Background: The function of the PSO4 complex in response to DNA damage remains unclear.
Results: The PSO4 complex is required for efficient accumulation of ATRIP at DNA damage sites and the subsequent CHK1 activation and RPA2 phosphorylation.
Conclusion: The PSO4 complex modulates ATR activation through interaction with RPA.
Significance: We reveal a role for RNA processing factor PSO4 in ATR activation.
Keywords: Cell Biology, Checkpoint Control, DNA Damage Response, DNA Repair, Signal Transduction
Abstract
The PSO4 core complex is composed of PSO4/PRP19/SNEV, CDC5L, PLRG1, and BCAS2/SPF27. Besides its well defined functions in pre-mRNA splicing, the PSO4 complex has been shown recently to participate in the DNA damage response. However, the specific role for the PSO4 complex in the DNA damage response pathways is still not clear. Here we show that both the BCAS2 and PSO4 subunits of the PSO4 complex directly interact and colocalize with replication protein A (RPA). Depletion of BCAS2 or PSO4 impairs the recruitment of ATR-interacting protein (ATRIP) to DNA damage sites and compromises CHK1 activation and RPA2 phosphorylation. Moreover, we demonstrate that both the RPA1-binding ability of BCAS2 and the E3 ligase activity of PSO4 are required for efficient accumulation of ATRIP at DNA damage sites and the subsequent CHK1 activation and RPA2 phosphorylation. Our results suggest that the PSO4 complex functionally interacts with RPA and plays an important role in the DNA damage response.
Introduction
The integrity of genomic DNA is continuously challenged by both exogenous and endogenous DNA-damaging agents. To maintain genetic information, cells have evolved many intricate DNA damage response mechanisms to detect and repair the various types of damage that can occur to DNA. One of the proteins essential for the accurate maintenance of genetic information is the single-stranded DNA-binding protein replication protein A (RPA). RPA is a heterotrimeric complex composed of RPA1, RPA2, and RPA3 subunits (1, 2). Each of the RPA subunits has at least one domain containing an oligonucleotide/oligosaccharide-binding fold (1, 2). In RPA, these oligonucleotide/oligosaccharide-binding folds are commonly referred to as DNA-binding domains (DBDs)2 (1, 2). RPA1 contains four oligonucleotide/oligosaccharide-binding folds, referred to as DBD-F, DBD-A, DBD-B, and DBD-C, following their arrangement from the N to the C terminus, and RPA2 and RPA3 have one oligonucleotide/oligosaccharide-binding fold each (1, 2). Binding of RPA to single-stranded regions of DNA is critical for the recruitment of ATRIP/ATR to sites of DNA damage, where checkpoint activation leads to the phosphorylation of CHK1 and several other downstream effectors (3, 4).
The PSO4 core complex is composed of PSO4/PRP19/SNEV, CDC5L, PLRG1, and BCAS2/SPF27 (5–11). This complex has been shown to have a role in pre-mRNA splicing from yeast to humans (12–17). The only identified catalytic center in the subunits of this complex is a U-box domain located in the N terminus of PSO4. U-box domains have been shown to have E3 ubiquitin ligase activity (18, 19). Several studies have shown that the E3 ubiquitin ligase activity of PSO4 is crucial for its function in pre-mRNA splicing (20–22). Besides its well defined roles in pre-mRNA splicing, the PSO4 complex has been found recently to play an important role in the DNA damage response (5, 14, 19, 23–26). In fact, cells harboring a mutant PSO4 in budding yeast showed broad hypersensitivity to DNA damage-inducing agents, suggesting that PSO4 is essential for mediating the DNA damage response (14, 26). Human PSO4 has also been shown to be a DNA-binding protein and plays a role in DNA repair through its interaction with terminal deoxynucleotidyl transferase (24). In addition, human PSO4 is required for the recruitment of the DNA repair protein Metnase to DNA damage sites (25). Moreover, CDC5L was found to directly interact with ATR and is required for the S phase cell-cycle checkpoint (27). Although these findings clearly indicate that the PSO4 complex participates in the DNA damage response, the nature of its function in these pathways is still not clear.
In this study, we adopted the tandem affinity purification approach to isolate an RPA-containing protein complex and identified the PSO4 complex as a component involved in RPA-mediated DNA damage response. We demonstrate that both the BCAS2 and PSO4 subunits of the PSO4 complex directly interact with RPA1 and are recruited to DNA damage sites. Depletion of BCAS2 or PSO4 leads to defects in the recruitment of ATRIP to DNA damage sites, CHK1 activation, and RPA2 phosphorylation. We further show that both the RPA1-binding ability of BCAS2 and the E3 ligase activity of PSO4 are required for efficient accumulation of ATRIP at DNA damage sites and the subsequent CHK1 and RPA2 phosphorylation. We propose that the PSO4 complex, via its interaction with RPA, modulates the DNA damage response.
EXPERIMENTAL PROCUDURES
Plasmids
All cDNAs were subcloned into pDONR201 (Invitrogen) as entry clones and were subsequently transferred to gateway-compatible destination vectors for the expression of N- or C-terminal-tagged fusion protein. All deletion mutants were generated using the QuikChange site-directed mutagenesis kit (Stratagene) and verified by sequencing.
Cell Cultures, Transfection, and shRNAs
HEK293T and HeLa Cells were maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. Sf9 insect cells were maintained in Grace's medium supplemented with 2% fetal bovine serum. Human cell lines were maintained in a 37 °C incubator with 5% CO2, whereas insect cells were maintained at 27 °C. Cell transfection was performed using Lipofectamine 2000 (Invitrogen) following the protocol of the manufacturer. Lentiviral nonsilencing control shRNA and shRNA target sets were purchased from Open Biosystems. The BCSA2 targeting sequences were as follows: 1, 5′-CCCGGATTATTCTGCCTTTG A-3′; 2, 5′-GCTGTGGTAATTCTATTTGTA-3′. The PSO4 targeting sequences were as follows: 1, 5′-GAACGGATGTGGAAGGAAGAA-3′; 2, 5′-CCTGTCTCTAATCATGTTTAT-3′. The RPA1 targeting sequences were as follows: 1, 5′-GCTACAAAGCGTTTCTTTA-3′; 2, 5′-GAGTCAGATGGGTCTGATA-3′. The nonsilencing control sequence was as follows: 5′-CCCATAAGAGTAATAATAT-3′. The shRNA-resistant wild-type and mutant PSO4 constructs were generated by changing seven nucleotides in the shRNA 2 targeting region (T57C, C60G, T63C, T66C, T69C, T72G, and T75C substitutions). The shRNAs were packaged into lentiviruses by cotransfection with the packaging plasmids pMD2G and pSPAX2 (provided by Songyang Zhou, Baylor College of Medicine) into HEK293T cells. Forty-eight hours after transfection, the supernatant was collected for infection of HeLa cells. Infected cells were selected with medium containing puromycin (2 μg/ml).
Antibodies
Polyclonal anti-PSO4, anti-BCAS2, or anti-ATRIP antibody was generated by immunizing rabbits with MBP-PSO4, MBP-BCAS2, or GST-ATRIP (residues 1–107) fusion proteins expressed and purified from Escherichia coli. Antisera were affinity-purified using the AminoLink Plus immobilization and purification kit (Pierce). Monoclonal anti-RPA2 (catalog no. ab2175) and polyclonal anti-phospho-RPA2 (Ser-4/Ser-8) (catalog no. A300-245A) antibodies were purchased from Abcam and Bethyl Laboratories, respectively. Anti-CHK1 and anti-CHK2 antibodies were purchased from Santa Cruz Biotechnology. Anti-CHK1 pS317 and anti-CHK2 T68 antibodies were purchased from Cell Signaling Technology. Anti-Myc (9E10) antibody was purchased from Covance. Anti-GAPDH and anti-FLAG (M2) antibodies were purchased from Millipore and Sigma, respectively.
The Establishment of Stable Cell Lines and Affinity Purification of S-Peptide, FLAG, and Streptavidin-binding Peptide (SFB)-tagged Protein Complexes
HEK293T cells were transfected with plasmids encoding SFB-tagged RPA1 or RPA3. A cell line stably expressing tagged RPA1 or RPA3 proteins was selected by culturing in medium containing puromycin (2 μg/ml) and confirmed by immunoblotting and immunostaining. For affinity purification, HEK293T cells stably expressing tagged proteins were lysed with NETN buffer (20 mm Tris-HCl (pH 8.0), 100 mm NaCl, 1 mm EDTA, and 0.5% Nonidet P-40) for 20 min. Crude lysates were removed by centrifugation at 14,000 rpm at 4 °C for 10 min, and the pellet was sonicated for 40 s in high-salt solution (20 mm HEPES (pH 7.8), 0.4 m NaCl, 1 mm EDTA, 1 mm EGTA, and protease inhibitor) to extract chromatin-bound protein fractions. The supernatants were cleared at 14,000 rpm to remove debris and then incubated with streptavidin-conjugated beads (Amersham Biosciences) for 2 h at 4 °C. The beads were washed three times with NETN buffer, and then bead-bound proteins were eluted with NETN buffer containing 1 mg/ml biotin (Sigma). The elutes were incubated with S protein beads (Novagen) for 2 h at 4 °C. The beads were again washed three times with NETN buffer and subjected to SDS-PAGE. Protein bands were excised and digested, and the peptides were analyzed by mass spectrometry.
Coimmunoprecipitation and Western Blotting
For whole-cell extracts, the cells were solubilized in NETN lysis buffer supplemented with 50 units/μl benzonase (Novagen), protease inhibitors, and phosphatase inhibitors. After removal of cell debris by centrifugation, the soluble fractions were collected. For FLAG immunoprecipitations, a 0.8-ml aliquot of lysate was incubated with 1 μg of the FLAG monoclonal antibody and 25 μl of a 1:1 slurry of protein A-Sepharose for 2 h at 4 °C. For endogenous immunoprecipitations, 1 mg of the whole-cell extract was incubated with 25 μl of a 1:1 slurry of protein A-Sepharose coupled with 2 μl of the indicated antibodies for 2 h at 4 °C. The Sepharose beads were washed three times with NTEN buffer, boiled in 2× SDS loading buffer, and resolved on SDS-PAGE. Membranes were blocked in 5% milk in TBST (50 mm Tris, 150 mm NaCl, and 0.05% Tween 20) buffer and then probed with antibodies as indicated.
Immunofluorescence Staining
Indirect immunofluorescence was carried out as described previously (28, 29). HEK293T or HeLa cells cultured on coverslips were treated with CPT (1 μm) for 1 h or IR (10 gray) for 1 h, respectively. Cells were then washed with PBS, pre-extracted with buffer containing 0.5% Triton X-100 for 5 min, and fixed with 3% paraformaldehyde for 10 min at room temperature. Cells were incubated in primary antibody for 20 min at room temperature. Following three 5-min washes with PBS, secondary antibody was added at room temperature for 20 min. Cells were then stained with DAPI to visualize nuclear DNA. The coverslips were mounted onto glass slides with antifade solution and visualized using a Nikon Eclipse i80 fluorescence microscope with a Nikon Plan Fluor ×60 oil objective lens.
Protein Purification
Full-length RPA1 was cloned into MBP-His-tagged vector for the expression of MBP-RPA1-His protein in insect cells. Transposition occurred in DH10Bac-competent cells, and correct bacmids confirmed by PCR were transfected into Sf9 cells for baculovirus production. After viral amplification, Sf9 cells were infected with baculovirus stocks expressing MBP-RPA1-His for 48 h. Cells were harvested and washed with 1× PBS and resuspended in lysis buffer (20 mm Tris-HCl; 300 mm NaCl; 1% Triton X-100; and 1 μg/ml each of leupeptin, aprotinin, and pepstatin). The extract was centrifuged at 18,000 rpm for 40 min. The supernatant was collected and loaded onto a pre-equilibrated nickel-nitrilotriacetic acid-agarose and washed with lysis buffer plus 20 mm imidazole and protease inhibitor. The bound protein was then eluted with lysis buffer containing 200 mm imidazole and protease inhibitor. Peak fractions were pooled and incubated with amylose resins for 2 h at 4 °C. After washing the beads with 100 ml of washing buffer (20 mm Tris-HCl; 500 mm NaCl; 0.5% Nonidet P-40; 1 mm DTT; and 1 μg/ml each of leupeptin, aprotinin, and pepstatin), the bound protein was used for a pulldown assay. Full-length PSO4 and BCAS2 were cloned into the PGEX-6P-1 vector for the expression of GST-tagged fusion proteins in E. coli. Cells were grown at 37 °C until log phase and induced with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside at 17 °C for 16 h. Cells were then harvested and resuspended in lysis buffer (20 mm Tris-HCl; 300 mm NaCl; 1% Triton X-100; 1 mm DTT; and 1 μg/ml each of leupeptin, aprotinin, and pepstatin). After sonication, the extract was centrifuged at 18,000 rpm for 40 min. The supernatant was collected and incubated with glutathione-Sepharose resin for 4 h at 4 °C. After washing the beads with washing buffer (20 mm Tris-HCl; 500 mm NaCl; 0.5% Nonidet P-40; 1 mm DTT; and 1 μg/ml each of leupeptin, aprotinin, and pepstatin), the bound proteins were eluted with washing buffer containing 20 mm glutathione and used for pulldown assays.
Lentivirus Packaging and Infection
The Tet-On-inducible, SFB-tagged lentiviral vector and packaging plasmids (pMD2G and pSPAX2) were provided by Prof. Songyang Zhou (Baylor College of Medicine). BCAS2 and PSO4 entry constructs were transferred into the Gateway-compatible, SFB-tagged lentiviral vector. Virus supernatant was collected 48 h after the cotransfection of lentiviral vectors and packaging plasmids (pMD2G and pSPAX2) into HEK293T cells. Cells were infected with viral supernatants with the addition of 8 μg/ml Polybrene (Sigma), and stable pools were selected with medium containing 500 μg/ml G418 (Calbiochem). The expression of the indicated genes in the stable pools was induced by the addition of 1 μg/ml doxycycline (Sigma) for 48 h for the experiments presented in this work.
RESULTS AND DISCUSSION
Both BCAS2 and PSO4 Interact and Colocalize with RPA
RPA plays a central role in DNA replication, repair, and recombination (1, 2). In an attempt to search for previously undetected proteins present in the RPA-containing complex, we performed tandem affinity purification using HEK293T cells stably expressing triple epitope (S-peptide, FLAG, and streptavidin-binding peptide)-tagged wild-type RPA1 or RPA3 for the identification of RPA-interacting proteins. Mass spectrometry analysis revealed a number of known RPA-associated proteins, including hPrimpol1/Primpol/CCDC111, BLM, and RAD52 (supplemental Tables 1 and 2) (30–35). Interestingly, we also identified that all four subunits of PSO4 exist in a complex with RPA (supplemental Tables 1 and 2).
To determine whether the PSO4 complex indeed interacts with RPA, we performed transient transfection and coimmunoprecipitation experiments. The results demonstrated that both the PSO4 and BCAS2 subunits could interact with RPA1 (Fig. 1A). In the same experiments, a weak interaction between PSO4 and RPA2 and RPA3 was also found to take place (Fig. 1A). These results indicate that the PSO4 complex associates with the RPA complex majorly through RPA1.
FIGURE 1.
The PSO4 complex associates and colocalizes with RPA. A, RPA1 interacts with PSO4 and BCAS2. HEK293T cells were transiently transfected with plasmids encoding SFB-tagged RPA1, RPA2, or RPA3 together with plasmids encoding Myc-tagged PSO4, PLRG1, CDC5L, or BCAS2. Cell lysates were immunoprecipitated (IP) with anti-FLAG antibody, and Western blot analysis was performed with anti-FLAG and anti-Myc antibodies. B, direct binding between recombinant MBP-tagged-RPA1 and GST-tagged-BCAS2 or GST-tagged-PSO4. Top panel, BCAS2 and PSO4 were detected by immunoblotting. Bottom panel, purified proteins visualized by Coomassie staining. C and D, association of endogenous PSO4 complex with RPA in HeLa cells was performed by coimmunoprecipitation using anti-RPA2 (C) or anti-BCAS2 (D) antibody. HeLa cells treated with CPT (1 μm) for 1 h were lysed in the presence of benzonase. Cell lysates were then incubated with protein A-agarose beads conjugated with the indicated antibodies, and Western blot analysis was carried out as indicated. E, BCAS2 colocalizes with RPA2. 293T cells were either mock-treated or treated with CPT (1 μm) or IR (10 gray) for 1 h before fixing and processing for BCAS2 and RPA2 immunofluorescence. A merged image shows colocalization. F, PSO4 colocalizes with RPA2. SFB-tagged PSO4 was expressed in HEK293T cells. Foci assembled by this fusion protein and by RPA2 following exposure to CPT (1 μm) or IR (10 gray) for 1 h were detected by immunofluorescence using anti-FLAG and anti-RPA2 antibodies, respectively. SFB-PSO4 foci were detected in green, whereas RPA2 foci were detected in red. A merged image shows colocalization.
We next tested the possibility that there could be a direct protein-protein interaction between RPA1 and PSO4 and BCAS2. Pulldown assays using recombinant MBP-tagged RPA1 and GST-tagged PSO4 or BCAS2 purified from E. coli demonstrated that RPA1 interacts strongly with BCAS2 and weakly with PSO4 in vitro (Fig. 1B).
To examine the interaction between endogenous RPA and the PSO4 complex, HeLa cell extracts were prepared and subjected to immunoprecipitation assays in the presence of either control IgG or anti-RPA2 antibody (Fig. 1C). Western blot analysis revealed that both PSO4 and BCAS2 were clearly detected in the immunoprecipitations obtained with the anti-RPA2 antibody but not with the control IgG (Fig. 1C). We also performed a reciprocal coimmunoprecipitation assay. As shown in Fig. 1D, the endogenous RPA1 and RPA2 were readily immunoprecipitated with the BCAS2-specific antibody but not with the control IgG. Moreover, treatment with the topoisomerase I inhibitor CPT slightly enhanced the interaction between BCAS2/PSO4 and RPA (Fig. 1, C and D). In these experiments, benzonase was included in the lysis buffer to exclude the possibility that the interaction occurs indirectly via DNA bridging (Fig. 1, C and D).
Upon occurrence of DNA damage, RPA and several other proteins involved in the DNA damage response could form large nuclear foci. A physical interaction between RPA and BCAS2 and PSO4, as demonstrated above, raises the possibility that BCAS2 and PSO4 may colocalize with RPA at DNA damage sites. Indeed, discrete foci of BCAS2 and PSO4 were readily detected in cells following CPT or IR treatment (Fig. 1, E and F). Moreover, these foci colocalize with RPA2 foci (more than 90%), indicating that the localization of BCAS2 and PSO4, like that of RPA, is regulated in response to DNA damage (Fig. 1, E and F).
The N Terminus of BCAS2 Binds to the C Terminus of RPA1
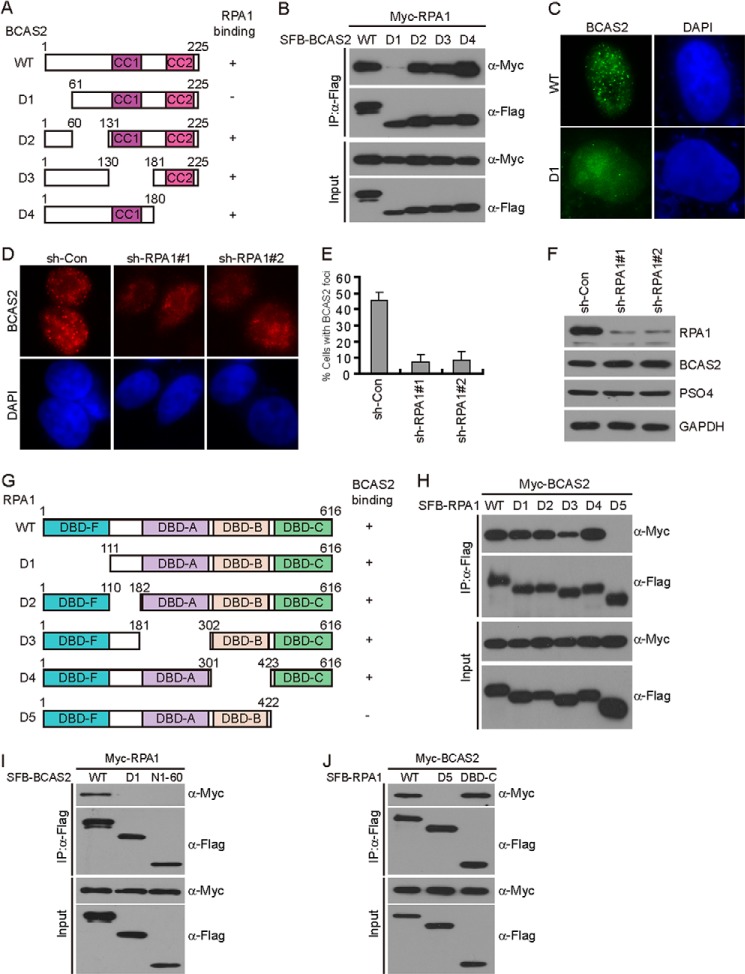
BCAS2 is a small subunit of the PSO4 complex that contains two coiled coil motifs (10, 11). Because BCAS2 directly interacts with RPA1, we next mapped the RPA1-binding domain on BCAS2. A series of deletion mutants that span the entire coding region of BCAS2 were generated (Fig. 2A). Coimmunoprecipitation experiments revealed that BCAS2 associated with RPA1 via its N terminus because the deletion mutant lacking the N-terminal 60 amino acids (D1) failed to coprecipitate with RPA1 (Fig. 2B). Interestingly, the D1 mutant, which does not bind to RPA1, failed to relocalize to DNA damage sites, suggesting that the binding of BCAS2 to RPA1 is important for its proper localization in response to DNA damage (Fig. 2C). In support of this conclusion, RPA1 depletion abolished BCAS2 focus formation after DNA damage (Fig. 2, D–F).
FIGURE 2.
BCAS2 localizes to DNA damage sites via an interaction with RPA1. A, schematic representation of wild-type and deletion mutants of BCAS2 used in this study. Their ability to bind to RPA1 is indicated. CC, coiled coil domain. B, the 60 amino acids at the N terminus of BCAS2 are required for RPA1 binding. HEK293T cells were transfected with plasmids encoding Myc-tagged RPA1 together with plasmids encoding SFB-tagged wild-type BCAS2 or the mutants. Cell lysates were immunoprecipitated (IP) with anti-FLAG antibody, and Western blot analysis was performed with the indicated antibodies. C, the N terminus of BCAS2 is required for its focus formation. HeLa cells were infected with lentivirus expressing SFB-tagged wild-type BCAS2 or the D1 mutant. 48 h later, cells were treated with CPT (1 μm) for 1 h before fixing and processing for immunofluorescence. D–F, RPA1 depletion impairs BCAS2 focus formation. HEK293T cells infected with non-target or RPA1-specific lentiviral shRNAs for 48 h were treated with CPT (1 μm) for 1 h before fixing and processing for BCAS2 immunofluorescence. Representative BCAS2 foci are shown (D). Quantification results were the average of three independent experiments and are presented as mean ± S.E. sh-Con, control shRNA. E, more than 100 cells were counted in each experiment. The knockdown efficiency of RPA1 using specific shRNAs was confirmed by immunoblotting of lysates prepared from HEK293T cells expressing the indicated shRNA (F). G, schematic of full-length RPA1 and the mutants used in this study. The ability to bind to BCAS2 is indicated. H, RPA1 with the DBD-C domain deletion could not bind to BCAS2. HEK293T cells were transfected with plasmids encoding Myc-tagged wild-type or mutant RPA1 together with a plasmid encoding SFB-tagged BCAS2. Cell lysates were immunoprecipitated with anti-FLAG antibody, and Western blot analysis was performed with the indicated antibodies. I, the N-terminal region of BCAS2 (N1–60, residues 1–60) is not sufficient for its interaction with RPA1. Coimmunoprecipitation experiments were carried out as indicated. J, the RPA1 DBD-C domain (residues 423–616) is not only essential but also sufficient for its interaction with BCAS2. Coimmunoprecipitation experiments were carried out as indicated.
To understand how RPA1 interacts with BCAS2 and facilitates its recruitment to sites of DNA damage, we used a panel of RPA1 constructs (Fig. 2G). The results indicated that RPA1 associated with BCAS2 via its DBD-C domain because the mutant that lacks the DBD-C domain (D5) failed to interact with BCAS2 (Fig. 2H).
To further test whether the N-terminal region of BCAS2 is sufficient to bind RPA1, we generated a construct encoding the N-terminal region alone (N1–60, residues 1–60). As shown in Fig. 2I, the N-terminal region alone failed to interact with RPA1. In contrast, the RPA1 DBD-C domain (residues 423–616) is not only essential but also sufficient for its interaction with BCAS2 (Fig. 2J).
BCAS2 and PSO4 Are Required for Efficient Accumulation of ATRIP at DNA Damage Sites and the Subsequent CHK1 Activation and RPA2 Phosphorylation
RPA is the major single-stranded DNA-binding protein and plays essential roles in DNA replication, DNA repair, and the initiation of DNA damage checkpoint (1, 2). Because BCAS2 and PSO4 exist in a complex with RPA and localize to DNA damage sites, we sought to determine whether BCAS2 and PSO4 also play important roles in the DNA damage response. As shown in Fig. 3, A and B, BCAS2 or PSO4 depletion led to impaired CHK1 and RPA2 phosphorylation after CPT or IR treatment. By contrast, BCAS2 or PSO4 depletion did not markedly affect phosphorylation of CHK2 (Fig. 3, A and B). Moreover, BCAS2 or PSO4 depletion severely reduced the recruitment of phospho-RPA2 and ATRIP to sites of DNA damage (Fig. 3, C–F). Taken together, these results indicate that BCAS2 or PSO4 depletion may specifically impair ATR- but not ATM-dependent signaling after DNA damage. Interestingly, depletion of BCAS2 caused a reduction in PSO4 protein levels, and vice versa, indicating that the stabilities of BCAS2 and PSO4 are interdependent (Fig. 3, A and B).
FIGURE 3.
BCAS2 and PSO4 are required for the recruitment of ATRIP to DNA damage sites and the subsequent CHK1 activation and RPA2 phosphorylation. A and B, BCAS2 or PSO4 depletion impairs CHK1 activation and RPA2 phosphorylation after DNA damage. HeLa cells infected with the indicated shRNAs were either mock-treated or treated with CPT (1 μm) (A) or IR (10 gray) (B) for 1 h. Cell lysates were immunoblotted with the indicated antibodies. The asterisk indicates a nonspecific band. sh-Con, control shRNA. C and D, BCAS2 or PSO4 depletion impairs CPT-induced phospho-RPA2 focus formation. BCAS2- or PSO4-depleted HeLa cells were treated with CPT (1 μm) for 1 h before fixing and processing for phospho-RPA2 immunofluorescence. Representative phospho-RPA2 foci are shown (C). Quantification results were the average of three independent experiments and are presented as mean ± S.E. (D). More than one hundred cells were counted in each experiment. E and F, BCAS2 or PSO4 depletion impairs CPT-induced ATRIP focus formation. BCAS2- or PSO4-depleted HeLa cells were treated with CPT (1 μm) for 1 h before fixing and processing for ATRIP immunofluorescence. Representative ATRIP foci are shown (E). Quantification results were the average of three independent experiments and are presented as mean ± S.E. (F). More than 100 cells were counted in each experiment.
The Ability of BCAS2 to Function in the DNA Damage Response Correlates with Its Association with RPA1
To further decipher the biological significance of the BCAS2-RPA1 interaction, we performed rescue experiments to test whether the RPA1-binding region on BCAS2 is required for efficient accumulation of ATRIP at DNA damage sites and the subsequent CHK1 activation and RPA2 phosphorylation. We generated HeLa cell lines to express wild-type BCAS2 or its deletion mutant defective in RPA1 binding under the control of a tetracycline-inducible promoter. By treating these cell lines with shRNA targeting the 3′ UTR of the BCAS2 transcript (shRNA 2), we can specifically knock down the endogenous, but not exogenous, BCAS2. The expression of wild-type and mutated BCAS2 was induced in BCAS2 knockdown cells when the cells were treated with doxycycline (Fig. 4, A and B). Interestingly, whereas wild-type BCAS2 successfully restored CHK1 activation and RPA2 phosphorylation to levels comparable with that of control cells, BCAS2 deletion mutant defective in RPA1 binding failed to do so (Fig. 4B). Consistently, the defects in ATRIP focus formation after DNA damage could be reversed by the expression of wild-type BCAS2 but not the D1 mutant (Fig. 4, C and D).
FIGURE 4.
Both the RPA1-binding ability of BCAS2 and the E3 ligase activity of PSO4 are required for the recruitment of ATRIP to DNA damage sites and the subsequent CHK1 and RPA2 phosphorylation. A–D, the mutant defective in RPA1 binding could not restore BCAS2 function in vivo. HeLa cell lines to express wild-type BCAS2 (WT) or its deletion mutant defective in RPA1 binding (D1) under the control of a tetracycline-inducible promoter were generated. The resulting cell lines were then infected with shRNA targeting the 3′ UTR of BCAS2 transcript (shRNA#2). 48 h after infection, cells were induced by doxycycline addition for 24 h prior to CPT (1 μm) treatment. 1 h later, cells were collected, and lysates were immunoblotted with the indicated antibodies (A and B) or subjected to immunostaining using ATRIP antibody (C and D). Representative ATRIP foci are shown (C). Quantification results were the average of three independent experiments and are presented as mean ± S.E. (D). More than 100 cells were counted in each experiment. The asterisk indicates a nonspecific band. sh-Con, control shRNA. E–H, the U-box deletion mutant of PSO4 could not restore PSO4 function in vivo. HeLa cell lines to express shRNA 2-resistant wild-type PSO4 (WT) or its U-box deletion mutant (ΔU) under the control of a tetracycline-inducible promoter were generated. The resulting cell lines were then infected with PSO4 shRNA 2. 48 h after infection, cells were induced by doxycycline addition for 24 h prior to CPT (1 mm) treatment. 1 h later, cells were collected, and lysates were immunoblotted with the indicated antibodies (E and F) or subjected to immunostaining using ATRIP antibody (G and H). Representative ATRIP foci are shown (G). Quantification results were the average of three independent experiments and are presented as mean ± S.E. (H). More than 100 cells were counted in each experiment. The asterisk indicates a nonspecific band.
The E3 Ligase Activity of PSO4 Is Indispensable for the Recruitment of ATRIP to DNA Damage Sites and the Subsequent CHK1 and RPA2 Phosphorylation
PSO4 is a U-box-containing E3 ubiquitin ligase (20–22). To explore the physiological relevance of this E3 ligase activity of PSO4 in ATRIP recruitment, CHK1 activation, and RPA2 phosphorylation, we took advantage of the inducible expression system to express the shRNA 2-resistant full-length PSO4 or the U-box domain deletion mutant (ΔU, lacking the 75 N-terminal amino acids) of PSO4 in PSO4-depleted HeLa cells (Fig. 4, E and F). Notably, the defects in ATRIP focus formation, CHK1 activation, and RPA2 phosphorylation in PSO4-depleted cells after DNA damage could be restored by the expression of wild-type PSO4,but not the ΔU mutant (Fig. 4, F–H). These results indicate that PSO4 may act as an E3 ubiquitin ligase and regulate certain unknown substrates at DNA damage sites that are critical for efficient recruitment of ATRIP to sites of DNA damage and the subsequent CHK1 activation and RPA2 phosphorylation. Interestingly, while our manuscript was in preparation, an independent study reported that PSO4 promotes RPA ubiquitination and facilitates the accumulation of ATRIP at sites of DNA damage (36).
In summary, we discovered a functional interaction between the PSO4 complex and RPA. We propose that the PSO4 complex, through its interaction with RPA, is recruited to sites of DNA damage and exerts its E3 ligase activity to ubiquitinate RPA and, thus, to modulate the cellular response to DNA damage.
Supplementary Material
Acknowledgments
We thank all of our colleagues in the Huang laboratory for insightful discussions.
This work was supported by National Program for Special Support of Eminent Professionals, National Basic Research Program of China Grants 2012CB944402 and 2013CB911003, by National Natural Science Funds for Distinguished Young Scholars, by Zhejiang University K.P. Chao's High Technology Development Foundation, and by China Fundamental Research Funds for the Central Universities.

This article contains supplemental Tables 1 and 2.
- DBD
- DNA-binding domain
- SFB
- S-peptide, FLAG, and streptavidin-binding peptide
- CPT
- camptothecin
- IR
- ionizing radiation
- ATR
- ataxia telangiectasia-mutated and Rad3-related
- ATRIP
- ATR-interacting protein
- MBP
- maltose-binding protein.
REFERENCES
- 1. Wold M. S. (1997) Replication protein A. A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 [DOI] [PubMed] [Google Scholar]
- 2. Zou Y., Liu Y., Wu X., Shell S. M. (2006) Functions of human replication protein A (RPA). From DNA replication to DNA damage and stress responses. J. Cell. Physiol. 208, 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zou L., Elledge S. J. (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 4. Ball H. L., Myers J. S., Cortez D. (2005) ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol. Biol. Cell 16, 2372–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Legerski R. J. (2009) The Pso4 complex splices into the DNA damage response. Cell Cycle 8, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 6. Ajuh P., Sleeman J., Chusainow J., Lamond A. I. (2001) A direct interaction between the carboxyl-terminal region of CDC5L and the WD40 domain of PLRG1 is essential for pre-mRNA splicing. J. Biol. Chem. 276, 42370–42381 [DOI] [PubMed] [Google Scholar]
- 7. Kleinridders A., Pogoda H. M., Irlenbusch S., Smyth N., Koncz C., Hammerschmidt M., Brüning J. C. (2009) PLRG1 is an essential regulator of cell proliferation and apoptosis during vertebrate development and tissue homeostasis. Mol. Cell. Biol. 29, 3173–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Groenen P. M., Vanderlinden G., Devriendt K., Fryns J. P., Van de Ven W. J. (1998) Rearrangement of the human CDC5L gene by a t(6;19)(p21;q13.1) in a patient with multicystic renal dysplasia. Genomics 49, 218–229 [DOI] [PubMed] [Google Scholar]
- 9. Ajuh P., Kuster B., Panov K., Zomerdijk J. C., Mann M., Lamond A. I. (2000) Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J. 19, 6569–6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maass N., Rösel F., Schem C., Hitomi J., Jonat W., Nagasaki K. (2002) Amplification of the BCAS2 gene at chromosome 1p13.3–21 in human primary breast cancer. Cancer Lett. 185, 219–223 [DOI] [PubMed] [Google Scholar]
- 11. Chen P. H., Lee C. I., Weng Y. T., Tarn W. Y., Tsao Y. P., Kuo P. C., Hsu P. H., Huang C. W., Huang C. S., Lee H. H., Wu J. T., Chen S. L. (2013) BCAS2 is essential for Drosophila viability and functions in pre-mRNA splicing. RNA 19, 208–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng S. C., Tarn W. Y., Tsao T. Y., Abelson J. (1993) PRP19. A novel spliceosomal component. Mol. Cell. Biol. 13, 1876–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tarn W. Y., Lee K. R., Cheng S. C. (1993) The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol. Cell. Biol. 13, 1883–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grey M., Düsterhöft A., Henriques J. A., Brendel M. (1996) Allelism of PSO4 and PRP19 links pre-mRNA processing with recombination and error-prone DNA repair in Saccharomyces cerevisiae. Nucleic Acids Res. 24, 4009–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan S. P., Cheng S. C. (2005) The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J. Biol. Chem. 280, 31190–31199 [DOI] [PubMed] [Google Scholar]
- 16. Chan S. P., Kao D. I., Tsai W. Y., Cheng S. C. (2003) The Prp19p-associated complex in spliceosome activation. Science 302, 279–282 [DOI] [PubMed] [Google Scholar]
- 17. Chen C. H., Kao D. I., Chan S. P., Kao T. C., Lin J. Y., Cheng S. C. (2006) Functional links between the Prp19-associated complex, U4/U6 biogenesis, and spliceosome recycling. RNA 12, 765–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hatakeyama S., Yada M., Matsumoto M., Ishida N., Nakayama K. I. (2001) U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276, 33111–33120 [DOI] [PubMed] [Google Scholar]
- 19. Zhang N., Kaur R., Lu X., Shen X., Li L., Legerski R. J. (2005) The Pso4 mRNA splicing and DNA repair complex interacts with WRN for processing of DNA interstrand cross-links. J. Biol. Chem. 280, 40559–40567 [DOI] [PubMed] [Google Scholar]
- 20. Ohi M. D., Vander Kooi C. W., Rosenberg J. A., Chazin W. J., Gould K. L. (2003) Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 10, 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Löscher M., Fortschegger K., Ritter G., Wostry M., Voglauer R., Schmid J. A., Watters S., Rivett A. J., Ajuh P., Lamond A. I., Katinger H., Grillari J. (2005) Interaction of U-box E3 ligase SNEV with PSMB4, the β7 subunit of the 20 S proteasome. Biochem. J. 388, 593–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vander Kooi C. W., Ohi M. D., Rosenberg J. A., Oldham M. L., Newcomer M. E., Gould K. L., Chazin W. J. (2006) The Prp19 U-box crystal structure suggests a common dimeric architecture for a class of oligomeric E3 ubiquitin ligases. Biochemistry 45, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu X., Legerski R. J. (2007) The Prp19/Pso4 core complex undergoes ubiquitylation and structural alterations in response to DNA damage. Biochem. Biophys. Res. Commun. 354, 968–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mahajan K. N., Mitchell B. S. (2003) Role of human Pso4 in mammalian DNA repair and association with terminal deoxynucleotidyl transferase. Proc. Natl. Acad. Sci. U.S.A. 100, 10746–10751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beck B. D., Park S. J., Lee Y. J., Roman Y., Hromas R. A., Lee S. H. (2008) Human Pso4 is a Metnase (SETMAR)-binding partner that regulates Metnase function in DNA repair. J. Biol. Chem. 283, 9023–9030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Henriques J. A., Vicente E. J., Leandro da Silva K. V., Schenberg A. C. (1989) PSO4. A novel gene involved in error-prone repair in Saccharomyces cerevisiae. Mutat. Res. 218, 111–124 [DOI] [PubMed] [Google Scholar]
- 27. Zhang N., Kaur R., Akhter S., Legerski R. J. (2009) Cdc5L interacts with ATR and is required for the S-phase cell-cycle checkpoint. EMBO Rep. 10, 1029–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu T., Ghosal G., Yuan J., Chen J., Huang J. (2010) FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science 329, 693–696 [DOI] [PubMed] [Google Scholar]
- 29. Wan L., Han J., Liu T., Dong S., Xie F., Chen H., Huang J. (2013) Scaffolding protein SPIDR/KIAA0146 connects the Bloom syndrome helicase with homologous recombination repair. Proc. Natl. Acad. Sci. U.S.A. 110, 10646–10651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wan L., Lou J., Xia Y., Su B., Liu T., Cui J., Sun Y., Lou H., Huang J. (2013) hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. EMBO Rep. 14, 1104–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Helleday T. (2013) PrimPol breaks replication barriers. Nat. Struct. Mol. Biol. 20, 1348–1350 [DOI] [PubMed] [Google Scholar]
- 32. Bianchi J., Rudd S. G., Jozwiakowski S. K., Bailey L. J., Soura V., Taylor E., Stevanovic I., Green A. J., Stracker T. H., Lindsay H. D., Doherty A. J. (2013) PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol. Cell 52, 566–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. García-Gómez S., Reyes A., Martínez-Jiménez M. I., Chocrón E. S., Mourón S., Terrados G., Powell C., Salido E., Méndez J., Holt I. J., Blanco L. (2013) PrimPol, an archaic primase/polymerase operating in human cells. Mol. Cell 52, 541–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mourón S., Rodriguez-Acebes S., Martínez-Jiménez M. I., García-Gómez S., Chocrón S., Blanco L., Méndez J. (2013) Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol. 20, 1383–1389 [DOI] [PubMed] [Google Scholar]
- 35. Im J. S., Lee K. Y., Dillon L. W., Dutta A. (2013) Human Primpol1. A novel guardian of stalled replication forks. EMBO Rep. 14, 1032–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marechal A., Li J.M., Ji X.Y., Wu C.S., Yazinski S.A., Nguyen H.D., Liu S., Jimenez A.E., Jin J., Zou L. (2014) PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol. Cell 10.1016/j.molcel.2013.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.