Background: SmgGDS-607 and SmgGDS-558 regulate GTPase movement through the prenylation pathway.
Results: The specificity of SmgGDS for GTPases depends on the GTPase CAAX sequence and the cellular context.
Conclusion: SmgGDS-607 binds to nonprenylated GTPases that end in a leucine and enter the geranylgeranylation pathway.
Significance: The identification of SmgGDS-607 as a novel CAAX-binding protein will accelerate the development of more effective cancer therapeutics.
Keywords: Isoprenoid, Protein Complexes, Protein Farnesylation, Protein Isoprenylation, Small GTPases, FTI, GGTI, K-Ras, Rap1, SmgGDS
Abstract
Ras family small GTPases localize at the plasma membrane, where they can activate oncogenic signaling pathways. Understanding the mechanisms that promote membrane localization of GTPases will aid development of new therapies to inhibit oncogenic signaling. We previously reported that SmgGDS splice variants promote prenylation and trafficking of GTPases containing a C-terminal polybasic region and demonstrated that SmgGDS-607 interacts with nonprenylated GTPases, whereas SmgGDS-558 interacts with prenylated GTPases in cells. The mechanism that SmgGDS-607 and SmgGDS-558 use to differentiate between prenylated and nonprenylated GTPases has not been characterized. Here, we provide evidence that SmgGDS-607 associates with GTPases through recognition of the last amino acid in the CAAX motif. We show that SmgGDS-607 forms more stable complexes in cells with nonprenylated GTPases that will become geranylgeranylated than with nonprenylated GTPases that will become farnesylated. These binding relationships similarly occur with nonprenylated SAAX mutants. Intriguingly, farnesyltransferase inhibitors increase the binding of WT K-Ras to SmgGDS-607, indicating that the pharmacological shunting of K-Ras into the geranylgeranylation pathway promotes K-Ras association with SmgGDS-607. Using recombinant proteins and prenylated peptides corresponding to the C-terminal sequences of K-Ras and Rap1B, we found that both SmgGDS-607 and SmgGDS-558 directly bind the GTPase C-terminal region, but the specificity of the SmgGDS splice variants for prenylated versus nonprenylated GTPases is diminished in vitro. Finally, we present structural homology models and data from functional prediction software to define both similar and unique features of SmgGDS-607 when compared with SmgGDS-558.
Introduction
Mutations and altered regulation of members of the Ras family of GTPases can lead to uncontrollable proliferation (1, 2), metastasis (3, 4), and evasion of apoptosis (5, 6) that occur in the development and progression of cancer (7). Oncogenic signaling by GTPases occurs at the cell membrane and is generally caused by increased expression of the GTPase (8), altered functions of GTPase-activating proteins or guanine nucleotide exchange proteins (9), or an activating mutation within the GTPase itself that interferes with GTP hydrolysis (10, 11). Because of the myriad of causes that can lead to uncontrollable signaling by a GTPase at cell membranes, research has been focused on blocking the prenylation of a GTPase and thus blocking it from reaching the plasma membrane. This therapeutic strategy has met with moderate success (12, 13).
Prenylation promotes the membrane localization of GTPases, and the addition of this hydrophobic group is the only necessary modification for GTPases that contain a C-terminal PBR to associate with membranes (14). The C-terminal region of GTPases such as K-Ras and Rap1B is known as the hypervariable region, which consists of a string of basic amino acids that make up the PBR followed by a CAAX motif (15). The addition of the prenyl moiety occurs on the cysteine of the CAAX motif (where A represents an aliphatic amino acid), which is then further processed by cleavage of the -AAX by Rce1 and carboxyl methylation by ICMT1 (16). The type of prenylation that a GTPase will undergo is decided primarily by the last amino acid of the CAAX motif; GTPases such as Rap1B that have a CAAL motif will be geranylgeranylated, whereas GTPases such as K-Ras that have a CAAM motif will be farnesylated (17). The mechanism by which GTPases containing a PBR traffic to the plasma membrane is not well defined, with limited evidence showing direct binding of a GTPase to microtubule adaptor proteins (18) and another report showing that K-Ras uses a unique, nonexocytic pathway (19). Recently, we found that the splice variants of SmgGDS, SmgGDS-607 and SmgGDS-558, play a role in promoting prenylation and trafficking of GTPases to the cell membrane (20). However, the molecular interactions that occur during these events have not yet been characterized.
SmgGDS has been found to be a key promoter in the malignancy of multiple cancers including non-small cell lung cancer (21), prostate cancer (22), and breast cancer (23). Prior to our report in 2010 (20), virtually all studies regarding SmgGDS (known as Rap1GDS1) focused on the 558-amino acid splice variant. SmgGDS-558 was originally reported as a weak guanine nucleotide exchange factor for multiple small GTPases that contain a PBR including Rap1 (24, 25), K-Ras (24), Rac1 (26), and RhoA (27) but more recently has been found to be a true guanine nucleotide exchange factor for only RhoA and RhoC (28). Structurally SmgGDS is comprised of armadillo (ARM)2 repeats similar to β-catenin or karyopherin α (29) and therefore has been hypothesized to act as a scaffold protein for multiple protein interactions (30). The ability of SmgGDS to interact with multiple GTPases and affect their nucleotide activity is most likely due to the binding of other guanine nucleotide exchange factors to the scaffold-like SmgGDS, because scaffolds form a platform for protein interactions (20, 31). The C-terminal region of a GTPase is needed for interaction with SmgGDS (24, 32), and the ablation of the PBR of a GTPase will diminish its ability to complex with SmgGDS (33).
Interestingly, it was reported that SmgGDS-607 will only associate with nonprenylated GTPases, whereas SmgGDS-558 will only associate with prenylated GTPases, yet the mechanism for the recognition of prenylated versus nonprenylated GTPases has not been characterized (20). We previously found that SmgGDS-607 more effectively regulates the prenylation of GTPases that will become geranylgeranylated, such as Rap1, RhoA, and Rac1, compared with GTPases that will become farnesylated, such as K-Ras (20). This effect that SmgGDS-607 has on GTPases that will become geranylgeranylated, but not on GTPases that will become farnesylated, provides insight into its mechanism of interaction.
In this study we use K-Ras and Rap1B as model GTPases that become farnesylated and geranylgeranylated, respectively, to define the parameters that promote their physical interactions with SmgGDS splice variants. We report that SmgGDS-607 recognizes the last amino acid of a GTPase for interaction and is also involved primarily in the geranylgeranylation pathway. Our results provide evidence that SmgGDS-607 and SmgGDS-558 structurally interact with GTPases similarly in vitro, and this is in contrast to their interactions in cells. Furthermore, we provide evidence that SmgGDS-607 is involved in the alternate prenylation pathway of K-Ras in cells treated with farnesyl transferase inhibitors (FTIs).
EXPERIMENTAL PROCEDURES
Cell Culture, cDNA Transfection, and Drug Treatment
The HEK-293T cell line was obtained from the American Type Tissue Collection (Manassas, VA). HEK-293T cells were cultured in DMEM with 10% heat-inactivated fetal bovine serum and antibiotics. All cDNAs were transfected into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. To inhibit prenylation, 10 μm FTI-277 (CAT344555; EMD Millipore) and GGTI-298 (CAT344555; EMD Millipore) or 15 μm mevastatin (M2537; Sigma) was added 90 min post-transfection to the cells.
cDNA Constructs
Human SmgGDS-558-HA and SmgGDS-607-HA constructs were generated as described previously (20). The majority of Myc-tagged GTPase constructs were generated as described previously (33). Myc-K-Ras CVIM was mutated by site-directed mutagenesis to change the methionine to a leucine (CVIL) and the cysteine to a serine (SVIM or SVIL) (see Fig. 1A). Site-directed mutagenesis was performed using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's protocols, using primers purchased from Eurofins MWG/Operon. The CAAX mutant Myc-Rap1B (CQLM) was generated by site directed mutagenesis through Top Gene Technologies (Montreal, Canada).
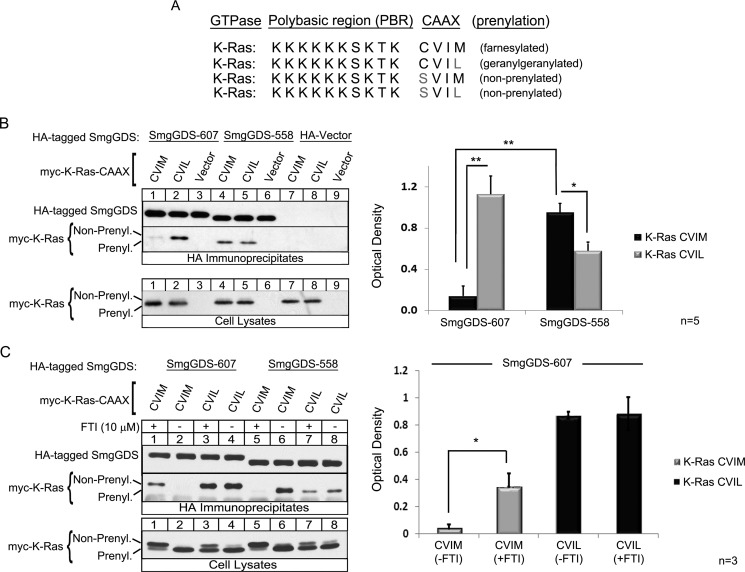
FIGURE 1.
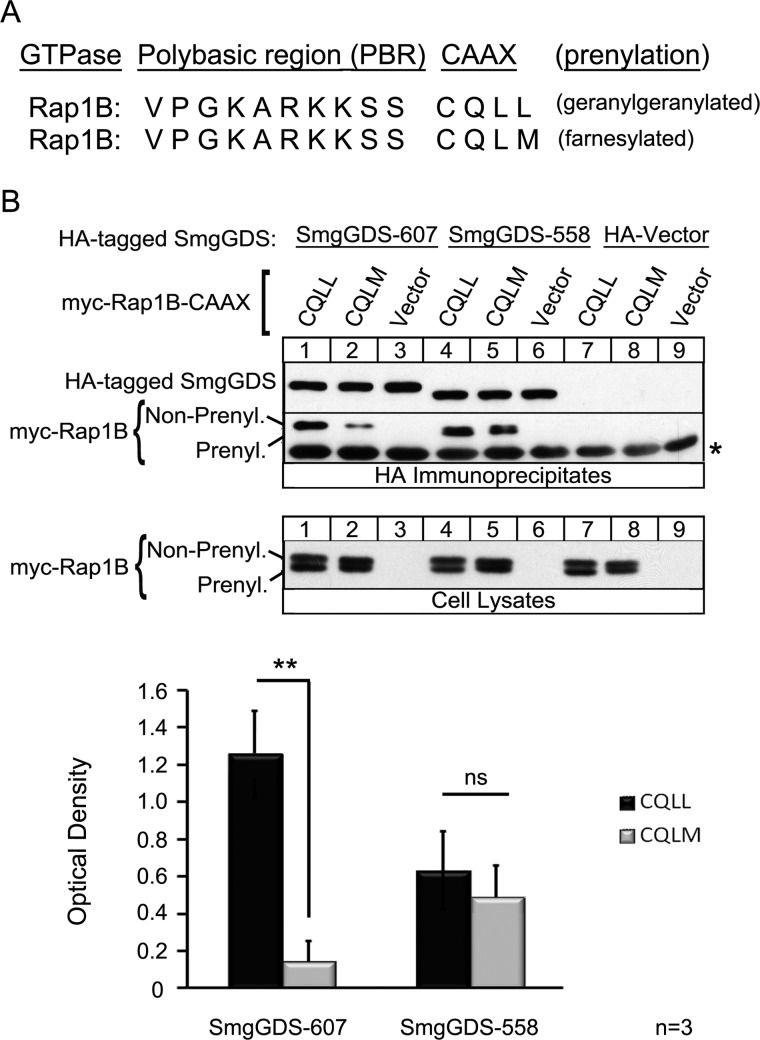
SmgGDS-607 is a novel CAAX-binding protein and associates with nonprenylated small GTPases that will become geranylgeranylated. A, WT K-Ras 4B has a CAAX motif of CVIM that allows for the protein to become farnesylated. The last amino acid of K-Ras was mutated to a leucine to promote geranylgeranylation (40). The cysteine of the CAAX motif was mutated to a serine to inhibit prenylation of K-Ras (20). B and C, HEK-293T cells were transfected with a cDNA encoding SmgGDS-607-HA, SmgGDS-558-HA, or the HA vector plus a cDNA encoding the Myc-tagged WT K-Ras CVIM, CVIL, or a Myc vector. 90 min post-transfection, the cells were treated with 10 μm FTI-277 (C, odd lanes) or vehicle (C, even lanes). After 24 h, cells were lysed, and an aliquot of each lysate was subjected to ECL-Western blotting using Myc antibody (Cell Lysates). The remaining volume of each cell lysate was immunoprecipitated with HA antibody, followed by ECL-Western blotting using HA antibody and Myc antibody (HA Immunoprecipitates). The results are shown as the optical density of IP Myc-K-Ras divided by the optical density of IP SmgGDS-HA and are the means ± S.E. of four (B) and three (C) independent experiments. *, p < 0.05; **, p < 0.01 by Student's t test.
Synthesis of K-Ras Peptides
Peptide synthesis was carried out using an automated solid phase peptide synthesizer (PS3; Protein Technologies Inc., Memphis, TN) employing standard Fmoc/1H-benzotriazolium1-[bis(dimethyl-amino)metheylene]-5-chloro-hexafluorophosphate (1-),3-oxide-based chemistry. Synthesis began on either preloaded Fmoc-M-Wang resin or Fmoc-L-Wang resin (0.25 mmol), and the peptide chain was elongated using 1H-benzotriazolium1-[bis(dimethyl-amino)metheylene]-5-chloro-hexafluorophosphate (1-),3-oxide/N-methylmorpholine-catalyzed, single coupling steps with 4 eq of both protected amino acids and HTCU for 30 min. Following complete chain elongation, the N terminus of the peptide was deprotected with 10% piperidine in DMF (v/v), and the presence of the resulting free amine was confirmed by ninhydrin analysis. The resin containing the peptide was washed with CH2Cl2, dried in vacuo overnight, weighed, and divided into two portions for further synthesis on a reduced scale. Using 0.13 mmol of peptide on resin, the free N terminus was modified with a PEG-based linker in DMF (5 ml) using Fmoc-8-amino-3,6-dioxaoctanoic acid (77 mg, 0.20 mmol, 1.5 eq) catalyzed by N,N-diisopropylethylamine (16.4 μl, 13.0 μmol) for 12 h. After installation of the linker and subsequent Fmoc deprotection, the resulting N terminus was biotinylated in DMF (5 ml) using biotin (49.0 mg, 0.20 mmol, 1.5 eq) and 1H-benzotriazolium1-[bis(dimethyl-amino)metheylene]-5-chloro-hexafluorophosphate (1-),3-oxide (82.7 mg, 0.20 mmol, 1.5 eq) catalyzed by N,N-diisopropylethylamine (16.4 μl, 13.0 μmol) for 16 h. After verifying the biotinylation was complete by ninhydrin analysis, the peptide was cleaved from the resin along with simultaneous side chain deprotection by treatment with reagent K containing TFA (10 ml), crystalline phenol (0.5 g), 1,2-ethanedithiol (0.25 ml), thioanisole (0.5 ml), and H2O (0.5 ml) for 2 h at room temperature. The released peptide was collected and combined with TFA washes of the resin before precipitation of the peptide in chilled Et2O (100 ml). The crude solid peptide was collected by centrifugation, the supernatant was removed, and the resulting pellet was washed two times with cold Et2O (50 ml), repeating the centrifugation and supernatant removal steps each time. The crude peptide (100 mg) was dissolved in a DMF/H2O solution (1:5, v/v, 25 ml), applied to a semipreparative C18 reverse phase HPLC column equilibrated in Solvent A, and washed with 10% Solvent B for 15 min. The peptide was eluted using a linear gradient (15–65% Solvent B over 1.5 h at a flow rate of 5 ml/min). Fractions were analyzed using an analytical C18 reverse phase HPLC column employing a linear gradient (0–100% Solvent B over 60 min at a flow rate of 1 ml/min) and detected at 214 nm. Fractions containing peptide product of at least 90% purity were pooled and concentrated by lyophilization to yield 35 mg (27% yield) of white peptide. The [M + 3H]3+ values for ESI-MS for Biotin-PEG-KKKKKKSKTKCVIM were 683.66 (calculated) and 683.73 (found), and those for ESI-MS for Biotin-PEG-KKKKKKSKTKCVIL were 677.56 (calculated) and 677.75 (found).
Synthesis of Rap1B (Free Thiol) Peptide
Solid phase peptide synthesis and purification was carried out in the same fashion as described above starting with Fmoc-L-Wang resin (0.25 mmol). No additional derivatizations were performed after peptide chain elongation was complete. After reagent K cleavage and HPLC purification, the free thiol-containing peptide resulted in 60 mg (37% yield) of white peptide. The [M + 2H]2+ values for ESI-MS were 943.51 (calculated) and 943.68 (found).
Synthesis of Rap1B (Farnesylated) Peptide
The starting free thiol peptide (20 mg, 10.6 μmol, 1 eq) was dissolved in DMF/n-butanol/H2O (0.10% TFA) (3:1:1, v/v/v, 6 ml). Farnesyl bromide (15 mg, 53.5 μmol, 5 eq) was dissolved in 0.50 ml of DMF and added directly into the reaction flask that contained the dissolved peptide. Zn(OAc)2·2H2O (11.7 mg, 53.5 μmol, 5 eq) was then added to initiate the alkylation reaction. After 4 h, the reaction was monitored by analytical reverse phase HPLC, purified by semipreparative C18 reverse phase HPLC, and identified via ESI-TOF MS. This reaction yielded 3.4 mg (19%) of the desired alkylated peptide. The [M + 2H]2+ values for ESI-MS were 1045.96 (calculated) and 1046.04 (found).
Synthesis of Rap1B (Geranylgeranylated) Peptide
Synthesis followed the same prenylation conditions as described above using geranylgeranyl bromide (19 mg, 53.5 μmol, 5 eq). This reaction yielded 2.6 mg (12%) of the desired alkylated peptide. The [M + 2H]2+ values for ESI-MS were 1079.64 (calculated) and 1079.73 (found).
Synthesis of Rap1B (C10-m-Bp) Peptide
Synthesis followed the same prenylation conditions as described above using C10-meta-Bp-Br (22.8 mg, 53.5 μmol, 5 eq) that was prepared as previously described (34–37). This reaction yielded 3.4 mg (14%) of the desired alkylated peptide. The [M + 2H]2+ values for ESI-MS were 1117.37 (calculated) and 1117.48 (found).
Photo-Cross-linking
Biotin-Rap1B (C10) peptides containing a benzophenone (synthesized as described above) were cross-linked to SmgGDS-607-HA by UV light irradiation utilizing a UVGL-25 Minerallight lamp (366 nm). Peptides and HEK-293T cell lysates overexpressing SmgGDS-607-HA or HA vector were combined and incubated on ice for 10 min. Mixtures were transferred to quartz tubes and placed ∼3 cm above the UV light for increasing time points from 0–60 min. Photo-cross-linking reaction was quenched immediately after the indicated time points by placing the sample in an Eppendorf tube, which was then placed in liquid nitrogen. A control sample was prepared as described above, and the tube was wrapped in tin foil before exposure to UV light. After the cross-linking procedure, the peptides were immunoprecipitated using streptavidin-Sepharose 4B beads (434341; Invitrogen) and subjected to ECL-Western blotting.
Expression of Recombinant Proteins
GST-K-Ras CVIM, GST-K-Ras CVIL, His-SmgGDS-607, and His-SmgGDS-588 were purified as described previously (38). Briefly, the coding sequence of K-Ras CVIM or K-Ras CVIL was cloned into pGEX-2T (GE Healthcare), or the coding sequence of SmgGDS-607 or SmgGDS-558 was cloned into pET-28a. The constructs were expressed in Escherichia coli BL21 (DE3). Bacteria were plated overnight at 37 °C with ampicillin (K-Ras) or kanamycin (SmgGDS) (250 μg/ml). Cells were added to 400 ml of LB broth supplemented with either ampicillin or kanamycin and cultured for 2 h at 30 °C when 1 mm isopropyl β-d-thiogalactopyranoside was added, and the cells were cultured for an additional 2 h at 30 °C.
GST Affinity Chromatography
Bacterial cells expressing GST-K-Ras were harvested, suspended in glutathione-binding buffer (20 mm Tris, pH 7.6, 150 mm NaCl, 1% Triton X-100, 1 mm PMSF, bacterial protease inhibitor mixture (Sigma), 20 μg/ml RNase, and 20 μg/ml DNase) and lysed in a French pressure cell. The lysate was centrifuged (30,000 × g for 20 min), and the soluble fraction was passed through a 0.45-μm filter. Bacterial lysates of GST-K-Ras CVIM or CVIL were incubated with 1.5 ml of glutathione-Sepharose-4B beads (GE Healthcare) for 30 min on a nutator at 4 °C. The slurry was centrifuged (1500 × g), and the resin was washed four times with 25 ml of binding buffer. Proteins were batch eluted three times with 1 ml of elution buffer (20 mm reduced glutathione, 100 mm Tris-HCl, pH 8.0, and 1% Triton X-100). GST-K-Ras was dialyzed into 10 mm Tris-HCl (pH 8.0), 20 mm NaCl, and 30% glycerol, and then stored at −80 °C.
Nitrilotriacetic Acid/Ni2+ Affinity Chromatography
Bacterial cells expressing His-SmgGDS were suspended in a binding buffer (20 mm Tris, pH 7.9, 0.5 m NaCl, 5 mm imidazole, bacterial protease inhibitor mixture (Sigma), 20 μg/ml RNase, and 20 μg/ml DNase) and lysed in a French pressure cell. The lysate was centrifuged (30,000 × g for 20 min), and the soluble fraction was passed through a 0.45-μm filter. The filtrate was applied to an equilibrated 2 ml of nitrilotriacetic acid/Ni2+-agarose affinity matrix (Qiagen). The column was washed with 20 ml of binding buffer. SmgGDS proteins were eluted with the addition of binding buffer containing 250 mm imidazole. Collecting 1-ml fractions, SmgGDS-607 and SmgGDS-558 elutes in fractions 3–4.
ECL-Western Blotting
Equal numbers of transfected cells were heated in Laemmli sample buffer and subjected to SDS-PAGE. The proteins were transferred as described previously (20) and immunoblotted using an antibody to SmgGDS (612511; BD Transduction Laboratories), GAPDH (sc-32233; Santa Cruz Biotechnology), mouse HA antibody (MMS-101P; Covance), mouse Myc antibody (PRB-150P; Covance), rabbit Myc antibody (C2845; Sigma), rabbit HA antibody (PRB-101P; Covance), and Gβ1 (C-16) antibody (sc-379; Santa Cruz). Bound antibodies were visualized as described previously (20).
Analysis of Immunoprecipitates from Cultured Cells
HEK-293T cells were transiently transfected with cDNAs encoding HA-tagged SmgGDS splice variants and Myc-tagged GTPases. After 24 h, equal numbers of cells were lysed in buffer containing 0.5% Nonidet P-40 with protease and phosphatase inhibitors, and the lysates were centrifuged (2500 × g, 5 min, 4 °C). A portion of the resulting supernatants was reserved for total cell lysates, and the remainder was immunoprecipitated using mouse monoclonal HA antibody-conjugated beads (A2095; Sigma), or streptavidin-Sepharose 4B beads (434341; Invitrogen) prebound to the indicated C-terminal K-Ras and Rap1B peptides. Immunoprecipitates, total cell lysate, and excess protein not bound to beads were subjected to ECL-Western blotting as described above.
Optical density values were measured by scanning the immunoblots utilizing an HP Scanjet (4850) at 600 dpi, and analyzing the scanned images using ImageJ software. Optical density values from Figs. 3A and 5C represent raw densitometry values. All other optical density values are presented as ratios of the immunoprecipitated GTPase compared with the amount of immunoprecipitated SmgGDS.
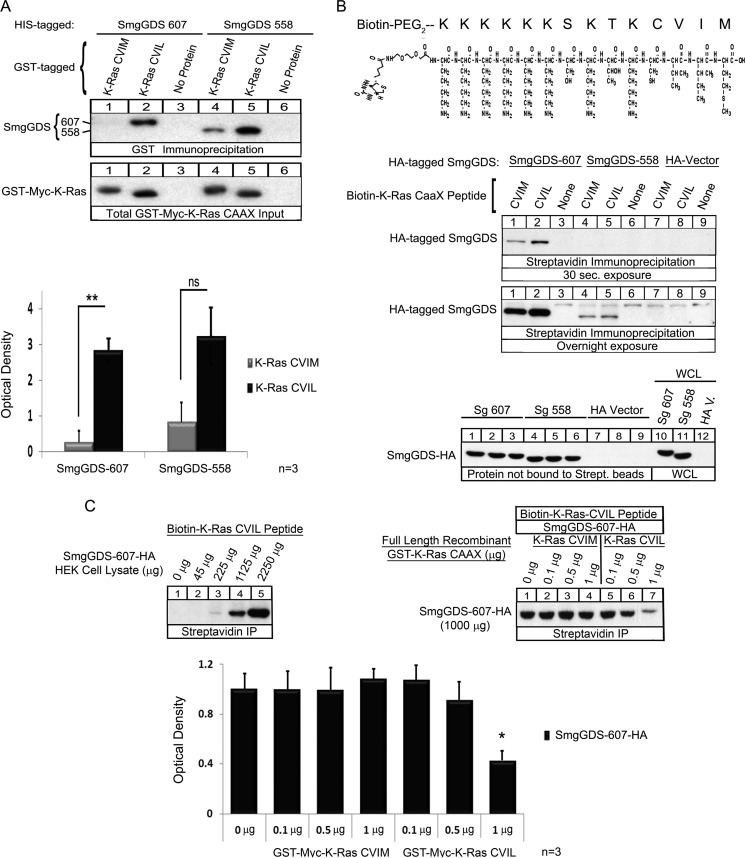
FIGURE 3.
SmgGDS-607 binds directly and specifically to K-Ras CVIL, and SmgGDS-558 binds unprenylated WT K-Ras CVIM or CVIL in vitro. A, 100 ng of recombinant His-tagged SmgGDS-607 and SmgGDS-558 were allowed to interact with 250 ng of recombinant GST-Myc-tagged full-length K-Ras CVIM or K-Ras CVIL proteins prebound to glutathione-Sepharose 4B beads. The complexes were isolated by pelleting the beads and subjected to ECL-Western blotting using a SmgGDS antibody. The results are shown as the optical density of the SmgGDS protein signal in the immunoblots and are the means ± S.E. of three independent experiments. ns, not significant; **, p < 0.01 by Student's t test. B, Biotin-PEG2-K-Ras CVIM or CVIL peptides with the sequence KKKKKKSKTKCVI(M/L) (top panel) were prebound (10 μg) to streptavidin beads and allowed to interact with HEK-293T cell lysates expressing either SmgGDS-607-HA, SmgGDS 558-HA, or HA vector. The complexes were isolated by pelleting the beads and subjected to ECL-Western blotting using HA antibody. The results are representative of three (30 sec. exposure) or two (Overnight exposure) independent experiments. C, lysates of HEK-293T cells expressing SmgGDS-607-HA were prepared as increasing concentrations of total cellular protein (45 μg to 2250 μg) (top left panel) or 1000 μg of total cellular protein (top right panel) and allowed to interact with the biotin-K-Ras CVIL peptide (10 μg) that was prebound to streptavidin beads. Full-length recombinant GST-K-Ras-CVIM or -CVIL proteins were added at increasing concentrations (0.1–1 μg) to the reaction mixtures (top right panel). SmgGDS-607-HA that was bound to the biotin-K-Ras CVIL peptide was isolated by pelleting the streptavidin beads and then subjected to ECL-Western blotting using HA antibody. The results are representative of two (top left panel) or three (top right panel) independent experiments. The graph shows the optical density (means ± S.E.) of the immunoblotted SmgGDS-607-HA protein that was pulled down by the biotin-K-Ras CVIL peptide in the presence of the indicated concentrations of full-length recombinant GST-K-Ras-CVIM or -CVIL proteins. *, p < 0.05 by one-way analysis of variance with Dunnett's post-test compared with the control value from the sample that did not receive full-length recombinant GST-K-Ras protein.
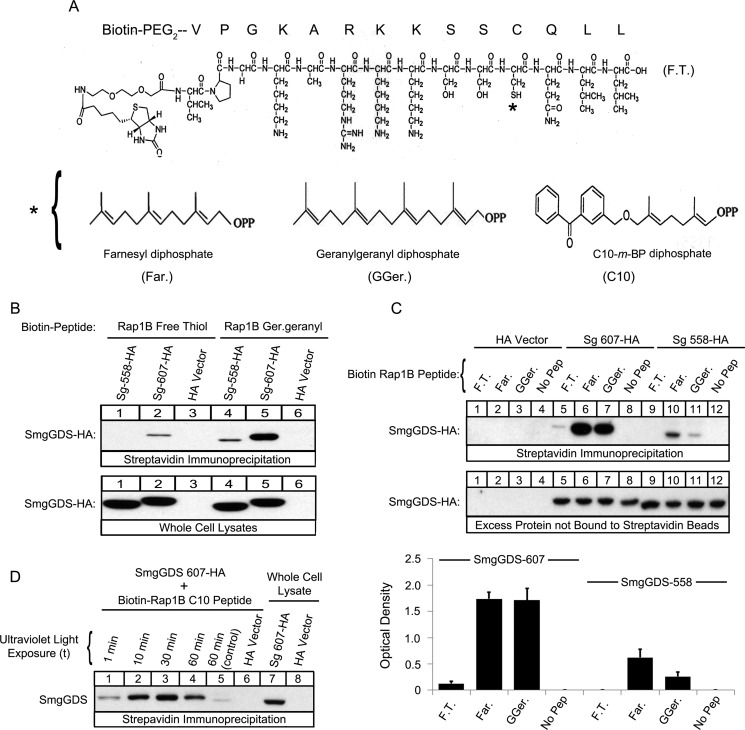
FIGURE 5.
SmgGDS-607 binds directly to both the prenylated and nonprenylated forms of a Rap1B C-terminal peptide, and SmgGDS-558 binds to the prenylated form of the peptide. A, the structure of the free thiol Rap1B peptide is shown, where the asterisk indicates the farnesyl (left panel), geranylgeranyl (middle panel), or C10-m-BP (right panel) group. The C10 group contains the photoactive benzophenone-containing residue, which allows for photo-cross-linking to a substrate. B and C, the indicated Biotin-PEG-Rap1B peptides in the free thiol (F.T.), farnesylated (Far.), or geranylgeranylated (GGer.) forms were prebound to streptavidin beads and allowed to interact with lysates of HEK-293T cells expressing either SmgGDS-607-HA, SmgGDS-558-HA, or HA vector. The bound complexes, whole cell lysates (B), and excess protein not bound to the streptavidin beads (C) were subjected to ECL-Western blotting using HA antibody. The results are representative of three independent experiments. D, Biotin-PEG-Rap1B C10-m-BP peptides were allowed to interact with HEK-293T cell lysates containing either SmgGDS-607-HA or HA vector and exposed to increasing times of UV light (1, 10, 30, and 60 min). The control sample was wrapped in tin foil and exposed to UV light for 60 min. Streptavidin beads were used to pull down the complexes of SmgGDS-607-HA cross-linked to the biotinylated Rap1B peptide, and then the complexes and whole cell lysates were subjected to ECL-Western blotting using HA antibody. The results are representative of three independent experiments.
Triton X-114 Fractionation
HEK-293T cells were lysed as described previously (39) in 1% Triton X-114 in TBS (50 mm Tris, 150 mm NaCl, pH 7.5), incubated 15 min on ice, and then centrifuged at 25,000 × g at 4 °C to remove insoluble debris. An aliquot of the cleared lysate was retained as total cell lysate. The remaining lysate was separated into aqueous and detergent fractions and examined by ECL-Western blotting as described previously (20). Proteins were detected with mouse Myc antibody to detect Myc-tagged K-Ras constructs, SmgGDS antibody (an aqueous phase loading control), and Gβ1 antibody (a detergent phase loading control).
Homology Modeling
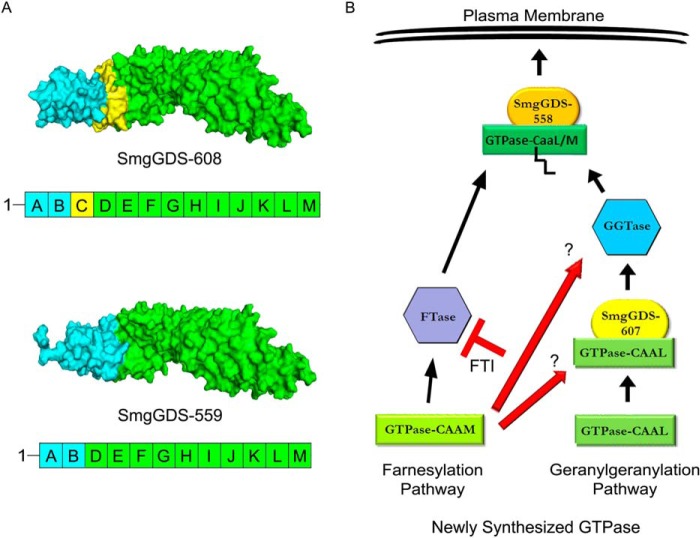
Homology models of SmgGDS-608 (NCBI accession number NP_001093896) and SmgGDS-559 (NCBI accession number NP_001093893) were generated by Dr. John Sondek's laboratory (University of North Carolina, Chapel Hill) as described previously (28). Models are displayed using PyMOL and are colored to represent the indicated armadillo (ARM) domains. An alanine is present at amino acid position 2 in both SmgGDS-608 and SmgGDS-559, and this alanine is lacking in SmgGDS-607 (NCBI accession number NP_001093897) and SmgGDS-558 (NCBI accession number NP_001093899). The presence of the alanine at position 2 is not expected to significantly change the modeled structure of the proteins.
Statistical Analyses
The means ± S.E. were measured for each value. Symbols above a column indicate a statistical comparison between the bracketed samples by one-way analysis of variance with Dunnett's post hoc multiple comparison's test or by Student's t test (two-tailed), as indicated in the figure legends. p values less than 0.05 were considered significant.
RESULTS
SmgGDS-607 Is a Novel CAAX-binding Protein and Selectively Associates with Nonprenylated K-Ras That Becomes Geranylgeranylated
To test the hypothesis that SmgGDS-607 selectively binds to GTPases that enter the geranylgeranylation pathway, K-Ras 4B (K-Ras) was used as a model. We made the indicated mutations to WT K-Ras (Fig. 1A) and then tested the ability of SmgGDS-607 and SmgGDS-558 to co-precipitate the proteins.
We found that SmgGDS-607 co-precipitates the slower migrating (nonprenylated) form of K-Ras CVIM and CVIL, and SmgGDS-558 co-precipitates the faster migrating (prenylated) form of K-Ras CVIM and CVIL (Fig. 1B, HA Immunoprecipitates, lanes 1 and 2 compared with lanes 4 and 5), consistent with the previous report that SmgGDS-607 associates with nonprenylated GTPases and SmgGDS-558 associates with prenylated GTPases (20). SmgGDS-607 co-precipitates significantly more with the mutant K-Ras CVIL than with WT K-Ras CVIM (Fig. 1B, HA Immunoprecipitates, lane 2 compared with lane 1). Interestingly, SmgGDS-558 associated with both K-Ras CVIM and K-Ras CVIL but exhibited significantly greater association with K-Ras CVIM (Fig. 1B, HA Immunoprecipitates, lanes 4 and 5 and graph). These results show that SmgGDS-607 and SmgGDS-558 are novel CAAX-specific binding proteins for K-Ras and indicate that the recognition of a GTPase by SmgGDS-607 or SmgGDS-558 requires more than just the PBR.
To test the hypothesis that SmgGDS-607 can bind WT K-Ras CVIM when the farnesylation pathway is blocked, we utilized a farnesyl-transferase inhibitor (FTI-277), which is a CAAX-peptidomimetic (12). As expected, we found that SmgGDS-558 did not co-precipitate K-Ras CVIM in cells treated with a FTI (Fig. 1C, HA Immunoprecipitates, lane 5 compared with lane 6), which further indicates that SmgGDS-558 will associate with a GTPase only after prenylation. Treatment of the HEK-293T cells with a FTI did not affect the ability of SmgGDS-607 or SmgGDS-558 to co-precipitate K-Ras CVIL (Fig. 1C, HA Immunoprecipitates, lanes 3 compared with lane 4 and lane 7 compared with lane 8, respectively), consistent with the FTI not affecting GTPases that will become geranylgeranylated. Interestingly, SmgGDS-607 associated with K-Ras CVIM significantly more in cells treated with a FTI (Fig. 1C, HA Immunoprecipitates, lane 1 compared with lane 2), indicating that SmgGDS-607 can interact with a GTPase ending in a methionine but only if it will traffic through the geranylgeranylation pathway. The ability of K-Ras CVIM to interact with SmgGDS-607 in cells treated with a FTI could result from the FTI causing a larger pool of unprenylated K-Ras CVIM (Fig. 1C, Cell Lysates, lane 1 compared with lane 2). Alternatively, SmgGDS-607 might interact more with nonprenylated K-Ras CVIM in FTI-treated cells because the FTI blocks K-Ras from interacting with the farnesyltransferase, thereby forcing K-Ras CVIM to interact with proteins in the geranylgeranylation pathway (40), which includes the geranylgeranyltransferase as well as SmgGDS-607.
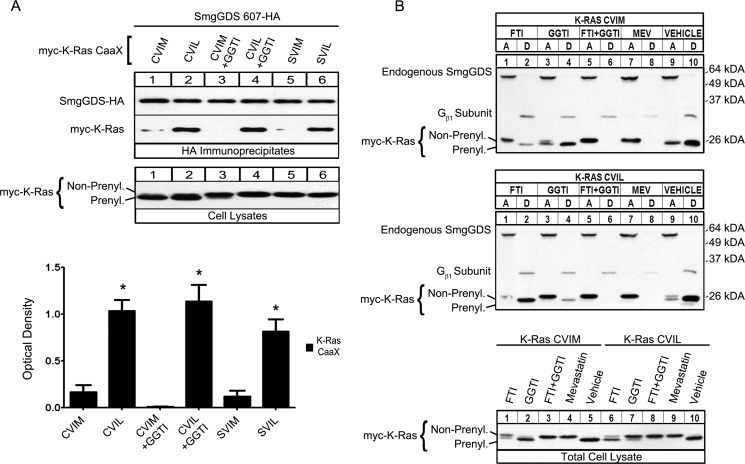
SmgGDS-607 Recognizes the Last Amino Acid of K-Ras for Preferential Binding
To test the hypothesis that SmgGDS-607 associates with K-Ras CVIM in cells treated with a FTI only when the GTPase can move through the geranylgeranylation pathway and not because there is a larger pool of K-Ras that is unprenylated, we utilized the nonprenylatable mutants K-Ras SVIM and SVIL in immunoprecipitation assays (Figs. 1A and 2A). We found that SmgGDS-607 only minimally co-precipitated both K-Ras CVIM and K-Ras SVIM but strongly co-precipitated both K-Ras CVIL and K-Ras SVIL (Fig. 2A, HA Immunoprecipitates lanes 1 and 5 compared with lanes 2 and 6, respectively). These results support the model that SmgGDS-607 maintains a preference for K-Ras ending in leucine even when presented with a large pool of nonprenylated K-Ras ending in methionine. Treating the cells with a geranylgeranyl transferase inhibitor (GGTI-298) did not significantly change the ability of K-Ras CVIM or K-Ras CVIL to interact with SmgGDS-607 (Fig. 2A, HA immunoprecipitates, lanes 3 and 4 compared with lanes 1 and lane 2, respectively). Taken together, these data indicate that SmgGDS-607 specifically recognizes the leucine at the end of the CAAX motif for binding. Furthermore, the association of SmgGDS-607 with K-Ras CVIM after FTI treatment indicates that regardless of the CAAX sequence, SmgGDS-607 prefers to interact with nonprenylated GTPases that are destined to interact with the geranylgeranyltransferase rather than the farnesyltransferase.
FIGURE 2.
K-Ras CVIM is farnesylated, whereas K-Ras CVIL is geranylgeranylated, and SmgGDS-607 recognizes the last amino acid in the CAAX motif for binding. A, SmgGDS-607-HA was expressed in HEK-293T cells with Myc-tagged WT K-Ras CVIM, CVIL, or their nonprenylatable mutants K-Ras SVIM and K-Ras SVIL, or a Myc vector. 90 min post-transfection, the cells were treated with either GGTI (15 μm) or vehicle. After 24 h, cells were lysed, and an aliquot of each lysate was subjected to ECL-Western blotting using Myc antibody (Cell Lysates). The remaining volume of each cell lysate was immunoprecipitated with HA antibody, and the immunoprecipitates were immunoblotted using HA and Myc antibodies (HA Immunoprecipitates). The results are shown as the optical density of IP Myc-K-Ras divided by the optical density of IP SmgGDS-HA and are the means ± S.E. of three independent experiments. *, p < 0.01 by one-way analysis of variance with Dunnett's post-test compared with the control value of Myc-K-Ras CVIM pulled down (lane 1). B, HEK-293T cells were transfected with a cDNA encoding either Myc-tagged WT K-Ras CVIM (top panel) or mutant K-Ras CVIL (bottom panel). 90 min post-transfection, cells were treated with either FTI-277 (10 μm), GGTI-298 (10 μm), both FTI and GGTI, mevastatin (10 μm), or vehicle control. After 24 h, the cells were lysed, and an aliquot of each lysate was subjected to ECL-Western blotting using Myc antibody (Total Cell Lysate). The remaining volume of each cell lysate was subjected to Triton X-114 fractionation, and equal volumes of the aqueous phase, detergent phase, or total cell lysate were immunoblotted using Myc antibody, antibody to SmgGDS (aqueous phase marker), or antibody to Gβ1 subunit (detergent phase marker).
K-Ras CVIM Is Farnesylated and K-Ras CVIL Is Geranylgeranylated
To verify that the K-Ras mutant CVIL does indeed become geranylgeranylated and not farnesylated, we utilized a fractionation assay that separates proteins based on hydrophobicity (39). The Triton X-114 fractionation assay separates proteins from a cell lysate into an aqueous (A) and a detergent (D) phase. Membrane-bound or hydrophobic proteins separate into the detergent phase, and cytosolic or hydrophilic proteins separate into the aqueous phase. Using cells transfected with K-Ras CVIM, we found that the majority of K-Ras CVIM fractionates into the detergent phase (Fig. 2B, top panel, lanes 9 and 10, Total Cell Lysate, lane 5). Treatment of these cells with either a FTI, both a FTI and a GGTI simultaneously, or mevastatin causes K-Ras CVIM to fractionate into the aqueous phase (Fig. 2B, top panel, lanes 1, 2, 5, 6, 7, and 8, and Total Cell Lysate, lanes 1, 3, and 4). In contrast, treatment of these cells with a GGTI did not affect the fractionation of K-Ras CVIM (Fig. 2B, top panel, lanes 3 and 4, and Total Cell Lysate, lane 2). Using cells transfected with K-Ras CVIL, we found that the majority of K-Ras CVIL also fractionates into the detergent phase (Fig. 2B, middle panel, lanes 9 and 10, and Total Cell Lysate, lane 10). Treatment of these cells with either a GGTI, both a FTI and a GGTI simultaneously, or mevastatin causes K-Ras CVIL to fractionate into the aqueous phase (Fig. 2B, middle panel, lanes 3–8, and Total Cell Lysate, lane 7–9). In contrast, treatment of these cells with a FTI does not affect the fractionation of K-Ras CVIL (Fig. 2B, middle panel, lanes 1 and 2, and Total Cell Lysate, lane 6).
The detergent phase loading control, Gβ1 subunit, fractionated primarily in the detergent phase regardless of FTI, GGTI, or mevastatin treatment, indicating that a large pool of this protein already pre-existed in the cells with minimal synthesis of new Gβ1 protein during treatment of the cells (Fig. 2B). A longer exposure of the same immunoblots demonstrates a partial block of Gβ1 prenylation in cells treated with FTI and GGTI simultaneously as well as in cells treated with mevastatin (data not shown).
Recognition of the Last Amino Acid in K-Ras by SmgGDS-607 Is Direct
The unique ability of SmgGDS-607 to recognize the last amino acid of a GTPase could be mediated by another protein that is known to participate in the prenylation pathway, such as the prenyltransferase, or perhaps an unknown protein. To identify whether the interaction between K-Ras and SmgGDS is direct, we made unprenylated recombinant GST-Myc-tagged K-Ras CVIM and GST-Myc-tagged K-Ras CVIL, as well as recombinant His-tagged SmgGDS-607 and His-tagged SmgGDS-558 to assess their interactions. Similar to the interactions that occur in cells, we found that recombinant K-Ras CVIM did not associate with SmgGDS-607, whereas recombinant K-Ras CVIL significantly and strongly interacts with SmgGDS-607 (Fig. 3A, GST Immunoprecipitation, lane 1 compared with lane 2). We next wanted to assess the importance of the body of K-Ras in its association with SmgGDS-607; therefore we prepared biotin-labeled peptides comprised of the C-terminal PBR and CAAX motif of K-Ras CVIM or K-Ras CVIL (Fig. 3B, top panel). Similar to the results we obtained with the recombinant full-length K-Ras, we found that the K-Ras CVIM peptide had a weaker association than the K-Ras CVIL peptide with SmgGDS-607 (Fig. 3B, Streptavidin Immunoprecipitation, 30 sec. exposure, lane 1 compared with lane 2). These data show that the association between K-Ras CVIL and SmgGDS-607 is direct and that the isolated C-terminal region of K-Ras is able to bind to SmgGDS-607.
Unprenylated K-Ras Can Bind to SmgGDS-558
Surprisingly, we found that full-length unprenylated recombinant K-Ras CVIM and K-Ras CVIL bound to SmgGDS-558 in vitro (Fig. 3A, GST immunoprecipitation, lanes 4 and 5). This is a novel interaction between an unprenylated GTPase and SmgGDS-558 because our previous data indicated that only prenylated GTPases will associate with SmgGDS-558 in cells (20). Furthermore, we observed that the unprenylated peptides of K-Ras CVIM and CVIL also (to a lesser degree) bound to SmgGDS-558 overexpressed in HEK-293T cells (Fig. 3B, Streptavidin Immunoprecipitation, Overnight exposure, lanes 4 and 5). Taken together, these data suggest that there are cellular protein(s) that bind to either the body of K-Ras or to SmgGDS-558 that help mediate the specificity of SmgGDS-558 for prenylated K-Ras. Alternatively, post-translational modifications that occur only in eukaryotic cells may promote the specificity of SmgGDS-558 for prenylated K-Ras.
The Association between SmgGDS-607 and a K-Ras CVIL Peptide Can Be Competed by Full-length K-Ras CVIL but Not K-Ras CVIM
We demonstrated that SmgGDS-607 associates well with both full-length K-Ras CVIL and the K-Ras CVIL peptide, but SmgGDS-607 associates poorly with full-length K-Ras CVIM and the K-Ras CVIM peptide. To confirm that these interactions are specifically defined by the last amino acid in K-Ras, we tested the prediction that the binding of SmgGDS-607 to the K-Ras CVIL peptide will be competitively inhibited by the presence of full-length K-Ras CVIL, but not by full-length K-Ras CVIM. To test this prediction, we utilized the biotin-K-Ras CVIL peptide in a competition assay with full-length K-Ras CVIM or K-Ras CVIL. To define the optimal conditions for this assay, we first determined that SmgGDS-607-HA associates with the K-Ras CVIL peptide in a dose-dependent manner (Fig. 3C, top left panel). We determined that 1000 μg of total protein from lysates of HEK-293T cells expressing SmgGDS-607-HA was an appropriate concentration to use for the competition assay, based on our dose-response curve (Fig. 3C, top left and top right panels). We found that the addition of full-length recombinant K-Ras CVIL protein, but not the addition of full-length recombinant K-Ras CVIM protein, significantly prevented the association of SmgGDS-607-HA with the K-Ras CVIL peptide (Fig. 3C, top right panel and graph). Taken together, these data demonstrate a dose-dependent association of K-Ras CVIL peptide to SmgGDS-607 that can be competitively blocked by full-length K-Ras CVIL protein.
SmgGDS-607 Recognizes the Last Amino Acid in Rap1B for Association
We tested whether the observed interactions of SmgGDS with specific CAAX motifs are unique to K-Ras or conserved among multiple GTPases using Rap1B as a model. Previous reports show that SmgGDS-607 binds to Rap1B (41), Rap1A, and RhoA (20), which are GTPases that contain a PBR and a CAAX motif ending in leucine and undergo geranylgeranylation. We tested the ability of SmgGDS-607 to co-precipitate Rap1B after mutating the last amino acid, changing the sequence from CQLL to CQLM (Fig. 4A). SmgGDS-607 co-precipitates Rap1B CQLL strongly and Rap1B CQLM significantly less so (Fig. 4B, HA Immunoprecipitates, lane 1 compared with lane 2). There was no significant difference in the co-precipitation of SmgGDS-558 with either Rap1B CQLL or Rap1B CQLM (Fig. 4B, HA Immunoprecipitates, lane 4 compared with lane 5), which demonstrates a difference in the ability of SmgGDS-558 to recognize K-Ras CAAX variants versus Rap1B CAAX variants. These data also show that SmgGDS-607 preferentially binds CAAX motifs ending in leucine in multiple GTPases.
FIGURE 4.
SmgGDS-607 recognizes the last amino acid of Rap1B for binding. A, the C-terminal region of Rap1B is depicted in two segments: the PBR identified by the basic lysines and arginines, followed by the CAAX motif. WT Rap1B has a CAAX sequence of CQLL, which allows for the protein to become geranylgeranylated. The last amino acid of Rap1B was mutated to a methionine for this study. B, HEK-293T cells were transfected with a cDNA encoding SmgGDS-607-HA, SmgGDS-558-HA, or the HA vector plus a cDNA encoding the Myc-tagged WT Rap1B CQLL or Rap1B CQLM mutant, or a Myc vector. After 24 h, the cells were lysed, and an aliquot of each lysate was subjected to ECL-Western blotting using Myc antibody (Cell Lysates). The remaining volume of each cell lysate was immunoprecipitated with HA antibody, followed by ECL-Western blotting using HA antibody and Myc antibody (HA Immunoprecipitates). The results are shown as the optical density of IP Myc-Rap1B divided by the optical density of IP SmgGDS-HA and are the means ± S.E. of three independent experiments. ns, not significant; **, p < 0.01 by Student's t test. A nonspecific background band in the immunoblot is indicated by the asterisk.
SmgGDS-607 Can Bind Prenylated Rap1B Peptides
We next assessed the role of prenylation in the interaction of SmgGDS with a GTPase. It was previously reported that a geranylgeranylated Rap1B peptide (PGKARKKSSC), but not a un-prenylated Rap1B peptide, binds to SmgGDS-558 (32, 42). We utilized a GTPase known to be geranylgeranylated, based on our hypothesis that SmgGDS-607 will preferentially interact with a GTPase that will become geranylgeranylated. Therefore, we synthesized biotin-labeled Rap1B peptides that were either in a free thiol form or conjugated to farnesyl diphosphate, geranylgeranyl diphosphate, or a photo-active C10-m-BP (C10) geranylgeranyl mimic (Fig. 5A).
Similar to data obtained using full-length GTPases in cells, we found that the nonprenylated Rap1B (free thiol) peptide co-precipitates SmgGDS-607 but not SmgGDS-558 (Fig. 5B, Streptavidin Immunoprecipitation, lane 2 compared with lane 1). As expected, the geranylgeranylated Rap1B (geranylgeranyl) peptide co-precipitates SmgGDS-558 (Fig. 5B, Streptavidin Immunoprecipitation, lane 4), but surprisingly geranylgeranylated Rap1B (geranylgeranyl) also co-precipitates SmgGDS-607 (Fig. 5B, Streptavidin Immunoprecipitation, lane 5). We next assessed whether a farnesyl or geranylgeranyl moiety conjugated to Rap1B affects association to SmgGDS. SmgGDS-607 strongly co-precipitates with both the farnesylated Rap1B (farnesyl) and geranylgeranylated Rap1B (geranylgeranyl) peptides, whereas interestingly, SmgGDS-558 co-precipitates less strongly with both prenylated peptides (Fig. 5C, Streptavidin Immunoprecipitation, lanes 6 and 7 compared with lanes 10 and 11). These are the first findings that identify a prenylated GTPase peptide binding to SmgGDS-607.
To test the hypothesis that a prenylated Rap1B peptide is directly binding to SmgGDS-607, we utilized a photo-activatable geranylgeranyl mimic (C10), which was conjugated to a Rap1B peptide (Fig. 5A). The benzophenone-containing prenyl-group is a photophore that is chemically stable and activated by UV light that cross-links the conjugated species (Rap1B peptide) to proteins that directly interact with the conjugated substrate (SmgGDS-607) (43–46). We found that the Rap1B (C10) peptide cross-linking to SmgGDS-607 increased with greater UV exposure (Fig. 5D). The control lane shows a weak association between SmgGDS-607 and the C10 peptide at a lower molecular weight comparable with the SmgGDS-607 whole cell lysate input (Fig. 5D, lane 5 compared with lane 7), which indicates binding but not cross-linking. Taken together, these results indicate a novel interaction between a prenylated GTPase peptide and SmgGDS-607, which differs from our previous data that SmgGDS-607 will only recognize a nonprenylated GTPase in cells (20).
SmgGDS-607 and SmgGDS-558 Share Similar Predicted Protein Structures as Well as Similar Functional Domains with Some Unique Differences
SmgGDS-607 and SmgGDS-558 are armadillo proteins that we previously described as consisting of ARM domains A–M (20). SmgGDS-558 is a splice variant of SmgGDS-607 and is lacking the “C” ARM domain (Fig. 6A, yellow shaded area). We analyzed homology models of the 608- and 559-amino acid forms of SmgGDS. These forms of SmgGDS contain an additional alanine at position 2 compared with the 607- and 558-amino acid forms of SmgGDS. The insertion of this alanine should have minimal or no effect on the structure of SmgGDS-608 compared with SmgGDS-607 and of SmgGDS-559 compared with SmgGDS-558. Thus, it is reasonable to assume that the predicted structures of SmgGDS-608 and SmgGDS-559 are similar to those of SmgGDS-607 and SmgGDS-558, respectively. These homology models predict that the N-terminal region consisting of ARMs A and B is structurally very different in SmgGDS-608 compared with SmgGDS-559 (Fig. 6A, blue shaded area). In contrast, SmgGDS-608 and SmgGDS-559 are predicted to share very similar structures in the region consisting of ARMs D-M (Fig. 6A, green shaded area). This region contains the electronegative patch in SmgGDS that interacts with the positively charged C-terminal PBR of a GTPase (28).
FIGURE 6.
SmgGDS-608/607 and SmgGDS-559/558 share similar structural domains with differences in the N-terminal region. A, PyMOL homology models of SmgGDS-608 and SmgGDS-559 are presented based on a previous published model of SmgGDS-608 (28). SmgGDS-608 corresponds to SmgGDS-607, whereas SmgGDS-559 corresponds to SmgGDS-558, as described under “Experimental Procedures.” The ARM domains in the proteins are designated A–M, with ARM C (yellow) being spliced out of SmgGDS-559/558. Both SmgGDS-608/607 and SmgGDS-559/558 have similar structures in ARMs D-M (green), but SmgGDS-608/607 differs significantly from SmgGDS-559/558 in the structure of ARMs A and B (blue). B, our results support a model in which a GTPase ending with a methionine enters the farnesylation pathway without the aid of SmgGDS-607. The farnesylated GTPase then interacts with SmgGDS-558 and traffics to the plasma membrane. In contrast, a small GTPase ending with a leucine interacts with SmgGDS-607 prior to becoming prenylated by the geranylgeranyltransferase. The geranylgeranylated GTPase then interacts with SmgGDS-558 and traffics to the plasma membrane. A FTI promotes the alternate geranylgeranylation of a GTPase that is normally farnesylated (e.g., K-Ras), and this leads to an increased association between a GTPase ending in a methionine and SmgGDS-607. More analyses are needed to determine whether the interaction of WT K-Ras with SmgGDS-607 is needed for the alternate geranylgeranylation of this GTPase in cells treated with FTI (red arrows).
Utilizing the I-TASSER server (47–49), we analyzed the function of SmgGDS-607 and SmgGDS-558 through COFACTOR software. The COFACTOR software utilizes functional libraries to assess the predicted protein structure match to functional sites and homologies of other known protein complexes. Table 1 shows the top five predicted enzymatic homologs of the SmgGDS splice variants using known enzyme-substrate complexes. Similar predicted functions between SmgGDS-607 and SmgGDS-558 include the functions of the V-type ATPase subunit H, PME-1, and PP2A core enzyme complex, and immunoglobulin A1 protease (Table 1). Interestingly, SmgGDS-558, but not SmgGDS-607, shared an enzymatic homology with a protein farnesyltransferase bound to a caged TCVIM peptide and farnesyl diphosphate. This difference between SmgGDS-558 and SmgGDS-607 is consistent with SmgGDS-558 uniquely recognizing prenylated GTPases.
TABLE 1.
Structure-based functional predictions of SmgGDS-607 and SmgGDS-558
The table shows analysis of SmgGDS-607 and SmgGDS-558 utilizing the I-TASSER structural and functional prediction software, which reports COFACTOR: structure-based functional predictions. The top five proteins that are shown share similar structural active sites to either SmgGDS-607 or SmgGDS-558. CscoreEC is the confidence score for the Enzyme Classification (EC) number prediction. CscoreEC values range between 0 and 1; a higher score indicates a more reliable EC number prediction. TM score is a measure of global structural similarity between query and template protein where scores <0.17 correspond to a random similarity. PDB Hit is the Protein Data Bank signifier.
| Rank | Name of enzyme homolog | CscoreEC | PDB hit | TM score | Predicted active site residues |
|---|---|---|---|---|---|
| SmgGDS-607 | |||||
| 1 | Vacuolar ATP synthase subunit H | 0.177 | 1ho8A | 0.387 | NA |
| 2 | PME-1 and PP2A core enzyme | 0.157 | 3c5wA | 0.279 | 372 |
| 3 | Immunoglobulin A1 protease | 0.15 | 3h09B | 0.411 | 364 |
| 4 | 1,4-α-Glucan-branching enzyme | 0.136 | 3k1dA | 0.388 | 335 and 384 |
| 5 | Fatty acid synthase subunit α | 0.133 | 2vkzG | 0.362 | 225 |
| SmgGDS-558 | |||||
| 1 | Vacuolar ATP synthase subunit H | 0.25 | 1ho8A | 0.427 | 282, 284, 289, 291, and 334 |
| 2 | Protein farnesyltransferase | 0.191 | 3dpyA | 0.38 | NA |
| 3 | PME-1 and PP2A core enzyme | 0.181 | 3c5wA | 0.29 | 239 and 325 |
| 4 | O-linked GlcNAc transferase | 0.163 | 1w3bA | 0.426 | 311 and 344 |
| 5 | Immunoglobulin A1 protease | 0.162 | 3h09B | 0.418 | 298 and 315 |
DISCUSSION
Our results indicate that the binding of a GTPase to SmgGDS-607 is based not only on the PBR of the GTPase (30) but also by the last amino acid of the GTPase. This finding indicates that SmgGDS-607 is a novel CAAX-binding protein and provides new insights into the regulation of GTPases before their entry into the prenylation pathway. Furthermore, our results show that SmgGDS-607 preferentially binds GTPases that will become geranylgeranylated rather than GTPases that will become farnesylated. These findings identify SmgGDS-607 as a target for cancer therapeutics, especially therapies aimed at inhibiting protein prenylation.
Our proposed model shows that SmgGDS-607 will associate with GTPases that end in a leucine and will move through the geranylgeranylation pathway, but not with a GTPase that ends in a methionine and will move through the farnesylation pathway (Fig. 6B). Previous data (20) along with the data presented in this study support the idea that SmgGDS-607 acts as a storage protein that binds the nonprenylated GTPase until the geranylgeranyltransferase (GGTase) can process it. Storage of a nonprenylated GTPase by SmgGDS-607 is an ideal way to regulate the entrance of a GTPase into the geranylgeranylation pathway. SmgGDS-607 may promote the trafficking of a GTPase to the GGTase similar to the interaction of the Rab escort protein-1 with Rab proteins (50, 51). In this case, SmgGDS-607 would recognize a newly synthesized GTPase ending in a leucine and present it to the GGTase for prenylation. Our previous studies indicate that certain signals such as nucleotide exchange (20) or phosphorylation (41) of a GTPase will determine the ability of a GTPase to bind SmgGDS-607 and/or be released to the GGTase.
We found that mutating the C-terminal residue of K-Ras from a methionine to a leucine significantly increases the binding of K-Ras to SmgGDS-607. This leucine substitution causes K-Ras to become geranylgeranylated instead of farnesylated, supporting our model that SmgGDS-607 prefers binding small GTPases that have a C-terminal leucine and will become geranylgeranylated.
Quite surprisingly, we found that treatment with a FTI increased the co-precipitation of complexes consisting of WT K-Ras and SmgGDS-607 from cells. This result might seem perplexing, because we would not expect SmgGDS-607 to bind WT K-Ras because of the presence of the C-terminal methionine. However, it is well known that the presence of an FTI causes WT K-Ras to have reduced interactions with the farnesyltransferase and increased interactions with the GGTase, resulting in the geranylgeranylation of K-Ras. It is intriguing to speculate that complexes of K-Ras and SmgGDS-607 can be isolated from FTI-treated cells because K-Ras has increased interaction with the GGTase in these cells. If SmgGDS-607 acts as a scaffold that binds the GGTase, then SmgGDS-607 would be expected to bind the GGTase when it is interacting with K-Ras in FTI-treated cells. In this case, SmgGDS-607 will not directly bind K-Ras (because of the presence of the C-terminal methionine), but instead SmgGDS-607 will bind the complex consisting of the GGTase and K-Ras.
Taken together, our findings support the model that SmgGDS-607 directly binds GTPases that have the CAAL motif (as indicated by our studies using recombinant proteins), but SmgGDS-607 can also indirectly bind GTPases that have the CAAM motif if they are associated with the GGTase (as in the case of K-Ras in FTI-treated cells). This model predicts that SmgGDS-607 will physically interact with both the GTPase and the GGTase. However, our initial studies trying to immunoprecipitate the GGTase I-β subunit with SmgGDS-607 or SmgGDS-558 either in the presence or absence of a GTPase have yielded no associations (data not shown). We have also tried to probe our immunoblots of immunoprecipitated complexes using antibodies that recognize the farnesyltransferase or GGTase with no success.
The interesting finding that K-Ras will associate with SmgGDS-607 in cells treated with a FTI provides novel insight into the mechanism of alternate prenylation and has potential therapeutic implications. FTIs were developed to block farnesylation of oncogenic Ras and prevent it from reaching the plasma membrane where its downstream target proteins are localized (52, 53). Although promising initially, the main limitation of FTIs was the lack of effectiveness in clinical trials (54, 55). Interestingly, it was found that in cells treated with FTIs, K-Ras could become alternatively geranylgeranylated and still continue to promote oncogenic signaling from the membrane (40). More success came from utilizing FTIs simultaneously with GGTIs; however, toxicity then became an issue (56, 57).
The association of K-Ras with SmgGDS-607 in cells treated with FTIs suggests that SmgGDS-607 might participate in the alternative geranylgeranylation of K-Ras in the presence of FTIs. If SmgGDS-607 regulates the entry of K-Ras into the geranylgeranylation pathway in FTI-treated cells, then inhibiting interactions of SmgGDS-607 with K-Ras (either through down-regulation or inactivation of SmgGDS-607) could provide a novel way to inhibit K-Ras geranylgeranylation when FTIs are administered. Thus, SmgGDS-607 could be a novel target for combination therapeutics, because initial studies show that knockdown of SmgGDS-607 alone is not deleterious in breast and lung cancers (20, 23). However, it is important to note that the association of K-Ras with SmgGDS-607 in FTI-treated cells could simply be an ancillary result, whereby K-Ras can become geranylgeranylated regardless of its association with SmgGDS-607. More tests are needed to define the function of SmgGDS-607 in the alternative geranylgeranylation of K-Ras when farnesylation is inhibited.
SmgGDS-558 differs from SmgGDS-607 because SmgGDS-558 is restricted to binding prenylated GTPases in cells (20). Interestingly, the analysis of predicted functional properties between SmgGDS-558 and SmgGDS-607 identified that the structure of SmgGDS-558 (but not SmgGDS-607) has enzymatic homology with a farnesyltransferase that has a caged K-Ras peptide and farnesyl pyrophosphate bound, which supports the observation that SmgGDS-558 will functionally bind prenylated GTPases (Table 1).
We found that in cells, SmgGDS-558 binds prenylated K-Ras that has either a methionine or leucine as the last amino acid, but interestingly SmgGDS-558 co-precipitates significantly more K-Ras CVIM than K-Ras CVIL. The most likely explanation for this finding is that the prenylated form of K-Ras is generated more from K-Ras CVIM than from K-Ras CVIL in the cells, thereby providing more prenylated K-Ras for SmgGDS-558 to co-precipitate from the cells. This possibility is supported by our finding that a higher ratio of prenylated to nonprenylated K-Ras is generated when K-Ras CVIM is expressed than when K-Ras CVIL is expressed (Fig. 1C, Cell Lysates, lane 6 compared with lane 8). A less likely explanation for SmgGDS-558 co-precipitating significantly more K-Ras CVIM than K-Ras CVIL is that SmgGDS-558 prefers to associate more with farnesylated K-Ras (which is generated from K-Ras CVIM) than with geranylgeranylated K-Ras (which is generated from K-Ras CVIL). Finally, a third potential explanation for the preference of SmgGDS-558 for K-Ras CVIM is that SmgGDS-558 interacts with prenylated K-Ras before the -AAX is cleaved from the CAAX motif by Rce1. In this speculative model, SmgGDS-558 binds prenylated GTPases that still retain their CAAX motif, with a preference for a CAAX motif ending in methionine rather than leucine. If these last two explanations are correct, we would expect SmgGDS-558 to exhibit the same binding preferences for the Rap1B CAAX variants as those shown for the K-Ras CAAX variants. However, we found that SmgGDS-558 co-precipitates equal amounts of Rap1B CQLM and Rap1B CQLL from cells (Fig. 4), suggesting that neither the type of isoprenoid modification (farnesyl versus geranylgeranyl) nor the last amino acid in the CAAX motif (methionine versus leucine) is dictating the specificity of SmgGDS-558 for the prenylated GTPases. Instead, the ratio of prenylated to nonprenylated GTPase seems to be the most likely indicator for interaction with SmgGDS-558. This conclusion is supported by our finding that Rap1B CQLM and Rap1B CQLL generate the same ratios of prenylated to nonprenylated Rap1B (Fig. 4B, Cell Lysates, lane 4 compared with lane 5) and are equally co-precipitated by SmgGDS-558.
The ability of SmgGDS-558 to associate with both farnesylated and geranylgeranylated GTPases in cells supports the idea that this splice variant may play a greater role than SmgGDS-607 in the trafficking of oncogenic GTPases. Indeed, previous studies indicate that the malignant phenotype of non-small cell lung carcinoma and breast cancer is decreased more by the RNAi-mediated knockdown of SmgGDS-558 than by SmgGDS-607 (20, 23). However, it is possible that the RNAi-mediated knockdown of SmgGDS-607 did not deplete SmgGDS-607 protein levels enough in these experiments to produce a functional effect. The development of small molecule inhibitors for both SmgGDS splice variants will aid further studies of their unique functions (20, 23).
We made the unexpected discovery that the preference of SmgGDS-558 for prenylated GTPases is lost when SmgGDS-558 is allowed to interact with GTPases in vitro. This conclusion is supported by our observation that even though SmgGDS-558 co-precipitates only the prenylated forms of WT K-Ras and K-Ras-CVIL in cells, SmgGDS-558 will bind to nonprenylated WT K-Ras and K-Ras-CVIL in recombinant systems in vitro. Additionally, we show that SmgGDS-558 will bind a nonprenylated C-terminal K-Ras peptide. Consistent with these findings, it was previously reported that SmgGDS-558 can induce guanine nucleotide exchange by the nonprenylated form of RhoA in a recombinant system but promotes guanine nucleotide exchange by only prenylated RhoA in cells (28). These findings indicate that SmgGDS-558 prefers prenylated GTPases only when interacting with the GTPases in a cellular system. Post-translational modifications or unknown protein partners may mediate the interaction of SmgGDS-558 with prenylated GTPases in cells, and the loss of these post-translational modifications or protein partners in recombinant systems may allow SmgGDS-558 to interact with nonprenylated GTPases in vitro.
Another interesting finding was the strong association of a prenylated Rap1B peptide to SmgGDS-607, which is surprising considering our previous model that SmgGDS-607 only binds nonprenylated GTPases. This novel discovery led us to assess whether this interaction was direct, and indeed using cross-linking we show that a prenylated Rap1B peptide covalently binds to SmgGDS-607. It is not clear why SmgGDS-607 directly binds the prenylated Rap1B C-terminal peptide in vitro, when SmgGDS-607 is restricted to binding only nonprenylated full-length Rap1B in cells. The prenylated peptide might bind to SmgGDS-607 through nonspecific hydrophobic interactions that arise in the in vitro system. Alternatively, the prenylated peptide might specifically bind a hydrophobic pocket in SmgGDS-607 that is accessible to the prenylated peptide in vitro but is inaccessible to full-length prenylated Rap1B in a cellular context. Further testing will be needed to define the reasons for this surprising result.
The homology models help explain several of the interactions we have observed in this study. According to these models, SmgGDS-608 and SmgGDS-559 are structurally very similar in ARMs D-M, which contains the electronegative patch that binds the PBR of a GTPase (28). We propose that when SmgGDS and a GTPase are allowed to interact in vitro without the presence of other proteins, the GTPases that are nonprenylated can associate with both SmgGDS splice variants through an electrostatic interaction between the PBR and the electronegative patch in ARMs D-M. This prediction is supported by our finding that both SmgGDS-607 and SmgGDS-558 bind nonprenylated K-Ras in recombinant systems.
The very different structure predicted for ARMs A and B in SmgGDS-608 compared with SmgGDS-559 might explain why the two SmgGDS splice variants differ in their ability to bind nonprenylated versus prenylated GTPases in cells. We propose that the different N-terminal structures of the two SmgGDS splice variants might cause them to have unique post-translational modifications or to interact with different protein partners in cells. These post-translational modifications and/or interactions with cellular protein partners may define the specificity of SmgGDS-607 for nonprenylated GTPases, and SmgGDS-558 for prenylated GTPases that is observed only in cellular systems. Thus, in vitro with no other proteins available, the splice variants would lose their ability to differentiate between prenylated and nonprenylated GTPases, and their interactions with GTPases in vitro would be dictated mainly by the electronegative patch in ARMs D-M.
This study is the first report that SmgGDS-607 binds GTPases by recognizing the last amino acid of the GTPase. SmgGDS-607 is shown to be involved specifically in the geranylgeranylation pathway for GTPases. Although direct, the associations of SmgGDS-607 and SmgGDS-558 with GTPases differ between cells and in vitro, suggesting that these interactions are regulated by processes that uniquely occur in intact cellular systems. Further studies should be focused on developing small molecule inhibitors for both splice variants of SmgGDS and assessing their potential therapeutic use.
Acknowledgments
We thank Dr. John Sondek (University of North Carolina-Chapel Hill) and Dr. Brant Hamel (Duke University) for the homology models of SmgGDS-608 and SmgGDS-559.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA136799 (to C. L. W.), R01 GM084152 and GM106082 (to M. D. D.), and R01 AI30162 and U54 AI057153 (to J. T. B.). This work was also supported by funds from the Medical College of Wisconsin Cancer Center (to C. L. W.), the Wisconsin Breast Cancer Showhouse (to C. L. W.), and the Rock River Cancer Research Foundation (to C. L. W.).
- ARM
- armadillo
- PBR
- poly-basic region
- FTI
- farnesyltransferase inhibitor
- GGTI
- geranylgeranyltransferase inhibitor
- GGTase
- geranylgeranyltransferase
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- DMF
- N,N-dimethylformamide.
REFERENCES
- 1. Lloyd A. C. (1998) Ras versus cyclin-dependent kinase inhibitors. Curr. Opin. Genet. Dev. 8, 43–48 [DOI] [PubMed] [Google Scholar]
- 2. Yoshida Y., Kawata M., Miura Y., Musha T., Sasaki T., Kikuchi A., Takai Y. (1992) Microinjection of smg/rap1/krev-1 p21 into Swiss 3T3 cells induces DNA synthesis and morphological changes. Mol. Cell. Biol. 12, 3407–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Janda E., Lehmann K., Killisch I., Jechlinger M., Herzig M., Downward J., Beug H., Grünert S. (2002) Ras and TGFβ cooperatively regulate epithelial cell plasticity and metastasis dissection of ras signaling pathways. J. Cell Biol. 156, 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bailey C. L., Kelly P., Casey P. J. (2009) Activation of Rap1 promotes prostate cancer metastasis. Cancer Res. 69, 4962–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mayo M. W., Wang C. Y., Cogswell P. C., Rogers-Graham K. S., Lowe S. W., Der C. J., Baldwin A. S., Jr. (1997) Requirement of NF-κB activation to suppress p53-independent apoptosis induced by oncogenic ras. Science 278, 1812–1815 [DOI] [PubMed] [Google Scholar]
- 6. Downward J. (1998) Ras signalling and apoptosis. Curr. Opin. Genet. Dev. 8, 49–54 [DOI] [PubMed] [Google Scholar]
- 7. Malumbres M., Barbacid M. (2003) RAS oncogenes. The first 30 years. Nat. Rev. Cancer 3, 459–465 [DOI] [PubMed] [Google Scholar]
- 8. Sahai E., Marshall C. J. (2002) RHO-GTPases and cancer. Nat. Rev. Cancer 2, 133–142 [DOI] [PubMed] [Google Scholar]
- 9. Vigil D., Cherfils J., Rossman K. L., Der C. J. (2010) Ras superfamily GEFs and GAPs. Validated and tractable targets for cancer therapy? Nat. Rev. Cancer 10, 842–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Viola M. V., Fromowitz F., Oravez S., Deb S., Finkel G., Lundy J., Hand P., Thor A., Schlom J. (1986) Expression of ras oncogene p21 in prostate cancer. N. Engl. J. Med. 314, 133–137 [DOI] [PubMed] [Google Scholar]
- 11. Boguski M. S., McCormick F. (1993) Proteins regulating ras and its relatives. Nature. 366, 643–654 [DOI] [PubMed] [Google Scholar]
- 12. Lerner E. C., Qian Y., Blaskovich M. A., Fossum R. D., Vogt A., Sun J., Cox A. D., Der C. J., Hamilton A. D., Sebti SM. (1995) Ras CAAX peptidomimetic FTI-277 selectively blocks oncogenic ras signaling by inducing cytoplasmic accumulation of inactive ras-raf complexes. J. Biol. Chem. 270, 26802–26806 [DOI] [PubMed] [Google Scholar]
- 13. Konstantinopoulos P. A., Karamouzis M. V., Papavassiliou A. G. (2007) Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat. Rev. Drug Discov. 6, 541–555 [DOI] [PubMed] [Google Scholar]
- 14. Hancock J. F., Paterson H., Marshall C. J. (1990) A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21 ras to the plasma membrane. Cell 63, 133–139 [DOI] [PubMed] [Google Scholar]
- 15. Cadwallader K. A., Paterson H., Macdonald S. G., Hancock J. F. (1994) N-terminally myristoylated ras proteins require palmitoylation or a polybasic domain for plasma membrane localization. Mol. Cell. Biol. 14, 4722–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Winter-Vann A. M., Casey P. J. (2005) Post-prenylation-processing enzymes as new targets in oncogenesis. Nat. Rev. Cancer 5, 405–412 [DOI] [PubMed] [Google Scholar]
- 17. Moores S. L., Schaber M. D., Mosser S. D., Rands E., O'Hara M. B., Garsky V. M., Marshall M. S., Pompliano D. L., Gibbs J. B. (1991) Sequence dependence of protein isoprenylation. J. Biol. Chem. 266, 14603–14610 [PubMed] [Google Scholar]
- 18. Thissen J. A., Gross J. M., Subramanian K., Meyer T., Casey P. J. (1997) Prenylation-dependent association of ki-ras with microtubules. Evidence for a role in subcellular trafficking. J. Biol. Chem. 272, 30362–30370 [DOI] [PubMed] [Google Scholar]
- 19. Apolloni A., Prior I. A., Lindsay M., Parton R. G., Hancock J. F. (2000) H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 20, 2475–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berg T. J., Gastonguay A. J., Lorimer E. L., Kuhnmuench J. R., Li R., Fields A. P., Williams C. L. (2010) Splice variants of SmgGDS control small GTPase prenylation and membrane localization. J. Biol. Chem. 285, 35255–35266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tew G. W., Lorimer E. L., Berg T. J., Zhi H., Li R., Williams C. L. (2008) SmgGDS regulates cell proliferation, migration, and NF-κB transcriptional activity in non-small cell lung carcinoma. J. Biol. Chem. 283, 963–976 [DOI] [PubMed] [Google Scholar]
- 22. Zhi H., Yang X. J., Kuhnmuench J., Berg T., Thill R., Yang H., See W. A., Becker C. G., Williams C. L., Li R. (2009) SmgGDS is up-regulated in prostate carcinoma and promotes tumour phenotypes in prostate cancer cells. J. Pathol. 217, 389–397 [DOI] [PubMed] [Google Scholar]
- 23. Hauser A. D., Bergom C., Schuld N. J., Chen X., Lorimer E. L., Huang J., Mackinnon A. C., Williams C. L. (2013) The SmgGDS splice variant SmgGDS-558 is a key promoter of tumor growth and RhoA activation in breast cancer. Mol. Cancer Res. 12, 130–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mizuno T., Kaibuchi K., Yamamoto T., Kawamura M., Sakoda T., Fujioka H., Matsuura Y., Takai Y. (1991) A stimulatory GDP/GTP exchange protein for smg p21 is active on the post-translationally processed form of c-ki-ras p21 and rhoA p21. Proc. Natl. Acad. Sci. U.S.A. 88, 6442–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yamamoto T., Kaibuchi K., Mizuno T., Hiroyoshi M., Shirataki H., Takai Y. (1990) Purification and characterization from bovine brain cytosol of proteins that regulate the GDP/GTP exchange reaction of smg p21s, ras p21-like GTP-binding proteins. J. Biol. Chem. 265, 16626–16634 [PubMed] [Google Scholar]
- 26. Hiraoka K., Kaibuchi K., Ando S., Musha T., Takaishi K., Mizuno T., Asada M., Ménard L., Tomhave E., Didsbury J. (1992) Both stimulatory and inhibitory GDPGTP exchange proteins, smg GDS and rho GDI, are active on multiple small GTP-binding proteins. Biochem. Biophys. Res. Commun. 182, 921–930 [DOI] [PubMed] [Google Scholar]
- 27. Isomura M., Kikuchi A., Ohga N., Takai Y. (1991) Regulation of binding of rhoB p20 to membranes by its specific regulatory protein, GDP dissociation inhibitor. Oncogene 6, 119–124 [PubMed] [Google Scholar]
- 28. Hamel B., Monaghan-Benson E., Rojas R. J., Temple B. R., Marston D. J., Burridge K., Sondek J. (2011) SmgGDS is a guanine nucleotide exchange factor that specifically activates RhoA and RhoC. J. Biol. Chem. 286, 12141–12148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peifer M., Berg S., Reynolds A. B. (1994) A repeating amino acid motif shared by proteins with diverse cellular roles. Cell. 76, 789–791 [DOI] [PubMed] [Google Scholar]
- 30. Williams C. L. (2003) The polybasic region of ras and rho family small GTPases. A regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cell Signal. 15, 1071–1080 [DOI] [PubMed] [Google Scholar]
- 31. Shin H. J., Lee C. H., Cho I. H., Kim Y. J., Lee Y. J., Kim I. A., Park K. D., Yui N., Shin J. W. (2006) Electrospun PLGA nanofiber scaffolds for articular cartilage reconstruction. Mechanical stability, degradation and cellular responses under mechanical stimulation in vitro. J. Biomater. Sci. Polym. Ed. 17, 103–119 [DOI] [PubMed] [Google Scholar]
- 32. Kotani K., Kikuchi A., Doi K., Kishida S., Sakoda T., Kishi K., Takai Y. (1992) The functional domain of the stimulatory GDP/GTP exchange protein (smg GDS) which interacts with the C-terminal geranylgeranylated region of rap1/krev-1/smg p21. Oncogene 7, 1699–1704 [PubMed] [Google Scholar]
- 33. Lanning C. C., Ruiz-Velasco R., Williams C. L. (2003) Novel mechanism of the co-regulation of nuclear transport of SmgGDS and rac1. J. Biol. Chem. 278, 12495–12506 [DOI] [PubMed] [Google Scholar]
- 34. Turek T. C., Gaon I., Gamache D., Distefano M. D. (1997) Synthesis and evaluation of benzophenone-based photoaffinity labeling analogs of prenyl pyrophosphates containing stable amide linkages. Bioorg. Med. Chem. Lett. 7, 2125–2130 [Google Scholar]
- 35. Turek T. C., Gaon I., Distefano M. D., Strickland C. L. (2001) Synthesis of farnesyl diphosphate analogues containing ether-linked photoactive benzophenones and their application in studies of protein prenyltransferases. J. Org. Chem. 66, 3253–3264 [DOI] [PubMed] [Google Scholar]
- 36. Kale T. A., Raab C., Yu N., Dean D. C., Distefano M. D. (2001) A photoactivatable prenylated cysteine designed to study isoprenoid recognition. J. Am. Chem. Soc. 123, 4373–4381 [DOI] [PubMed] [Google Scholar]
- 37. Turek T. C., Gaon I., Distefano M. D. (1997) Synthesis and rapid purification of 32P-labeled photoactive analogs of farnesyl pyrophosphate. J. Labelled Comp. Radiopharm. 39, 139–146 [Google Scholar]
- 38. Maresso A. W., Barbieri J. T. (2002) Expression and purification of two recombinant forms of the type-III cytotoxin, pseudomonas aeruginosa ExoS. Protein Expr. Purif. 26, 432–437 [DOI] [PubMed] [Google Scholar]
- 39. Hancock J. F. (1995) Prenylation and palmitoylation analysis. Methods Enzymol. 255, 237–245 [DOI] [PubMed] [Google Scholar]
- 40. Whyte D. B., Kirschmeier P., Hockenberry T. N., Nunez-Oliva I., James L., Catino J. J., Bishop W. R., Pai J. K. (1997) K- and N-ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem. 272, 14459–14464 [DOI] [PubMed] [Google Scholar]
- 41. Ntantie E., Gonyo P., Lorimer E. L., Hauser A. D., Schuld N., McAllister D., Kalyanaraman B., Dwinell M. B., Auchampach J. A., Williams C. L. (2013) An adenosine-mediated signaling pathway suppresses prenylation of the GTPase Rap1B and promotes cell scattering. Sci. Signal. 6, ra39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shirataki H., Kaibuchi K., Hiroyoshi M., Isomura M., Araki S., Sasaki T., Takai Y. (1991) Inhibition of the action of the stimulatory GDP/GTP exchange protein for smg p21 by the geranylgeranylated synthetic peptides designed from its C-terminal region. J. Biol. Chem. 266, 20672–20677 [PubMed] [Google Scholar]
- 43. Kyro K., Manandhar S. P., Mullen D., Schmidt W. K., Distefano M. D. (2011) Photoaffinity labeling of ras converting enzyme using peptide substrates that incorporate benzoylphenylalanine (bpa) residues. Improved labeling and structural implications. Bioorg. Med. Chem. 19, 7559–7569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dormán G., Prestwich G. D. (1994) Benzophenone photophores in biochemistry. Biochemistry 33, 5661–5673 [DOI] [PubMed] [Google Scholar]
- 45. Chowdhry V., Westheimer F. (1979) Photoaffinity labeling of biological systems. Annu. Rev. Biochem. 48, 293–325 [DOI] [PubMed] [Google Scholar]
- 46. Vervacke J. S., Wang Y. C., Distefano M. D. (2013) Photoactive analogs of farnesyl diphosphate and related isoprenoids. Design and applications in studies of medicinally important isoprenoid-utilizing enzymes. Curr. Med. Chem. 20, 1585–1594 [DOI] [PubMed] [Google Scholar]
- 47. Roy A., Yang J., Zhang Y. (2012) COFACTOR. An accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 40, W471–W477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER. A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Alexandrov K., Horiuchi H., Steele-Mortimer O., Seabra M. C., Zerial M. (1994) Rab escort protein-1 is a multifunctional protein that accompanies newly prenylated Rab proteins to their target membranes. EMBO J. 13, 5262–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stenmark H., Olkkonen V. M. (2001) The Rab GTPase family. Genome Biol. 2, REVIEWS3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Basso A. D., Kirschmeier P., Bishop W. R. (2006) Thematic review series. Lipid posttranslational modifications. farnesyl transferase inhibitors. J. Lipid Res. 47, 15–31 [DOI] [PubMed] [Google Scholar]
- 53. Law B. K., Norgaard P., Moses H. L. (2000) Farnesyltransferase inhibitor induces rapid growth arrest and blocks p70s6k activation by multiple stimuli. J. Biol. Chem. 275, 10796–10801 [DOI] [PubMed] [Google Scholar]
- 54. Rao S., Cunningham D., de Gramont A., Scheithauer W., Smakal M., Humblet Y., Kourteva G., Iveson T., Andre T., Dostalova J., Illes A., Belly R., Perez-Ruixo J. J., Park Y. C., Palmer P. A. (2004) Phase III double-blind placebo-controlled study of farnesyl transferase inhibitor R115777 in patients with refractory advanced colorectal cancer. J. Clin. Oncol. 22, 3950–3957 [DOI] [PubMed] [Google Scholar]
- 55. Macdonald J. S., McCoy S., Whitehead R. P., Iqbal S., Wade J. L., 3rd, Giguere J. K., Abbruzzese J. L. (2005) A phase II study of farnesyl transferase inhibitor R115777 in pancreatic cancer. A southwest oncology group (SWOG 9924) study. Invest. New Drugs. 23, 485–487 [DOI] [PubMed] [Google Scholar]
- 56. Sun J., Qian Y., Hamilton A. D., Sebti S. M. (1998) Both farnesyltransferase and geranylgeranyltransferase I inhibitors are required for inhibition of oncogenic K-ras prenylation but each alone is sufficient to suppress human tumor growth in nude mouse xenografts. Oncogene 16, 1467–1473 [DOI] [PubMed] [Google Scholar]
- 57. Lobell R. B., Omer C. A., Abrams M. T., Bhimnathwala H. G., Brucker M. J., Buser C. A., Davide J. P., deSolms S. J., Dinsmore C. J., Ellis-Hutchings M. S., Kral A. M., Liu D., Lumma W. C., Machotka S. V., Rands E., Williams T. M., Graham S. L., Hartman G. D., Oliff A. I., Heimbrook D. C., Kohl N. E. (2001) Evaluation of farnesyl:protein transferase and geranylgeranyl:protein transferase inhibitor combinations in preclinical models. Cancer Res. 61, 8758–8768 [PubMed] [Google Scholar]