Background: The precise roles for Rictor/Sin1 complexes in IFN signaling remain to be defined.
Results: Targeted disruption Rictor/Sin1 results in defects in activation of elements of Stat pathways. These proteins are required for IFN antineoplastic effects on malignant erythroid precursors.
Conclusion: Rictor/Sin1 play critical roles in IFN signaling.
Significance: This study provides evidence for a key mechanism for gene regulation associated with generation of IFN antineoplastic responses.
Keywords: Akt PKB, Antiviral Agents, Cell Signaling, Cytokines/Interferon, Gene Regulation, Interferon, Signal Transduction
Abstract
We provide evidence that type I IFN-induced STAT activation is diminished in cells with targeted disruption of the Rictor gene, whose protein product is a key element of mTOR complex 2. Our studies show that transient or stable knockdown of Rictor or Sin1 results in defects in activation of elements of the STAT pathway and reduced STAT-DNA binding complexes. This leads to decreased expression of several IFN-inducible genes that mediate important biological functions. Our studies also demonstrate that Rictor and Sin1 play essential roles in the generation of the suppressive effects of IFNα on malignant erythroid precursors from patients with myeloproliferative neoplasms. Altogether, these findings provide evidence for critical functions for Rictor/Sin1 complexes in type I IFN signaling and the generation of type I IFN antineoplastic responses.
Introduction
IFNs are cytokines with key and central roles in innate immunity, immune modulation, and immune surveillance against neoplasia (1, 2). These cytokines exhibit important antiviral, growth inhibitory, and pro-apoptotic properties and have been used extensively over the years in clinical settings for the treatment of leukemias, neurologic disorders, and viral infections (3–7). Despite the fact that new effective targeted therapies have replaced IFNs in the management of certain diseases, these cytokines still play key roles in the treatment of certain malignancies, such as myeloproliferative neoplasms (8, 9). Notably, there has recently been renewed enthusiasm for the clinical use of IFNs in malignancies, taking advantage of the emerging better understanding of their biological functions and the pathophysiological mechanisms in which IFNs are involved (7, 10).
The IFNs are classified into three major classes, types I, II, and III (3, 7, 10–12). The different classes of IFNs bind to distinct cell surface receptors, classified as type I, II and III IFN receptors (3, 7, 10–12). The binding of IFNs to their corresponding receptors induces conformational changes in the receptor structures, leading to activation of associated JAK kinases and downstream engagement of STAT transcription factors, which form homo- or heterodimers and translocate to the nucleus (13–17). STAT complexes then bind to the promoters of interferon-stimulated genes (ISGs)2 to initiate transcription (13–17). The activation of STATs has served as an important paradigm for the transduction of IFN signals and initiation of transcription of ISGs. The activities of different STATs are regulated by various post-translational modifications like tyrosine/serine phosphorylation, acetylation, and sumoylation (16, 17) and/or by interactions with other proteins like PIAS proteins (18). IFN-dependent nuclear translocation of STATs requires tyrosine phosphorylation, and the nuclear import is facilitated by importins (19), whereas the transcriptional activity of STATs may also depend on chromatin binding (20).
In addition to classical Jak-STAT pathways, there are several cellular signaling cascades engaged by IFN receptors, whose coordinated functions are required for optimal production of ISG products, including the p38 MAPK pathway (21), the PI3K-Akt pathway (22, 23), mTORC1 and mTORC2 cascades (24, 25), and pathways involving members of the PKC family of proteins (26, 27). Engagement of these pathways complements the function of Jak-STAT pathways, either by providing accessory signals for optimal transcriptional activation of ISGs (21, 26, 27) or by promoting mRNA translation of these genes and ultimate production of their protein products (22–25).
There is recent evidence implicating mTORC2 complexes in mRNA translation of ISGs and the generation of the antiviral effects of type I IFNs (25). In efforts to better define the role of the mTOR pathway and, in particular mTORC2 complexes in the induction of IFN-responses, we performed studies to examine whether there is cross-talk between components of the mTORC2 cascade and functional activation of IFN-engaged STAT proteins. Our studies demonstrate that in cells with targeted disruption of the Rictor gene, there is a reduction in IFN-inducible STAT2 tyrosine phosphorylation and ISGF3-DNA complex formation, as well as diminished phosphorylation of STAT1 on serine 727, an event required for the full transcriptional activity of STAT1 (4, 13, 15). Using gene microarray studies, we identified several genes involved in the generation of antiviral and antiproliferative responses, whose expression is reduced in the absence of Rictor. We also found that knockdown of Rictor or Sin1 results in reversal of the inhibitory effects of IFNα on malignant hematopoietic precursors from patients with polycythemia vera, establishing an unexpected critical role for these proteins in the generation of the antineoplastic effects of type I IFNs.
MATERIALS AND METHODS
Cells and Reagents
Immortalized Rictor+/+ (Rictorex3cond/w) and Rictor−/− (Rictorex3del/ex3del) MEFs provided by Dr. Mark Magnuson (28) and immortalized Sin1+/+ and Sin1−/− MEFs (29) were maintained in DMEM supplemented with 10% FBS and gentamycin. U937 cells were from ATCC. Control shRNA or Rictor shRNA lentivirus infected U937 cells have been described previously (25) and were maintained in RPMI supplemented with 10% FBS, gentamycin, and puromycin. Rictor and Sin1 specific siRNAs were purchased from Dharmacon. Microarray chips were from Illumina. The antibodies against phosphorylated forms of STAT1 on serine 727, tyrosine 701, and phosphotyrosine-Tyk2 were from Cell Signaling (Danvers, MA). Antibodies against STAT1, human STAT2, tubulin, and Hsp90 were from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-mouse STAT2 antibody was from Abcam, and the anti-Tyk2 antibody was from BD Biosciences. An antibody against Rictor was from Bethyl Laboratories. Antibodies against Tyr(P)689 STAT2, Sin1, and GAPDH were from Millipore (Temecula, MA).
Immunoblotting
MEFs were starved overnight in DMEM supplemented with 0.5% FBS, followed by treatment with 5–10 × 103 IU/ml of mouse IFNα, as indicated. U937 cells were treated with 5–10 × 103 IU/ml of human IFNα. Following treatment, cells were washed in PBS, and lysed in phosphorylation lysis buffer supplemented with protease and phosphatase inhibitors. Equal protein aliquots were resolved by SDS-PAGE and processed for immunoblotting, as in our previous studies (22–25).
Electrophoretic Mobility Shift Assays
Gel shift and supershift assays were performed as in previous studies (27). Briefly, Rictor+/+ and Rictor−/− MEFs were either left untreated or treated with mouse IFNα for 15 min. The nuclear extracts were incubated with 32P-labeled synthetic ISRE (5′-AGCTTCCCTTCTGAGGAAACGAAACCA) oligonucleotides, and the protein DNA complexes were resolved by native PAGE. For supershift experiments, the extracts were incubated with an anti-STAT1 antibody (Millipore) or nonimmune rabbit IgG (Jackson ImmunoResearch Laboratories), used as control. The DNA-protein complexes were visualized by autoradiography.
Gene Expression Microarrays and Data Analysis
Rictor+/+ and Rictor−/− MEFs were treated with 2.5 × 103 IU/ml of mouse IFNα for 24 h in DMEM supplemented with 0.5% FBS, as indicated. RNA was isolated using RNAeasy RNA isolation kit from Qiagen. The quality of RNA was analyzed using Agilent 2100 Bioanalyzer. cRNA synthesis, MouseWG-6 v2.0 Expression BeadChip hybridization (Illumina), washing, and staining were performed as per the manufacturer instructions. Arrays were scanned on Illumina BeadStation 500. All array data were deposited in the GEO database (GEO identifier GSE47896). Probe average intensity signal was calculated with BeadStudio without background correction. Raw data were analyzed with Bioconductor using the one ChannelGUI package (30). Average probe intensities were log2-transformed and normalized by the Lowess method (31). All experimental groups were filtered to have an interquartile range for each probe ≥0.25. Differential expression of wild-type versus wild-type + IFN and knock-out versus knock-out + IFN were assessed by using an empirical Bayes method (32) together with a false discovery rate correction of the p value ≤ 0.05 (33). Hierarchical clustering was done using MeV v4.4.1 software. Functional analysis was performed using IPA 2014.
Quantitative RT-PCR
Rictor+/+ and Rictor−/− or Sin1+/+ and Sin1−/− MEFs were treated with 2.5 × 103 IU/ml of mouse IFNα for 24 h, and RNA was isolated as described above. 1 μg of total RNA was reverse transcribed using an Omniscript RT-PCR kit from Qiagen and oligo(dT) from Invitrogen. Real time quantitative PCR was carried using FAM-labeled primers and probes for ISG54, Daxx, Pyhin1, Slfn2, OAS2, Mx1, and PHF11 from Applied Biosystems (Foster City, CA). β-Actin was used for normalization. The mRNA amplification was calculated using ΔΔCt method as described previously (23, 24), and data were plotted as fold change over untreated samples.
Hematopoietic Colony Forming Assays
Peripheral blood was collected from patients with polycythemia vera after obtaining informed consent, approved by the Northwestern University Institutional Review Board. Hematopoietic colony formation of early erythroid (burst forming unit-erythroid) progenitors was assessed by colonogenic assays in methylcellulose, as in our previous studies (34).
RESULTS
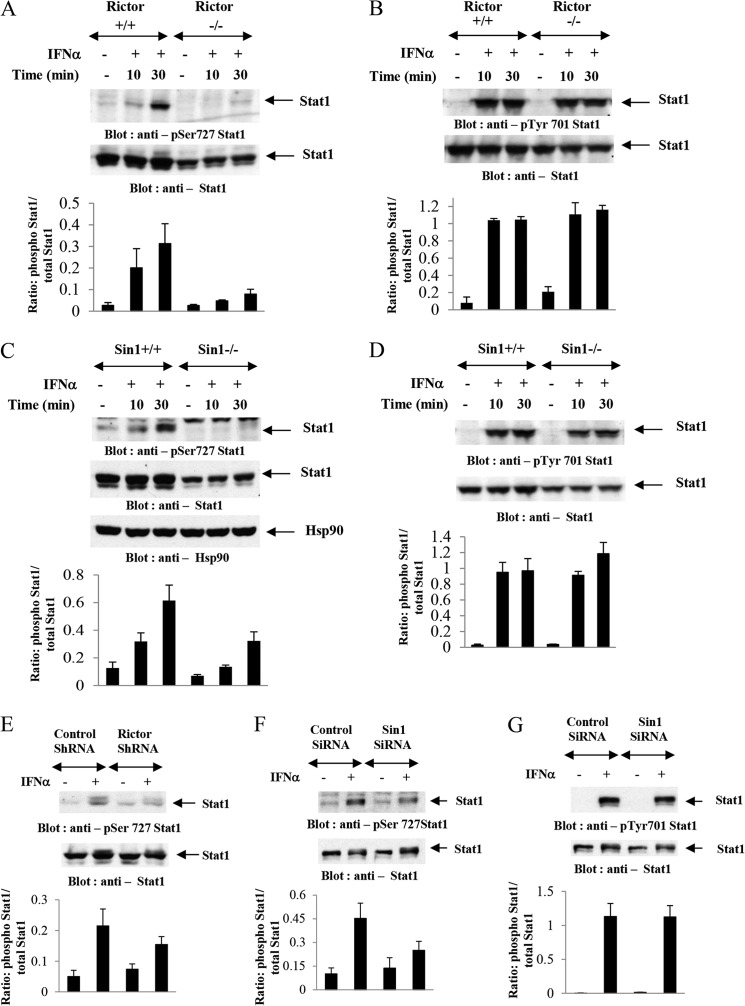
In previous work we provided evidence that targeted knock-out of Rictor or mLST8 genes results in defective ISG expression and diminished antiviral responses (25). Because our findings demonstrated that IFNα-inducible ISRE promoter activity requires Rictor and mLST8 expression (25), we performed studies to define potential mechanisms by which mTORC2 complexes may modulate IFN-dependent transcriptional activation of ISGs. At the outset we examined the effects of targeted disruption of the Rictor gene on phosphorylation of STAT1. Extensive previous work has established that phosphorylation of STAT1 on tyrosine 701 is required for STAT dimerization (15, 35), whereas phosphorylation on serine 727 is required for full transcriptional activation of the protein (4, 13, 15). As expected, treatment of wild-type MEFs with IFNα resulted in phosphorylation of STAT1 on serine 727 (Fig. 1A). This phosphorylation was decreased in Rictor−/− MEFs (Fig. 1A), suggesting a defect in STAT1 serine kinase activity in the absence of the Rictor gene. It should be noted that some decrease in the protein levels of STAT1 was noticeable in Rictor−/− cells, but this relatively modest decrease does not account for the substantial reduction in Ser727 STAT1 phosphorylation (Fig. 1A). Notably, phosphorylation of STAT1 on Tyr701 was intact in the absence of Rictor (Fig. 1B). Similarly, IFNα inducible phosphorylation of STAT1 on Ser727 was diminished in Sin1−/− MEFs (Fig. 1C), whereas phosphorylation of STAT1 on Tyr701 was IFN-inducible (Fig. 1D).
FIGURE 1.
Type 1 IFN-induced phosphorylation of Ser STAT1 is Rictor and Sin1 dependent. A and B, Rictor+/+ or Rictor−/− MEFs were treated with IFNα for the indicated times, and equal amounts of protein were processed for Western blotting with anti-Ser727-STAT1 (A) or anti-Tyr701-STAT1 (B) antibodies. The blots in respective top panels were stripped and probed with an anti-STAT1 antibody. The signals for phospho-STAT1 and total STAT1 from three independent experiments (including the blots shown) were quantitated by densitometry, and the intensity of phospho-STAT1 relative to STAT1 was calculated. The data are expressed as means of ratios of phospho-STAT1/STAT1 ± S.E. for each experimental condition. C and D, Sin1+/+ or Sin1−/− MEFs were treated with IFNα for the indicated times, and equal amounts of protein were processed for Western blotting with anti-Ser727-STAT1 (C) or anti-Tyr701-STAT1 (D) antibodies. The blots in respective top panels were stripped and probed with an anti-STAT1 antibody or an anti-HSP90 antibody, as indicated. The signals for phospho-STAT1 and total STAT1 from three independent experiments (including the blots shown) were quantitated by densitometry and the intensity of phospho-STAT1 relative to STAT1 was calculated. The data are expressed as means of ratios of phospho-STAT1/STAT1 ± S.E. for each experimental condition. E, U937 cells stably infected with control shRNA or Rictor shRNA were treated with human IFNα as indicated. Equal protein aliquots were processed for immunoblotting with anti-Ser727-STAT1 antibody (top panel). The same blot was stripped and probed with an anti-STAT1 antibody (middle panel). The signals for phospho-STAT1 and total STAT1 from three independent experiments (including the blots shown) were quantitated by densitometry, and the intensity of phospho-STAT1 relative to STAT1 was calculated. The data are expressed as means of ratios of phospho-STAT1/STAT1 ± S.E. for each experimental condition (bottom panel). F and G, U937 cells transiently transfected with control siRNA or Sin1 siRNA were treated with human IFNα as indicated. Equal protein aliquots were processed for immunoblotting with anti-Ser727-STAT1 (F, top panel) or anti-Tyr701-STAT1 (G, top panel) antibody, as indicated. The respective blots were stripped and probed with an anti-STAT1 antibody (F and G, middle panels), as indicated. The signals for phospho-STAT1 and total STAT1 from three independent experiments (including the blots shown) were quantitated by densitometry, and the intensity of phospho-STAT1 relative to STAT1 was calculated. The data are expressed as means of ratios of phospho-STAT1/STAT1 ± S.E. for each experimental condition (F and G, bottom panels).
In subsequent studies we sought to determine whether shRNA-mediated knockdown of Rictor in malignant hematopoietic cells also results in diminished STAT1-Ser727 phosphorylation. U937 myelomonocytic leukemia cells were stably infected with either control shRNA or Rictor shRNA, using lentiviral infection. IFN induction of Ser727 STAT1 phosphorylation was found to be selectively impaired in Rictor shRNA-expressing, but not control shRNA-expressing, cells (Fig. 1E). Similarly, Sin1 knockdown also resulted in decreased IFN-inducible STAT1 Ser727 phosphorylation (Fig. 1F). In contrast, IFN-inducible tyrosine 701 STAT1 phosphorylation was unaffected by Sin1 knockdown in U937 cells (Fig. 1G).
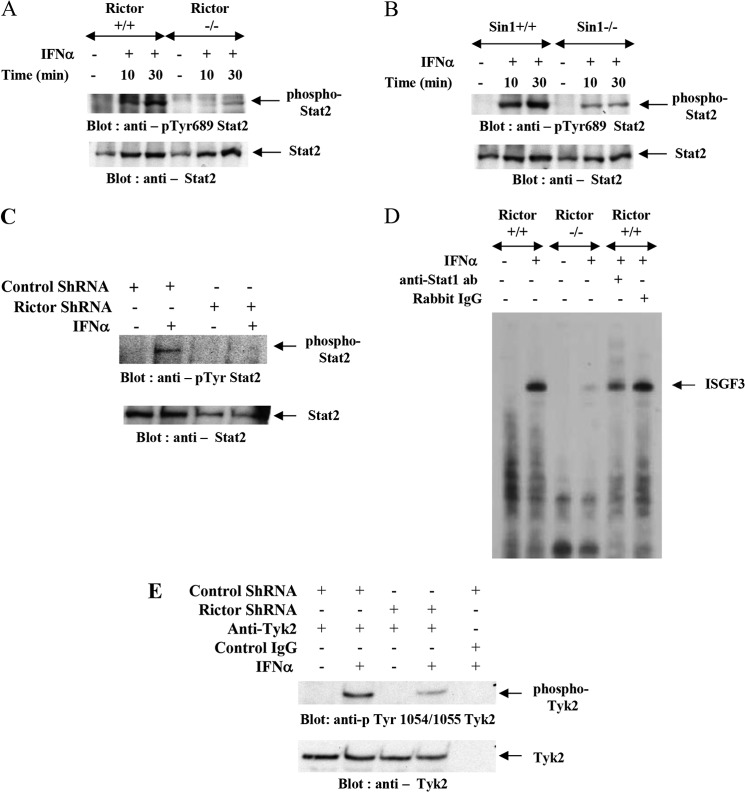
We also assessed the effect of Rictor or Sin1 knock-out on IFN-induced STAT2 tyrosine phosphorylation. Phosphorylation of STAT2 on tyrosine 689 plays a role in formation of the ISGF3 transcription complex and IFN-induced transcription of genes (36, 37). Treatment with IFN-induced STAT2 tyrosine phosphorylation in Rictor+/+ or Sin1+/+ MEFs, but this induction of STAT2 phosphorylation was diminished in Rictor−/− and Sin1−/− MEFs (Fig. 2, A and B). Similarly, U937 cells stably transfected with Rictor shRNA exhibit a decrease in IFN-induced phosphorylation of STAT2 on tyrosine (Fig. 2C). We further assessed the effects of targeted disruption of Rictor on IFN-dependent STAT-DNA complex formation. Rictor+/+ and Rictor−/− MEFs were treated with IFNα, and the formation of ISGF3-DNA complexes was analyzed by EMSAs. As expected, we observed IFN-induced ISGF3 binding to ISRE in Rictor+/+ cells, but no such complexes were evident in Rictor−/− cells (Fig. 2D). It should be noted that IRF9 protein levels were not affected by Rictor knock-out (data not shown). In U937 cells in which Rictor was knocked down, there were also diminished levels of IFN-induced Tyk2 tyrosine phosphorylation when compared with control cells (Fig. 2E).
FIGURE 2.
IFN-induced STAT2 phosphorylation and ISGF3 complex formation is Rictor- and Sin1-dependent. A, Rictor+/+ or Rictor−/− MEFs were treated with IFNα for the indicated times, and equal amounts of protein were processed for Western blotting with anti-Tyr689-STAT2 antibody (top panel). Equal amounts of protein were resolved in parallel on the same gel and processed for immunoblotting with anti-STAT2 antibody (bottom panel). B, Sin1+/+ or Sin1−/− MEFs were treated with IFNα for the indicated times, and equal amounts of protein were processed for Western blotting with anti-Tyr689-STAT2 antibody (top panel). Equal amounts of protein were resolved in parallel on the same gel and processed for immunoblotting with anti-STAT2 antibody (bottom panel). C, U937 cells stably infected with control shRNA or Rictor shRNA were treated with human IFNα as indicated. Equal protein aliquots were processed for immunoblotting with anti-Tyr-STAT2 antibody (top panel). The same blot was stripped and probed with an anti-STAT2 antibody (bottom panel). D, nuclear extracts were prepared from untreated or IFNα-treated Rictor+/+ and Rictor−/− MEFs and incubated with 32P-labeled synthetic ISRE. The protein-DNA complexes were resolved on a native PAGE, and complexes were detected by autoradiography. For antibody supershift experiments, protein extracts were incubated with the specified antibody or control nonimmune rabbit IgG, as indicated. E, U937 cells stably infected with control shRNA or Rictor shRNA were treated with human IFNα for 10 min. Equal protein aliquots were processed for immunoprecipitation with anti-Tyk2 or control IgG antibody as indicated. The immunoprecipitated proteins were processed for immunoblotting with anti-phospho-Tyk2 antibody (top panel). The same blot was stripped and probed with anti-Tyk2 antibody (bottom panel).
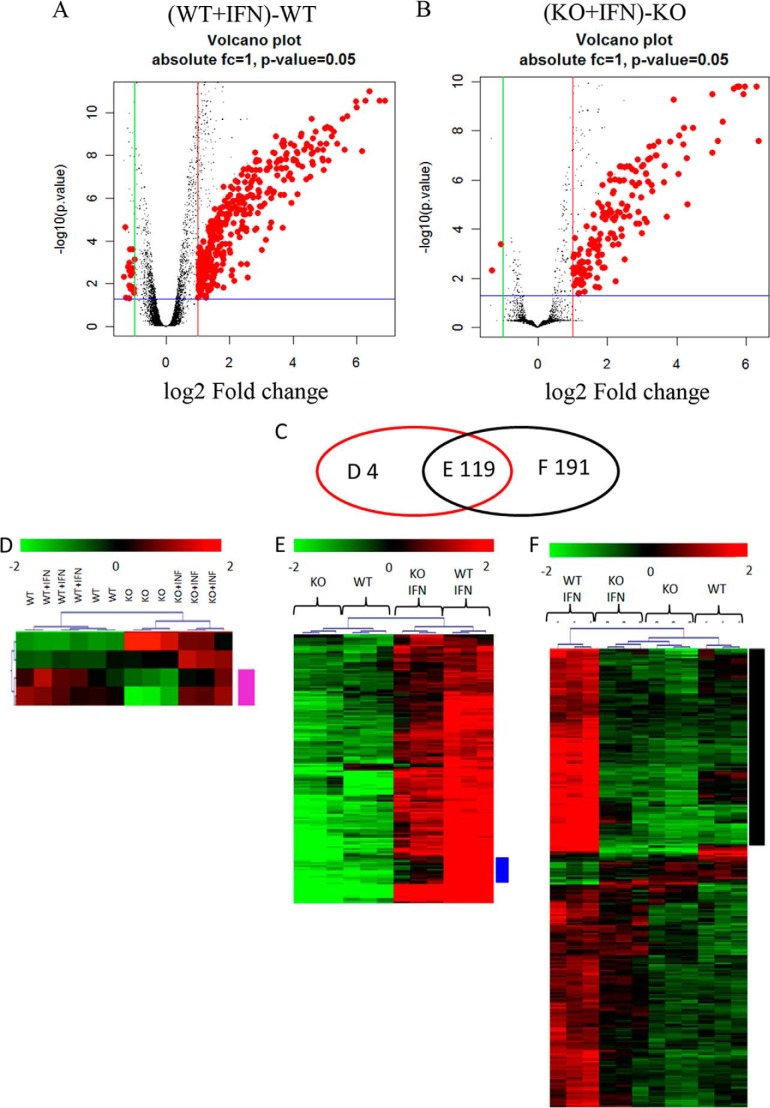
Altogether, our studies indicate that Rictor/Sin1 complexes are required for serine phosphorylation of STAT1 and the formation of type I IFN-inducible STAT-DNA-binding complexes to initiate ISG transcription. To define the changes in ISG expression in the absence of Rictor, we next used gene expression microarrays to identify genes differentially induced by IFNs in Rictor+/+ and Rictor−/− cells. Rictor+/+ and Rictor−/− MEFs were either left untreated or treated with IFNα for 24 h, and gene expression analysis was then carried out by binding cRNA to mouse WG6 v2.0 Illumina bead chips. Analysis of data from three independent experiments revealed that 310 genes were differentially expressed in IFN-treated Rictor+/+ cells (Fig. 3A), and 123 genes were differentially expressed in IFN-treated Rictor−/− cells (Fig. 3B). Of these, 81 genes were found to be induced only in Rictor+/+ MEFs, but not in Rictor−/− cells (Fig. 3F, black cluster). Furthermore, 13 distinct genes were induced in both Rictor+/+ and Rictor−/− MEFs (log2 fold change ≥ 1), but the induction levels were much higher in Rictor+/+ MEFs (Fig. 3E, blue cluster); i.e., 94 IFN-inducible genes are differentially expressed in Rictor+/+ MEFs (supplemental Table S1). Most of the genes that were differentially up-regulated by IFN treatment in Rictor+/+ MEFs could be classified as genes that play important roles in innate immunity, antiviral, and antimicrobial responses, as well as mediators of growth inhibitory and/or pro-apoptotic effects.
FIGURE 3.
Differential expression of IFN regulated genes in the presence or absence of Rictor. Rictor+/+ (WT) and Rictor−/− MEFs were either left untreated or treated with IFNα for 24 h. The transcription profiles of Rictor+/+ and Rictor−/− cells were compared with transcription profiles of IFN-treated Rictor+/+ and Rictor−/− cells in three independent experiments, using MouseWG-6 v2.0 Illumina bead chips. Following normalization and removal of genes whose expression was absent or unchanged, differential gene expression was assessed with the regularized t test by comparing WT versus WT + IFN and KO versus KO + IFN, using a false discovery rate of ≤0.05 (33), together with an absolute log2 fold change threshold of ≥1. A and B, volcano plots of probes found differentially expressed after IFN treatment are shown. 404 probes (310 genes) were differentially expressed in IFN-treated Rictor+/+ cells (A). 163 probes (123 genes) were differentially expressed in IFN-treated Rictor−/− cells (B). Blue horizontal lines refer to a p value threshold of 0.05. Green and red vertical lines refer respectively to a −/+ log2 fold change threshold of 1. C, Venn diagram of the overlaps existing between genes found differentially expressed in Rictor−/− (red ellipse) and in Rictor+/+ (black ellipse) upon treatment with IFN. D, hierarchical clustering of probes differentially expressed only in Rictor−/− cells. The basal level of gene expression in Rictor−/− cells is different from that of Rictor+/+ cells, and in two of four genes, the treatment with IFN brings the expression level in Rictor−/− cells comparable to that observed in Rictor+/+ cell in the absence of IFN (violet gene cluster). E, hierarchical clustering of probes differentially expressed in both Rictor−/− and Rictor+/+ cells upon IFN treatment. The differences in the effects of IFN treatment are not dramatic between Rictor−/− and Rictor+/+ cells, except for the set of transcripts in the blue gene cluster, which are characterized by a less efficient IFN-driven expression in Rictor−/− cells (supplemental Table S1, 13 genes). F, hierarchical clustering of probes differentially expressed only in IFN-treated Rictor+/+ cells. The black gene cluster contains only genes that cannot be activated in Rictor−/− cells upon IFN treatment (supplemental Table S1, 81 genes).
We also constructed two functional networks, using IPA 2014 software, to identify relationships that may exist among the 94 genes that are up-regulated upon IFN treatment in Rictor+/+ MEFs. To the networks we also added STAT1 (supplemental Fig. S1) or STAT2 (supplemental Fig. S2). STAT1 and 2 were included to observe the effects of their functional association with Rictor-dependent genes. Notably, STAT1 shows a higher connectivity to genes selectively activated by IFN treatment in Rictor+/+ MEFs compared with STAT2.
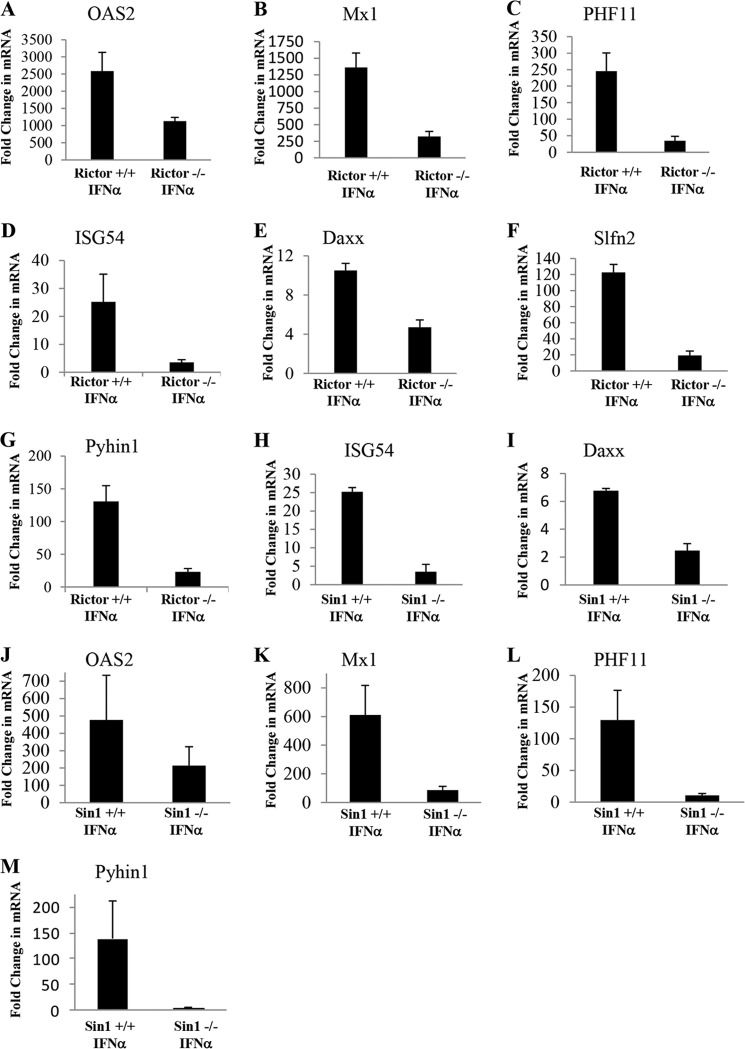
The expression levels of several genes identified as differentially expressed in Rictor+/+ cells and involved in antiviral responses, such as OAS2, Mx1 (38), and PHF11 (39), were confirmed by quantitative real time RT-PCR (Fig. 4, A–C). The identification of Rictor dependence of their expression is consistent with our previous studies, demonstrating that Rictor expression is essential for the generation of IFN-dependent antiviral responses (25). Importantly, several genes involved in IFN-inducible growth inhibitory and/or pro-apoptotic responses, such as ISG54 (40), Daxx (41), and Slfn2 (42), were also induced by IFNα to a greater extent in Rictor+/+ MEFs as compared with Rictor−/− MEFs (Fig. 4, D–F). Similarly, Pyhin1, a gene belonging to the HIN200 family of IFN-inducible proteins that have roles in the control of cell cycle progression, cell differentiation, apoptosis, and tumor suppression (43), was preferentially expressed in Rictor+/+ cells, as compared with Rictor−/− MEFs (Fig. 4G).
FIGURE 4.
Requirement of Rictor and/or Sin1 for OAS2, Mx1, Phf11, Isg54, Daxx, Slfn2, and Pyhin1 expression. A–G, Rictor+/+ or Rictor−/− MEFs were treated with IFNα as indicated, and total RNA was isolated. Expression levels of the indicated genes were determined by real time RT-PCR, using β-actin for normalization. The data are expressed as fold change over untreated samples and represent means ± S.E. of six independent experiments for B–G and four experiments for A. H–M, Sin1+/+ or Sin1−/− MEFs were treated with IFNα for 24 h as indicated, and total RNA was isolated. Expression of Isg54, Daxx, OAS2, Mx1, PHF11, and Pyhin1 genes was determined by real time RT-PCR, using β-actin for normalization. The data are expressed as fold change over untreated samples and represent means ± S.E. of three experiments for Isg54 and Daxx and two experiments for OAS2, Mx1, PHF11, and Pyhin1.
To determine whether targeted disruption of the Sin1 gene has similar effects on transcription of IFN-induced genes, Sin1+/+ and Sin1−/− MEFs were treated with IFNα for 24 h, and mRNA expression of ISG54, Daxx, OAS2, Mx1, PHF11, and Pyhin1 was assessed by quantitative real time RT-PCR. As in the case of Rictor−/− cells, there was decreased IFN-dependent induction of ISG54, Daxx, OAS2, Mx1, PHF11, and Pyhin1 mRNA in Sin1−/− MEFs, as compared with Sin1+/+ MEFs (Fig. 4, H–M), definitively establishing a requirement for Rictor/Sin1 complexes in transcriptional activation of key ISGs involved in pro-apoptotic/growth inhibitory responses.
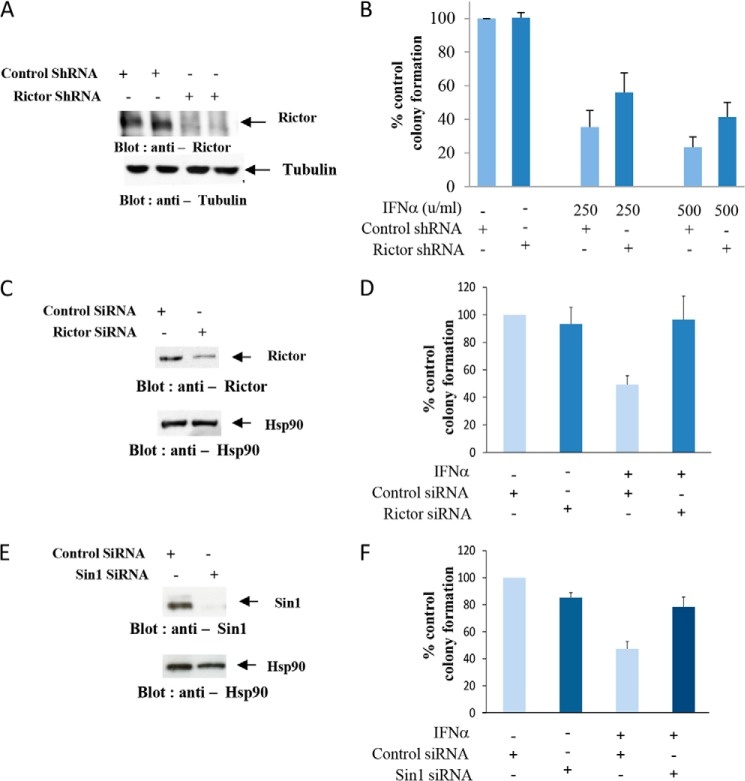
In subsequent studies, we sought to directly define the functional significance of Rictor/Sin1 complexes in the generation of IFN-dependent anti-leukemic responses. For this series of experiments, the effects of Rictor knockdown on IFN-dependent, U937-derived, leukemic progenitor colony formation was assessed. Rictor expression was knocked down in U937 cells by lentiviral transduced Rictor shRNA (Fig. 5A). The inhibitory effects of IFNα against primitive leukemic progenitors (CFU-L) were partially reversed in cells with diminished Rictor expression (Fig. 5B). Studies were also carried out to determine the effects of Rictor/Sin1 complexes in the generation of IFN-responses in primary malignant precursors from patients with polycythemia vera. To this end, we used transient transfection with Rictor- or Sin1-validated siRNAs. As shown in studies using U937 cells, these siRNAs clearly decreased expression of Rictor and Sin1 proteins, respectively (Fig. 5, C and E). As anticipated, treatment with IFNα suppressed malignant erythroid (BFU-E) colony formation from primary polycythemia vera patient samples (Fig. 5, D and F). These inhibitory effects were partially reversed by knockdown of Rictor (Fig. 5D) or Sin1 (Fig. 5F), establishing critical and essential roles for these components of mTORC2 complexes in the generation of the antineoplastic effects of IFNα in myeloproliferative neoplasms.
FIGURE 5.
Essential roles for Rictor and Sin1 in the generation of the antineoplastic effects of IFNα. A, cell lysates from U937 cells stably infected with lentiviral control shRNA or Rictor shRNA were resolved by SDS-PAGE and then immunoblotted with an anti-Rictor antibody or anti-tubulin antibody as control. B, U937 cells stably infected with control shRNA or Rictor shRNA were processed for clonogenic assays in methylcellulose in the presence or absence of different doses of human IFNα, and leukemic CFU-L colonies were scored. The data are expressed as percentages of control untreated cells and represent means ± S.E. from four independent experiments. C, cell lysates from U937 cells transiently transfected with control siRNA or Rictor siRNA were resolved by SDS-PAGE and then immunoblotted with an anti-Rictor antibody (upper panel). The same blot was stripped and reprobed with an anti-Hsp90 antibody, as a loading control (lower panel). D, PBMCs from polycythemia vera patients were transfected with either control siRNA or Rictor siRNA. These cells were incubated in clonogenic assays in methylcellulose in the presence or absence of human IFNα. Malignant erythroid BFU-E progenitors were scored after 14 days in culture, and the data are expressed as percentages of untreated control siRNA derived colony formation and represent means ± S.E. of four independent experiments. E, cell lysates from U937 cells transiently transfected with control siRNA or Sin1 siRNA were resolved by SDS-PAGE then immunoblotted with an anti-Sin1 antibody (upper panel). The same blot was stripped and reprobed with an anti-Hsp90 antibody as a loading control, as indicated (lower panel). F, PBMCs from polycythemia vera patients were transfected with either control siRNA or Sin1 siRNA. The cells were incubated in clonogenic assays in methylcellulose in the presence or absence of human IFNα. Μalignant BFU-E erythroid progenitors were scored after 14 days in culture, and the data are expressed as percentages of untreated control siRNA derived colony formation and represent means ± S.E. of three independent experiments.
DISCUSSION
Although Jak-STAT pathways in IFN signaling were the first to be discovered and precisely defined, in recent years it has become clear that IFNs activate additional signaling cascades in addition to Jak-STATs (13). These pathways are activated in parallel or shortly after type I IFN receptor engagement of Jak-STAT pathways and complement the functions and activities of Jak-STAT generated signals (13). There continues to be emerging evidence that this parallel engagement of complementary pathways either facilitates transcriptional activation of ISGs by modulating/optimizing the activities of STAT transcription factors and/or promotes mRNA translation of target genes after completion of the promoter-transcriptional activation process. For instance, engagement of the p38 MAPK pathway is essential for ISG-transcriptional activation and complements the function of STAT proteins (44). At the same time, activation of the p38 MAP kinase pathway by certain external stimuli can lead to ISG induction (45), suggesting the presence of a positive feedback regulatory loop in the IFN system, involving the p38 MAP kinase pathway (21). Similarly, members of the PKC family of proteins play important complementary roles in IFN-activated, STAT-mediated, transcription of ISGs (26, 27) and the generation of the pro-apoptotic and growth inhibitory effects of type I IFNs (46, 47). Moreover, the PI3K-Akt-mTOR signaling cascade has been shown to play a critical and essential role in mRNA translation of ISGs, and the generation of specific ISG products required for the induction of the biological effects of IFNs (22–25, 48, 49). Beyond mTOR pathways, signals downstream of MAPK pathways, specifically Mnk-mediated phosphorylation of the eukaryotic initiation factor 4E on serine 209, are also essential for mRNA translation of certain ISGs (50).
Two distinct mTOR complexes with distinguishing components, functions, and downstream effector elements exist, mTORC1 and mTORC2 (51). Both complexes share the mTOR kinase as their central catalytic subunit, the mTOR inhibitor Deptor, and the scaffold proteins tti1 and tel2 (51). mLST8 is also present in both mTORC1 and mTORC2, although its expression is only required for mTORC2 activity (51). Specific elements of mTORC1 complexes are Raptor, a unique scaffold protein whose function is required for mTORC1 assembly and substrate binding, and the protein PRAS40 (51, 52). Rictor and Sin1 are present in mTORC2 complexes, and they are both scaffold proteins important for the integrity and function of mTORC2 complexes, whereas Sin1 is also involved in the interaction of mTORC2 with its downstream effector kinase SGK1 (51).
In previous studies, we had provided evidence implicating both mTORC1 (24) and mTORC2 (25) in the expression of ISGs. Importantly, our studies had suggested specific roles for mTORC2 complexes in IFN signaling, as compared with growth factor/oncogene signaling, associated with regulatory effects on the AKT/mTORC1 axis (25). Notably, beyond reduced mRNA levels in polysomal fractions from IFN-treated cells, we had also found defective ISRE-driven transcription in Rictor knock-out cells, raising the possibility that mTORC2 complexes, or elements of mTORC2 complexes, play dual roles in IFN signaling, both by facilitating transcription and by promoting mRNA translation of ISGs (25). The demonstration of diminished antiviral responses in Rictor and mLST8 knock-out cells underscored the importance of Rictor-mediated signals in the biological properties of type I IFNs (25).
In the present study, we sought to define the requirement of Rictor complexes in ISG expression and to define the mechanisms involved in the process. Gene expression analysis demonstrated that a large group of ISGs with antiviral, antiproliferative, and/or pro-apoptotic properties require an intact Rictor-dependent signaling pathway for optimal expression. Importantly, our studies established that in Rictor and Sin1 knock-out MEFs, there are defects in activation of STAT pathways, including tyrosine phosphorylation of STAT2 on tyrosine 689, which is required for ISGF3 activation and also IFN-induced STAT1 phosphorylation on serine 727, a cellular event required for optimal STAT1-dependent ISG transcription (53). Such negative effects on the activation of elements of STAT pathways were also seen in MEFs with targeted deletion of the Sin1 gene and also in experiments in which either Rictor or Sin1 was transiently knocked down in malignant hematopoietic cells. Notably, Rictor or Sin1 expression was not essential for phosphorylation of STAT1 on Tyr701.
It remains to be determined whether the observed decreased STAT-DNA binding reflects a direct involvement of Rictor/Sin1 complexes or an indirect requirement reflecting altered mRNA translation of genes and production of proteins whose functions are essential for STAT transcriptional activity. It should be noted that there is previous evidence that genetic disruption of Rictor and Sin1 can result in decreased stability and expression of certain proteins such as Akt kinase and PKCα (54, 55), and our data also suggest a decrease in the total levels of STAT1 protein, although this is not sufficient to account for the decreased levels of STAT1 serine phosphorylation in response to IFNα treatment. It is possible that defective expression of a serine kinase(s) implicated in phosphorylation of STAT1 on serine 727 may account for the diminished IFN-inducible phosphorylation in the absence of Rictor or Sin1. There is evidence that PKCδ (27, 56), PKCϵ (57), and CDK8 (58) can act as STAT1 serine kinases during engagement of type I and/or II IFN receptors in different cell types. Although PKCα and PKCϵ protein levels are diminished in Rictor knock-out MEFS, PKCδ expression levels are unchanged (54, 55). It is therefore possible that changes in PKCα and PKCϵ expression associated with the absence of intact Rictor/Sin1 complexes may account for impaired serine 727 STAT1 phosphorylation. However, because there may be cross-compensation among different PKC isoforms, this remains to be precisely determined in detailed future studies. There are other reports that show that chromatin binding of STAT1 is required for phosphorylation of STAT1 on serine 727 (20), raising the possibility of additional mechanisms that may be involved. Because tyrosine phosphorylation of STAT2 and ISGF3 binding is defective in these cells, the observed decreased STAT1 serine phosphorylation may be a consequence of decreased chromatin binding. It should be noted that diminished STAT-DNA binding complex formation and altered ISG expression observed in the absence of Rictor and Sin1 reflect early defective engagement of IFN signals downstream of type I IFN receptor activation. Sin1 has been shown to interact with IFNAR2 (59–61). It is therefore possible that its localization at the type I IFN receptor level has a role in the functional activation of the receptor complex during IFN treatment and engagement of Jak kinases, required for tyrosine phosphorylation of STAT2 and subsequent formation of ISGF3 complexes.
Importantly, our studies provide the first direct evidence implicating Rictor/Sin1 complexes in the generation of the antineoplastic effects of IFNs. We observed that Rictor expression is essential for type I IFN-dependent expression of several genes with pro-apoptotic and antineoplastic properties, whereas knockdown of Rictor reversed the inhibitory effects of IFNα on U937-derived leukemic CFU-L precursors. Importantly, knockdown of either Rictor or Sin1 reversed the suppressive effects of IFNα on malignant erythroid progenitors from patients with polycythemia vera, a myeloproliferative neoplasm against which IFN exhibits major clinical activity (8, 9). This finding is somewhat surprising, because the mTOR pathway is perceived as an oncogenic pathway, because of its ability to influence initiation of translation of mRNAs encoding for pro-tumorigenic proteins that promote cell survival, angiogenesis, and metastasis and/or exhibit regulatory effects on cell cycle progression and metabolism (51). Our findings provide evidence for unique roles for elements of mTORC2 complexes in the IFN system, acting as novel mediators and essential components of transcriptional and translational regulation of genes that mediate IFN-inducible antineoplastic responses. Thus, depending on the stimulus and/or cellular context, engagement of Rictor/Sin1 complexes can result in specific and sometimes opposing effects as they relate to tumorigenesis.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA77816, CA155566, and CA161796. This work was also supported by a merit review grant from the Department of Veterans Affairs and by funds made available to ENF as a Tier 1 Canada Research Chair.

This article contains supplemental Table S1 and Figs. S1 and S2.
- ISG
- interferon-stimulated gene
- MEF
- mouse embryonic fibroblast
- mTORC
- mTOR complex.
REFERENCES
- 1. Dunn G. P., Koebel C. M., Schreiber R. D. (2006) Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 6, 836–848 [DOI] [PubMed] [Google Scholar]
- 2. Hervas-Stubbs S., Perez-Gracia J. L., Rouzaut A., Sanmamed M. F., Le Bon A., Melero I. (2011) Direct effects of type I interferons on cells of the immune system. Clin. Cancer Res. 17, 2619–2627 [DOI] [PubMed] [Google Scholar]
- 3. Borden E. C., Sen G. C., Uze G., Silverman R. H., Ransohoff R. M., Foster G. R., Stark G. R. (2007) Interferons at age 50. Past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6, 975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stark G. R. (2007) How cells respond to interferons revisited. From early history to current complexity. Cytokine Growth Factor Rev. 18, 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Platanias L. C. (2013) Interferons and their antitumor properties. J. Interferon Cytokine Res. 33, 143–144 [DOI] [PubMed] [Google Scholar]
- 6. Kotredes K. P., Gamero A. M. (2013) Interferons as inducers of apoptosis in malignant cells. J. Interferon Cytokine Res. 33, 162–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bracarda S., Eggermont A. M., Samuelsson J. (2010) Redefining the role of interferon in the treatment of malignant diseases. Eur. J. Cancer 46, 284–297 [DOI] [PubMed] [Google Scholar]
- 8. Tefferi A., Vainchenker W. (2011) Myeloproliferative neoplasms. Molecular pathophysiology, essential clinical understanding, and treatment strategies. J. Clin. Oncol. 29, 573–582 [DOI] [PubMed] [Google Scholar]
- 9. Kiladjian J. J., Mesa R. A., Hoffman R. (2011) The renaissance of interferon therapy for the treatment of myeloid malignancies. Blood 117, 4706–4715 [DOI] [PubMed] [Google Scholar]
- 10. Fuchs S. Y. (2013) Hope and fear for interferon. The receptor-centric outlook on the future of interferon therapy. J. Interferon Cytokine Res. 33, 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Levy D. E., Marié I. J., Durbin J. E. (2011) Induction and function of type I and III interferon in response to viral infection. Curr. Opin Virol 1, 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kotenko S. V. (2011) IFN-λs. Curr. Opin Immunol. 23, 583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Platanias L. C. (2005) Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386 [DOI] [PubMed] [Google Scholar]
- 14. Piehler J., Thomas C., Garcia K. C., Schreiber G. (2012) Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation. Immunol. Rev. 250, 317–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stark G. R., Darnell J. E., Jr. (2012) The JAK-STAT pathway at twenty. Immunity 36, 503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lim C. P., Cao X. (2006) Structure, function, and regulation of STAT proteins. Mol. Biosyst 2, 536–550 [DOI] [PubMed] [Google Scholar]
- 17. Decker T., Kovarik P. (1999) Transcription factor activity of STAT proteins. Structural requirements and regulation by phosphorylation and interacting proteins. Cell Mol. Life Sci. 55, 1535–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tahk S., Liu B., Chernishof V., Wong K. A., Wu H., Shuai K. (2007) Control of specificity and magnitude of NF-κB and STAT1-mediated gene activation through PIASy and PIAS1 cooperation. Proc. Natl. Acad. Sci. U.S.A. 104, 11643–11648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fagerlund R., Mélen K., Kinnunen L., Julkunen I. (2002) Arginine/lysine-rich nuclear localization signals mediate interactions between dimeric STATs and importin α5. J. Biol. Chem. 277, 30072–30078 [DOI] [PubMed] [Google Scholar]
- 20. Sadzak I., Schiff M., Gattermeier I., Glinitzer R., Sauer I., Saalmüller A., Yang E., Schaljo B., Kovarik P. (2008) Recruitment of Stat1 to chromatin is required for interferon-induced serine phosphorylation of Stat1 transactivation domain. Proc. Natl. Acad. Sci. U.S.A. 105, 8944–8949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Platanias L. C. (2003) Map kinase signaling pathways and hematologic malignancies. Blood 101, 4667–4679 [DOI] [PubMed] [Google Scholar]
- 22. Kaur S., Sassano A., Joseph A. M., Majchrzak-Kita B., Eklund E. A., Verma A., Brachmann S. M., Fish E. N., Platanias L. C. (2008) Dual regulatory roles of phosphatidylinositol 3-kinase in IFN signaling. J. Immunol. 181, 7316–7323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaur S., Sassano A., Dolniak B., Joshi S., Majchrzak-Kita B., Baker D. P., Hay N., Fish E. N., Platanias L. C. (2008) Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proc. Natl. Acad. Sci. U.S.A. 105, 4808–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaur S., Lal L., Sassano A., Majchrzak-Kita B., Srikanth M., Baker D. P., Petroulakis E., Hay N., Sonenberg N., Fish E. N., Platanias L. C. (2007) Regulatory effects of mammalian target of rapamycin-activated pathways in type I and II interferon signaling. J. Biol. Chem. 282, 1757–1768 [DOI] [PubMed] [Google Scholar]
- 25. Kaur S., Sassano A., Majchrzak-Kita B., Baker D. P., Su B., Fish E. N., Platanias L. C. (2012) Regulatory effects of mTORC2 complexes in type I IFN signaling and in the generation of IFN responses. Proc. Natl. Acad. Sci. U.S.A. 109, 7723–7728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Redig A. J., Sassano A., Majchrzak-Kita B., Katsoulidis E., Liu H., Altman J. K., Fish E. N., Wickrema A., Platanias L. C. (2009) Activation of protein kinase C (PKC) η by type I interferons. J. Biol. Chem. 284, 10301–10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Uddin S., Sassano A., Deb D. K., Verma A., Majchrzak B., Rahman A., Malik A. B., Fish E. N., Platanias L. C. (2002) Protein kinase Cδ (PKCδ) is activated by type I interferons and mediates phosphorylation of Stat1 on serine 727. J. Biol. Chem. 277, 14408–14416 [DOI] [PubMed] [Google Scholar]
- 28. Shiota C., Woo J. T., Lindner J., Shelton K. D., Magnuson M. A. (2006) Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev. Cell 11, 583–589 [DOI] [PubMed] [Google Scholar]
- 29. Jacinto E., Facchinetti V., Liu D., Soto N., Wei S., Jung S. Y., Huang Q., Qin J., Su B. (2006) SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates AKT phosphorylation and substrate specificity. Cell 127, 125–137 [DOI] [PubMed] [Google Scholar]
- 30. Sanges R., Cordero F., Calogero R. A. (2007) oneChannelGUI. A graphical interface to Bioconductor tools, designed for life scientists who are not familiar with R language. Bioinformatics 23, 3406–3408 [DOI] [PubMed] [Google Scholar]
- 31. Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- 32. Smyth G. K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- 33. Reiner A., Yekutieli D., Benjamini Y. (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19, 368–375 [DOI] [PubMed] [Google Scholar]
- 34. Sharma B., Joshi S., Sassano A., Majchrzak B., Kaur S., Aggarwal P., Nabet B., Bulic M., Stein B. L., McMahon B., Baker D. P., Fukunaga R., Altman J. K., Licht J. D., Fish E. N., Platanias L. C. (2012) Sprouty proteins are negative regulators of interferon (IFN) signaling and IFN-inducible biological responses. J. Biol. Chem. 287, 42352–42360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Darnell J. E., Jr., Kerr I. M., Stark G. R. (1994) Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264, 1415–14121 [DOI] [PubMed] [Google Scholar]
- 36. Qureshi S. A., Salditt-Georgieff M., Darnell J. E., Jr. (1995) Tyrosine-phosphorylated Stat1 and Stat2 plus a 48-kDa protein all contact DNA in forming interferon-stimulated-gene factor 3. Proc. Natl. Acad. Sci. U.S.A. 92, 3829–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qureshi S. A., Leung S., Kerr I. M., Stark G. R., Darnell J. E., Jr. (1996) Function of Stat2 protein in transcriptional activation by α interferon. Mol. Cell. Biol. 16, 288–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haller O., Kochs G., Weber F. (2007) Interferon, Mx, and viral countermeasures. Cytokine Growth Factor Rev. 18, 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rahman N., Stewart G., Jones G. (2010) A role for the atopy-associated gene PHF11 in T-cell activation and viability. Immunol. Cell Biol. 88, 817–824 [DOI] [PubMed] [Google Scholar]
- 40. Reich N. C. (2013) A death-promoting role for ISG54/IFIT2. J. Interferon Cytokine Res. 33, 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dionne K. R., Zhuang Y., Leser J. S., Tyler K. L., Clarke P. (2013) Daxx upregulation within the cytoplasm of reovirus-infected cells is mediated by interferon and contributes to apoptosis. J. Virol. 87, 3447–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Katsoulidis E., Carayol N., Woodard J., Konieczna I., Majchrzak-Kita B., Jordan A., Sassano A., Eklund E. A., Fish E. N., Platanias L. C. (2009) Role of Schlafen 2 (SLFN2) in the generation of interferon α-induced growth inhibitory responses. J. Biol. Chem. 284, 25051–25064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schattgen S. A., Fitzgerald K. A. (2011) The PYHIN protein family as mediators of host defenses. Immunol. Rev. 243, 109–118 [DOI] [PubMed] [Google Scholar]
- 44. Uddin S., Majchrzak B., Woodson J., Arunkumar P., Alsayed Y., Pine R., Young P. R., Fish E. N., Platanias L. C. (1999) Activation of the p38 mitogen-activated protein kinase by type I interferons. J. Biol. Chem. 274, 30127–30131 [DOI] [PubMed] [Google Scholar]
- 45. Takauji R., Iho S., Takatsuka H., Yamamoto S., Takahashi T., Kitagawa H., Iwasaki H., Iida R., Yokochi T., Matsuki T. (2002) CpG-DNA-induced IFN-α production involves p38 MAPK-dependent STAT1 phosphorylation in human plasmacytoid dendritic cell precursors. J. Leukocyte Biol. 72, 1011–1019 [PubMed] [Google Scholar]
- 46. Yanase N., Hayashida M., Kanetaka-Naka Y., Hoshika A., Mizuguchi J. (2012) PKC-δ mediates interferon-α-induced apoptosis through c-Jun NH2-terminal kinase activation. BMC Cell Biol. 13, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kaur S., Parmar S., Smith J., Katsoulidis E., Li Y., Sassano A., Majchrzak B., Uddin S., Tallman M. S., Fish E. N., Platanias L. C. (2005) Role of protein kinase Cδ (PKCδ) in the generation of the effects of IFN-α in chronic myelogenous leukemia cells. Exp. Hematol. 33, 550–557 [DOI] [PubMed] [Google Scholar]
- 48. Kroczynska B., Kaur S., Katsoulidis E., Majchrzak-Kita B., Sassano A., Kozma S. C., Fish E. N., Platanias L. C. (2009) Interferon-dependent engagement of eukaryotic initiation factor 4B via S6 kinase (S6K)- and ribosomal protein S6K-mediated signals. Mol. Cell. Biol. 29, 2865–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kroczynska B., Sharma B., Eklund E. A., Fish E. N., Platanias L. C. (2012) Regulatory effects of programmed cell death 4 (PDCD4) protein in interferon (IFN)-stimulated gene expression and generation of type I IFN responses. Mol. Cell. Biol. 32, 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Joshi S., Kaur S., Redig A. J., Goldsborough K., David K., Ueda T., Watanabe-Fukunaga R., Baker D. P., Fish E. N., Fukunaga R., Platanias L. C. (2009) Type I interferon (IFN)-dependent activation of Mnk1 and its role in the generation of growth inhibitory responses. Proc. Natl. Acad. Sci. U.S.A. 106, 12097–12102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Laplante M., Sabatini D. M. (2012) mTOR signaling in growth control and disease. Cell 149, 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Beauchamp E. M., Platanias L. C. (2013) The evolution of the TOR pathway and its role in cancer. Oncogene 32, 3923–3932 [DOI] [PubMed] [Google Scholar]
- 53. Decker T., Kovarik P. (2000) Serine phosphorylation of STATs. Oncogene 19, 2628–2637 [DOI] [PubMed] [Google Scholar]
- 54. Facchinetti V., Ouyang W., Wei H., Soto N., Lazorchak A., Gould C., Lowry C., Newton A. C., Mao Y., Miao R. Q., Sessa W. C., Qin J., Zhang P., Su B., Jacinto E. (2008) The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 27, 1932–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ikenoue T., Inoki K., Yang Q., Zhou X., Guan K. L. (2008) Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 27, 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Deb D. K., Sassano A., Lekmine F., Majchrzak B., Verma A., Kambhampati S., Uddin S., Rahman A., Fish E. N., Platanias L. C. (2003) Activation of protein kinase Cδ by IFN-γ. J. Immunol. 171, 267–273 [DOI] [PubMed] [Google Scholar]
- 57. Choudhury G. G. (2004) A linear signal transduction pathway involving phosphatidylinositol 3-kinase, protein kinase Cepsilon, and MAPK in mesangial cells regulates interferon-γ-induced STAT1α transcriptional activation. J. Biol. Chem. 279, 27399–27409 [DOI] [PubMed] [Google Scholar]
- 58. Bancerek J., Poss Z. C., Steinparzer I., Sedlyarov V., Pfaffenwimmer T., Mikulic I., Dölken L., Strobl B., Müller M., Taatjes D. J., Kovarik P. (2013) CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 38, 250–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ghosh D., Srivastava G. P., Xu D., Schulz L. C., Roberts R. M. (2008) A link between SIN1 (MAPKAP1) and poly(rC) binding protein 2 (PCBP2) in counteracting environmental stress. Proc. Natl Acad. Sci. U.S.A. 105, 11673–11678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang S. Z., Roberts R. M. (2004) Interaction of stress-activated protein kinase-interacting protein-1 with the interferon receptor subunit IFNAR2 in uterine endometrium. Endocrinology 145, 5820–5831 [DOI] [PubMed] [Google Scholar]
- 61. Wang S. Z., Roberts R. M. (2005) The evolution of the Sin1 gene product, a little known protein implicated in stress responses and type I interferon signaling in vertebrates. BMC Evol. Biol. 5, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.