Background: The ribosome is a highly charged complex comprising RNAs and proteins.
Results: Ribosomal proteins exhibit low hydrophobicity and a significant degree of intramolecular charge segregation.
Conclusion: The majority of ribosomal proteins from all organisms, particularly halophiles, use intramolecular charge segregation to minimize electrostatic repulsion with rRNA.
Significance: The electrostatic properties of ribosomal proteins are important for ribosome stability, assembly, and interaction with translation factors and nascent proteins.
Keywords: Archaea, Bacteria, Protein Folding, Protein Stability, Ribosomes, Electrostatics, Hydrophobicity, Net Charge, Ribosomal Proteins
Abstract
Ribosomes are large and highly charged macromolecular complexes consisting of RNA and proteins. Here, we address the electrostatic and nonpolar properties of ribosomal proteins that are important for ribosome assembly and interaction with other cellular components and may influence protein folding on the ribosome. We examined 50 S ribosomal subunits from 10 species and found a clear distinction between the net charge of ribosomal proteins from halophilic and non-halophilic organisms. We found that ∼67% ribosomal proteins from halophiles are negatively charged, whereas only up to ∼15% of ribosomal proteins from non-halophiles share this property. Conversely, hydrophobicity tends to be lower for ribosomal proteins from halophiles than for the corresponding proteins from non-halophiles. Importantly, the surface electrostatic potential of ribosomal proteins from all organisms, especially halophiles, has distinct positive and negative regions across all the examined species. Positively and negatively charged residues of ribosomal proteins tend to be clustered in buried and solvent-exposed regions, respectively. Hence, the majority of ribosomal proteins is characterized by a significant degree of intramolecular charge segregation, regardless of the organism of origin. This key property enables the ribosome to accommodate proteins within its complex scaffold regardless of their overall net charge.
Introduction
Several crystal structures of large and small ribosomal subunits became available over the past decade (1–4), providing the opportunity to examine the peculiar features of ribosomal RNA and proteins at atomic resolution. As a result, the last few years (5, 6) have witnessed unprecedented progress on the structural and functional analysis of ribosomal proteins (7, 8) and the mechanism of ribosome assembly (9, 10).
The electrostatic properties of ribosomal proteins, however, have not been explored in detail despite their fundamental importance for ribosome assembly, stabilization, and interaction with cellular proteins, molecular chaperones, co-translational modification agents the newly synthesized nascent chain during translation, and ions of different nature (9, 11–13).
Here, we focus on the electrostatic properties of ribosomal proteins from 10 organisms: three from Bacteria (Escherichia coli, Thermus thermophilus, and Deinococcus radiodurans), two from Eukaryota (Saccharomyces cerevisiae and Tetrahymena thermophila), two from non-halobacterial archaea (Methanothermobacter thermautotrophicus and Sulfolobus solfataricus), and three from the Halobacteria class of the Archaea domain (Haloarcula marismortui, Halalkalicoccus jeotgali, and halophilic archaeon DL31). The physiological medium of ribosomes from the former three groups includes moderate salt content (i.e. ∼150 mm), whereas the environment of archaeal ribosomes from halophilic organisms is characterized by much higher salt concentrations up to 2–5 m (14).
Because of the high salt content of the physiological environment, the isoelectric point (pI) of proteins from halophiles is known to be generally lower than the pI of proteins from non-halophilic organisms (15, 16). In other words, the proteome of halophiles is dominated by biomolecules with a net negative charge at physiological pH. This property is manifested in the high abundance of Asp and/or Glu in proteins from halophilic organisms (14). These residues are most effective at capturing hydration water in solution (17). Hence, high populations of Asp and Glu enable proteins from halophiles to successfully compete with the high bulk salt content for protein hydration.
Surprisingly, it is not known whether the above trend toward negatively charged proteins is also specifically followed by the corresponding ribosomal proteins from halophiles. Given that the function of ribosomal proteins relies on a close interaction with the highly negatively charged rRNA, it is not clear how a stable assembly could result from the interaction between highly negatively charged particles. In addition, the general electrostatic features of ribosomal proteins from all organisms are not well understood, and it is not known whether they share any common properties.
Our motivation to address the above questions is 3-fold. First, the electrostatic properties of the ribosome, its surface, and its proteins affect how the ribosome interacts with other proteins and various co-solvents and ions in solutions (11, 12). Second, electrostatic properties of the ribosomal surface and of surfaces of ribosomal proteins may help select appropriate nascent chains for co-translational protein folding studies. Third, knowledge on the physical features of ribosomal proteins contributes to our understanding of RNA-protein interactions within the ribosome.
We found that ribosomal proteins from halophilic bacteria have a higher percent of low pI, acidic proteins than ribosomal proteins from non-halophilic organisms. We also show that the majority of ribosomal proteins from a variety of organisms exhibit a large degree of intramolecular charge segregation, and the extent of this charge segregation is larger in the case of ribosomal proteins from halophiles. This property supports tight binding to ribosomal RNA and better hydration of the solvent-exposed side of ribosomal proteins. Better hydration is anticipated because a high density of solvent-exposed negative charges is known to effectively compete for water synergistically with the RNA phosphate groups on the ribosomal surface (14, 17, 18). Importantly, our analysis shows that charge segregation is a general property of ribosomal proteins from all species, although this effect is particularly pronounced in ribosomal proteins from halophiles.
EXPERIMENTAL PROCEDURES
Organisms Studied in This Work
Several organisms were chosen for the analysis of ribosomal proteins. Table 1 summarizes the organism names and database resources. Protein sequences and structures were obtained from the UniProt Knowledgebase (19) and the Protein Data Bank (20), respectively.
TABLE 1.
List of source information for the cell strains and ribosomal protein Protein Data Bank files of all organisms analyzed in this work
N.A., not applicable.
| Organism | Strain | UniProt | Protein Data Bank files |
|---|---|---|---|
| Bacteria | |||
| E. coli | K12 | ECOLI | 50 S, 2AW4 (56); 30 S, 2AVY (56); L7/L12, 1RQU (58); L9, 1DIV (59) |
| T. thermophilus | HB8/ATCC27634/DSM579 | THET8 | 50 S, 3I8I (60) |
| 30 S, 3I8H (60) | |||
| D. radiodurans | ATCC13939/DSM20539/JCM16871/LMG4051/NBRC15346/NCIMB9279/R1/VKMB-1422 | DEIRA | 50 S, 2ZJR and 2ZJQ (with L7/L12) (61) |
| Eukaryota | |||
| S. cerevisiae | ATCC204508/S288c | YEAST | 60 S, 3U5E (proteins) and 3U5D (RNA) (62); 40 S, 3U5C (proteins) and 3U5B (RNA) (62); P0, chain q in 4B6A; P1α, chain r in 4B6A; P2β, chain s in 4B6A (42) |
| T. thermophila | SB210 | TETTS | 60 S, 4A1D and 4A1E (45); 40 S, 2XZM (29) |
| Archaea | |||
| Other classes | |||
| M. thermautotrophicus | ATCC29096/DSM1053/JCM10044/NBRC100330/ΔH | METTH | N.A. |
| S. solfataricus | ATCC35092/DSM1617/JCM11322/P2 | SULSO | N.A. |
| Halobacteria | |||
| H. marismortui | ATCC43049/DSM3752/JCM8966/VKMB-1809 | HALMA | 50 S, 2QA4 (57); 30 S, N.A. |
| H. jeotgali | DSM18796/CECT7217/JCM14584/KCTC4019/B3 | HALJB | N.A. |
| Halophilic archaeon DL31 | DL31 | 9ARCH | N.A. |
Determination of Charge- and Hydrophobicity-related Parameters
pI values of each ribosomal protein were determined with the ProtParam tool (21). The percentage of negatively charged (i.e. low pI) proteins in various proteomes was estimated based on the bimodal distribution of the pI (15) of all species except for E. coli. The fraction of negatively charged proteins in the E. coli proteome was calculated from the “pI bias” value of −27% (15). The relationship between pI bias and percentage of negatively charged proteins is defined by Equation 1.
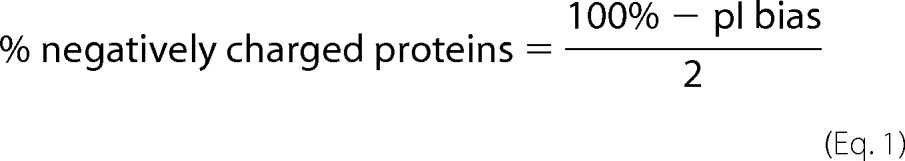 |
The mean net charge per residue (MNC)3 of each protein was determined by dividing the total net charge by the number of amino acids. Charges of +1, +1, −1, and −1 were assigned to Arg, Lys, Asp, and Glu, respectively. The charges of all other amino acids were regarded as negligible at the physiological pH of 7.4.
The hydrophobicity of individual residues was estimated according to Kyte and Doolittle (22) from hydropathy values, which are based upon water vapor transfer free energies and the interior-exterior distribution of side chains in proteins. Hydropathies were normalized on a scale from 0 to 1. The mean hydrophobicity per residue (MH) of each protein is defined as the sum of the normalized hydropathies of all amino acids divided by the total number of residues.
Following the criteria established by Uversky at el. (23), proteins can be grouped into natively folded and intrinsically disordered (IDP) proteins upon plotting the dependence of MNC on MH. The line separating the natively folded from the natively unfolded classes is defined according to the following expression (24).
To take into account the inherent uncertainties of the above predictions, Oldfield et al. (24) proposed to use the two relations below that take into account the presence of boundary regions with uncertain structural classification.
and
Equation 3 separates IDPs from partially ordered proteins, and Equation 4 separates partially ordered from natively folded proteins. MNC and MH values determined for each protein plotted in Fig. 2 were rounded to two decimal places.
FIGURE 2.
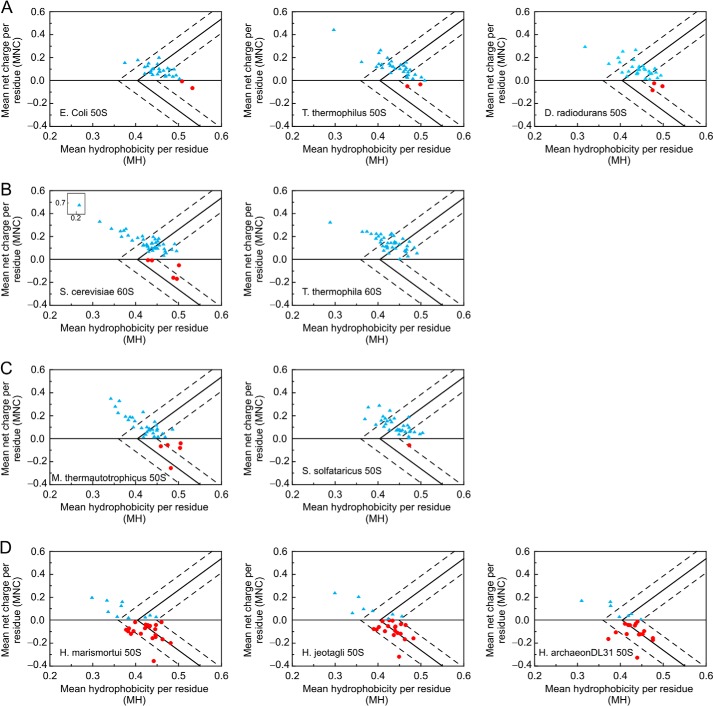
Plots illustrating the MNC versus MH of proteins from the large ribosomal subunits of different organisms. Data are shown for bacteria (A), Eukaryota (B), non-halophilic archaea (C), and halophilic archaea (D). Light blue triangles and red circles denote proteins with positive and negative MNC, respectively. The solid line separates IDPs (on the left) from independently folded proteins (on the right). The regions on the left, right, and in between the dashed lines host IDPs, folded, and partially ordered proteins, respectively. Calculations were performed based on the UniProt Knowledgebase sequence information (see “Experimental Procedures”). H. archaeon, halophilic archaeon.
Determination of Amino Acid Composition
Variations in amino acid composition were calculated using Equation 5.
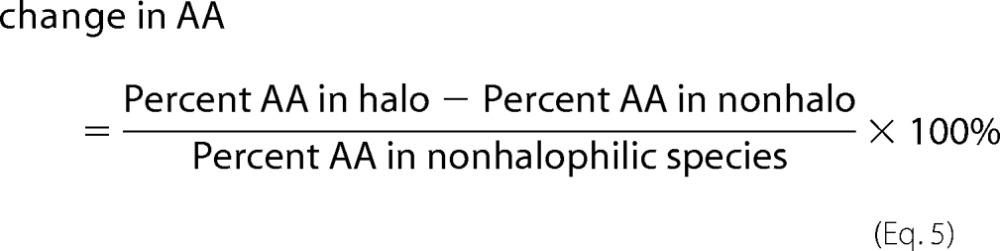 |
where AA denotes an individual amino acid. Changes in AA were assessed for both ribosomal and cellular proteins. The %AA data for the cellular proteins were taken from Rao and Argos (25).
Computation of Electrostatic Surface Potential
Protein Data Bank files were converted to PQR files with PDB2PQR software (26, 27). PQR files were used as input for electrostatic potential calculations using the Adaptive Poisson-Boltzmann Solver (APBS) software package developed by Baker et al. (12). Electrostatic surface potential calculations were performed by solving the linearized Poisson-Boltzmann equation (28). In general, the linearized Poisson-Boltzmann equation is not valid for very highly charged systems, and presumably the ribosome belongs to this class given its large surface charge density. However, a rigorous solution of the nonlinear Poisson-Boltzmann equation is challenging in the case of the ribosome due to, for instance, uncertainties in the local concentration of divalent cations like Mg2+ and putrescine2+ and the variable dielectric constant at the ribosomal surface (11, 30). Nonetheless, we also performed electrostatic surface potential calculations with the nonlinear Poisson-Boltzmann equation (28), and we obtained results equivalent to those of the linearized Poisson-Boltzmann equation calculations (data not shown). The ribosome was treated as a dielectric continuum with dielectric constant 2.0 embedded in a solvent of dielectric constant 78.0. All calculations used a 150 mm sodium chloride concentration. In Figs. 3 and 6, surfaces are colored so that blue denotes regions of positive potential (>+1 kBT/e unless otherwise stated) and red denotes regions of negative potential (<−1 kBT/e unless otherwise stated). kBT is 4.11 × 10−21 J at room temperature where kB is the Boltzmann constant in J/Kelvin (K), T is the temperature in K, and e is the charge of a single electron in coulombs. Table 2 lists the parameters used in the electrostatic calculations. Electrostatic surface potentials were viewed with the PyMOL software package (31) equipped with the APBS plug-in.
FIGURE 3.
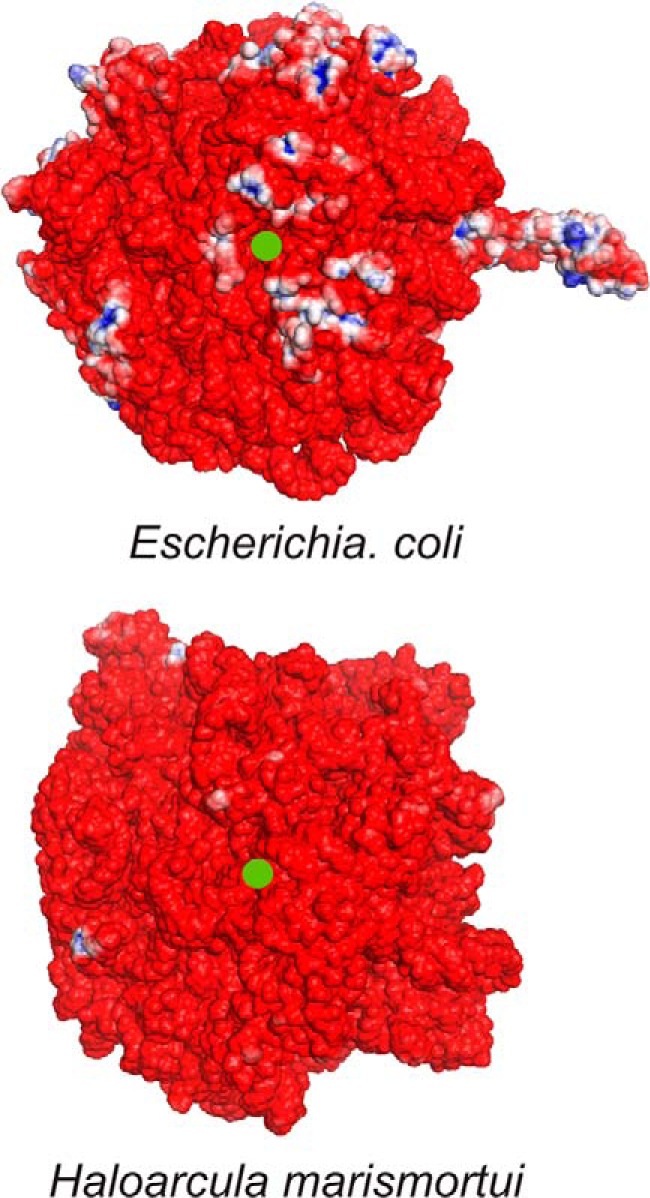
Electrostatic surface potential maps of the 50 S subunit of ribosomes from E. coli and H. marismortui in proximity to the exit tunnel. The ribosomal exit tunnel is denoted by a green dot. Electrostatic potentials were obtained with APBS (150 mm ionic strength, solute dielectric of 2.0, and solvent dielectric of 78.0) using three-dimensional structures with Protein Data Bank codes 2AW4 (56) and 2QA4 (57) for E. coli and H. marismortui, respectively. Regions with positive (>+3 kBT/e) and negative potential (<−3 kBT/e) are shown in blue and red, respectively. kBT denotes an energy unit of 4.11 × 10−21 J at room temperature (where kB is the Boltzmann constant and T is temperature), and e denotes the electric charge in coulombs.
FIGURE 6.
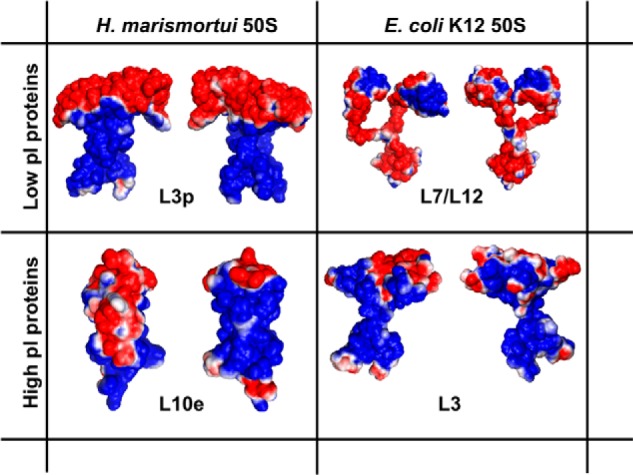
Electrostatic surface potential map of four representative 50 S ribosomal proteins. Front and back views are shown for L3p and L10e from H. marismortui and L7/L12 and L3 from E. coli. Protein Data Bank code information is as follows: L3p, chain B of 2QA4 (57); L7/L12, 1RQU (58); L10e, chain H of 2QA4; L3, chain D of 2AW4 (56).
TABLE 2.
Summary of parameters used to compute electrostatic surface potentials with the APBS software package
| Parameter | Value |
|---|---|
| Resolution (Å) | ∼0.5 |
| Dielectric constant of species | 2.0 |
| Dielectric constant of solvent | 78.0 |
| Ion concentration in solvent (mm) | 150 |
| Number of grid points | Protein Data Bank file-specific |
| Solvent radius (Å) | 1.4 |
| Temperature (K) | 310.0 |
| Vacuum sphere density (grid points/Å2) | 10.0 |
Computation of Extent of Charge Segregation
The average extent of charge segregation in ribosomal proteins was quantitatively assessed by first determining the solvent-accessible surface area (ASA) of the charged functional groups of Asp and Glu (COO−), Lys (NH3+), and Arg (NH-C+(NH2)2) of each of the 50 S ribosomal proteins embedded in the ribosome. The total surface area (Total SAi) due to either positively or negatively charged (i = +, −) groups and the corresponding percentage of solvent-accessible charged surface area (%ASAi) of each ribosomal protein were assessed as follows.
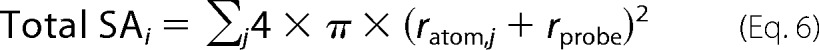 |
and
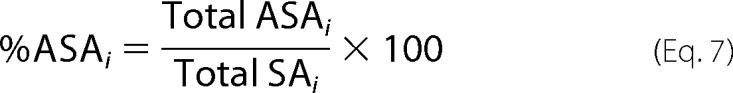 |
where j denotes each of the positively or negatively charged groups, rprobe denotes the radius of a rolling sphere probe, and the parameter Total ASAi denotes the total positively or negatively charged surface area that is exposed to the solvent in each ribosome-embedded protein. The resulting %ASAi values were averaged over all ribosomal proteins of any given species to yield %ASAi. For each ribosomal protein, SAi and ASAi values were computed with the NACCESS 2.1.1 software package (32). Simple Python scripts were written to eliminate incompatibilities between the APBS and NACCESS output files. The NACCESS algorithm uses the rolling ball approach of Lee and Richards (33) to calculate ASA. The radius of the rolling sphere probe rprobe was set to 1.4 Å.
RESULTS AND DISCUSSION
Role of Net Charge
We analyzed all available proteins from the large ribosomal subunit of the ribosomes from E. coli, T. thermophilus, D. radiodurans, S. cerevisiae, T. thermophila, M. thermautotrophicus, S. solfataricus, H. marismortui, H. jeotgali, and halophilic archaeon DL31 in terms of pI and MNC. The UniProt Knowledgebase (19) served as a source of sequence information. MNC values were then computed (see details under “Experimental Procedures”). Our work focuses on proteins from the large ribosome subunit because data from the small subunit were too scarce, i.e. often entirely unavailable or missing large fractions of proteins.
As shown in Table 3, ribosomal proteins from non-halophiles and extreme halophiles have distinct characteristics. Specifically, ribosomal proteins from non-halophilic organisms have a large percentage of high pI proteins, ranging from 86 to 100% (Table 3). High pI proteins are positively charged at physiological pH. Hence, this trend is consistent with the fact that ribosomal proteins experience strong electrostatic interactions with the highly negatively charged ribosomal RNA (6). This is a peculiarity of ribosomal proteins given that the percentage of high pI proteins in the corresponding entire proteomes of non-halophiles fluctuates between 27 and 60%, leaning toward a high abundance in acidic (i.e. low pI) proteins.
TABLE 3.
Electrostatic properties of proteins from 10 different organisms
The table illustrates the percentage of proteins with high and low pI (i.e. with pI >7 or <7, respectively) and the MNC.
| Organism | Percentage of high pI proteins in large ribosomal subunit | Percentage of low pI proteins in large ribosomal subunit | Average MNC of proteins in large ribosomal subunita | Percentage of high pI proteins within the entire proteome (15, 36, 37) | Percentage of low pI proteins within the entire proteome (15, 36, 37) |
|---|---|---|---|---|---|
| % | % | % | % | ||
| E. coli | 93 | 7 | 0.08 | 36 | 64 |
| T. thermophilus | 94 | 6 | 0.12 | ∼42 | ∼58 |
| D. radiodurans | 91 | 9 | 0.10 | ∼35 | ∼65 |
| S. cerevisiae | 89 | 11 | 0.13 | ∼50 | ∼50 |
| T. thermophila | 100 | 0 | 0.15 | ∼60 | ∼40 |
| S. solfataricus | 97 | 3 | 0.12 | ∼57 | ∼43 |
| M. thermautotrophicus | 86 | 14 | 0.09 | ∼27 | ∼73 |
| H. marismortuib | 34 | 66 | −0.04 | ∼3 | ∼97 |
| H. jeotgalib | 33 | 67 | −0.03 | ∼8 | ∼92 |
| Halophilic archaeon DL31b | 30 | 70 | −0.05 | ∼7 | ∼93 |
a The average MNC is defined as the MNC averaged over all proteins belonging to the large ribosomal subunit of each species.
b Halophile.
Conversely, ribosomal proteins from extreme halophiles have on average a much lower pI; i.e. they are much more acidic than ribosomal proteins from non-halophilic organisms. The ratio of low pI to high pI ribosomal proteins from halophiles is ∼2:1. This trend is consistent with the general properties of the proteomes of halophilic organisms, which are dramatically shifted toward low pI values (15).
MNC values averaged over all the ribosomal proteins in the large ribosomal subunit of each species are also shown in Table 3 and are consistent with the pI trends. MNC values are positive for ribosomal proteins from non-halophiles and negative for those from halophiles.
The high degree of acidity of proteins from halophiles arises as a natural response to the high salt concentration of the physiological medium (14, 34). Acidic proteins have a larger fraction of negatively charged amino acids at the physiological pH of 7.4, i.e. Asp and Glu. Proteins with a high Asp and Glu content are more hydrated (17) than proteins from non-halophilic organisms, which have a lower Asp and Glu content (14). The presence of highly hydration-prone residues in proteins from halophiles results from the need to compete with the high salt content of the medium to maintain an effective hydration shell. Conversely, proteins from organisms that live under conventional salt concentrations (i.e. ∼150 mm) are not as pressed to compete for hydration water (14, 17, 25, 35).
Considering the above arguments, the presence of a substantial percentage of positively charged high pI ribosomal proteins (30–34%) in high salt environments is intriguing. This value is considerably larger than the percent of high-pI positively charged proteins from the corresponding entire organism, which is only 3–8% (36, 37) (Table 3). Indeed, the pI distribution of the entire proteome of extreme halophiles, averaged over several exponents of this class, is known to be dramatically shifted toward low pI values (15). We suggest that the relatively high abundance of positively charged ribosomal proteins in halophiles results from the necessity to ensure effective binding to rRNA together with the need to prevent excessive protein destabilization caused by electrostatic repulsion in negatively charged proteins (38).
At this juncture, it is not clear how the high pI positively charged ribosomal proteins from halophiles can be successfully biosynthesized and survive in the high salt medium before being incorporated into the ribosome given the difficulties in being properly hydrated. It is possible that these proteins may remain associated with the ribosome during biosynthesis before being assembled within a new ribosome. Alternatively, they may become co- or post-translationally associated with a specific negatively charged molecular chaperone before participating in ribosome assembly. More research is needed to address this topic experimentally.
Role of Hydrophobicity
To explore the nonpolar content of ribosomal proteins, we plotted the MH as a function of pI for ribosomal proteins from different organisms. Fig. 1 shows that ribosomal proteins from halophiles exhibit a dramatic shift toward a low pI, consistent with Table 3. Fig. 1 also shows that, regardless of organism type, the hydrophobicity of high pI ribosomal proteins is generally lower than that of low pI proteins. This trend may have evolved in response to the need to effectively penetrate the rRNA during ribosome assembly (9, 18, 39, 40). Remarkably, the nonpolar content of ribosomal proteins from halophiles is lower than that of ribosomal proteins from non-halophilic organisms even at low pI values, suggesting a general role of low hydrophobicity in ribosomal proteins from halophiles.
FIGURE 1.
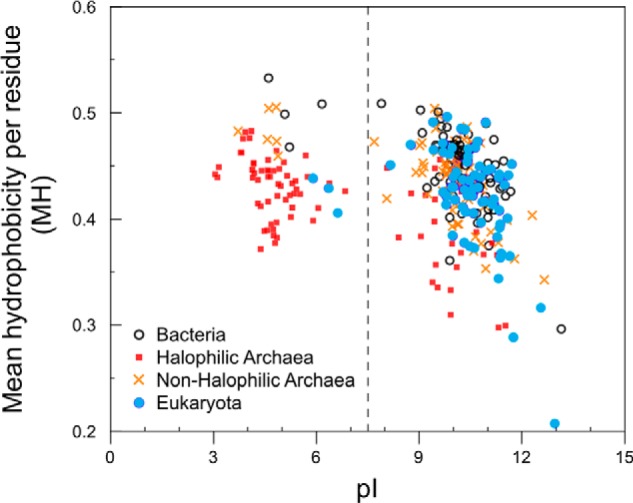
Hydrophobicity and pI of ribosomal proteins in the large subunit of the ribosome from nine organisms. Red solid squares, 50 S ribosomal proteins from halophilic archaea H. marismortui, H. jeotgali, and halophilic archaeon DL31; blue solid circles, eukaryotic 60 S ribosomal proteins from S. cerevisiae and T. thermophila; black open circles, bacterial 50 S ribosomal proteins from E. coli, T. thermophilus, and D. radiodurans; orange crosses, 50 S ribosomal proteins from non-halophilic archaea S. solfataricus and M. thermautotrophicus. The vertical dotted line denotes the physiological pH of 7.4.
We further explored the role of hydrophobicity by plotting the MNC versus MH of ribosomal proteins from 10 different species in Fig. 2. Relative MNC and MH values are excellent predictors of the most highly populated state of proteins (ordered, partially ordered, or fully disordered) under physiological conditions (23). For instance, many IDPs have a high net charge (leading to strong charge-charge repulsion) and low hydrophobicity, resulting in an insufficient driving force for autonomous folding (28). Chen et al. (41) suggested that ribosomal proteins contain conserved regions of predicted disorder. Fig. 2, A–C, show Uversky-type plots of ribosomal proteins from seven non-halophilic organisms, and D shows the corresponding plots for proteins from halophiles. The solid line represents the putative discriminating edge between IDPs (to the left) and independently folded (to the right) proteins based on prior work from Uversky and co-workers (23, 24). Regions enclosed within the dashed lines correspond to proteins from either structural class (24).
Inspection of Fig. 2 and Table 4 reveals a number of trends. First, the fraction of low MH ribosomal proteins from halophiles is somewhat lower than that of non-halophiles. This observation parallels the fact that the MH of the entire proteome of halophilic organisms is lower than that of non-halophilic species (34).
TABLE 4.
Hydrophobicity and degree of independent folding of proteins from the large ribosomal subunit of 10 organisms
| Organism | Fraction of proteins with MH <0.36 | Average MH | Fraction of IDPsa | Fraction of independently folded proteinsa |
|---|---|---|---|---|
| % | % | % | ||
| Non-halophiles | ||||
| Bacteria | ||||
| E. coli | 0.0 | 0.46 | 6.7 | 30 |
| T. thermophilus | 3.0 | 0.44 | 21 | 27 |
| D. radiodurans | 3.0 | 0.44 | 18 | 21 |
| Eukaryota | ||||
| S. cerevisiae | 6.5 | 0.43 | 26 | 11 |
| T. thermophila | 2.6 | 0.43 | 34 | 5.3 |
| Non-halophilic archaea | ||||
| S. solfataricus | 0.0 | 0.44 | 23 | 20 |
| M. thermautotrophicus | 8.6 | 0.43 | 31 | 14 |
| Halophiles | ||||
| Halophilic archaea | ||||
| H. marismortui | 13 | 0.41 | 28 | 3.1 |
| H. jeotgali | 11 | 0.42 | 22 | 0.0 |
| Halophilic archaeon DL31 | 4.3 | 0.42 | 26 | 0.0 |
a Note that the values reported in this column refer only to the ribosomal proteins falling outside of the error bar regions (dashed lines) of Fig. 2.
Second, the third column of Table 4 shows that all ribosomes have a significant fraction of IDPs, confirming the general importance of disorder in ribosomal structure and assembly (41). Furthermore, the fourth column of Table 4 shows that, whereas the ribosomal proteins falling within the independently folded region vary between ∼5 (T. thermophila) and 30% (E. coli) for non-halophiles, only ∼0–3% of the proteins from the halophiles are independently folded.
Third, ribosomal proteins from non-halophiles are largely positively charged (MNC > 0), whereas ribosomal proteins from halophiles are comparably distributed between negatively and positively charged (Fig. 2 and Table 4).
Fourth, Fig. 2 shows that negatively charged ribosomal proteins, predominantly found in halophiles, display fewer IDPs as well as fewer independently folded proteins than positively charged ribosomal proteins. This result is consistent with the previously reported coexistence of ordered and disordered regions with negatively and positively charged character, respectively, in the ribosomal proteins of the H. marismortui halophile (6, 18, 41).
The latter observation suggests that positively charged disordered ribosomal proteins or protein regions may prefer to be embedded within the negatively charged rRNA-rich core of the ribosome. This led us to hypothesize that there may be some degree of surface charge segregation in ribosomal proteins. To test this hypothesis, we calculated and visualized the electrostatic surface potential of the E. coli and H. marismortui 50 S ribosomal subunits with the APBS software package. Fig. 3 illustrates the results. Ribosomal proteins make up ∼40% of the molecular weight of the ribosome (5). In non-halophilic organisms, these proteins have a pI ranging from 9 to 12. Thus, if the surface charge of the 50 S ribosomal proteins were randomly distributed, then E. coli ribosomes would be expected to display a much larger fraction of positively charged blue regions arising from the protein portion of the 50 S subunit. Moreover, the E. coli 50 subunit would be expected to be much more positively charged than the corresponding subunit of the H. marismortui ribosome given the previously discussed pI of the respective proteins. However, we only observed moderate differences in the surface charge of the ribosomes from the two species (Fig. 3). This finding supports the hypothesis that charge is ubiquitously segregated in ribosomal proteins from different organisms and that the bulk of the positive charge due to proteins may be buried inside the ribosome. This concept is pictorially represented in Fig. 4 and will be further explored below.
FIGURE 4.
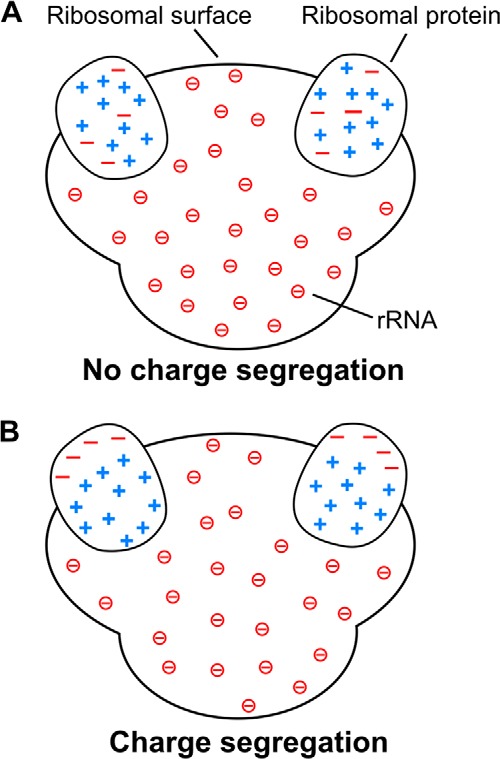
Schematic models for the charge distribution of ribosomal proteins illustrating the charge segregation concept. A, ribosomal proteins with no intramolecular charge segregation. B, ribosomal proteins with intramolecular charge segregation supported by this study. The negative charges enclosed in circles are from the phosphate groups of rRNA. For simplicity, only two ribosomal proteins are shown embedded in each ribosome.
Amino Acid Content of Ribosomal Proteins
To better understand the origin of the shifted MNC and MH values discussed before, we studied the specific amino acid composition of ribosomal proteins from halophilic and non-halophilic organisms. Fig. 5 shows that the increased acidity of the ribosomal proteins from halophiles is due to an increase in the percentage of aspartates and glutamates and to a decrease in the percentage of lysines. Interestingly, the percentage of arginines increases slightly. Fukuchi et al. (34) showed that, in the entire proteome of halophiles, the probability of finding solvent-exposed (as opposed to buried) negatively charged residues is higher by 1.7-fold for halophiles than for non-halophiles. In contrast, the corresponding probability for positively charged residues is only 1.1-fold higher for halophiles (34). This observation points to the importance of solvent-exposed negatively charged residues in halophiles. The abundance of these residues and their contribution to surface hydration have already been discussed in previous sections.
FIGURE 5.
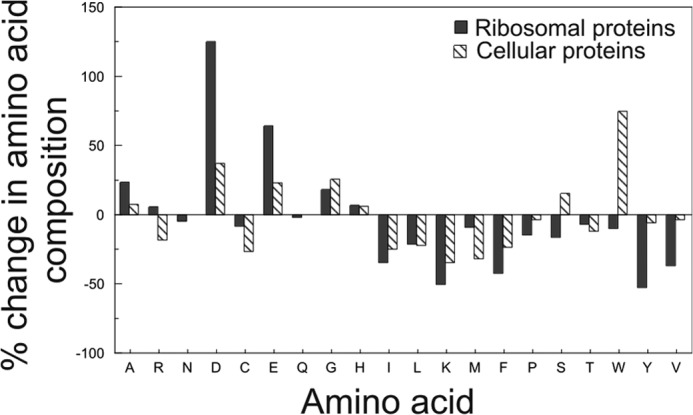
Percent change in amino acid composition of ribosomal and cellular proteins from non-halophilic to halophilic organisms. The black bars denote ribosomal proteins. The striped bars denote selected cellular proteins according to Rao and Argos (25). No data are available for the asparagine (N) and glutamine (Q) content of cellular proteins. Data were generated from the amino acid sequences of ribosomal proteins from seven non-halophilic and three halophilic organisms (details are given under “Experimental Procedures”).
Leucine, isoleucine, and valine contribute the most to the overall hydrophobicity of a protein (22). The fraction of these residues in ribosomal proteins is lower than in non-ribosomal proteins (Fig. 5). At high concentration of the salts that are abundant in the halophilic intracellular medium (e.g. K+ salts (11)), Hofmeister effects cause an increase protein stability (43) in part due to a strengthening of the hydrophobic effect (14, 44). The lower content in nonpolar residues of ribosomal proteins from halophiles may therefore be dictated by the need to bring the thermodynamic stability of these proteins down to non-halophilic values. The large decrease in Lys content (relative to a slight increase in Arg) may be a consequence of the higher hydrophobicity of the Lys side chain (46). Conversely, the moderate increase in the Arg content of ribosomal proteins from halophiles may be due to the low nonpolar character of this residue (14, 25). The overall trends in amino acid composition of ribosomal proteins of halophiles and non-halophiles are similar to those of the corresponding entire proteomes (14).
Charge Segregation of Ribosomal Proteins
Very little is known about the charge distribution of ribosomal proteins and its relation with the highly negatively charged rRNA. This topic is particularly intriguing in the case of ribosomal proteins from halophiles because of their high abundance in negative charges.
Given that the charge density of rRNA is nearly identical across halophilic and non-halophilic organisms, rRNAs from different organisms are likely to interact similarly with ribosomal proteins. However, how do negatively charged ribosomal proteins cope with such a highly negative rRNA charge density in their proximity? We hypothesize that a major characteristic feature of ribosomal proteins from all organisms is the presence of charge segregation. This property serves the purpose of preserving effective binding to rRNA despite their difference in overall net charge. For instance, it is known that some ribosomal proteins of H. marismortui have long positively charged extensions penetrating deep into the RNA during ribosomal assembly (18, 40).
As an initial qualitative estimate of the extent of charge segregation in the large ribosomal subunits, we inspected the surface electrostatic potential of ribosomal proteins for the presence of distinct patches of positive and negative charge density. Fig. 6 provides four representative examples of the electrostatic surface potential of low and high pI proteins from halophilic (H. marismortui) and non-halophilic (E. coli) organisms. The presence of significant charge segregation is evident in the images. Furthermore, as shown in Table 5, charge segregation is detected in the majority (62–100%) of ribosomal proteins from both halophiles and non-halophiles. Importantly, this property is not limited to the nonglobular extensions, as had been proposed by Klein et al. (18), but is also manifested by the presence of extensive negatively and positively charged patches on the surface of globular proteins. The small number of available proteins that could be used to generate the data in Table 5 (high resolution three-dimensional structures were required) prevents a quantitative evaluation of the statistical significance of charge separation across different organisms. Despite this limitation, the data in the last column of Table 5 suggest that proteins from halophiles may exhibit more charge segregation than proteins from non-halophiles.
TABLE 5.
Qualitative assessment of the fraction of ribosomal proteins exhibiting charge segregation
This analysis was performed upon visual inspection of the electrostatic surface area of 193 ribosomal proteins. N.A., not applicable.
| Organism | Protein Data Bank file | Proteins in large ribosomal subunita |
|||
|---|---|---|---|---|---|
| Low pI |
High pI |
||||
| Total number of proteins | Fraction of proteins exhibiting charge segregation | Total number of proteins | Fraction of proteins exhibiting charge segregation | ||
| % | % | ||||
| Non-halophiles | |||||
| Bacteria | |||||
| E. coli | 2AW4 | 2 | 100b | 27 | 85 |
| T. thermophilus | 3I8I | 2 | 100b | 29 | 90 |
| D. radiodurans | 2ZJR | 1 | 100b | 24 | 83 |
| Eukaryota | |||||
| S. cerevisiae | 3U5E | 5c | 80 | 37 | 70 |
| T. thermophila | 4A1D 4A1E | 0 | N.A. | 39 | 62 |
| Halophiles | |||||
| Halophilic archaea | |||||
| H. marismortui | 2QA4 | 18 | 100 | 9 | 100 |
A more quantitative characterization of the extent of charge segregation was carried out by computing the fraction of ASA of the charged groups of Asp and Glu (COO−), Lys (NH3+), and Arg (NH-C+(NH2)2) of 50 S ribosomal proteins embedded in the ribosome. Table 6 shows the results for proteins from five organisms: E. coli, T. thermophilus, D. radiodurans, S. cerevisiae, and H. marismortui. It is clear that, for all species examined, the fraction of negatively charged ASA is much higher (average value, 75–86%) than the fraction of positively charged ASA (average value, 49–59%). The difference is especially pronounced in proteins from the halophilic H. marismortui. This result confirms the hypothesis that charge segregation is a property of ribosomal proteins.
TABLE 6.
Quantitative assessment of the average extent of charge segregation of ribosomal proteins
Data were evaluated as percentage of positively and negatively charged solvent-accessible surface area of ribosomal proteins embedded within the ribosome, averaged over the ribosomal proteins of each species. See explicit definitions of %ASAi+ and %ASAi− under “Experimental Procedures.” %ASA+ and %ASA− values of each individual protein are also provided (see supplemental data).
| Organisma | %ASAi+ | %ASAi− |
|---|---|---|
| % | % | |
| E. coli | 57 | 75 |
| T. thermophilus | 59 | 81 |
| D. radiodurans | 57 | 84 |
| S. cerevisiae | 51 | 83 |
| H. marismortui | 49 | 86 |
a Data were evaluated for the ribosomal proteins of the large ribosomal subunit of each organism.
In summary, the data in Table 6 show that the negative charge of ribosomal proteins preferentially points toward the solvent-accessible region, consistent with the schematic in Fig. 4B. In addition, ribosomal proteins from halophiles exhibit a stronger degree of charge segregation than ribosomal proteins from non-halophiles, consistent with the model in Fig. 7.
FIGURE 7.
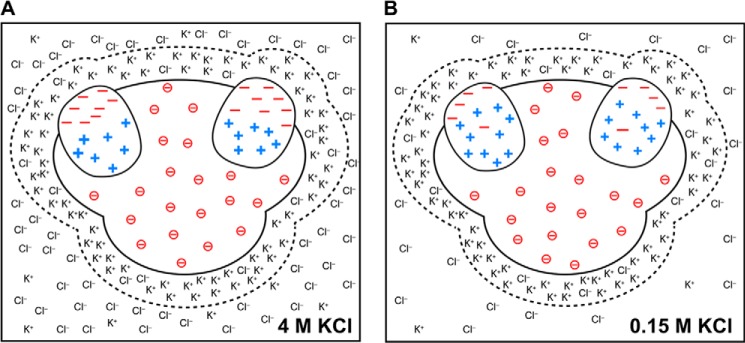
Schematic model illustrating the proposed charge distribution in ribosomes from halophilic and non-halophilic organisms. A, ribosome from a halophile in 4 m KCl. B, ribosome from a non-halophile in 150 mm KCl. The dashed lines around the ribosomal surfaces enclose a simplified view of counterion layers and emphasize the postulated similarity of these layers in halophilic and non-halophilic organisms. For simplicity, only two ribosomal proteins are shown embedded in each ribosome.
How Does the Ribosome Cope with Surface Negative Charges Contributed by Both rRNA and Ribosomal Proteins?
Given the presence of charge segregation (leading to high solvent exposure of negative charges) in ribosomal proteins, it is important to ask how the ribosome may cope with the high surface density of negative charges contributed by both rRNA and ribosomal proteins.
A high salt environment often leads to an increase in the pKa of aspartate and glutamate from ∼3 and ∼4 at 5 mm NaCl to 4.9 and 5.3 at 5 m NaCl, respectively (48, 49). This effect contributes to decrease the destabilizing effect of charge repulsion among negatively charged residues (38) within solvent-exposed regions of ribosomal proteins. In addition, the pKa of both acidic and basic residues may increase due to RNA-protein proximity (28). This prediction leads to an expected lowering in the negative charge near aspartates and glutamates and increasing positive charge near lysines and arginines, leading to additional stabilizing effects.
In addition, just like all nucleic acids (e.g. double-stranded DNA), the ribosomal surface is surrounded by a counterion layer whose concentration is very high (given the high surface charge density) regardless of the bulk salt concentration (50). This phenomenon is known as the polyelectrolyte effect (51–54). The layer of counterions shields repulsive interactions via coulombic interactions and, at the high bulk salt concentrations of the halophilic media, exerts an additional stabilizing role via Hofmeister effects (43).
Hence, based on the above information on the known behavior of nucleic acids and charged residues in proteins, we propose that ribosomes from non-halophilic organisms have layers of counterions of a composition fairly similar to that of ribosomes from halophiles. Hence, regardless of the bulk salt content, the ribosomal surface may maintain an overall similar surface microenvironment to properly sustain all interactions essential for biological activity (54). This concept is pictorially illustrated in Fig. 7.
Finally, the L7/L12 stalk ribosomal proteins (55) are to be regarded as a special case. These proteins are not embedded within the ribosome, and they are not directly bound to rRNA. They associate with the ribosome via rRNA-bound accessory proteins. The L7/L12 proteins are also highly dynamic because of their peculiar functional role in translation, i.e. to bind translation factors as they approach the ribosome and to promote GTP hydrolysis. As shown in Fig. 6, the L7/L12 proteins exhibit some degree of charge segregation. However, supplemental Tables 3–5 show that the solvent-exposed regions are not always preferentially exposing negative charges in different organisms. The meaning of charge segregation in the L7/L12 proteins is not well understood yet, and it likely does not fall within the trends discussed in this work.
Conclusion
In summary, ribosomal proteins are generally characterized by a high pI and a net positive charge at physiological pH. However, ribosomal proteins from extreme halophilic organisms are overall more negatively charged, less hydrophobic, and have an overall slightly less ordered character than those from non-halophiles. In other words, the overall MNC and MH values are lower for ribosomal proteins from halophiles than those from non-halophiles. The latter findings for halophiles parallel the hydrophobicity trends of the corresponding halophilic proteomes. On the other hand, ribosomal proteins from halophiles have a higher percentage of high-pI proteins than the corresponding proteomes. This trend suggests that, despite their largely negatively-charged proteome, halophiles retain some positively charged proteins in the ribosome to preserve its integrity.
How can the highly negatively charged ribosomal proteins of halophiles be compatible with close interactions with the highly negatively charged rRNA? Importantly, we found that a large fraction of ribosomal proteins experiences charge segregation with positively charged regions buried within the rRNA and negatively charged regions exposed to the solvent. Interestingly, while halophiles have a higher fraction of charge-segregated proteins than non-halophilic organisms, intramolecular charge segregation is a common property of ribosomal proteins from a variety of species.
Supplementary Material
Acknowledgments
We thank Prof. Julie Mitchell for help with using NACCESS; Newsha Ardalani and Sharad Brahma Aksha Punuganti for help with electrostatic potential and accessible surface area calculations; Rudy Clausen for writing a Python script for MNC and MH calculations; Yue Liu for technical assistance; Paweł Mackiewicz for providing pI distribution data on the entire proteome of H. Jeotgali and H. Archaeon DL31; Thomas Record, Jr., Irina Shkel, and Sarah Woodson for helpful discussions; and Dr. Nathan Baker for consultations regarding the APBS software.
This work was supported in part by National Science Foundation Grant MCB 0951209 (to S. C.).

This article contains supplemental Tables 1–5.
- MNC
- mean net charge per residue
- MH
- mean hydrophobicity per residue
- APBS
- Adaptive Poisson-Boltzmann Solver
- IDP
- intrinsically disordered protein
- ASA
- solvent-accessible surface area.
REFERENCES
- 1. Harms J., Schluenzen F., Zarivach R., Bashan A., Gat S., Agmon I., Bartels H., Franceschi F., Yonath A. (2001) High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107, 679–688 [DOI] [PubMed] [Google Scholar]
- 2. Selmer M., Dunham C. M., Murphy F. V., 4th, Weixlbaumer A., Petry S., Kelley A. C., Weir J. R., Ramakrishnan V. (2006) Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 [DOI] [PubMed] [Google Scholar]
- 3. Yusupov M. M., Yusupova G. Z., Baucom A., Lieberman K., Earnest T. N., Cate J. H., Noller H. F. (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science 292, 883–896 [DOI] [PubMed] [Google Scholar]
- 4. Ramakrishnan V. (2011) Molecular biology. The eukaryotic ribosome. Science 331, 681–682 [DOI] [PubMed] [Google Scholar]
- 5. Melnikov S., Ben-Shem A., Garreau de Loubresse N., Jenner L., Yusupova G., Yusupov M. (2012) One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 19, 560–567 [DOI] [PubMed] [Google Scholar]
- 6. Wilson D. N., Nierhaus K. H. (2005) Ribosomal proteins in the spotlight. Crit. Rev. Biochem. Mol. Biol. 40, 243–267 [DOI] [PubMed] [Google Scholar]
- 7. Korobeinikova A. V., Garber M. B., Gongadze G. M. (2012) Ribosomal proteins: structure, function, and evolution. Biochemistry 77, 562–574 [DOI] [PubMed] [Google Scholar]
- 8. Brodersen D. E., Nissen P. (2005) The social life of ribosomal proteins. FEBS J. 272, 2098–2108 [DOI] [PubMed] [Google Scholar]
- 9. Woodson S. A. (2011) RNA folding pathways and the self-assembly of ribosomes. Acc. Chem. Res. 44, 1312–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shajani Z., Sykes M. T., Williamson J. R. (2011) Assembly of bacterial ribosomes. Annu. Rev. Biochem. 80, 501–526 [DOI] [PubMed] [Google Scholar]
- 11. Guttman H. J., Cayley S., Li M., Anderson C. F., Record M. T., Jr. (1995) K+-ribosome interactions determine the large enhancements of 39K NMR transverse relaxation rates in the cytoplasm of Escherichia coli K-12. Biochemistry 34, 1393–1404 [DOI] [PubMed] [Google Scholar]
- 12. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Culviner P. H., Knight A. M., Kurt N., Zou T., Ozkan S. B., Cavagnero S. (2013) Electrostatic effect of the ribosomal surface on nascent polypeptide dynamics. ACS Chem. Biol. 8, 1195–1204 [DOI] [PubMed] [Google Scholar]
- 14. Lanyi J. (1974) Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol. Rev. 38, 272–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kiraga J., Mackiewicz P., Mackiewicz D., Kowalczuk M., Biecek P., Polak N., Smolarczyk K., Dudek M. R., Cebrat S. (2007) The relationships between the isoelectric point and: length of proteins, taxonomy and ecology of organisms. BMC Genomics 8, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Elevi Bardavid R., Oren A. (2012) Acid-shifted isoelectric point profiles of the proteins in a hypersaline microbial mat: an adaptation to life at high salt concentrations? Extremophiles 16, 787–792 [DOI] [PubMed] [Google Scholar]
- 17. Kuntz I. D. (1971) Hydration of macromolecules. 3. Hydration of polypeptides. J. Am. Chem. Soc. 93, 514–516 [DOI] [PubMed] [Google Scholar]
- 18. Klein D. J., Moore P. B., Steitz T. A. (2004) The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 340, 141–177 [DOI] [PubMed] [Google Scholar]
- 19. UniProt Consortium (2007) The Universal Protein Resource (UniProt). Nucleic Acids Res. 35, D193–D197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) The Protein Data Bank. Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gasteiger E., Hoogland C., Gattiker A., Duvaud S. E., Wilkins M., Appel R., Bairoch A. (2005) in The Proteomics Protocols Handbook (Walker J., ed), pp. 571–607, Humana Press, Totowa, NJ [Google Scholar]
- 22. Kyte J., Doolittle R. F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 23. Uversky V. N., Gillespie J. R., Fink A. L. (2000) Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 41, 415–427 [DOI] [PubMed] [Google Scholar]
- 24. Oldfield C. J., Cheng Y., Cortese M. S., Brown C. J., Uversky V. N., Dunker A. K. (2005) Comparing and combining predictors of mostly disordered proteins. Biochemistry 44, 1989–2000 [DOI] [PubMed] [Google Scholar]
- 25. Rao J. K., Argos P. (1981) Structural stability of halophilic proteins. Biochemistry 20, 6536–6543 [DOI] [PubMed] [Google Scholar]
- 26. Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., Baker N. A. (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A. (2004) PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dill K. A., Bromberg S. (2010) Molecular Driving Forces: Statistical Thermodynamics in Chemistry and Biology, 2nd Ed., Garland Science, New York [Google Scholar]
- 29. Rabl J., Leibundgut M., Ataide S. F., Haag A., Ban N. (2011) Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331, 730–736 [DOI] [PubMed] [Google Scholar]
- 30. Ni H., Anderson C. F., Record M. T. (1999) Quantifying the thermodynamic consequences of cation (M2+, M+) accumulation and anion (X−) exclusion in mixed salt solutions of polyanionic DNA using Monte Carlo and Poisson-Boltzmann calculations of ion-polyion preferential interaction coefficients J. Phys. Chem. B 103, 3489–3504 [Google Scholar]
- 31. DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 32. Hubbard S. J., Thornton J. M. (1993) NACCESS, Department of Biochemistry and Molecular Biology, University College London, London [Google Scholar]
- 33. Lee B., Richards F. M. (1971) The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 55, 379–400 [DOI] [PubMed] [Google Scholar]
- 34. Fukuchi S., Yoshimune K., Wakayama M., Moriguchi M., Nishikawa K. (2003) Unique amino acid composition of proteins in halophilic bacteria. J. Mol. Biol. 327, 347–357 [DOI] [PubMed] [Google Scholar]
- 35. Kuntz I. D., Jr., Brassfield T. S., Law G. D., Purcell G. V. (1969) Hydration of macromolecules. Science 163, 1329–1331 [DOI] [PubMed] [Google Scholar]
- 36. Mongodin E. F., Nelson K. E., Daugherty S., Deboy R. T., Wister J., Khouri H., Weidman J., Walsh D. A., Papke R. T., Sanchez Perez G., Sharma A. K., Nesbø C. L., MacLeod D., Bapteste E., Doolittle W. F., Charlebois R. L., Legault B., Rodriguez-Valera F. (2005) The genome of Salinibacter ruber: convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc. Natl. Acad. Sci. U.S.A. 102, 18147–18152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goo Y. A., Roach J., Glusman G., Baliga N. S., Deutsch K., Pan M., Kennedy S., DasSarma S., Ng W. V., Hood L. (2004) Low-pass sequencing for microbial comparative genomics. BMC Genomics 5, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Elcock A. H., McCammon J. A. (1998) Electrostatic contributions to the stability of halophilic proteins. J. Mol. Biol. 280, 731–748 [DOI] [PubMed] [Google Scholar]
- 39. Sayers E. W., Gerstner R. B., Draper D. E., Torchia D. A. (2000) Structural preordering in the N-terminal region of ribosomal protein S4 revealed by heteronuclear NMR spectroscopy. Biochemistry 39, 13602–13613 [DOI] [PubMed] [Google Scholar]
- 40. Timsit Y., Acosta Z., Allemand F., Chiaruttini C., Springer M. (2009) The role of disordered ribosomal protein extensions in the early steps of eubacterial 50 S ribosomal subunit assembly. Int. J. Mol. Sci. 10, 817–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen J. W., Romero P., Uversky V. N., Dunker A. K. (2006) Conservation of intrinsic disorder in protein domains and families: II. functions of conserved disorder. J. Proteome Res. 5, 888–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Greber B. J., Boehringer D., Montellese C., Ban N. (2012) Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat. Struct. Mol. Biol. 19, 1228–1233 [DOI] [PubMed] [Google Scholar]
- 43. Record M. T., Jr., Guinn E., Pegram L., Capp M. (2013) Introductory lecture: interpreting and predicting Hofmeister salt ion and solute effects on biopolymer and model processes using the solute partitioning model. Faraday Discuss. 160, 9–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Date M. S., Dominy B. N. (2013) Modeling the influence of salt on the hydrophobic effect and protein fold stability. Commun. Comput. Phys. 13, 90–106 [Google Scholar]
- 45. Klinge S., Voigts-Hoffmann F., Leibundgut M., Arpagaus S., Ban N. (2011) Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334, 941–948 [DOI] [PubMed] [Google Scholar]
- 46. Tadeo X., López-Méndez B., Trigueros T., Laín A., Castaño D., Millet O. (2009) Structural basis for the aminoacid composition of proteins from halophilic archea. PLoS Biol. 7, e1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dassarma S., Capes M. D., Karan R., Dassarma P. (2013) Amino acid substitutions in cold-adapted proteins from Halorubrum lacusprofundi, an extremely halophilic microbe from antarctica. PLoS One 8, e58587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Abe Y., Ueda T., Iwashita H., Hashimoto Y., Motoshima H., Tanaka Y., Imoto T. (1995) Effect of salt concentration on the pKa of acidic residues in lysozyme. J. Biochem. 118, 946–952 [DOI] [PubMed] [Google Scholar]
- 49. Nozaki Y., Tanford C. (1967) in Methods in Enzymology (Hirs C. H. W., ed) Vol. 11, pp. 715–734, Academic Press, New York [Google Scholar]
- 50. Anderson C. F., Record M. T. (1995) Salt-nucleic acid interactions. Annu. Rev. Phys. Chem. 46, 657–700 [DOI] [PubMed] [Google Scholar]
- 51. Leipply D., Lambert D., Draper D. E. (2009) in Methods in Enzymology (Herschlag D., ed) Vol. 469, pp. 433–463, Elsevier, Waltham, MA: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Draper D. E., Grilley D., Soto A. M. (2005) Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 34, 221–243 [DOI] [PubMed] [Google Scholar]
- 53. Draper D. E. (2004) A guide to ions and RNA structure. RNA 10, 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Draper D. E. (1995) Protein-RNA recognition. Annu. Rev. Biochem. 64, 593–620 [DOI] [PubMed] [Google Scholar]
- 55. Wahl M. C., Möller W. (2002) Structure and function of the acidic ribosomal stalk proteins. Curr. Protein Pept. Sci. 3, 93–106 [DOI] [PubMed] [Google Scholar]
- 56. Schuwirth B. S., Borovinskaya M. A., Hau C. W., Zhang W., Vila-Sanjurjo A., Holton J. M., Cate J. H. (2005) Structures of the bacterial ribosome at 3.5 Å resolution. Science 310, 827–834 [DOI] [PubMed] [Google Scholar]
- 57. Kavran J. M., Steitz T. A. (2007) Structure of the base of the L7/L12 stalk of the Haloarcula marismortui large ribosomal subunit: analysis of L11 movements. J. Mol. Biol. 371, 1047–1059 [DOI] [PubMed] [Google Scholar]
- 58. Bocharov E. V., Sobol A. G., Pavlov K. V., Korzhnev D. M., Jaravine V. A., Gudkov A. T., Arseniev A. S. (2004) From structure and dynamics of protein L7/L12 to molecular switching in ribosome. J. Biol. Chem. 279, 17697–17706 [DOI] [PubMed] [Google Scholar]
- 59. Hoffman D. W., Davies C., Gerchman S. E., Kycia J. H., Porter S. J., White S. W., Ramakrishnan V. (1994) Crystal structure of prokaryotic ribosomal protein L9: a bi-lobed RNA-binding protein. EMBO J. 13, 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jenner L. B., Demeshkina N., Yusupova G., Yusupov M. (2010) Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 17, 555–560 [DOI] [PubMed] [Google Scholar]
- 61. Harms J. M., Wilson D. N., Schluenzen F., Connell S. R., Stachelhaus T., Zaborowska Z., Spahn C. M., Fucini P. (2008) Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol. Cell 30, 26–38 [DOI] [PubMed] [Google Scholar]
- 62. Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. (2011) The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334, 1524–1529 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.