Background: Several cross-talk mechanisms between autophagy and apoptosis have been identified, where the same protein plays a central role in the opposing processes.
Results: This study identified a novel cross-talk mechanism involving an Atg7·caspase-9 complex.
Conclusion: Atg7 and caspase-9 mutually regulate each other's activity.
Significance: The Atg7·caspase-9 complex may determine the balance between autophagy and apoptosis in response to stress.
Keywords: Apoptosis, Autophagy, Caspase, Cell Death, Cellular Regulation
Abstract
Several cross-talk mechanisms between autophagy and apoptosis have been identified, in which certain co-regulators are shared, allowing the same protein to participate in these opposing processes. Our studies suggest that caspase-9 is a novel co-regulator of apoptosis and autophagy and that its caspase catalytic activity is dispensable for its autophagic role. We provide evidence that caspase-9 facilitates the early events leading to autophagosome formation; that it forms a complex with Atg7; that Atg7 is not a direct substrate for caspase-9 proteolytic activity; and that, depending on the cellular context, Atg7 represses the apoptotic capability of caspase-9, whereas the latter enhances the Atg7-mediated formation of light chain 3-II. The repression of caspase-9 apoptotic activity is mediated by its direct interaction with Atg7, and it is not related to the autophagic function of Atg7. We propose that the Atg7·caspase-9 complex performs a dual function of linking caspase-9 to the autophagic process while keeping in check its apoptotic activity.
Introduction
Autophagy is a multistep homeostatic and stress-induced process that mediates the degradation of cytoplasmic components through a lysosomal pathway (1). Autophagy involves the coordinated function of multiple Atg proteins that choreograph the various phases of the degradation process from the generation of the autophagosome to its fusion with an endocytic organelle and/or lysosome. Two ubiquitin-like reactions are required for autophagosome formation: (i) the ubiquitination-like conjugation of activated Atg12 to Atg5 and (ii) the post-translational lipidation of Atg8/LC33 with phosphatidylethanolamine (2). Atg7 is an E1-like enzyme that is required for both reactions, utilizing Atg10 as the E2 enzyme for the Atg12·Atg5 conjugation and Atg3 as the E2 enzyme for the lipidation of LC3 (3–5). The preformed Atg12·Atg5 conjugate is also required for the formation of lipidated LC3. Its phosphatidylethanolamine lipid moiety mediates membrane tethering and hemifusion of LC3; therefore, it serves as a reliable marker for a nascent autophagosome (6).
Autophagy is implicated in multiple human diseases that can be characterized by an imbalance in protein, organelle, or cellular homeostasis (7). In cancer, autophagy is involved in tumor biogenesis as well as in its response to therapy (8, 9). Although autophagy may function under certain circumstances as a Type II mechanism of cell death, there is increasing evidence that in advanced cancer, it improves the fitness of the metabolically stressed cancer cells. This survival function is mediated via the elimination of damaged organelles and cytoplasmic stress by-products, the recycling of basic building blocks, and the utilization of recycled metabolites as an internal source of energy (10, 11). As a survival mechanism, autophagy is expected to exhibit complete polarity to apoptosis. Such polarity has been detected in tumor cells, where either autophagy or a programmed cell death mechanism prevails (12). Reported cross-regulation mechanisms between autophagy and apoptosis hinge on shared proteins with distinct functions in the opposing processes (13). Such co-regulators include Bcl-2, which interacts with proapoptotic Bcl-2 family members as well as with Beclin-1 and Atg12 (14, 15); Atg5, an essential autophagy player (16) that promotes mitochondrial apoptosis following its cleavage by calpains (17); FLIP, which inhibits caspase-8 activity, but also binds to Atg3, which blocks its conjugation to LC3 (18); and Atg12, which is essential for autophagy and also binds to Bcl-2 or Mcl-1 to promote apoptosis (15). In addition, caspases were found to cleave and inactivate Beclin-1 and Atg4D (19–22), whereas autophagy degrades active caspase-8 (23).
In the current study, we present evidence for a novel cross-talk mechanism between apoptosis and autophagy: autophagic involvement of caspase-9 (C9), which is independent of its caspase activity. Potential involvement of catalytically active caspases in autophagic degradation has been reported in Drosophila (24, 25). Although the critical function of caspases in apoptosis is firmly established, it has become evident that certain caspases mediate other cellular processes distinct from apoptotic cell death (26–29). Thus, non-apoptotic functions have been determined for caspase-1, -3, -7, -8, -11, and -14, which involve their catalytic activity (30–38). Two recent studies identified non-apoptotic activities for C9 that are dependent on its catalytic activity, including myotube formation in skeletal muscle (39) and the involvement of C9 enzymatic activity in lysosomal function (40). Our findings suggest that the Atg7·C9 complex can distinctly impact autophagy and apoptosis and may serve as a therapeutic target for the concomitant removal of apoptosis repression and the institution of autophagy inhibition.
EXPERIMENTAL PROCEDURES
Reagents
Abs against caspase-9 (sc-17784 for immunostaining and sc-8355 or sc-7885 for immunoblotting and immunoprecipitation), β-tubulin (sc-9104), beclin-1 (sc-11427), β-actin (sc-47778), and ubiquitin (sc-8017) were from Santa Cruz Biotechnology, Inc.; p62 (610497) was from BD Transduction; anti-Vps34 (ab5451) was from Abcam; anti-MAP-LC3 Abs for immunoblotting were from AnaSpec (San Jose, CA), and anti-MAP-LC3 Ab (sc-16756) for immunostaining was from Santa Cruz Biotechnology. Pepstatin A, 3MA, MG132, rapamycin, Hoechst, and DAPI were from Sigma; E64D was from Calbiochem; and Alexa Fluor 488- or 594-conjugated anti-rabbit or anti-mouse Ig and pCR3.1, pcDNA4/TO, pcDNA4/TO/lacZ, and pcDNA6/TR plasmids were from Invitrogen. Active recombinant human caspase-9 was purchased from BioVision, and in vitro translated purified GST-Atg7 was purchased from Abnova.
Cell Lines
The tumor cell lines, including Hct116, HeLa, MB-MDA-231, and RKO, were obtained from ATCC. VP-16-resistant MDA-MB-231 cells were selected by multiple cycles of etoposide treatments. Tumor cell lines and WT or engineered MEF were grown in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, 20 mm l-glutamine, and 100 units/ml each of penicillin and streptomycin.
Transfection
Transfections were carried out with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions using non-linearized plasmids for transient expression and linearized plasmids for stable expression. To generate caspase-9 KO MEF that stably express Tet-inducible C325A caspase-9 mutant, the cells were first stably transfected with linearized pcDNA6/TR that encodes the Tet repressor. Blasticidin selected clones with verified Tet repressor expression were further stably transfected with the linearized Tet-inducible plasmids pcDNA4/TO, pcDNA4/TO/lacZ (Invitrogen), or pcDNA4/TO/C325A caspase-9. All procedures involving linearization, transfection, and stable clone selection were carried out as described previously (41–43).
RNAi
Beclin-1, caspase-9, and Vps34 specific siRNAs were obtained from Dharmacon as the siGENOME SMARTpool, which consists of four nonoverlapping siRNAs. The matching negative control from Dharmacon was the siGENOME Non-targeting siRNA Pool 2. Additionally, three distinct caspase-9 siRNAs were obtained from Invitrogen as the Stealth Select RNAi CASP9 set along with their matching negative controls, the Stealth RNAi negative controls LO and Hi GC. All RNAi results obtained by the Dharmacon siGENOME SMARTpool were confirmed by at least an additional distinct caspase-9-specific siRNA. Hct116, MDA-MB231, RKO, MEF WT, and MEF caspase-9 KO cells (2.5 × 105) were plated in a 6-well plate and, following 16 h (at 20% confluence), were transfected with 25–100 nm siRNA in Opti-MEM medium (Invitrogen) without fetal calf serum using oligofectamine reagent (Invitrogen) according to the manufacturer's transfection protocol. After 4 h, fetal calf serum was added to a final concentration of 10%. Mouse caspase-9 shRNA-expressing pLKO.1 plasmids (set of five non-overlapping target sequences) were obtained from Open Biosystems (catalog no. RMM4534-NM_015733). The non-mammalian shRNA-expressing negative control pLKO.1 plasmid was obtained from Sigma-Aldrich (catalog no. SHC002).
Molecular Cloning of Tet-inducible Mouse C325A Caspase-9 Mutant Expression Plasmid
Total RNA was isolated from WT MEF cells using RNA STAT-60 reagent (Tel-Test “B”, Inc.). Reverse transcription of the MEF total RNA was carried out using SuperScript II RNase H− reverse transcriptase (Invitrogen), using an oligo(dT)12–18 primer. PCR was performed with the Expand High Fidelity PCR system (Roche Applied Science) following the manufacturer's protocol. An amplicon containing the catalytic mutant (C325A) mouse caspase-9 open reading frame (ORF), which extends 6 nucleotides (nt) into the 5′-untranslated region (UTR) and 39 nt into the 3′-UTR was generated in two steps using the following four primers by overlap extension using the PCR method (44): 5′-CGGGGTACCCTCGCCATGGACGAGGCG-3′ (forward; outside) and 5′-ACCACCCGCGGCCTGGATGAAGAAGAGCTT-3′ (reverse; inside C325A underlined) and 5′-TTCATCCAGGCCGCGGGTGGTGAGCAGAAAGACCAT-3′ (forward; inside-C325A underlined) and 5′-CCGGAATTCCTTCGGAGAGAATAATGAGGC-3′ (reverse; outside). The putative mutant caspase-9 amplicon was size-selected on a 1% agarose gel and extracted using the QIAquick gel extraction kit (Qiagen). It was then subjected to a second round of PCR with the forward and reverse outside primers and was size-selected and extracted as above. The purified mutant amplicon was digested with the restriction endonucleases KpnI and EcoRI and ligated (T4 ligase; New England Biolabs) into the Tet-inducible vector pcDNA4/TO (Invitrogen). Following transformation of DH5α cells (Invitrogen), randomly selected putative mouse C325A caspase-9 clones were sequenced by the University of Pittsburgh Genomics and Proteomics Core Laboratories.
Generation of Tet-inducible N-terminal 3× FLAG Mouse C325A Caspase-9 Mutant Expression Plasmid
We carried out PCR using the recombinant C325A mouse caspase-9 plasmid (above) as a template with the following primers: 5′-CGGGGTACCGCCATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGATGGACGAGGCGGACCGG-3′ (forward; 3× FLAG tag underlined) and the reverse outside primer described above for mouse C325A caspase-9. All other procedures for the processing of the N-terminal 3× FLAG mouse C325A amplicon, its ligation to pcDNA4/TO, and sequence verification were as above.
Generation of N-terminal 3× FLAG Mouse C325A Caspase-9 ΔCARD, ΔLarge Subunit, ΔLinker Domain and Small Subunit, and ΔA and ΔB Large Subunit Deletion Mutant Expression Plasmids
All deletion mutant plasmids were generated from the mouse caspase-9 C325A expression plasmid by PCR and overlap extension using PCR as described above using the following primers: ΔCARD, 5′-CGGGGTACCGCCATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGGTCTGTGTTCCAGGGAAG-3′ and the reverse outside primer described above for mouse C325A caspase-9; Δlarge subunit, 5′-CGGGGTACCGCCATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGATGGACGAGGCGGACCGG-3′ (forward; outside), 5′-ATAGGGGACAGCATCATGAGCTCCGCCAGAACC-3′ (reverse; inside), 5′-GGAGCTCATGATGCTGTCCCCTATCAGGAAGGC-3′ (forward; inside), and the reverse outside primer described above for mouse C325A caspase-9; Δlinker domain and small subunit, 5′-CGGGGTACCGCCATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGATGGACGAGGCGGACCGG-3′ and 5′-CCGGAATTCTCAATCTGGCTCAGAGTCACTGTC-3′; ΔA large subunit, 5′-CGGGGTACCGCCATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGATGGACGAGGCGGACCGG-3′ (forward; outside), 5′-GCAGTCCAGGGCATCATGAGCTCCGCCAGAACC-3′ (inside; reverse), 5′-GGAGCTCATGATGCCCTGGACTGCTTTGTGGTG-3′ (forward; inside), and the reverse outside primer described above for mouse C325A caspase-9; ΔB large subunit, 5′-CGGGGTACCGCCATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGATGGACGAGGCGGACCGG-3′ (forward; outside), 5′-ATAGGGGACAGCACGGTGGTTCCGGTGTGCCAT-3′ (reverse; inside), 5′-CGGAACCACCGTGCTGTCCCCTATCAGGAAGGC-3′ (forward; inside), and the reverse outside primer described above for mouse C325A caspase-9. All deletion mutant amplicons were processed as described for C325A mouse caspase-9. Sequences of randomly selected clones were verified by the University of Pittsburgh Genomics and Proteomics Core laboratories.
Molecular Cloning of Mouse N-terminal V5 Atg7
Total RNA was isolated from WT MEF cells and reverse transcribed as described above for mouse C325A caspase-9. PCR was carried out as above using the following primer pair: 5′-CGCGGATCCGCCATGAGAGGGCCCTTCGAAGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGCGTACCGGTATGGGGGACCCTGGACTG-3′ (forward; V5 tag underlined) and 5′-ACGCGTCGACTTGCTTGCTTCAGACAGTCTC-3′ (reverse). The putative V5-tagged Atg7 amplicon was size-selected on an agarose gel and extracted as described for caspase-9 above. It was subjected to a second round of PCR as above. The purified V5-Atg7 amplicon was digested with restriction enzymes BamHI and SalI and ligated to pcDNA4/TO, which had been digested with BamHI and XhoI. Following transformation of DH5α cells, randomly selected clones were sequenced by the University of Pittsburgh Genomics and Proteomics Core laboratories.
Generation of Human C287A Caspase-9 Mutant Expression Plasmid
A human full-length WT caspase-9 plasmid whose cDNA is derived from Jurkat RNA was used as a template in overlap extension using PCR with the following four primers: 5′-CGCGGATCCCTCGCCATGGACGAAGCGGAT-3′ (forward; outside), 5′-CCCACCCGCGGCCTGGATGAAAAAGAGCTT-3′ (reverse; inside C287A underlined), 5′-TTCATCCAGGCCGCGGGTGGGGAGCAGAAAGACCAT-3′ (forward; inside C287A underlined), and 5′-ACGCGTCGACTGCAAGATAAGGCAGGGTGAG-3′ (reverse; outside). The putative C287A caspase-9 amplicon, which extends 6 nt into the 5′-UTR and 32 nt into the 3′-UTR, was size-selected etc., as described above, and digested with restriction enzymes BamHI and SalI. It was ligated to the vector pCR3.1 (Invitrogen), which had been digested with BamHI and XhoI. Following transformation, sequence verification was carried out as described previously.
Molecular Cloning of Human ATG7 Expression Plasmid
Total RNA was isolated from MCF-7 breast cancer cells, and reverse transcription and PCR were all carried out as described above for the mouse C325A caspase-9 mutant. The following primers were utilized to generate a full-length ATG7 ORF that extends 6 nt into both the 5′- and 3′-UTRs: 5′-CGGGGTACCGAAATAATGGCGGCAGCTACG-3′ (forward) and 5′-CCGGAATTCGCCATCTCAGATGGTCTCATC-3′ (reverse). The putative full-length amplicon was processed as described for C325A caspase-9, including digestion by KpnI and EcoRI. It was ligated to the mammalian expression vector pCR3.1 that had also been digested with KpnI and EcoRI. Transformation and sequence verification were performed as described above.
In Vitro Transcription-Translation
Atg7 and caspase-9 cDNAs were expressed in the TNT T7 transcription-translation reticulocyte lysate system (Promega). Each coupled transcription-translation reaction contained 1 μg of plasmid DNA in a final volume of 50 μl in a reticulocyte lysate reaction mixture according to the manufacturer's instructions. After incubation at 30 °C for 90 min, the reaction products were immediately used or stored at −78 °C.
Apoptotic Assays
Apoptotic assays included long-term clonogenicity, flow cytometry of propidium iodide and Annexin V, and immunoblotting for processed caspases or their substrates, as described previously (41, 42, 45, 46). Caspase-9 activity was measured by the caspase-9 Glo Kit (Promega).
Autophagy Induction
Autophagy was induced by starvation (amino acids and serum, 0.5–24 h); rapamycin (2–10 μm, 5–24 h), MG132 (2 μm, 5–24 h), and TRAIL (50–200 ng/ml, 5–24 h).
Immunoblotting and Immunoprecipitation
Cell lysates were prepared with 1% Nonidet P-40, 20 mm Tris-HCl, pH 7.4, 137 mm NaCl, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, and 10 μg/ml aprotinin. The immunoblotting and immunoprecipitation procedures were described in our previous publications (41, 42, 45, 46). All immunoprecipitations were controlled by a sham procedure with nonspecific matching immunoglobulins. Quantification of scanned protein bands was performed by the US-SCAN-IT Gel software.
Cell Microscopy and Image Acquisition
Confocal images were obtained with an Olympus Fluoview 1000 confocal microscope and the companion software FV10-ASW1.6. Images were acquired with the use of the same setting at a resolution of 1020 × 1024 pixels. Morphometric measurements were performed using Metamorph (Universal Imaging) on at least 50 cells/condition. Physiologically expressed endogenous LC3 puncta were monitored by two measures: (i) counting of dot number/cell and (ii) determination of cumulative dot area/cell area. Cumulative dot area and cell area were determined by Metamorph on images where the set threshold eliminated the detection of low or non-puncta staining.
For electron microscopy, cells were fixed with 2% paraformaldehyde and 2% glutaraldehyde in 0.1 m phosphate buffer (pH 7.0), followed by 1% OsO4. After dehydration, thin sections were stained with uranyl acetate and lead citrate for observation under a JEM 1011CX electron microscope (JEOL, Peabody, MA). At least 50 cells/treatment were utilized for quantification of all digitally acquired microscopic images.
Statistical Analysis
All immunoblot analyses are representative of at least three experiments. Images are representative of ∼50 cells from at least three repeats. Quantification by Metamorph was performed on at least 50 cells/treatment, and the results were confirmed in three independent experiments. Statistical analysis was performed by GraphPad Prism V software, utilizing the nonparametric tests Mann-Whitney U (MWU) and Wilcoxon signed-rank, as appropriate.
RESULTS
Involvement of Non-apoptotic C9 in Autophagy
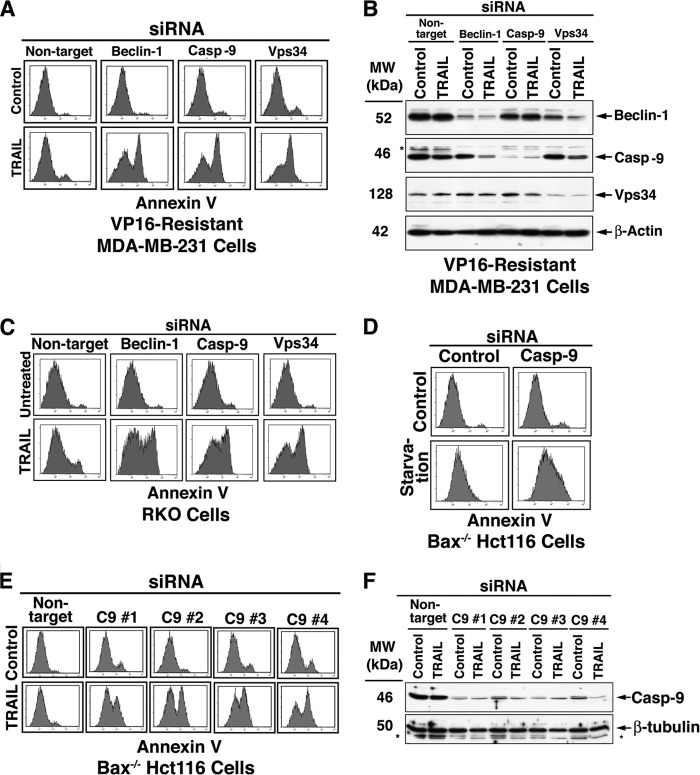
Recent studies have demonstrated the induction of cytoprotective autophagy by chemotherapeutic agents, including TRAIL, in apoptosis-resistant tumor cells (46–49). Apoptotic sensitization of such tumor cells could be achieved by pharmacological inhibition of autophagy or through RNAi of Atg genes, including Beclin-1, Vps34, and UVRAG (46, 48). Upon inhibition, 30–50% of the treated cells turn Annexin V-positive, and all such treated cells lose their clonogenic capability, as determined by a long term clonogenicity assay (46). In our efforts to elucidate the mechanism(s) of cell death induced by autophagy inhibition, we knocked down C9 with the expectation that it would inhibit the apoptotic shift. Surprisingly, C9 knockdown (KD) did not inhibit the execution of the autophagy-to-apoptosis shift but rather enhanced it significantly (data not shown). Utilizing tumor cell lines with significant TRAIL resistance (VP16-selected MDA-MB-231, RKO, and Bax−/− Hct116), we determined that comparable TRAIL sensitization was mediated by Beclin-1, Vps34, or C9 RNAi (Fig. 1, A–C). Likewise, C9 KD shifted to cell death the starvation response of Bax−/− Hct116 cells (Fig. 1D). To ascertain that these observations were not related to off-target effects, we performed the experiments with multiple distinct C9 siRNAs that demonstrated effective sensitization of Bax−/− Hct116 cells to TRAIL (Fig. 1, E and F). The conclusion from this initial series of experiments was that C9 performs a non-apoptotic function, which in the context of cellular adaptation to stress supersedes its apoptotic activity.
FIGURE 1.
C9 RNAi sensitizes apoptosis-deficient tumor cells to TRAIL or starvation. A–D, TRAIL-resistant tumor cells (including VP16-selected MDA-MB-231, Bax−/− Hct116, and RKO) were treated with the indicated siRNAs for 60 h and then by TRAIL (100 ng/ml, 6 h) or starvation (Hanks' balanced salt solution, 16 h). The cells were assessed by flow cytometry for Annexin V binding (A, C, and D). E and F, reversal of Bax−/− Hct116 TRAIL resistance by independent C9 siRNAs. (#1, a mixture of four distinct siRNAs from Dharmacon; #2, #3, and #4, distinct C9 siRNAs from Invitrogen, as detailed under “Experimental Procedures”). The control siRNAs from Invitrogen provided a similar pattern to that shown for the non-target siRNA pool from Dharmacon. Reduced expression of the siRNA-targeted proteins in the cells utilized in A and E are shown in B and F, respectively. The asterisks indicate unidentified protein bands. The Dharmacon siRNA pools (#1 and its matching non-target control) were utilized in experiments shown in A–D. Each of the shown experiments has been independently repeated with similar results at least three times.
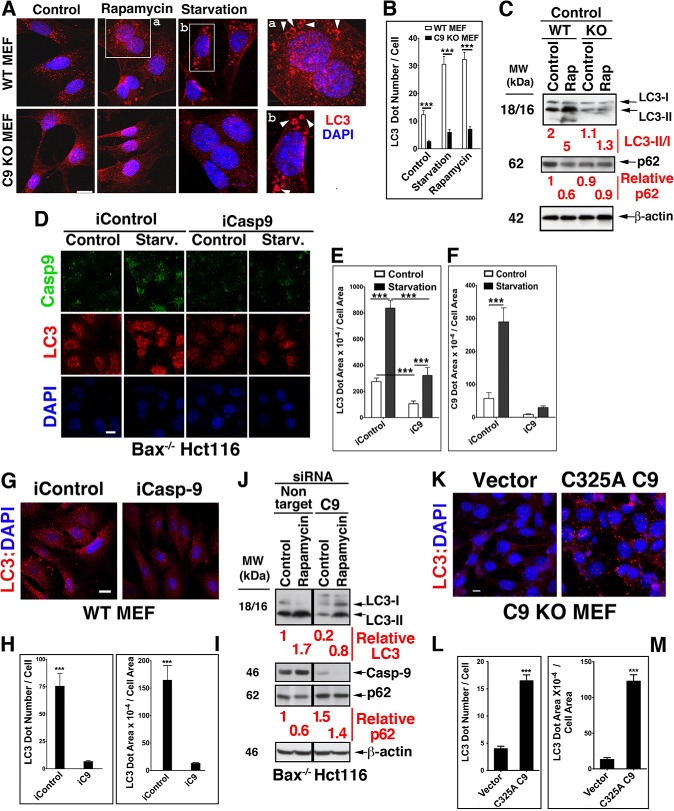
The impact of C9 on autophagy was assessed by modifications in its expression level in cells undergoing autophagy in response to rapamycin or starvation, classical inducers of autophagic flux. The modifications in C9 expression levels included the utilization of C9 KO MEF from two sources (50, 51), C9 KD, and transient and stable transfection of C9 KO MEF with the murine catalytically inactive C325A C9 mutant. Distinct and large endogenous LC3 puncta were detected in WT MEF but markedly less so in C9 KO MEF under basal conditions or following treatments by rapamycin or starvation (Fig. 2, A and B). Further, LC3 decorated vesicles were detected only in WT MEF treated with rapamycin or exposed to starvation and not in similarly treated C9 KO MEF. The expression of C9 also impacted the ratio of LC3-II/LC3-I in either control or rapamycin-treated MEF; whereas rapamycin treatment of WT MEF increased this ratio by enhanced generation of LC3-II, there was no change in the already reduced level of LC3-II formation in C9 KO MEF (Fig. 2C). Likewise, whereas rapamycin enhanced p62 degradation in WT MEF, it did not impact the p62 expression level in C9 KO MEF. Although both LC3-I and LC3-II were present in C9 KO MEF, the difference in the LC3-II/I ratio between the two cell lines suggests that C9 enhances the mechanism responsible for the conversion of LC3-I to LC3-II. This interpretation is supported by the impact of C9 KD on the autophagic response of Bax−/− Hct116 to starvation or rapamycin (Fig. 2, D–J). In particular, the LC3-I to LC3-II conversion was more efficient in rapamycin-treated cells in combination with non-target siRNA control than with C9 siRNA (Fig. 2J). Likewise, the p62 degradation levels mediated by rapamycin were reduced in the same cells in which C9 KD reduced the conversion of LC3-I to LC3-II. The impact of C9 on the autophagosome formation process was corroborated by the reduced expression of LC3 puncta in C9 KD Hct116 cells subjected to starvation (Fig. 2, D–F). The level of LC3 puncta under basal conditions was also impacted by the expression level of C9, as demonstrated by C9 KD in WT MEF (Fig. 2, G–I). Conversely, transfection of C9 KO MEF with the C325A C9 mutant, but not vector control, increased the basal expression of LC3 puncta (Fig. 2, K–M).
FIGURE 2.
The expression level of C9 impacts the pattern and expression level of LC3 under basal conditions or autophagy induced by rapamycin or starvation. A and B, reduced expression of LC3 vesicles in starved or rapamycin-treated C9 KO MEF (2–4 μm, 6–16 h) as compared with WT MEF. Scale bar (A), 30 μm. Increased magnifications of the boxed areas are shown in a and b. B, quantification of LC3 puncta in WT and C9 KO MEF was performed on at least 50 cells per treatment, and similar results were obtained in three independent experiments. Data are means ± S.E (error bars). ***, p < 0.0001 (MWU). C, differential LC3-II/LC3-I ratio and p62 degradation levels in C9 KO MEF as compared with WT MEF treated with rapamycin. Densitometric measurements of the bands were utilized for quantification. D–F, C9 KD in Bax−/− Hct116 cells reduces the LC3 puncta expression level under basal and starvation conditions. Scale bar (D), 50 μm. E and F, quantification of results shown in D. G–I, C9 KD in WT MEF reduces the basal expression level of LC3. Scale bar (G), 100 μm. H and I, quantification of results shown in G. J, increased accumulation of LC3-I, decreased LC3-II, and reduced degradation of p62 in rapamycin-treated Bax−/− Hct116 cells with reduced C9 expression. All samples were run in the same gel, and the two-component appearance of the immunoblots is due to the removal of an unrelated middle lane. Dharmacon siRNA pools were utilized in D, G, and J. K–M, the level of LC3 puncta is significantly enhanced in C9 KO MEF transfected with C325A C9 mutant. Scale bar (K), 100 μm. L and M, quantification of results shown in K.
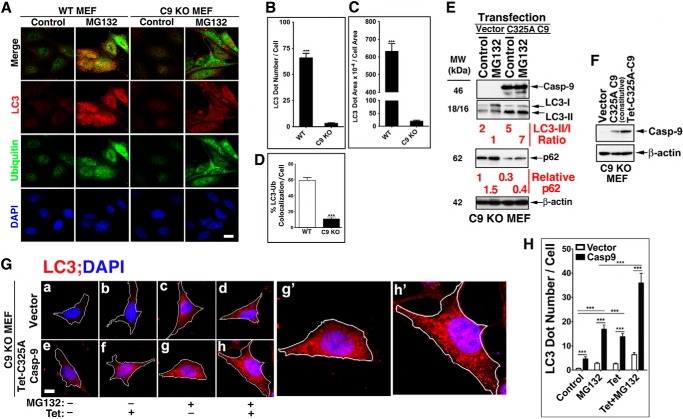
Because protein degradation is mediated by both proteasomal and lysosomal mechanisms, characterization of an autophagic effector(s) may be best conducted in cells with inhibited proteasomal activity. In support of such an approach, it has been documented that proteasomal inhibition induces an autophagic response as a compensatory mechanism for protein degradation (52, 53). In line with the compensatory notion, we observed a marked difference in the response of WT and C9 KO MEF to the proteasomal inhibitor MG132. A significant reduction in LC3 puncta formation in response to MG132 was detected in the absence of C9 (Fig. 3, A–D). The WT MEF LC3 puncta were highly co-localized with ubiquitin-decorated proteins that accumulated in response to MG132, but such co-localization was not observed in C9 KO MEF harboring a reduced number of LC3 puncta. The reduced formation of autophagosomes as detected by LC3 puncta or LC3-II/LC3-I ratio was corrected by transfection of the C9 KO MEF with the C325A C9 mutant (Fig. 3E). Thus, the ratio of LC3-II/LC3-I was increased 7-fold upon transient transfection of C325A C9 mutant as compared with MG132-treated vector control cells. This change in LC3-II/LC3-I ratio was accompanied by a reduction in the expression level of p62 as compared with vector-transfected C9 KO MEF. Because the C9 expression level in our transient transfection is not controllable, we generated C9 KO MEF that stably express Tet-inducible C325A C9. Low level constitutive expression of C9 (due to leakiness) was detected in the absence of Tet (Fig. 3F). Thus, we utilized both non-treated and Tet-treated C9 KO MEF that were stably transfected with Tet-inducible C325A C9 as cells expressing different incremental levels of C9. Stable expression of C325A C9 increased the LC3 expression signal above vector-transfected cells in both control and MG132-treated MEF (Fig. 3, G and H). Increased formation of autophagosomes was also documented by transmission electron microscopy in C9 KO MEF reconstituted with C325A C9 relative to those reconstituted with vector under basal conditions (Fig. 4, A and B) or in response to rapamycin (Fig. 4, C and D). Cumulatively, these findings suggest that non-apoptotic C9 enhances the autophagic response mediated by archetypical inducers of autophagy, including starvation, rapamycin, and proteasomal inhibition. The findings further suggest that C9 significantly affects the autophagosome formation process (as assessed by autophagosome morphology and LC3 puncta) and appears to specifically impact the LC3-I to LC3-II conversion process.
FIGURE 3.
The presence of C9 impacts MEF autophagic response to proteasomal inhibition. A–D, increased LC3 puncta and their co-localization with ubiquitin in WT, but not in C9 KO MEF, treated with MG132 (2 μm/ml, 6 h). A, representative images obtained with WT or C9 KO MEF. Scale bar, 40 μm. B and C, quantification of the LC3 dot number/cell (B) and LC3 dot area/cell area (C); data are means ± S.E. (error bars) of at least 50 cells; ***, p < 0.0001 (MWU). D, quantification of co-localization of LC3 puncta with ubiquitin, as performed by Metamorph; E, transfection of C325A C9 mutant into C9 KO MEF, but not vector control, increases the LC3-II/LC3-I ratio and decreases the p62 expression level in control and MG132-treated cells. F–H, constitutive or induced expression of non-catalytically active C325A C9 mutant increases LC3 puncta in C9 KO MEF at the basal level and in response to MG132. F, immunoblot assessment for the expression of C9 in C9 KO MEF stably transfected with the indicated plasmids. G, a representative cell for each treatment is shown (a–h); g′ and h′, enlargements of g and h, respectively. Scale bar, 10 μm. H, quantification of results shown in G; the data are means ± S.E. of LC3 dot number/cell of at least 50 cells/treatment in one experiment of at least three performed with similar results.
FIGURE 4.
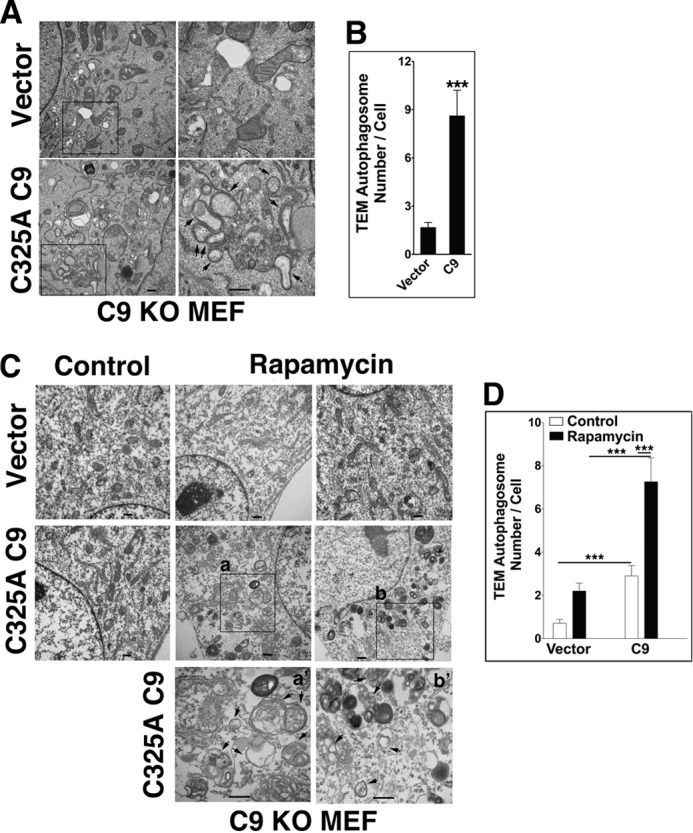
Increased C9 expression is associated with enhanced autophagosome formation under basal or autophagic conditions. A and B, transfection of C325A C9, but not vector control, increases the number of autophagosomes/cell in C9 KO MEF under basal conditions. The arrows indicate autophagosomes. Scale bars, 500 nm; right panels show increased magnification of the boxed areas. B, quantification was performed by counting autophagosomes in at least 50 cells/treatment in one of at least three experiments with similar results. Data are means ± S.E. (error bars); ***, p < 0.0001 (MWU). TEM, transmission electron microscopy. C and D, increased autophagosome formation in response to rapamycin in C9 KO MEF transfected with C325A C9 as compared with vector control. Scale bars, 500 nm; bottom panels a′ and b′ show increased magnification of the boxed areas in a and b, respectively. D, quantification was performed as described in B. Two independent experiments are shown in A and C.
Interaction of C9 with Atg7
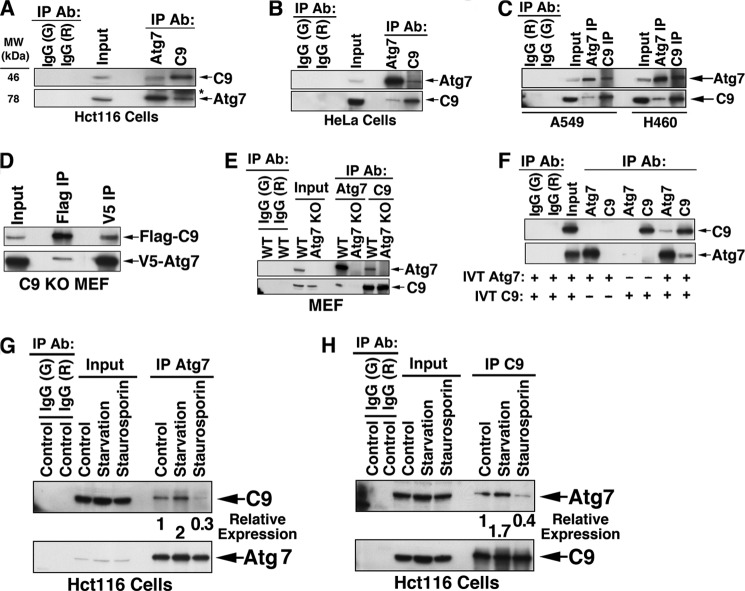
To expand our investigation of the newly identified autophagic function of C9, we screened C9 immunoprecipitated complexes for an array of autophagic proteins. We identified Atg7 as a C9-interacting protein. Thus, a complex between endogenous Atg7 and endogenous C9 was confirmed by a two-way immunoprecipitation in Hct116, HeLa, and two non-small cell lung cancer (A549 and H460) cell lines (Fig. 5, A–C). Two-way co-immunoprecipitation of Atg7 and C9 was also confirmed for C9 KO MEF that were transiently transfected with 3× FLAG-C325A C9 and V5-Atg7 (Fig. 5D). The specificity of the interaction was confirmed by a lack of C9 co-immunoprecipitation in Atg7 KO MEF (Fig. 5E). Further, a direct interaction between Atg7 and C9 was confirmed by co-immunoprecipitation of in vitro translated Atg7 and C9 proteins (Fig. 5F).
FIGURE 5.
Interaction of C9 with Atg7. A–C, co-immunoprecipitation (co-IP) of endogenous Atg7 with endogenous C9 in four tumor cell lines, including Hct116 (A), HeLa (B), and A549 and H460 (C). Sham IP by matching control IgG (goat (G) and rabbit (R)) is shown in the two left lanes. D, co-IP of transfected Atg7 and C9. C9 KO MEF were transiently transfected with murine 3× FLAG-C325A C9 and murine V5-Atg7 and subjected to IP by anti-FLAG or anti-V5 Abs. E, the Atg7·C9 complex is absent in Atg7 KO MEF. WT and Atg7 KO MEF cell lysates were subjected to IP by anti-Atg7 or anti-C9 Abs. WT MEF were also subjected to control IgG IP. F, co-IP of in vitro translated (IVT) Atg7 and in vitro translated C9. The ratio of input to pellet in all IP procedures was 1:3. G and H, the expression level of the Atg7·C9 complex is increased under starvation and decreased during apoptosis. Hct116 cells were subjected to starvation (Hanks' balanced salt solution, 6 h) or staurosporin (2 μm, 6 h) and subjected to IP of Atg7 (G) and C9 (H). The relative expression levels of C9 (G) or Atg7 (H), as determined by their optical density, are indicated.
We then assessed the impacts of starvation and apoptosis on the Atg7·C9 complex formation. As determined by either Atg7 immunoprecipitation (Fig. 5G) or C9 immunoprecipitation (Fig. 5H), the Atg7·C9 complex formation was enhanced by starvation and decreased by staurosporin-mediated apoptosis. The disruption of the Atg7·C9 complex in response to staurosporin may suggest that the apoptotic activity of C9 favors its dissociation from Atg7.
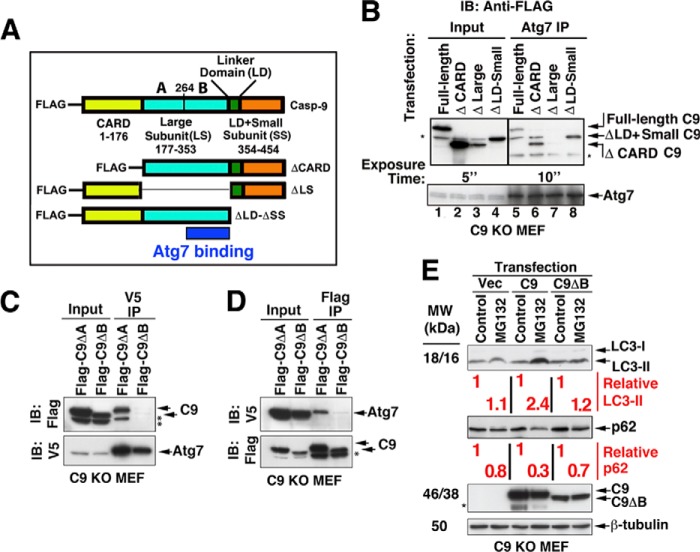
To start mapping the Atg7 binding site within C9, we generated, in addition to the 3× FLAG-tagged full-length C325A C9 construct, three deletion mutants (ΔCARD, Δlarge subunit (ΔLS), and Δlinker domain and small subunit (ΔLD + ΔSS)) (Fig. 6A). FLAG-tagged full-length C325A C9 and the indicated deletion mutants were transiently transfected into C9 KO MEF, which were subjected to immunoprecipitation of endogenous Atg7. The C9 large subunit was identified as the core of the binding area because Atg7 co-immunoprecipitated the full-length C9, ΔCARD C9, and ΔLD + ΔSS but not the ΔLS (Fig. 6B). Subsequently, we generated two additional C9 mutants, each with a partial deletion of the LS: 3× FLAG-C9ΔVal177–Arg263 (ΔA) and 3× FLAG-C9ΔAla264–Asp353 (ΔB). Each of these mutants was transfected with V5-Atg7 into C9 KO MEF and assessed for Atg7 binding. The 3× FLAG-C9ΔB mutant failed to co-precipitate with V5-Atg7 (Fig. 6, C and D), identifying the region that stretches from Ala264 to Asp353 within C9 LS as required for the interaction of C9 with Atg7.
FIGURE 6.
The C-terminal region of the C9 large subunit is required for its interaction with Atg7 and for its impact on LC3-II formation. A, a schema of C9 constructs. B, C9 large subunit is required for C9 interaction with Atg7. C9 KO MEF were transiently transfected with the indicated FLAG-tagged C9 constructs. The cells were then subjected to immunoprecipitation of endogenous Atg7 and probed with anti-FLAG Ab (top) and anti-Atg7 Ab (bottom). The proteins were run on one SDS gel, but different exposure times are shown for the input (top left) and IP (top right). The asterisks indicate unidentified protein bands (including those in lanes 5 and 6 that run parallel to the ΔLD + ΔSS). IB, immunoblot. C and D, Atg7 binds to the C-terminal portion of the C9 large subunit. C9 KO MEF were transiently transfected with V5-Atg7 and FLAG-C9ΔVal177-Arg263 (ΔA) or FLAG-C9 ΔAla264–Asp353 (ΔB). The cell lysates were subjected to V5 IP (C) and FLAG IP (D). E, LC3-II accumulation in response to MG132 is dependent on the C9 large subunit region Ala264–Asp353. C9 KO MEF were transfected with FLAG-C325A C9 or FLAG-C9 ΔAla264–Asp353 (ΔB) for 24 h and then treated with MG132 or vehicle control (6 h). The relative expression levels of LC3-II and p62 as compared between pairs of vehicle control and MG132 treatments were determined by their optical density. The vertical lines separate pairs.
To determine if the C9 interaction site with Atg7 is also required for C9 autophagic function, we assessed the LC3-II levels in C9 KO MEF transfected with either full-length C325A C9 or C9ΔAla264–Asp353 (C9ΔB) and treated with MG132 (Fig. 6E). MG132 treatment enhanced the LC3-II level in C9-transfected cells but not in those transfected with either vector or with the C9ΔB deletion mutant. These findings suggest that the C9 site, which is required for interaction with Atg7, is also required for its impact on LC3-II formation.
Mutual Impacts of Atg7 and C9 on Each Other
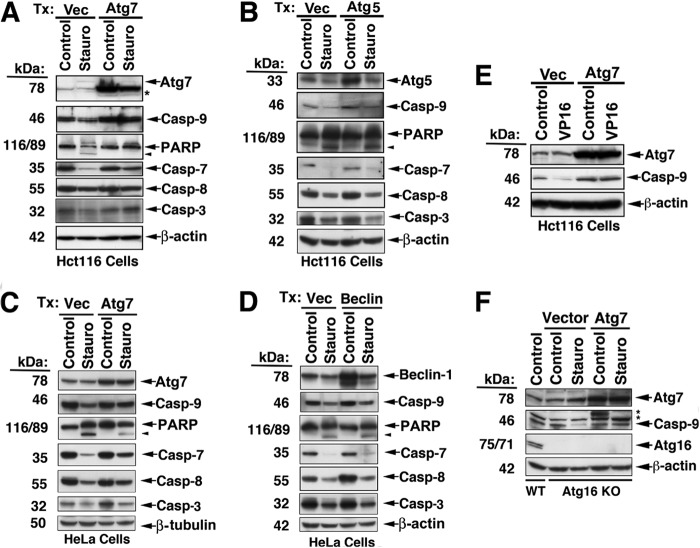
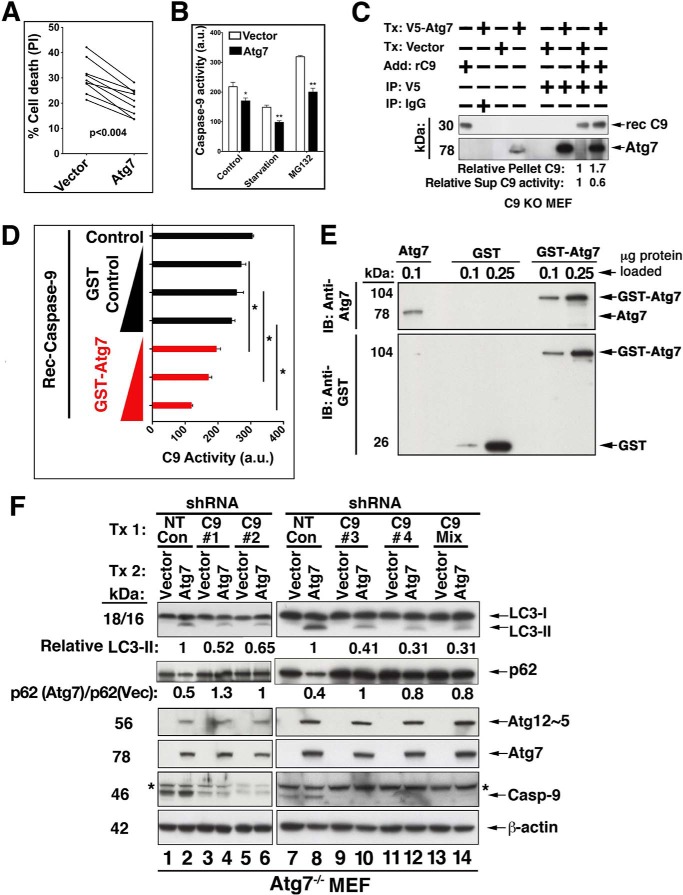
The possibility that Atg7 is a substrate for C9 proteolytic activity or C9-activated caspase-3/7 was excluded in tumor cell lines treated by staurosporin or etoposide. Thus, neither a change in the expression level of endogenous Atg7 nor the appearance of any Atg7 cleavage products was detected in staurosporin- or VP16-treated cells confirmed for the processing and activation of C9 (Fig. 7, A, C, and E). In line with the validation that Atg7 does not serve as an in vivo substrate of C9, the interaction of Atg7 with C9 appears to have an inhibitory effect on the processing of the C9 prodomain. Thus, overexpression of Atg7 repressed the processing of C9 under apoptotic conditions mediated by staurosporin in Hct116 and HeLa cells (Fig. 7, A, C, and E). In contrast to Atg7, transfection of either Atg5 or Beclin-1 (Fig. 7, B and D) did not inhibit the staurosporin-mediated processing of C9 or other downstream caspases. To investigate if the Atg7-mediated inhibition was dependent on the autophagic activity of Atg7, we transfected Atg7 into Atg16 KO MEF and assessed its impact on staurosporin-mediated processing of the C9 prodomain (Fig. 7F). The staurosporin-mediated processing of C9 was inhibited in Atg7-transfected, but not in vector-transfected Atg16 KO MEF. Because Atg16 is an essential autophagic effector and the Atg16 KO MEF are autophagy-deficient (54), the C9-targeted inhibitory activity of Atg7 appears to be independent of the autophagic function of Atg7. Although C9 processing was markedly inhibited in the presence of Atg7 (Fig. 7, A, C, and E), the processing of caspase-7, -8, or -3 or poly(ADP ribose) polymerase was only partially inhibited, suggesting that the apoptotic activity of staurosporin was not completely dependent on C9. Indeed, transfection of Atg7 KO MEF with Atg7, but not with vector control, significantly inhibited, but only partially (percentage of inhibition, 32.3 ± 3.3% (mean ± S.E.)), the staurosporin-mediated cell death of these cells (Fig. 8A). The observed partial inhibition may relate to the ability of staurosporin, a broad spectrum kinase inhibitor, to initiate multiple cell death pathways in which the activation of caspase-7 and -8 may be both upstream and downstream of C9. In support of this interpretation, benzyloxycarbonyl-VAD-fluoromethyl ketone, a pan-caspase inhibitor, arrested the cell death mediated by staurosporin treatment (24 h), whereas benzyloxycarbonyl-LEHD-fluoromethyl ketone, a C9-specific inhibitor, demonstrated only partial inhibition at a level of ∼25%. Reconstitution of Atg7 in Atg7 KO MEF also reduced the catalytic activity of C9, as assessed by the processing of a specific C9 fluorescence substrate in the response of these cells to starvation or MG132 (Fig. 8B). To investigate the capability of Atg7 to sequester C9, we transfected C9 KO MEF with V5-Atg7 or vector control, immunoprecipitated the transfected Atg7, and added active recombinant C9 to the pellet. The washed pellet and its supernatant (sup) were assessed by immunoblotting for the presence of Atg7 and C9, and the supernatant was further assessed for C9 enzymatic activity (Fig. 8C). An increased level of recombinant C9 was present in the Atg7 immunoprecipitate (1:1.7 ratio of vector/Atg7 transfection), and the C9 activity in the matching supernatant was correspondingly decreased (1:0.6 ratio). These experiments suggest that the Atg7·C9 complex could function by sequestering C9 from its activation site. The ability of Atg7 to inhibit C9 enzymatic activity was confirmed in a cell-free system, where active recombinant C9 was mixed with increasing doses of purified GST-Atg7. Each of the utilized doses of GST-Atg7 significantly inhibited recombinant C9 protease activity, as compared with the GST control (Fig. 8, D and E).
FIGURE 7.
C9 processing is inhibited by Atg7. A–E, inhibition of C9 processing in cells overexpressing Atg7 but not Atg5 or Beclin-1. Hct116 cells (A, B, and E) and HeLa cells (C and D) were transfected (Tx) as indicated for 48 h and then treated with staurosporin (2 μm, 16 h) or VP16 (20 μm, 16 h). F, C9 processing is inhibited by overexpressed Atg7 in cells with deficient autophagic capability. Atg16 KO MEF were transfected with either vector control or Atg7 and treated with vehicle control or staurosporin. The asterisks indicate unidentified protein bands.
FIGURE 8.
Reciprocal impacts of Atg7 and C9. A, increased expression of Atg7 attenuates staurosporin-mediated cell death. Atg7 KO MEF were transfected (Tx) with vector control or WT Atg7 and assessed for cell susceptibility to staurosporin by flow cytometry of Annexin V + propidium iodide. Nine independent experiments conducted with various staurosporin doses (0.5–2 μm) are shown. Inhibition range was 15.7–50%; inhibition mean ± S.E. was 32.3 ± 3.3%. Statistical significance was assessed by Wilcoxon signed rank test. B, during autophagy, Atg7 inhibits C9 apoptotic activity. Bax−/− Hct116 cells were transiently transfected with Atg7 for 48 h and then subjected to starvation or MG132 for 5 h. The cells were then assessed for C9 activity (C9 Glo). The results are means ± S.E. (error bars) of six replicates in one of three experiments with similar results; *, p < 0.05; **, p < 0.005 (MWU). a.u., arbitrary units. C, recombinant C9 is sequestered by its interaction with Atg7. C9 KO MEF were transfected with V5-Atg7 or vector control and subjected to V5 IP. Recombinant active C9 was then added to the protein A-Atg7 beads. Following washing, the pellet was assessed by immunoblotting, and the protein bands were quantified by densitometric measurements. The wash supernatant was assessed for C9 activity by the C9 Glo assay. D and E, the protease activity of recombinant C9 is inhibited by recombinant Atg7. Recombinant C9 was incubated alone or with wheat germ in vitro translated GST-Atg7 or the GST-control protein (1–5 μg/ml, 1 h, 37 °C). C9 enzymatic activity was determined by the C9 Glo assay. The data are means ± S.D. of triplicate determinations in one experiment of three performed with similar results; *, p < 0.05 (Wilcoxon signed rank test). E, immunoblotting of the proteins utilized in D. The doses of the loaded proteins are equivalent to the lowest and medium doses utilized in D. F, C9 KD reduces the Atg7 conjugation activity required for LC3-II formation and p62 degradation. Atg7 KO MEF were transfected with WT V5-Atg7 or matching vector control (24 h) and subsequently treated with non-target (NT) control shRNA (lanes 1, 2, 7, and 8) and four independent C9 shRNAs (lanes 3–6 and 9–12) or the shRNA mix (lanes 13 and 14) for 48 h. The cell lysates were assessed by immunoblotting. Lanes 1–6 and 7–14 represent two independent experiments. LC3-II was formed only in the presence of Atg7, and its level was reduced by the C9 shRNAs. Likewise, p62 degradation that was observed in Atg7-transfected cells (lanes 2 and 8) was blocked by the distinct C9 shRNAs (lanes 4, 6, 10, 12, and 14). Atg12·Atg5 complex was also produced only in the presence of Atg7, but its production level was not impacted by C9 KD. Densitometric quantification of LC3-II (in relative units) is indicated. Also, the ratios of p62 in Atg7 transfectants relative to p62 in their matching vector controls (lanes 2 versus 1, 4 versus 3, 6 versus 5, 8 versus 7, 10 versus 9, 12 versus 11, and 14 versus 13) are indicated.
To investigate the impact of C9 on the Atg7 autophagic function, we transfected Atg7 KO MEF with WT Atg7 concomitant with a C9 shRNA construct(s) (Fig. 8F). The transfection of Atg7, but not the vector control (lanes 1, 2, 7, and 8) repaired the cell's capability of producing the Atg12·Atg5 complex and of converting LC3-I to LC3-II. However, in the presence of each of four independent C9 shRNAs or their mix (lanes 4, 6, 10, 12, and 14), LC3-II formation was reduced, whereas the conjugation capability required for Atg12·Atg5 complex formation remained unchanged. In agreement with the LC3-II formation levels, degradation of p62 was reconstituted by Atg7 transfection and reversed to a reduced level by the C9 KD. These findings suggest that the autophagic function of C9 is mediated at least partly by its interaction with Atg7, it is localized to the molecular phase of LC3-II formation, and it significantly impacts the degradation of p62.
DISCUSSION
Our studies revealed an unexpected non-caspase function for C9 that is localized to the early phase of autophagosome formation and is mediated via its interaction with Atg7. Depending on the cellular context, Atg7 and C9 mutually impact each other; whereas Atg7 represses the apoptotic activity of C9, the latter facilitates the Atg7-dependent formation of autophagosomal LC3-II. Induced autophagy is associated with increased formation of the Atg7·C9 complex, which facilitates LC3-II formation while keeping C9 apoptotic activity in check. Reduced expression of the Atg7·C9 complex is associated with an apoptotic response, where C9 is relieved from sequestration and repression by Atg7.
The survival activity of C9 was initially discovered by an RNAi approach that was expected to block a shift from autophagy to apoptosis upon autophagy inhibition but was surprisingly found to exacerbate the ensuing apoptotic process. Whereas KD of caspase-8 inhibited apoptosis and elucidated its major role in the apoptotic process induced by inhibition of autophagy, C9 KD intensified the apoptotic response, suggesting the supersession of C9 survival activity over its apoptotic activity in the context of an adaptive response to cellular stress.
Our findings suggest that the processing of C9 is effectively inhibited in cells that overexpress Atg7 and that this inhibition is not dependent on the cytoprotective effect of autophagy because it occurs in Atg16 KO MEF, which are deficient in an essential autophagic protein. The Atg7 interaction with the C9 large subunit is consistent with the ability of Atg7 to interact with both pro-C9 and with (CARD-deficient) recombinant C9. Pro-C9 monomers are mainly cytosolic, and their interaction with Atg7 may sequester them from the mitochondria and apoptosome. Because C9 processing occurs mainly on the apoptosome, cytosolic sequestration of C9 by Atg7 may indirectly inhibit C9 activation and processing. Additionally, Atg7 may interact with apoptosome-associated C9, inhibiting its activation and/or processing. Unlike other caspases, whose activation requires prodomain processing (i.e. intrachain cleavage), the activation of C9 has been considered independent of its autocatalytic processing (55, 56). Although this issue has remained controversial, autoprocessing of C9 is either required for its activation (57) or for the molecular timer-mediated recruitment of pro-C9 to the apoptosome (55, 58). Thus, the Atg7-mediated inhibition of C9 processing could potentially inhibit the activation of C9 at the level of the apoptosome or interfere with its recruitment to the apoptosome. Additionally, apoptosome-independent activity of C9 has been described (59), and thus, Atg7 may be involved in the regulation of this cell death mechanism. These potential mechanisms of Atg7-mediated inhibition of C9 are now being intensely investigated.
Currently, it is not clear what determines the participation of C9 in apoptosis or autophagy. It is possible that a significant release of mitochondrial apoptogenic proteins would shift the balance toward apoptosome activation, whereas insufficient mitochondrial involvement would allow the opposing regulation of C9 by Atg7. Our findings suggest that the Atg7·C9 complex is involved in the determination of the balance between apoptosis and autophagy. Such toggling activity could potentially serve as a therapeutic target for the concomitant enhancement of apoptosis and the inhibition of autophagy. Although reciprocal regulation between Atg7 and C9 is a likely mechanism for the potential determination between participation of C9 in either autophagy or apoptosis, additional molecular scenarios should be considered. Because specific C9 phosphorylation sites are associated with inactivation of its enzymatic function, a phosphorylation mechanism may serve as a potential determinant in the involvement of C9 in either apoptosis or autophagy. Additionally, association of C9 with XIAP or Bruce or its potential ubiquitination by these E3 ligases (60, 61) may force C9 into an autophagic rather than apoptotic mode. Further studies should determine the exact molecular mechanisms that tie C9 to autophagosome formation and to the ultimate determination between its opposing functions within either apoptosis or autophagy.
Acknowledgments
We thank Dr. Richard Flavell (Yale University) and Dr. Tak W. Mak (University Health Network, Toronto, Ontario, Canada) for providing the caspase-9 knock-out MEF cell lines; Dr. Douglas Green and Dr. Brenda Schulman for the Atg7 knock-out MEF (St. Jude Children's Research Hospital, Memphis, TN); Dr. Shizuo Akira (Osaka University, Japan) for the Atg16 knock-out MEF; and Dr. Bert Vogelstein (Johns Hopkins University) for the Bax−/− Hct116 cell line.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 CA134776. This work was also supported by Department of Defense Grant W81XWH-12-1-0228 and the Pennsylvania Breast and Cervical Cancer Research Initiative (to H. R.).
- LC3
- light chain 3
- C9
- casapase-9
- IP
- immunoprecipitation
- KD
- knockdown
- MWU
- Mann-Whitney U
- Ab
- antibody
- MEF
- mouse embryo fibroblast(s)
- nt
- nucleotides.
REFERENCES
- 1. Yang Z., Klionsky D. J. (2009) An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol 335, 1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ohsumi Y. (2001) Molecular dissection of autophagy. Two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2, 211–216 [DOI] [PubMed] [Google Scholar]
- 3. Fujioka Y., Noda N. N., Fujii K., Yoshimoto K., Ohsumi Y., Inagaki F. (2008) In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. J. Biol. Chem. 283, 1921–1928 [DOI] [PubMed] [Google Scholar]
- 4. Taherbhoy A. M., Tait S. W., Kaiser S. E., Williams A. H., Deng A., Nourse A., Hammel M., Kurinov I., Rock C. O., Green D. R., Schulman B. A. (2011) Atg8 transfer from Atg7 to Atg3. A distinctive E1-E2 architecture and mechanism in the autophagy pathway. Mol. Cell 44, 451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Noda N. N., Satoo K., Fujioka Y., Kumeta H., Ogura K., Nakatogawa H., Ohsumi Y., Inagaki F. (2011) Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol. Cell 44, 462–475 [DOI] [PubMed] [Google Scholar]
- 6. Nakatogawa H., Ichimura Y., Ohsumi Y. (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178 [DOI] [PubMed] [Google Scholar]
- 7. Levine B., Kroemer G. (2008) Autophagy in the pathogenesis of disease. Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. White E. J., Martin V., Liu J. L., Klein S. R., Piya S., Gomez-Manzano C., Fueyo J., Jiang H. (2011) Autophagy regulation in cancer development and therapy. Am. J. Cancer Res. 1, 362–372 [PMC free article] [PubMed] [Google Scholar]
- 9. White E., DiPaola R. S. (2009) The double-edged sword of autophagy modulation in cancer. Clin. Cancer Res. 15, 5308–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shen S., Kepp O., Kroemer G. (2012) The end of autophagic cell death? Autophagy 8, 1–3 [DOI] [PubMed] [Google Scholar]
- 11. Kroemer G., Levine B. (2008) Autophagic cell death. The story of a misnomer. Nat. Rev. Mol. Cell Biol. 9, 1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kroemer G., Mariño G., Levine B. (2010) Autophagy and the integrated stress response. Mol Cell 40, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maiuri M. C., Zalckvar E., Kimchi A., Kroemer G. (2007) Self-eating and self-killing. Crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 8, 741–752 [DOI] [PubMed] [Google Scholar]
- 14. Pattingre S., Tassa A., Qu X., Garuti R., Liang X. H., Mizushima N., Packer M., Schneider M. D., Levine B. (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–939 [DOI] [PubMed] [Google Scholar]
- 15. Rubinstein A. D., Eisenstein M., Ber Y., Bialik S., Kimchi A. (2011) The autophagy protein atg12 associates with antiapoptotic bcl-2 family members to promote mitochondrial apoptosis. Mol. Cell 44, 698–709 [DOI] [PubMed] [Google Scholar]
- 16. Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 17. Yousefi S., Perozzo R., Schmid I., Ziemiecki A., Schaffner T., Scapozza L., Brunner T., Simon H. U. (2006) Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 8, 1124–1132 [DOI] [PubMed] [Google Scholar]
- 18. Lee J. S., Li Q., Lee J. Y., Lee S. H., Jeong J. H., Lee H. R., Chang H., Zhou F. C., Gao S. J., Liang C., Jung J. U. (2009) FLIP-mediated autophagy regulation in cell death control. Nat. Cell Biol. 11, 1355–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luo S., Rubinsztein D. C. (2010) Apoptosis blocks Beclin 1-dependent autophagosome synthesis. An effect rescued by Bcl-xL. Cell Death Differ. 17, 268–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cho D. H., Jo Y. K., Hwang J. J., Lee Y. M., Roh S. A., Kim J. C. (2009) Caspase-mediated cleavage of ATG6/Beclin-1 links apoptosis to autophagy in HeLa cells. Cancer Lett. 274, 95–100 [DOI] [PubMed] [Google Scholar]
- 21. Djavaheri-Mergny M., Maiuri M. C., Kroemer G. (2010) Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene 29, 1717–1719 [DOI] [PubMed] [Google Scholar]
- 22. Li H., Wang P., Sun Q., Ding W. X., Yin X. M., Sobol R. W., Stolz D. B., Yu J., Zhang L. (2011) Following cytochrome c release, autophagy is inhibited during chemotherapy-induced apoptosis by caspase-8-mediated cleavage of Beclin-1. Cancer Res. 71, 3625–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hou W., Han J., Lu C., Goldstein L. A., Rabinowich H. (2010) Autophagic degradation of active caspase-8. A crosstalk mechanism between autophagy and apoptosis. Autophagy 6, 891–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berry D. L., Baehrecke E. H. (2007) Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell 131, 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hou Y. C., Chittaranjan S., Barbosa S. G., McCall K., Gorski S. M. (2008) Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J. Cell Biol. 182, 1127–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yi C. H., Yuan J. (2009) The Jekyll and Hyde functions of caspases. Dev. Cell 16, 21–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Launay S., Hermine O., Fontenay M., Kroemer G., Solary E., Garrido C. (2005) Vital functions for lethal caspases. Oncogene 24, 5137–5148 [DOI] [PubMed] [Google Scholar]
- 28. Maelfait J., Beyaert R. (2008) Non-apoptotic functions of caspase-8. Biochem. Pharmacol. 76, 1365–1373 [DOI] [PubMed] [Google Scholar]
- 29. Burguillos M. A., Deierborg T., Kavanagh E., Persson A., Hajji N., Garcia-Quintanilla A., Cano J., Brundin P., Englund E., Venero J. L., Joseph B. (2011) Caspase signalling controls microglia activation and neurotoxicity. Nature 472, 319–324 [DOI] [PubMed] [Google Scholar]
- 30. Arama E., Bader M., Rieckhof G. E., Steller H. (2007) A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol. 5, e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li J., Brieher W. M., Scimone M. L., Kang S. J., Zhu H., Yin H., von Andrian U. H., Mitchison T., Yuan J. (2007) Caspase-11 regulates cell migration by promoting Aip1-Cofilin-mediated actin depolymerization. Nat. Cell Biol. 9, 276–286 [DOI] [PubMed] [Google Scholar]
- 32. Keller M., Rüegg A., Werner S., Beer H. D. (2008) Active caspase-1 is a regulator of unconventional protein secretion. Cell 132, 818–831 [DOI] [PubMed] [Google Scholar]
- 33. Kang T. B., Ben-Moshe T., Varfolomeev E. E., Pewzner-Jung Y., Yogev N., Jurewicz A., Waisman A., Brenner O., Haffner R., Gustafsson E., Ramakrishnan P., Lapidot T., Wallach D. (2004) Caspase-8 serves both apoptotic and nonapoptotic roles. J. Immunol. 173, 2976–2984 [DOI] [PubMed] [Google Scholar]
- 34. Huh J. R., Vernooy S. Y., Yu H., Yan N., Shi Y., Guo M., Hay B. A. (2004) Multiple apoptotic caspase cascades are required in nonapoptotic roles for Drosophila spermatid individualization. PLoS Biol. 2, E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Denecker G., Hoste E., Gilbert B., Hochepied T., Ovaere P., Lippens S., Van den Broecke C., Van Damme P., D'Herde K., Hachem J. P., Borgonie G., Presland R. B., Schoonjans L., Libert C., Vandekerckhove J., Gevaert K., Vandenabeele P., Declercq W. (2007) Caspase-14 protects against epidermal UVB photodamage and water loss. Nat Cell Biol. 9, 666–674 [DOI] [PubMed] [Google Scholar]
- 36. Carlile G. W., Smith D. H., Wiedmann M. (2004) Caspase-3 has a nonapoptotic function in erythroid maturation. Blood 103, 4310–4316 [DOI] [PubMed] [Google Scholar]
- 37. Ben Moshe T., Barash H., Kang T. B., Kim J. C., Kovalenko A., Gross E., Schuchmann M., Abramovitch R., Galun E., Wallach D. (2007) Role of caspase-8 in hepatocyte response to infection and injury in mice. Hepatology 45, 1014–1024 [DOI] [PubMed] [Google Scholar]
- 38. Leverrier S., Salvesen G. S., Walsh C. M. (2011) Enzymatically active single chain caspase-8 maintains T-cell survival during clonal expansion. Cell Death Differ. 18, 90–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murray T. V., McMahon J. M., Howley B. A., Stanley A., Ritter T., Mohr A., Zwacka R., Fearnhead H. O. (2008) A non-apoptotic role for caspase-9 in muscle differentiation. J. Cell Sci. 121, 3786–3793 [DOI] [PubMed] [Google Scholar]
- 40. Jeong H. S., Choi H. Y., Lee E. R., Kim J. H., Jeon K., Lee H. J., Cho S. G. (2011) Involvement of caspase-9 in autophagy-mediated cell survival pathway. Biochim. Biophys. Acta 1813, 80–90 [DOI] [PubMed] [Google Scholar]
- 41. Han J., Goldstein L. A., Hou W., Froelich C. J., Watkins S. C., Rabinowich H. (2010) Deregulation of mitochondrial membrane potential by mitochondrial insertion of granzyme B and direct Hax-1 cleavage. J. Biol. Chem. 285, 22461–22472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Han J., Goldstein L. A., Hou W., Rabinowich H. (2007) Functional linkage between NOXA and Bim in mitochondrial apoptotic events. J. Biol. Chem. 282, 16223–16231 [DOI] [PubMed] [Google Scholar]
- 43. Han J., Hou W., Lu C., Goldstein L. A., Stolz D. B., Watkins S. C., Rabinowich H. (2013) Interaction between Her2 and Beclin-1 proteins underlies a new mechanism of reciprocal regulation. J. Biol. Chem. 288, 20315–20325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 45. Han J., Goldstein L. A., Hou W., Gastman B. R., Rabinowich H. (2010) Regulation of mitochondrial apoptotic events by p53-mediated disruption of complexes between antiapoptotic Bcl-2 members and Bim. J. Biol. Chem. 285, 22473–22483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Han J., Hou W., Goldstein L. A., Lu C., Stolz D. B., Yin X. M., Rabinowich H. (2008) Involvement of protective autophagy in TRAIL resistance of apoptosis-defective tumor cells. J. Biol. Chem. 283, 19665–19677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Herrero-Martín G., Høyer-Hansen M., García-Garcia C., Fumarola C., Farkas T., López-Rivas A., Jäättelä M. (2009) TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 28, 677–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hou W., Han J., Lu C., Goldstein L. A., Rabinowich H. (2008) Enhancement of tumor-TRAIL susceptibility by modulation of autophagy. Autophagy 4, 940–943 [DOI] [PubMed] [Google Scholar]
- 49. Mills K. R., Reginato M., Debnath J., Queenan B., Brugge J. S. (2004) Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is required for induction of autophagy during lumen formation in vitro. Proc. Natl. Acad. Sci. U.S.A. 101, 3438–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hakem R., Hakem A., Duncan G. S., Henderson J. T., Woo M., Soengas M. S., Elia A., de la Pompa J. L., Kagi D., Khoo W., Potter J., Yoshida R., Kaufman S. A., Lowe S. W., Penninger J. M., Mak T. W. (1998) Differential requirement for caspase 9 in apoptotic pathways in vivo. Cell 94, 339–352 [DOI] [PubMed] [Google Scholar]
- 51. Kuida K., Haydar T. F., Kuan C. Y., Gu Y., Taya C., Karasuyama H., Su M. S., Rakic P., Flavell R. A. (1998) Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell 94, 325–337 [DOI] [PubMed] [Google Scholar]
- 52. Pandey U. B., Nie Z., Batlevi Y., McCray B. A., Ritson G. P., Nedelsky N. B., Schwartz S. L., DiProspero N. A., Knight M. A., Schuldiner O., Padmanabhan R., Hild M., Berry D. L., Garza D., Hubbert C. C., Yao T. P., Baehrecke E. H., Taylor J. P. (2007) HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 447, 859–863 [DOI] [PubMed] [Google Scholar]
- 53. Ding W. X., Ni H. M., Gao W., Yoshimori T., Stolz D. B., Ron D., Yin X. M. (2007) Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am. J. Pathol. 171, 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Saitoh T., Fujita N., Jang M. H., Uematsu S., Yang B. G., Satoh T., Omori H., Noda T., Yamamoto N., Komatsu M., Tanaka K., Kawai T., Tsujimura T., Takeuchi O., Yoshimori T., Akira S. (2008) Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 456, 264–268 [DOI] [PubMed] [Google Scholar]
- 55. Malladi S., Challa-Malladi M., Fearnhead H. O., Bratton S. B. (2009) The Apaf-1·procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J. 28, 1916–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Srinivasula S. M., Hegde R., Saleh A., Datta P., Shiozaki E., Chai J., Lee R. A., Robbins P. D., Fernandes-Alnemri T., Shi Y., Alnemri E. S. (2001) A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 410, 112–116 [DOI] [PubMed] [Google Scholar]
- 57. Hu Q., Wu D., Chen W., Yan Z., Shi Y. (2013) Proteolytic processing of the caspase-9 zymogen is required for apoptosome-mediated activation of caspase-9. J. Biol. Chem. 288, 15142–15147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bratton S. B., Salvesen G. S. (2010) Regulation of the Apaf-1-caspase-9 apoptosome. J. Cell Sci. 123, 3209–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gyrd-Hansen M., Farkas T., Fehrenbacher N., Bastholm L., Høyer-Hansen M., Elling F., Wallach D., Flavell R., Kroemer G., Nylandsted J., Jäättelä M. (2006) Apoptosome-independent activation of the lysosomal cell death pathway by caspase-9. Mol. Cell. Biol. 26, 7880–7891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hao Y., Sekine K., Kawabata A., Nakamura H., Ishioka T., Ohata H., Katayama R., Hashimoto C., Zhang X., Noda T., Tsuruo T., Naito M. (2004) Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat. Cell Biol. 6, 849–860 [DOI] [PubMed] [Google Scholar]
- 61. Morizane Y., Honda R., Fukami K., Yasuda H. (2005) X-linked inhibitor of apoptosis functions as ubiquitin ligase toward mature caspase-9 and cytosolic Smac/DIABLO. J. Biochem. 137, 125–132 [DOI] [PubMed] [Google Scholar]