Background: PinX1 interacts with TRF1 and hTERT, but the significance of the physical association is not fully understood.
Results: PinX1 is involved in chromosome stability by regulating TRF1 protein stability, and the function of PinX1 is linked to hTERT.
Conclusion: PinX1 is functionally connected with hTERT to maintain TRF1 stability.
Significance: These findings suggest that a TRF1 turnover mechanism is linked to telomerase.
Keywords: DNA Damage Response, Telomerase, Telomeres, Tumor Cell Biology, Tumor Suppressor Gene, DNA Damage Response, TERT, Chromosomal Instability, Telomere Deficiency, Telomeric Repeat-binding Factor 1
Abstract
TRF1, a telomere-binding protein, is important for telomere protection and homeostasis. PinX1 interacts with TRF1, but the physiological consequences of their interaction in telomere protection are not yet understood. Here we investigated PinX1 function on TRF1 stability in HeLa cells. PinX1 overexpression stabilized TRF1, but PinX1 depletion by siRNA led to TRF1 degradation, TRF1 ubiquitination, and less TRF1 telomere association. The depletion also induced DNA damage responses at telomeres and chromosome instability. These telomere dysfunctional phenotypes were in fact due to TRF1 deficiency. We also report that hTERT, a catalytic component of telomerase, plays dual roles in the TRF1 steady state pathway. PinX1-mediated TRF1 stability was not observed in hTERT-negative immortal cells, but was pronounced when hTERT was ectopically expressed in the cells, suggesting that hTERT may be needed in the PinX1-mediated TRF1 stability pathway. Interestingly, the knockdown of both PinX1 and hTERT in HeLa cells stabilized TRF1, suppressed DNA damage response activation, and restored chromosome stability. In summary, our findings suggested that PinX1 may maintain telomere integrity by regulating TRF1 stability and that hTERT may act as both a positive and a negative regulator of TRF1 homeostasis in a PinX1-dependent manner.
Introduction
Telomeres are specialized nucleoprotein complexes that cap the natural ends of chromosomes and protect them from end-to-end fusion. Human somatic cells show a progressive loss of telomeric DNA during successive rounds of cell division due to DNA end replication problems (1, 2). When telomeres shorten to a critical point, DNA damage responses (DDRs)3 are triggered at the chromosomal ends, forcing normal cells into senescence. In most human immortal cells, telomeres are maintained by telomerase, a ribonucleoprotein complex, which adds TTAGGG repeats to the telomeric ends. Telomerase, suppressed in normal somatic cells, is reactivated to counteract telomere shortening in most human cancer cells (3–5). Introduction of hTERT, a catalytic component of telomerase, into telomerase-silent somatic cells restores telomerase activity and leads to telomere maintenance and an indefinite life span of normal somatic cells (6–8).
Telomeric DNA is protected by a shelterin complex composed of six telomere-specific proteins, TRF1, TRF2, TIN2, RAP1, TPP1, and POT1 (9). Shelterin is involved in the formation of t-loops, protects telomeres from being recognized as double-strand breaks (9, 10), and controls telomerase-dependent telomere extension (11, 12). TRF1 binds to double-stranded telomeric DNA and plays critical roles in telomere length regulation and telomere protection. Long term overexpression of TRF1 in telomerase-positive human cancer cells results in progressive telomere shortening, and expression of a dominant negative mutant form of TRF1 designed to inhibit binding of endogenous TRF1 to telomeres was shown to lead to telomere elongation, indicating that TRF1 acts as a negative regulator of telomere length (12). Previous studies revealed that loss of TRF1 leads to embryonic lethality in mice, causes telomere end fusion, and retards the proliferation of murine embryonic stem cells, indicating that TRF1 is crucial for the maintenance of telomere integrity and cell viability (13, 14).
Genetic and molecular studies have revealed that telomere-TRF1 association is tightly regulated by a series of sequential posttranslational modifications, including ubiquitination, ADP-ribosylation, and phosphorylation (15–17). ADP-ribosylation of TRF1 by tankyrase releases TRF1 from telomeres (17), and phosphorylation of TRF1 by casein kinase 2 enhances the ability of TRF1 to bind to telomeres (16). Moreover, TRF1 released from telomeres is known to undergo protein degradation via the ubiquitin-mediated proteasomal degradation pathway (15, 16).
PinX1 was identified as a TRF1-interacting protein and acts as a telomerase inhibitor. In a previous study, overexpression of PinX1 or delivery of the carboxyl-terminal (functional) fragment of PinX1 in telomerase-positive cancer cells reduced telomerase activity, shortened telomeres, and induced cell crisis (18, 19), suggesting that PinX1 might be an intrinsic telomerase inhibitor. PinX1, located at 8p23, is frequently deleted in many tumors, and tumor tissues originating from different organs express low levels of PinX1 compared with adjacent normal tissues, indicating that PinX1 is a putative tumor suppressor gene in human cells. Deletion of PinX1 causes embryonic lethality in mice, in which PinX1 heterozygous knockout led to the development of a range of malignant tumors (20). PinX1, as a nucleolar protein, is known to allow TRF1 to accumulate in the nucleolus, but the physiological role of the nucleolar transit of the protein remains unclear (21).
Although the role of PinX1 in regulating telomere length by inhibiting telomerase activity has been characterized, the physiological role of PinX1-TRF1 interactions has not yet been elucidated. In this study, we show that PinX1 plays a critical role in the maintenance of telomere integrity by controlling TRF1 steady state protein levels. Also, hTERT was found to be involved in both PinX1-mediated TRF1 stability and PinX1 depletion-mediated TRF1 degradation.
EXPERIMENTAL PROCEDURES
Cell Culture
HeLa cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen), 10 μg/ml streptomycin, and penicillin (Invitrogen). GM847 cells were cultured in DMEM supplemented with 10% FBS and 0.1% gentamicin (Sigma). GM847/vector and GM847/hTERT cells were established by infection with Lenti-EGFP and Lenti-hTERT (G200; ABM, BC, Canada), respectively, followed by positive clone selection with 450 μg/ml G418 (Duchefa, Haarlem, The Netherlands) over 14 days (22).
Plasmid Construction
Ubiquitin C (UBC) constructs were generated by PCR amplification of the UBC gene using the primers 5′-CCGAATTCAAATGCAGATCTTCGTGAAG-3′ and 5′-AAGCGGCGCCTACCACCCTGAGACGGAG-3′, and the EcoRI and NotI sites are underlined. Amplified DNAs were gel-purified, digested with EcoRI and NotI, and ligated into the pCMV-HA. Site-directed mutagenesis was performed to create the PinX1L291A and TRF1T122A mutants according to the manufacturer's instructions (Stratagene). The primers used for the mutagenesis were as follows: TRF1T122A-F, 5′-CCAGTCTAACAGCTTGCCAGTTGAGAGCTATATACATATGTC-3′; TRF1T122A-R, 5′-GACATATGTATATAGCTCTCAACTGGCAAGCTGTTAGACTGG-3′; PinX1L291A-F, 5′-CCGGGACTTCACCGCGAAGCCCAAAAAGA-3′; PinX1L291A-R, 5′-TCTTTTTGGGCTTCGCGGTGAAGTCCCGG-3′. Positive clones were confirmed by DNA sequencing (Cosmogenetech, Seoul, Korea).
Transfection, siRNA, and Plasmids
Cells at 50–60% confluence were transfected with 1 μg of plasmid or 50 nm siRNA using JetPRIMETM transfection reagent (Polyplus, Illkirch, France). StealthTM siRNAs purchased from Invitrogen were as follows: PinX1 (HSS123667, HSS123668, HSS182773), hTERT (HSS144248, HSS144247, HSS144249), and control (catalog no. 2935–300). A mixture of three siRNAs was used for PinX1 and hTERT silencing. Some of the work was done with control siRNA purchased from Genolution (5′-ACGUGACACGUUCGGAGAAUU-3′; Genolution, Seoul, Korea). Plasmids encoding myc-PinX1, HA-PinX193–328, HA-PinX1149–268, HA-PinX1205–328, and GFP-PinX1 were described in a previous report (21). pcDNA3-hTERT-myc was generously provided by Dr. Ishikawa's group.
Immunoblotting and Antibodies
Cell lysates were prepared from passive lysis buffer (Promega) containing a mixture of protease inhibitors (Roche Applied Science) and incubated with the following primary antibodies: TRF1 (1:1,000, ab10579; Abcam, Cambridge, UK); PinX1 (1:3,000, H00054981-A01; Abnova, Taipei City, Taiwan); γ-H2AX (1:3,000, NB100-2280; Novus Biologicals, Littleton, CO); hTERT (1:5,000, 1531-1; Epitomics, Burlingame, CA); GFP (1:5000, 632381; Clontech, Sparks, MD); pT68-CHK2 (Thr-68) (1:3,000, 2197P), β-actin (1:5,000, 4967), and GAPDH (1:5,000, 2118) (Cell Signaling Technology); and c-myc (9E10) (1:5,000, sc-40) and HA-probe (Y-11) (1:500, sc-805; Santa Cruz Biotechnology). The secondary antibodies included horseradish peroxidase-conjugated anti-mouse (1:5,000) and anti-rabbit (1:5,000) from Cell Signaling Technology.
Protein Stability Assay
Cells were transfected with plasmids or siRNAs, and cycloheximide (CHX) (Sigma) was added 24–48 h later at 500 μg/ml for the times indicated in the figures. Cell lysates prepared from cells collected at different time points were subjected to immunoblotting. Signal intensity of the bands was semiquantified using Quantity One (Bio-Rad).
In Vivo Ubiquitination Assay
Cells were transfected with 50 nm siRNA against PinX1 or control. After 24 h, cells were then transfected with 1 μg of plasmids encoding myc-TRF1 and HA-ubiquitin (for 36 h and then treated with 1 μm MG132 (Calbiochem) for 8 h to inhibit proteasome function. Cells were lysed with passive lysis buffer. With gentle agitation, 500 μg of clarified cell lysates was incubated with protein G-agarose (Amersham Biosciences) for 1 h at 4 °C. The supernatant was added to 0.5 μg of anti-myc antibody. After 1 h of incubation at 4 °C, protein G-agarose was added, and the mixture was incubated for 1 h at 4 °C. The agarose beads were resuspended in SDS sample buffer (Cell Signaling Technology) and boiled for 5 min. The immunoprecipitates were then analyzed by Western blot analysis.
Immunoprecipitation
Immunoprecipitation (IP) assay was performed as described in the protocol of the IP assay kit (Sigma). HeLa cells transfected with plasmids were lysed in passive lysis buffer containing a 1× protease inhibitor mixture (Roche Applied Science). Then, 800 μg of clarified cell lysates was incubated with 2 μg of anti-TRF1 antibody at 4 °C overnight. For precipitation of myc-TRF1, 1 μg of anti-myc antibody (9E10) was added to 500 μg of lysates for 1 h. Protein G-agarose suspension was then added to each reaction and incubated for 4 h at 4 °C. After sequentially washing the beads, the proteins were subjected to immunoblot analysis.
Chromatin Immunoprecipitation (ChIP) Assay and Telomere Slot Blotting
ChIP assay was performed with an EZ ChIPTM assay kit (Upstate, Charlottesville, VA). In brief, cells were fixed in formaldehyde and lysed, followed by sonication to obtain chromatin fragments with an average size of 500 bp. Lysates were immunoprecipitated with anti-HA antibody and anti-myc antibody and supplemented with protein G-Sepharose beads. The chromatin was eluted from the beads, and cross-links were reversed and isolated using the column provided in the kit (Upstate). The isolated DNA molecules were denatured at 95 °C for 5 min and slot-blotted onto Hybond N+ membranes (Amersham Biosciences). For detection of telomeres, 80% of the isolated samples was loaded, while the remaining 20% was loaded for d(CAC)8 sequences. Hybridization was performed with d(TTAGGG)4 and d(CAC)8 labeled with a DIG oligonucleotide 3′ end labeling kit (Roche Applied Science) and found with a detection starter kit (Roche Applied Science).
Telomere Dysfunction-induced DNA Damage Foci (TIF) Detection
Telomere FISH was performed as described previously (21). In brief, cells were transfected with siRNAs for 78 h, extracted in Triton X-100 buffer (0.5% Triton X-100, 20 mm HEPES-KOH (pH 7.4), 50 mm NaCl, 3 mm MgCl2 and 300 mm sucrose), and fixed with 4% formaldehyde (w/v) (Sigma) in PBS. Dehydration was followed by soaking the cells in 70% ethanol for 2 h at 4 °C. The cells were then rehydrated in 2× SSC, 50% formamide, denatured at 80 °C for 5 min, and hybridized for 2 h at 30 °C with 20 μl of hybridization mixture (5 mm Tris-HCl (pH 7.4), 1 mm MgCl2, 0.45 mm citric acid, 4.1 mm NaHPO4, 70% formamide deionized, 0.1 μg of telomere probe of peptide nucleic acid (Cy3-(TTAGGG)4; Applied Biosystems), and 5% blocking reagent (Roche Applied Science). After hybridization, the cells were incubated with anti-γ-H2AX antibody (1:500) for 3 h at room temperature in the dark. After washing, cells were incubated with a mixture of Alexa Fluor 488 goat anti-rabbit IgG (1:500; Invitrogen) and DAPI (Sigma) at room temperature for 1 h in the dark and then washed and mounted in anti-fade medium (Invitrogen). Images were acquired using LSM 700 (Carl Zeiss) operating software with the ZEN2009 laser scanning system (Carl Zeiss).
Immunofluorescence
Immunofluorescence analysis was performed as described previously (21). In brief, cells were extracted in Triton X-100 buffer, fixed with 4% paraformaldehyde (w/v), and permeabilized with Triton X-100 buffer. After blocking with 2% bovine serum albumin in PBS, cells were incubated with anti-TRF1 (1:200; Abcam) or anti-hTERT (1:1,000; Epitomics) overnight at 4 °C. After washing, cells were incubated with Alexa Fluor 594 goat anti-mouse IgG (1:500; Molecular Probes) or Alexa Fluor 633 goat anti-rabbit IgG (1:500; Molecular Probes) and DAPI (Sigma).
Analysis of Anaphase Bridges and Multipolar Mitosis
Hematoxylin and eosin (H&E)-stained cells were examined under a light microscope at 400× magnification for multipolar mitosis and anaphase bridges. At least 100 high power fields were reviewed. Multipolar mitosis was classified for chromosomes in metaphase or anaphase showing three or more spindle poles. Anaphase bridging was defined as the presence of a chromatin bridge spanning anaphase poles.
Statistical Analysis
Statistical analysis was performed using SPSS version 12.0 statistical software (SPSS Corporation, Chicago, IL) and assessed using a Student's t test, Mann-Whitney U test, and the χ2 test as deemed appropriate. The level of statistical significant was set at p < 0.05.
RESULTS
Excess PinX1 Enhances TRF1 Protein Stability
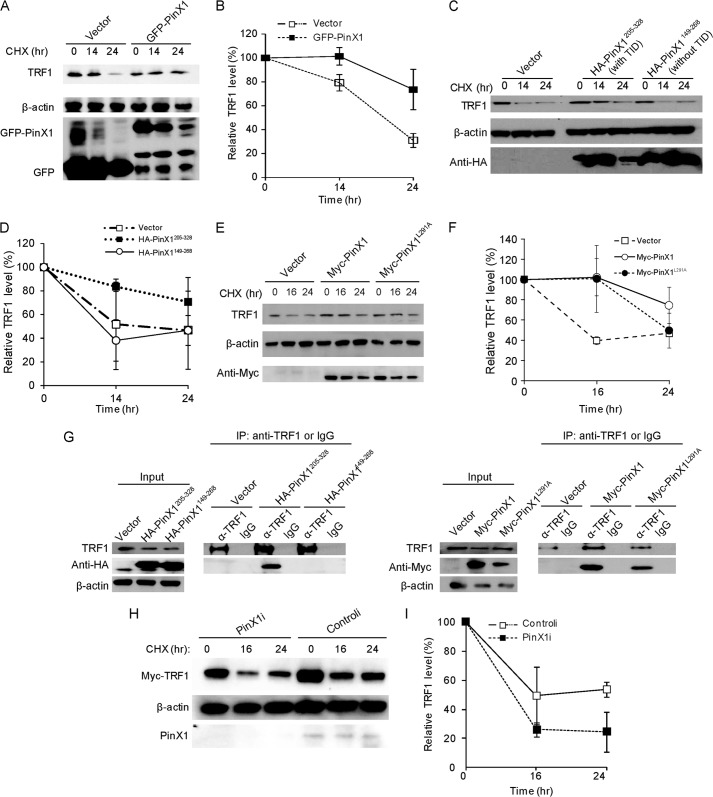
TRF1 protein stability is known to be important for the TRF1 binding to telomeres (16, 23, 24). In a previous report, we showed that overexpression of PinX1 increases TRF1 binding to telomeres in HeLa cells (21). In this study, we attempted to evaluate whether PinX1 exerts any influence on TRF1 protein stability. To address this question, protein stability assays were performed as follows. HeLa cells were transfected with GFP-PinX1, and 24 h later cells were treated with CHX for 0, 14, and 24 h to block protein biosynthesis, and endogenous TRF1 protein level was monitored by immunoblotting. As the exposure time with CHX increased, TRF1 protein was gradually decreased in vector-transfected cells whereas the level of TRF1 protein was maintained in GFP-PinX1-expressed cells (Fig. 1, A and B). TRF1 protein stability assay was further performed with PinX1 mutants. Cells transfected with a plasmid encoding HA-PinX1205–328, a small carboxyl-terminal fragment, which contains a TRF1-interacting domain (TID), retained the stability of TRF1 protein (Fig. 1, C and D). Interaction between HA-PinX1205–328 and TRF1 is shown in Fig. 1G. However, cells transfected with a plasmid encoding HA-PinX1149–268, in which a TID is disrupted, showed rapid degradation of the protein (Fig. 1, C and D). IP assay revealed a lack of interaction between TRF1 and HA-PinX1149–268 (Fig. 1G). These may indicate that the physical interaction of PinX1 with TRF1 is critical to PinX1-mediated TRF1 protein stability. Previous studies with PinX1 mutants revealed that leucine 291 in PinX1 is critical for mediating TRF1-PinX1 interaction (25). We tested TRF1 stability by expressing full-length PinX1 with a point mutation at 291, replacing leucine with alanine (25). Unexpectedly, TRF1 stability was maintained in myc-PinX1L291A-expressed cells (Fig. 1, E and F). IP assay showed that myc-PinX1L291A bound well to TRF1 in HeLa cells (Fig. 1G). Previous studies have utilized in vitro assay systems, such as peptide binding assay or GST pulldown assay, to identify PinX1 sites responsible for TRF1 interaction (25, 26). Differences in assay systems may account for the discrepancies between previous results and ours. According to our results, leucine 291 in PinX1 seems to be insignificant to TRF1 interaction in vivo, in which posttranslational modifications are active. Our data indicate that excess PinX1 stabilizes TRF1 protein, which may be mediated by binding between PinX1 and TRF1.
FIGURE 1.
PinX1 regulates TRF1 protein stability. A, stability of TRF1 protein increased by overexpression of GFP-PinX1. HeLa cells were transfected with plasmid expressing GFP-PinX1, and 24 h later cells were treated with CHX for the indicated times. Lysates were resolved on 4–12% gradient SDS-PAGE, and protein levels were monitored by immunoblotting. B, quantification of the data represented in A. C, TID in PinX1 important for TRF1 protein stability. PinX1 mutant forms with a TID (HA-PinX1205–328) and without a TID (HA-PinX1149–268) were expressed in HeLa cells, and a TRF1 protein stability assay was performed. D, quantification of the data represented in C. E, TRF1 also stabilized by PinX1L291A. Myc-PinX1L291A was transiently expressed in HeLa cells, and TRF1 protein stability assay was performed. F, quantification of the data represented in E. G, interaction between PinX1 mutants and TRF1. HeLa cells expressing HA-PinX1205–328, HA-PinX1149–268, myc-PinX1 or myc-PinX1L291A were subjected to immunoprecipitation with anti-TRF1 antibody or normal IgG, followed by immunoblotting. H, TRF1 degradation enhanced by depletion of PinX1. HeLa cells treated with siRNA against PinX1 (PinX1i) were transfected with pCMV-myc-TRF1, and 24 h later CHX was treated for the indicated times. I, quantification of the data represented in H. TRF1 level was semiquantified using β-actin as a loading control, and the TRF1 amount at the 0-h time point was defined as 100%. Error bars represent the S.D. from at least three independent experiments in B, D, F, and I.
PinX1 Depletion Decreases Telomeric Association of TRF1 and Increases TRF1 Ubiquitination
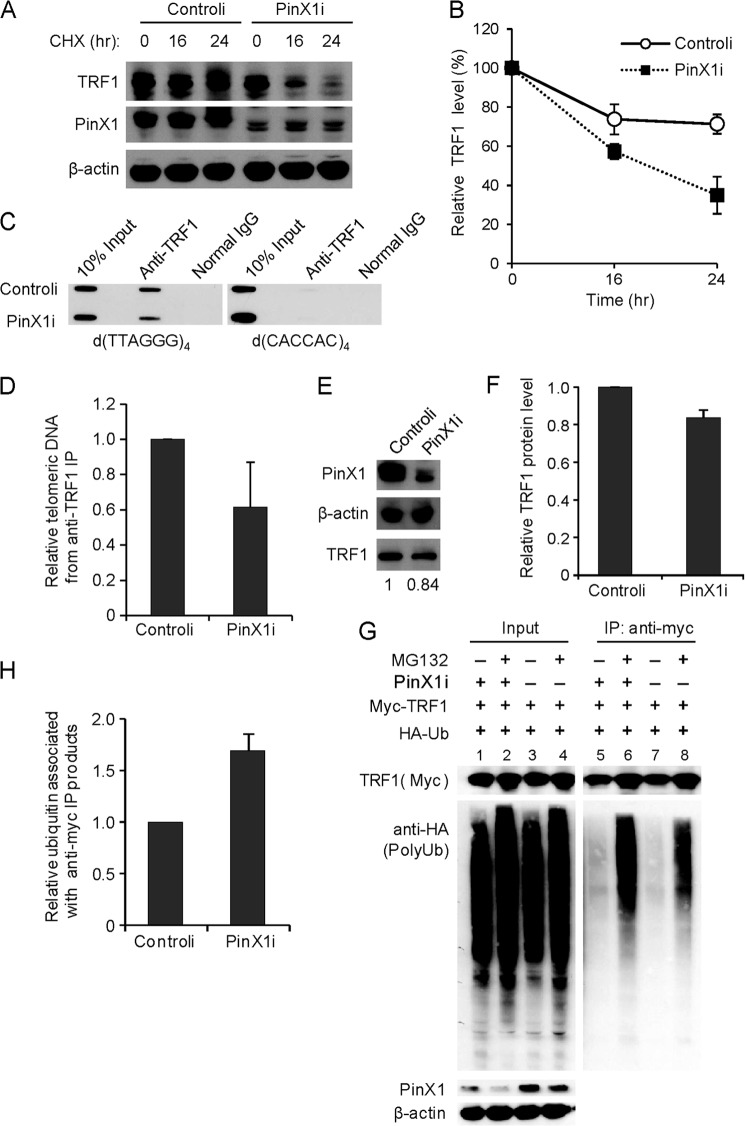
TRF1 protein stability was further examined in PinX1-depleted cells. To investigate this, we treated HeLa cells with siRNA targeting PinX1 (named as PinX1i), which allowed a reduction of PinX1 protein (Fig. 1H). To determine the specificity of PinX1 siRNA, we performed quantitative RT-PCR and confirmed reduced mRNA levels of PinX1, but not of TRF1, hTERT, or TGF-β, in the PinX1i-treated cells (data not shown). For the protein stability assay, HeLa cells were co-transfected with myc-TRF1 and PinX1i, followed by CHX chase. Myc-TRF1 protein was degraded more rapidly in the PinX1i-transfected cells compared with control siRNA (controli)-transfected cells (Fig. 1, H and I). Next, a similar experiment was performed to detect the stability of endogenous TRF1 in the PinX1i-treated cells, and a rapid decrease in endogenous TRF1 was indeed observed in the PinX1-depleted cells (Fig. 2, A and B). Consistent with the results for PinX1 overexpression, these findings demonstrated the involvement of PinX1 in TRF1 stability.
FIGURE 2.
Depletion of PinX1 reduces residence of TRF1 on telomeres and increases TRF1 ubiquitination. A, endogenous TRF1 degradation enhanced by depletion of PinX1. HeLa cells were treated with 50 nm PinX1i, and 48 h later CHX was treated for the indicated times. B, quantification of the data represented in A. TRF1 level was semiquantified using β-actin as a loading control, and the TRF1 amount at the 0-h time point was defined as 100%. Error bars represent the S.D. from four independent experiments. C, reduced TRF1 level on telomeres in PinX1-depleted cells. PinX1-depleted HeLa cells were subjected to ChIP with anti-TRF1 antibody. The precipitated DNA was probed for the presence of TTAGGG or CAC minisatellite repeats on slot blots. Normal IgG ChIPs were performed as a negative control. D, quantification of ChIPs in C. Telomere signals of TRF1 ChIPs were normalized with input signals and calculated as a ratio relative to that recovered from controli-expressing cells. Error bar represents the S.D. from three independent experiments. E, immunoblot of the samples used in C. Endogenous PinX1 and TRF1 proteins were detected by immunoblotting. The relative amounts of TRF1 normalized to β-actin are written below the gel. F, quantification of TRF1 amounts in E. Error bars represent the S.D. from three independent experiments. G, TRF1 ubiquitination in PinX1-depleted cells. HeLa cells were transfected with PinX1i, HA-ubiquitin (HA-Ub), and myc-TRF1, after which a proteasome inhibitor, MG132, was treated for 8 h. Immunoprecipitation was performed with anti-myc antibody, and immunoblotting was followed with anti-myc and anti-HA antibodies. H, quantification of the data represented in G. Ubiquitinated TRF1 (anti-HA) was normalized with anti-myc IP products and calculated as a ratio relative to that recovered from controli-treated cells. Error bars indicate the S.D. from three independent experiments.
To investigate whether depletion of PinX1 exerts any influence on TRF1 binding to telomeres, a telomeric ChIP assay with an anti-TRF1 antibody was performed in PinX1-depleted HeLa cells. The results of the ChIP assay revealed decreased telomeric association of TRF1 in the PinX1-depleted cells (Fig. 2, C and D). TRF1 amount was also measured in the ChIP samples, and it was slightly low in the PinX1-depleted cells (0.84 ± 0.04) compared with the controli-treated cells (Fig. 2, E and F). The TRF1 protein stability and ChIP assays may suggest that diminished telomeric association of TRF1 is partly caused by a reduction of TRF1 amount in PinX1-depleted cells.
TRF1 protein is known to be degraded by the ubiquitin-proteasome degradation pathway (15). To investigate whether the instability of TRF1 upon PinX1 depletion was due to the ubiquitination of TRF1, in vivo ubiquitination assay was performed as follows. HeLa cells were transfected with PinX1i, HA-ubiquitin, and myc-TRF1, followed by treatment of a proteasome inhibitor, MG132, and immunoprecipitation with anti-myc antibody. Immunoprecipitated myc-tagged proteins were then subjected to anti-HA immunoblotting to reveal ubiquitin-conjugated TRF1 products (Fig. 2, G and H); polyubiquitinated TRF1 products were more abundant in PinX1-depleted cells than controli-treated cells (Fig. 2G, lanes 6 and 8). Depletion of PinX1 seems to lead to increased TRF1 ubiquitination. Together with previous results, our findings suggest that PinX1 may stabilize TRF1 protein by inhibiting its ubiquitination.
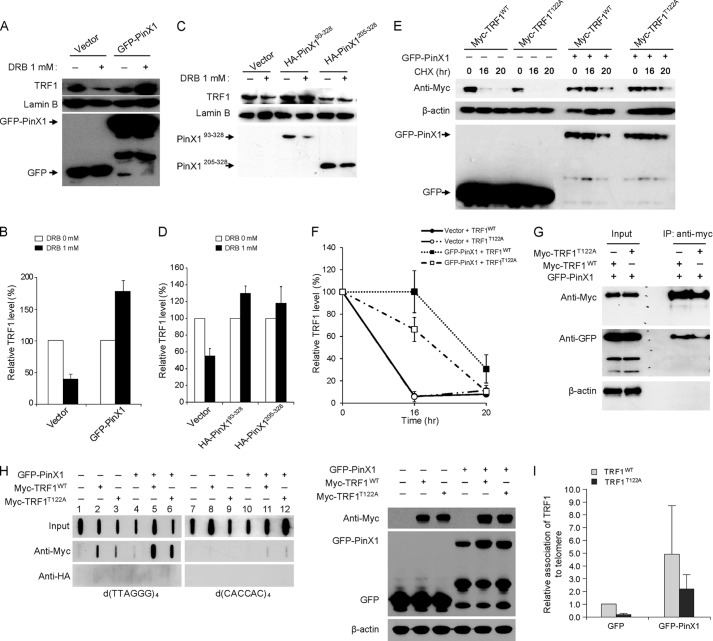
PinX1 Stabilizes TRF1T122A and Enhances Telomeric Binding of the Mutant
Treatment with 5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole (DRB), an inhibitor of casein kinase 2, is known to lead to ubiquitination and degradation of TRF1 (16). Casein kinase 2 is known to phosphorylate TRF1 on threonine at position 122, and this phosphorylation is suggested to be important for TRF1 stability and the efficient telomere binding of TRF1 (16). Accordingly, we attempted to evaluate whether PinX1-mediated TRF1 stability is associated with TRF1 phosphorylation. To address this possibility, HeLa cells transfected with GFP-PinX1 were treated with DRB, and TRF1 protein levels were assessed. DRB treatment indeed decreased TRF1 levels in vector-transfected cells (Fig. 3, A and B), consistent with a previous report (16). In GFP-PinX1-transfected cells, however, TRF1 protein remained abundant in the presence of DRB (Fig. 3, A and B). Similar results were also detected in HA-PinX193–328- and HA-PinX1205–328-expressing cells (Fig. 3, C and D). These may suggest that PinX1 stabilizes TRF1 regardless of casein kinase 2-mediated phosphorylation. This was further supported by protein stability assay with myc-TRF1T122A, a mutant of TRF1 incapable of phosphorylation at residue 122 (16), which showed that GFP-PinX1 maintained the stability of myc-TRF1T122A (Fig. 3, E and F). Co-IP assay further demonstrated that GFP-PinX1 bound well to myc-TRF1T122A (Fig. 3G). Next, we tested whether PinX1 is able to enhance the telomeric association of TRF1T122A. ChIP assay revealed that overexpression of GFP-PinX1 results in increased interaction between myc-TRF1T122A and telomeres (Fig. 3H, lanes 3 and 6, and I). Our data indicate that PinX1 stabilizes TRF1T122A and allows the mutant protein to bind to telomeres.
FIGURE 3.
PinX1 stabilizes TRF1T122A and allows the protein to bind to telomeres. A, increased stability of TRF1 in PinX1-expressing cells after treatment with DRB. HeLa cells transfected with GFP-PinX1 for 24 h were treated with 1 mm DRB for 8 h, and then nuclear lysates were resolved by SDS-PAGE, and proteins were detected by immunoblotting. B, graphical representation of relative TRF1 levels in A. The amount of TRF1 protein in DRB-treated cells was determined relative to those in DRB-untreated cells. Lamin B was used as a loading control, and error bars indicate S.D. from three independent experiments. C, PinX1 mutants containing TID domain also enhancing TRF1 stability. Nuclear lysates prepared from HeLa cells expressing HA-PinX1 constructs treated with DRB were resolved by SDS-PAGE, and proteins were detected by immunoblotting. D, quantification of data represented in C. The relative TRF1 levels were determined as described in B. Error bars indicate the S.D. from three independent experiments. E, PinX1 stabilizing TRF1T122A. HeLa cells co-transfected with GFP-PinX1 and myc-TRF1WT or myc-TRF1T122A were subjected to protein stability assays, and TRF1 was detected by anti-myc antibody. GFP vector was used as a control (−). F, quantification of data represented in E. TRF1 level was semiquantified using β-actin as a loading control, and the TRF1 amount at 0-h time point was defined as 100%. Error bars represent the S.D. of the mean from three independent experiments. G, interaction of PinX1 with TRF1T122A. HeLa cells expressing GFP-PinX1 and myc-TRF1WT or myc-TRF1T122A were subjected to immunoprecipitation with anti-myc antibody, followed by immunoblot with anti-myc and anti-GFP antibodies. H, increased TRF1T122A on telomeres in PinX1-overexpressed cells. HeLa cells co-transfected with GFP-PinX1 and myc-TRF1WT or myc-TRF1T122A were subjected to ChIP with anti-myc antibody. The presence of telomeres in the precipitated DNA was detected by hybridization with d(TTAGGG)4. HA ChIPs were performed as a negative control. Right panel shows immunoblots of the samples used in the ChIP assay. Expressions of GFP-PinX1, myc-TRF1WT, and myc-TRF1T122A were confirmed by immunoblotting. I, quantification of ChIPs in H. Telomere signals of myc ChIPs were normalized with input signals and calculated as a ratio relative to that recovered from myc-TRF1WT in GFP vector-expressing cells. Error bars indicate the S.D. from three independent experiments.
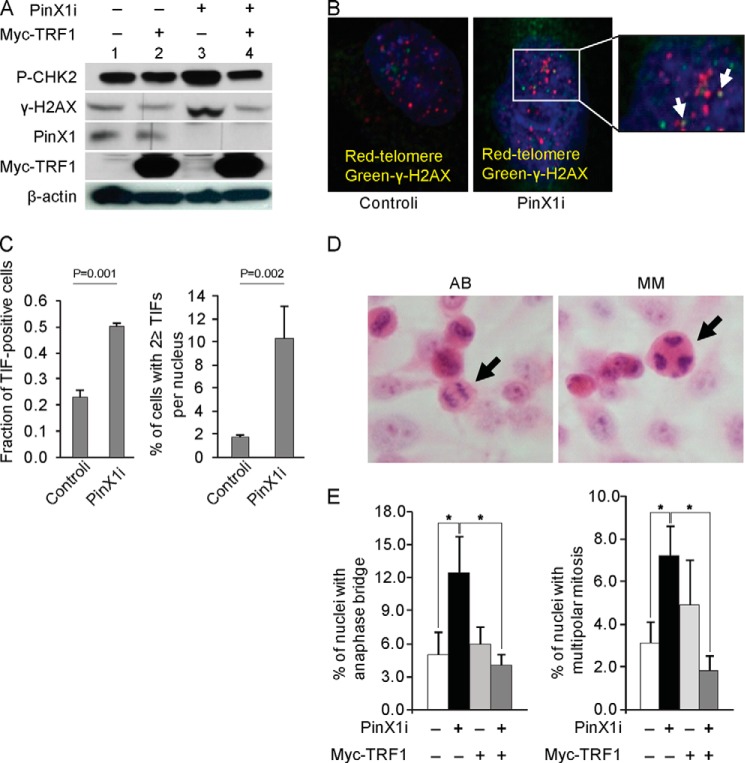
PinX1 Depletion Leads to DDR Activation and Mild Telomere Dysfunctional Phenotypes, and TRF1 Overexpression Restores Chromosome Stability in PinX1-depleted Cells
Shelterin complex functions to protect chromosomal ends from DNA damage surveillance and repair pathways (9), and TRF1, as a member of the shelterin complex, plays a critical role in telomere protection. According to our results, PinX1 seems to control TRF1 stability. Accordingly, we further investigated whether loss of PinX1 leads to any telomere damage. To do so, DDR proteins were monitored in PinX1-depleted cells (Fig. 4A). DDR proteins, which were measured by phosphorylation at H2AX (γ-H2AX) and CHK2 (27), accumulated more in the PinX1-depleted cells (Fig. 4A, lane 3) compared with the control cells (Fig. 4A, lane 1). Next, we monitored TIFs which are detected by co-localization of γ-H2AX and telomeres, representing activation of DDRs at telomeres (28). Under our experimental condition, spots of telomere and γ-H2AX co-localization were not markedly large in size. Although a robust DDR at telomeres was not detected in the PinX1-depleted cells, spots were slightly more frequent in the PinX1-depleted cells (Fig. 4, B and C). TIF-positive cells comprised ∼50% of the PinX1-depleted cells and 20% of the control cells (Fig. 4C). Approximately 10% (10.3 ± 2.83%) of the PinX1-depleted cells exhibited more than two TIFs per cell, whereas this was seen in approximately only 2% (1.8 ± 0.14%) of the control cells (Fig. 4C). We further examined the presence of anaphase bridges and multipolar mitosis that occurred as a result of telomere dysfunction (20, 24). Anaphase bridges were more frequent in the PinX1-depleted cells (12%) than the controli-transfected cells (5%, p < 0.005) (Fig. 4, D and E). The incidence of multipolar mitosis also increased approximately 2.4-fold in the PinX1-depleted cells (p < 0.001) (Fig. 4E).
FIGURE 4.
PinX1 depletion induces DDRs at telomeres, and TRF1 overexpression restores telomere function in PinX1-depleted cells. A, activation of DDRs in PinX1-depleted cells. Lysates from HeLa cells transfected with siRNA against PinX1 (PinX1i) and/or myc-TRF1 plasmid were prepared at 72 h after transfection, and then γ-H2AX and phosphorylated CHK2 (P-CHK2) were determined by immunoblotting. Scramble siRNA and empty vectors were used as controls (−). B, representative images of TIF. HeLa cells transfected with siRNA against PinX1 (PinX1i) or scramble siRNA (controli) were immunostained with anti-γ-H2AX antibody (green) followed by telomere-FISH (red). In total, 117 PinX1i-treated cells and 111 controli-treated cells were counted. TIF spots in the image are enlarged for better visualization and are indicated by arrows. C, quantification of the data represented in B. TIF-positive cells and cells with ≥2 TIFs/nucleus were counted. Mean values were derived from two independent experiments. p values were determined by χ2test. D, representative images of anaphase bridging (AB) and multipolar mitosis (MM) in PinX1-depleted cells. Cells stained with H&E were viewed at 400× magnification. Arrows indicate nuclei with anaphase bridging and multipolar mitosis. E, increased chromosomal instability in PinX1-depleted cells and rescue of stability in the cells after TRF1 overexpression. Anaphase bridging and multipolar mitosis were counted in PinX1-depleted cells and in PinX1-depleted and myc-TRF1-overexpressed cells. A minimum of 500 cells was examined, and mean values were derived from three to four independent experiments. *, p < 0.001 by χ2 test.
To discern whether PinX1 depletion-mediated DDR activation and mild telomere dysfunction are caused by short telomeres, telomere length was measured in the PinX1-depleted cells. Southern blotting revealed that telomere length remained unchanged in the PinX1-depleted cells (data not shown). A similar result was also observed in the PinX1-overexpressed cells (data not shown). Transient depletion or overexpression of PinX1 did not seem to change telomere length. Our findings suggest that DDR activation and TIF phenotypes in the PinX1-depleted cells seem unlikely to be due to defects caused by shortened telomeres. However, we cannot rule out the possibility that the telomeres within the TIF foci may be critically short, which was undetected by Southern blotting.
We further investigated whether PinX1 depletion-mediated DDR activation was compromised by an excess amount of TRF1. To do so, myc-TRF1 was expressed in PinX1i-treated cells. Myc-TRF1 overexpression in PinX1-depleted cells markedly reduced the amount of γ-H2AX and phosphorylated CHK2 (Fig. 4A, lanes 3 and 4). Similarly, when myc-TRF1 was ectopically expressed in PinX1-depleted cells, the incidences of anaphase bridges and multipolar mitosis decreased to the level seen in the control cells (Fig. 4E). These results suggest that DNA damage caused by PinX1 depletion is likely to be due to a deficiency in TRF1.
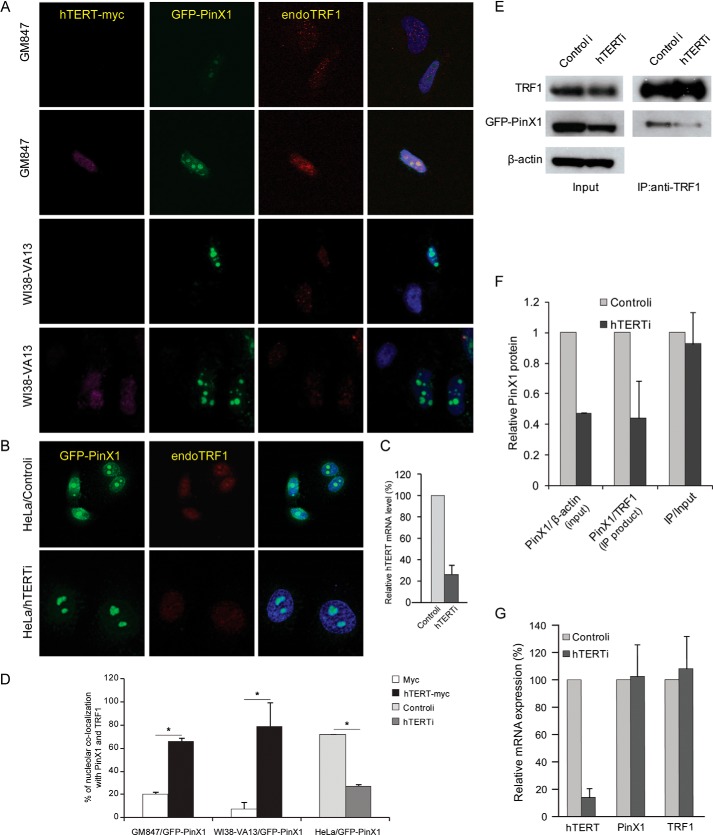
Depletion of hTERT Reduces PinX1-mediated TRF1 Nucleolar Localization
In a previous report, we showed accumulation of TRF1 in the nucleolus upon overexpression of nucleolar PinX1 in HeLa cells, but this phenomenon was not evident in ALT cells, which are hTERT-negative cells (21). To confirm whether this nucleolar localization of TRF1 is dependent on hTERT, we performed immunostaining with hTERT-expressed ALT cells. For the experiments, hTERT-myc and GFP-PinX1 were transiently expressed in GM847 cells, an hTERT-negative ALT cell line. Nucleolar co-localization of TRF1 and GFP-PinX1 was detected in 66% of hTERT-myc positive cells, whereas such co-localization was detected in only 21% of hTERT-myc negative cells (Fig. 5, A and D). A similar result was observed in WI38-VA13 cells, another hTERT-negative ALT cell line; nucleolar co-localization was observed in 79% of hTERT-myc positive cells and 7% of hTERT-myc negative cells (Fig. 5, A and D). These results were further confirmed in the hTERT-depleted HeLa cells (Fig. 5B). hTERT knockdown was confirmed in the siRNA-treated HeLa cells by quantitative RT-PCR (Fig. 5C). Nucleolar localization of TRF1, which was detected in 72% of controli-treated HeLa cells, was markedly reduced in the hTERT-depleted cells, of which only 26% showed the nucleolar co-localization (Fig. 5,B and 5D). Consistent with previous results (21), PinX1-mediated TRF1 nucleolar accumulation was more pronounced when cells expressed hTERT.
FIGURE 5.
PinX1-induced nucleolar localization of TRF1 is dependent on hTERT expression. A, nucleolar localization of TRF1 in hTERT-expressing ALT cells. GM847 and WI38-VA13 cells that transiently expressed hTERT-myc and GFP-PinX1 were subjected to immunostaining. Endogenous TRF1 (red) that co-localized with GFP-PinX1 (green) in nucleoli was counted in hTERT-myc expressing cells (pink) and in hTERT-myc negative cells. B, reduced nucleolar localization of TRF1 in hTERT-depleted HeLa cells. HeLa cells co-transfected with siRNA against hTERT (hTERTi) and plasmid containing GFP-PinX1 were examined for the presence of TRF1 in the nucleolus co-localized with GFP-PinX1. TRF1 (red) that co-localized with GFP-PinX1 (green) in nucleoli was counted in hTERTi- and controli-treated cells. C, RT-PCR of hTERT mRNA in hTERT-depleted cells. hTERT silencing was confirmed by quantitative RT-PCR. Mean values were derived from two independent experiments. Error bars, S.D. D, quantification of the data represented in A and B. A minimum of 100 cells was examined, and mean values were derived from two to five independent experiments. Error bars, S.D. *, p < 0.0001 by χ2 test. E, immunoprecipitation assay in HeLa cells. HeLa cells transfected with GFP-PinX1 and hTERTi or controli were immunoprecipitated with anti-TRF1 antibody, and TRF1 and GFP-PinX1 were detected by immunoblotting. F, quantification of the data represented in E. Input GFP-PinX1 was normalized to β-actin, and GFP-PinX1 co-precipitates were normalized to TRF1 precipitates, and then GFP-PinX1 in IP fraction were calculated as a ratio relative to the input amount. Error bars indicate S.D. from three independent experiments. G, specificity of siRNA against hTERT for target gene. cDNA was prepared from HeLa cells transfected with 50 nm siRNA against hTERT for 72 h. The expression of hTERT, PinX1, and TRF1 was measured by quantitative RT-PCR using Taqman gene expression system, and GAPDH was used as an internal control. The level of gene expression was described as a ratio relative to that obtained from controli-treated cells. Error bars indicate S.D. from three independent experiments.
Next, we investigated whether hTERT is necessary for PinX1-TRF1 interaction. IP assay was performed with cell extracts prepared from GFP-PinX1 expressed HeLa cells in which hTERT was depleted. We noted that hTERTi-treated cells expressed lower amounts of GFP-PinX1 protein, which led to smaller amounts of GFP-PinX1 co-immunoprecipitation along with TRF1 (Fig. 5, E and F). The relative amount of GFP-PinX1, namely IP PinX1 over input PinX1, was nearly the same for both hTERT depleted and control siRNA-treated cells (Fig. 5F). To examine whether reduced PinX1 protein was due to low specificity of the hTERTi, quantitative RT-PCR was performed. Treatment of hTERTi effectively silenced hTERT, but did not reduce the level of PinX1 or TRF1 (Fig. 5G). This indicates that reduction of GFP-PinX1 protein is unlikely to be due to an off-target effect of hTERTi. Further studies in this direction are in progress in our laboratory. Nevertheless, the results of the co-IP assays suggest that the interaction between PinX1 and TRF1 is maintained regardless of hTERT status. We thus speculated that the low incidence of nucleolar localization of TRF1 in the knockdown of hTERT might be partially due to a reduction in PinX1 protein, which, in turn, recruits less TRF1 into nucleoli. Further investigation is necessary to understand how nucleolar accumulation of TRF1 is related to hTERT.
Roles of hTERT on the TRF1 Steady State Pathway
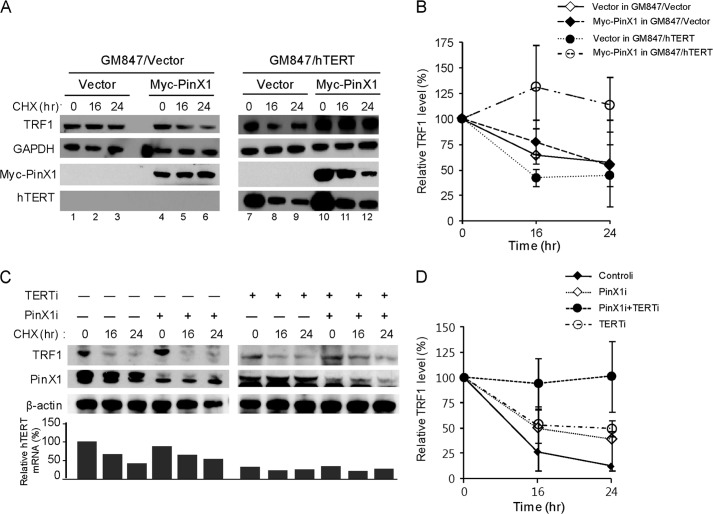
PinX1 physically interacts with both TRF1 and hTERT, and our findings revealed that PinX1 function is associated with TRF1 protein stability. However, role of hTERT on PinX1 function has not been discovered. Next, we investigated whether hTERT has any influence on PinX1-mediated TRF1 stability. Initially, TRF1 stability was examined in HeLa cells transfected with GFP-PinX1 and siRNA against hTERT, but the results were not conclusive because of a reduction in GFP-PinX1 in the hTERT depleted cells (Fig. 5E). Instead, the TRF1 stability assay was performed in GM847/hTERT cells, a stable cell line expressing hTERT, in which myc-PinX1 was transiently expressed (Fig. 6, A and B). In these cells, TRF1 protein appeared to be stable (Fig. 6A, lanes 10–12). This stability, however, was not observed in GM847/vector cells expressing myc-PinX1 (Fig. 6A, lanes 4-6). These results may suggest that hTERT plays a role in the PinX1-mediated TRF1 stability. However, our findings are yet inconclusive because of the use of ALT cells, and understanding the function of PinX1 in primary cells may provide clues to the functional connections between PinX1, hTERT and TRF1.
FIGURE 6.
Role of hTERT in TRF1 homeostasis. A, TRF1 protein stability in ALT cells expressing hTERT. Stable cell lines, GM847/hTERT and GM847/vector, were used for the protein stability assay after transfection with myc-PinX1 or an empty vector. Proteins were detected by immunoblotting. B, quantification of the data represented in A. Error bars represent S.D. from three independent experiments. C, TRF1 protein stability in hTERT and PinX1 co-depleted cells. Protein stability assay was performed with HeLa cells transfected with siRNA against hTERT (hTERTi) and PinX1 (PinX1i). PinX1 and TRF1 proteins were detected by immunoblotting. hTERT status was detected by quantitative RT-PCR, and the hTERT amount normalized with GAPDH in controli-treated cells at 0-h point was defined as 100%. D, quantification of the data represented in C. TRF1 level was semiquantified using β-actin as a loading control, and relative TRF1 levels at time 0 were defined as 100%. Error bars indicate the S.D. from three independent experiments.
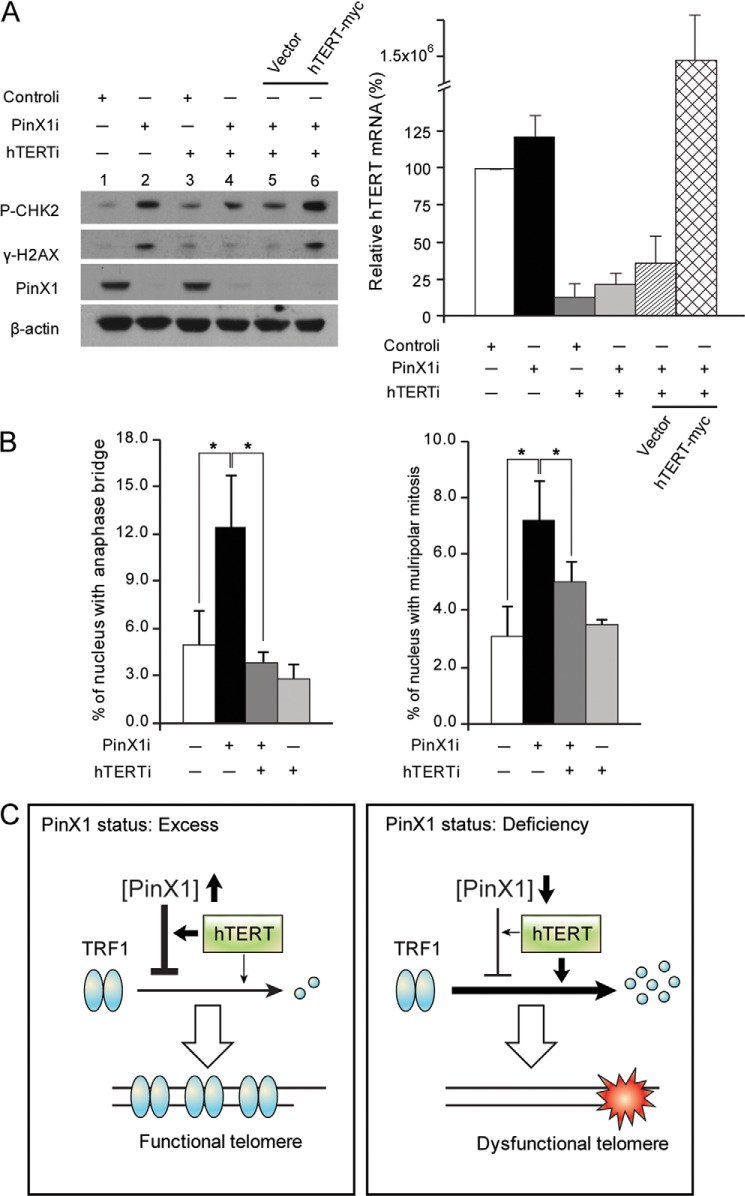
Next, the stability assay was performed in hTERT and PinX1 co-depleted HeLa cells. Depletion of PinX1 and hTERT was determined by immunoblotting and quantitative RT-PCR, respectively (Fig. 6C). Interestingly, TRF1 protein stability was maintained in the cells where both PinX1 and hTERT were depleted (Fig. 6, C and D). This, however, was not seen in the cases where either hTERT or PinX1 alone was depleted. We then examined DDR proteins in the co-depleted cells. Induction of γ-H2AX and phosphorylated CHK2, seen in the cells where PinX1 was depleted (Fig. 7A, lane 2), disappeared in the co-depleted cells (Fig. 7A, lane 4). Next, hTERT was ectopically expressed in the co-depleted cells to see whether excess hTERT compromises DDR activation. Indeed, the level of γ-H2AX and phosphorylated CHK2 was significantly induced in the co-depleted cells where hTERT-myc was overexpressed (Fig. 7A, lane 6). These results suggest that hTERT may play a critical role in PinX1 depletion-mediated DDR activation. Overexpression of hTERT in the co-depleted cells was confirmed by quantitative RT-PCR (Fig. 7A, right). hTERT mRNA was maintained at a high level in cells transfected with hTERTi, PinX1i, and a plasmid encoding hTERT-myc (Fig. 7A, right, 1.5 × 106 ± 7.5 × 105), although hTERTi reduced target hTERT mRNA levels to one tenth of that of controli- and pcDNA3-hTERT-myc-treated cells (1.5 × 107 ± 6.6 × 106).
FIGURE 7.
hTERT is involved in PinX1 depletion-mediated telomere dysfunction. A, suppressed DDR activation in hTERT- and PinX1-co-depleted HeLa cells and restoration of DDR activation by hTERT overexpression in the co-depleted cells. Cell lysates prepared from HeLa cells transfected with indicated siRNAs were subjected to immunoblotting for the detection of DDR proteins. For hTERT expression in the co-depleted cells, HeLa cells were transfected with siRNA against hTERT (hTERTi) and PinX1 (PinX1i), and 24 h later, transfected with pcDNA3-hTERT-myc for 48 h. γ-H2AX and phosphorylated CHK2 (P-CHK2) were determined with anti-γ-H2AX and anti-pT68-CHK2 antibodies, respectively. hTERT status was determined by quantitative RT-PCR, shown in the graph on the right. B, reduced PinX1 depletion-mediated telomere dysfunction phenotypes in hTERT-depleted cells. HeLa cells treated with indicated siRNAs were fixed and stained with H&E, and anaphase bridging and multipolar mitosis were scored. The results of controli- or PinX1i-treated cells were from Fig. 4E. A minimum of 2,000 cells was examined, and mean values were derived from three independent experiments. *, p < 0.001 by χ2 test. C, proposed model for the role of PinX1 in telomere protection. The left panel shows that excess PinX1 together with hTERT stabilizes TRF1 by protecting the protein from ubiquitin-mediated proteolysis. The stable TRF1 binds to telomeres, which eventually leads to telomere protection. The right panel shows that PinX1 shortage induces TRF1 degradation, which results in telomere dysfunction. In this pathway, in contrast to PinX1 excess, hTERT may actively participate in TRF1 degradation. This model shows that hTERT acts as a positive and a negative regulator of TRF1 stability depending on PinX1 content.
Furthermore, we examined the incidence of anaphase bridges and multipolar mitosis in the co-depleted cells (Fig. 7B). Compared with the PinX1 alone-depleted cells (12 ± 3.4%), the co-depleted cells showed a reduced incidence of anaphase bridges (4 ± 0.5%) (Fig. 7B). Similarly, the incidence of multipolar mitosis decreased in the co-depleted cells (5 ± 1.9%), compared with the PinX1-depleted cells (7 ± 1.6%) (Fig. 7B). Together, knockdown of hTERT in the PinX1-depleted cells suppressed DDR activation and restored TRF1 protein stability and chromosome integrity. These may indicate that upon knockdown of PinX1 hTERT may involve in TRF1 degradation pathway, which may lead to DDR activation and chromosome instability. In addition our results that TRF1 stability is influenced by hTERT status in ALT cells may suggest association of hTERT function in TRF1 stability when PinX1 is present. Our findings demonstrate the functional connections between hTERT and PinX1 on TRF1 stability although the mechanism is not clear yet.
DISCUSSION
In this study, we examined the role of PinX1 on TRF1 stability and telomere function. Our findings revealed that PinX1 stabilizes TRF1 and allows more TRF1 to bind to telomeres. PinX1 depletion leads to DDR activation and mild telomere dysfunction. Additionally, we also investigated the role of hTERT on TRF1 stability in connection with PinX1. PinX1 overexpression in ALT cells did not maintain TRF1 stability, whereas TRF1 stability was restored by expressing hTERT in the cells. Interestingly, the knockdown of both hTERT and PinX1 in HeLa led to the suppression of DDR activation and the restoration of TRF1 and chromosome stabilities. Our findings suggest that hTERT may be important to PinX1-mediated TRF1 stability, but upon PinX1 knockdown, hTERT may participate in TRF1 degradation.
The TRF1 steady state is tightly regulated by posttranslational modifications such as phosphorylation, poly(ADP-ribosyl)ation, and ubiquitination (15–17). Human TRF1 undergoes ubiquitin-mediated proteolysis after the protein dissociates from telomeres (15). Telomere-unbound TRF1 may have two fates, one for degradation and the other for telomeric association. Therefore, suppressing the TRF1 degradation pathway and maintaining TRF1 stability are important to preserving telomere integrity. Our results showed that PinX1 indeed stabilizes TRF1, which was similar to the report by Yonekawa et al. (29). The physical interaction between PinX1 and TRF1 seems to be critical for PinX1-mediated TRF1 stability which appeared to be achieved by protecting TRF1 from ubiquitin-mediated proteolysis. We also found that excess PinX1 was capable of stabilizing TRF1T122A and leading to increased telomeric association of this mutant. Notwithstanding, it would be interesting to discover whether telomere-bound TRF1T122A is capable of protecting telomeres in the same manner as wild type TRF1. Once again, our results emphasize that TRF1 stability is critical for TRF1 binding to telomeres and for telomere protection.
TRF1 is known to play a critical role in telomere protection. Mouse embryonic fibroblasts derived from a TRF1 knock-out mouse showed increased telomere fragility and fusions (30). Similarly, HeLa cells treated with siRNA targeting TRF1 exhibited TIF induction (31). In this study, we found that depletion of PinX1 resulted in DDR activation, mild TIF phenotypes, and chromosome instability, which was consistent with previous reports (20). These phenotypes seem to be caused by TRF1 shortage because overexpression of TRF1 restored the phenotypes in the PinX1-depleted cells. It is likely that TRF1 deficiency causes diminished TRF1 binding to telomeres and mild telomere dysfunctional phenotypes in the PinX1-depleted cells.
According to our results, hTERT seems to be needed for PinX1-mediated TRF1 stability. Because both hTERT and TRF1 are known to bind to the same region of PinX1 (19), these three proteins cannot simultaneously participate in the TRF1-stabilizing process. A possible scenario for this may be that PinX1, after interacting with hTERT, goes through conformational change or posttranslational modification, the form of which may protect TRF1 from degradation. Further studies are necessary to uncover the precise mechanism of how PinX1 and hTERT stabilize TRF1.
Our finding that co-depletion of PinX1 and hTERT leads to TRF1 stability may suggest that hTERT plays a positive role in the PinX1 depletion-mediated TRF1 degradation pathway. This finding was further confirmed by assays that showed hTERT overexpression in the co-depleted cells restored DDR activation. TRF1 overloading on telomeres inhibits telomerase accessibility to telomeres, which in turn shortens telomeres (12). However, a deficiency of TRF1 leads to telomere deprotection and cell growth defect (13, 31, 32). Thus, regulation of TRF1 loading on telomeres might be critical for telomere homeostasis and cell viability. hTERT may be involved in TRF1 homeostasis by participating in both TRF1 stability and TRF1 degradation (Fig. 7C). Although highly speculative, hTERT may act on TRF1 stability when telomere length needs to be shortened, whereas it may function in TRF1 degradation when telomeres are too short via TRF1 overloading. Thus, in telomerase-positive cancer cells, the amount of PinX1 might need to be tightly regulated. Overall, our results indicate that PinX1 may maintain telomere integrity by modulating TRF1 steady state and that hTERT is involved in TRF1 homeostasis in a PinX1-dependent manner. These findings provide a novel TRF1 turnover mechanism linked to telomerase and will aid in the discovery of a molecular mechanism for the role of hTERT on telomere length regulation.
Acknowledgment
We thank Yonsei-Carl Zeiss Advanced Imaging Center, Yonsei University College of Medicine, for technical assistance.
This work was supported by National Research Foundation of Korea Grants 2013R1A2A2A05005990, 2012M3A9B6055350, and 2011-0030086 (to Y. N. P.) and 2010-0008254 and 2011-0015638 (to B. K. O.) funded by the Ministry of Education, Science and Technology.
- DDR
- DNA damage response
- CHX
- cycloheximide
- controli
- control siRNA
- DRB
- 5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole
- IP
- immunoprecipitation
- TID
- TRF1 interacting domain
- TIF
- telomere dysfunction-induced DNA damage foci
- TRF
- telomere repeat-binding factor
- hTERT
- human telomerase reverse transcriptase.
REFERENCES
- 1. Blackburn E. H. (2000) Telomere states and cell fates. Nature 408, 53–56 [DOI] [PubMed] [Google Scholar]
- 2. de Lange T. (1998) Telomeres and senescence: ending the debate. Science 279, 334–335 [DOI] [PubMed] [Google Scholar]
- 3. Autexier C., Greider C. W. (1996) Telomerase and cancer: revisiting the telomere hypothesis. Trends Biochem. Sci. 21, 387–391 [PubMed] [Google Scholar]
- 4. Collins K. (2000) Mammalian telomeres and telomerase. Curr. Opin. Cell Biol. 12, 378–383 [DOI] [PubMed] [Google Scholar]
- 5. Kim N. W., Piatyszek M. A., Prowse K. R., Harley C. B., West M. D., Ho P. L., Coviello G. M., Wright W. E., Weinrich S. L., Shay J. W. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 6. Bodnar A. G., Ouellette M., Frolkis M., Holt S. E., Chiu C. P., Morin G. B., Harley C. B., Shay J. W., Lichtsteiner S., Wright W. E. (1998) Extension of life-span by introduction of telomerase into normal human cells. Science 279, 349–352 [DOI] [PubMed] [Google Scholar]
- 7. Morales C. P., Holt S. E., Ouellette M., Kaur K. J., Yan Y., Wilson K. S., White M. A., Wright W. E., Shay J. W. (1999) Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat. Genet. 21, 115–118 [DOI] [PubMed] [Google Scholar]
- 8. Nakayama J., Tahara H., Tahara E., Saito M., Ito K., Nakamura H., Nakanishi T., Tahara E., Ide T., Ishikawa F. (1998) Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nat. Genet. 18, 65–68 [DOI] [PubMed] [Google Scholar]
- 9. de Lange T. (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 [DOI] [PubMed] [Google Scholar]
- 10. van Steensel B., Smogorzewska A., de Lange T. (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92, 401–413 [DOI] [PubMed] [Google Scholar]
- 11. Ancelin K., Brunori M., Bauwens S., Koering C. E., Brun C., Ricoul M., Pommier J. P., Sabatier L., Gilson E. (2002) Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol. 22, 3474–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van Steensel B., de Lange T. (1997) Control of telomere length by the human telomeric protein TRF1. Nature 385, 740–743 [DOI] [PubMed] [Google Scholar]
- 13. Iwano T., Tachibana M., Reth M., Shinkai Y. (2004) Importance of TRF1 for functional telomere structure. J. Biol. Chem. 279, 1442–1448 [DOI] [PubMed] [Google Scholar]
- 14. Karlseder J., Kachatrian L., Takai H., Mercer K., Hingorani S., Jacks T., de Lange T. (2003) Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol. Cell. Biol. 23, 6533–6541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang W., Dynek J. N., Smith S. (2003) TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev. 17, 1328–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim M. K., Kang M. R., Nam H. W., Bae Y. S., Kim Y. S., Chung I. K. (2008) Regulation of telomeric repeat binding factor 1 binding to telomeres by casein kinase 2-mediated phosphorylation. J. Biol. Chem. 283, 14144–14152 [DOI] [PubMed] [Google Scholar]
- 17. Smith S., Giriat I., Schmitt A., de Lange T. (1998) Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282, 1484–1487 [DOI] [PubMed] [Google Scholar]
- 18. Chen G., Da L., Wang H., Xu Y., Chen G., Sun C., Wang L., Zhao J., Zhang F., Feng J., Wang Y., Tiollais P., Li T., Zhao M. (2011) HIV-Tat-mediated delivery of an LPTS functional fragment inhibits telomerase activity and tumorigenicity of hepatoma cells. Gastroenterology 140, 332–343 [DOI] [PubMed] [Google Scholar]
- 19. Zhou X. Z., Lu K. P. (2001) The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell 107, 347–359 [DOI] [PubMed] [Google Scholar]
- 20. Zhou X. Z., Huang P., Shi R., Lee T. H., Lu G., Zhang Z., Bronson R., Lu K. P. (2011) The telomerase inhibitor PinX1 is a major haploinsufficient tumor suppressor essential for chromosome stability in mice. J. Clin. Invest. 121, 1266–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yoo J. E., Oh B. K., Park Y. N. (2009) Human PinX1 mediates TRF1 accumulation in nucleolus and enhances TRF1 binding to telomeres. J. Mol. Biol. 388, 928–940 [DOI] [PubMed] [Google Scholar]
- 22. Jung A. R., Yoo J. E., Shim Y. H., Choi Y. N., Jeung H. C., Chung H. C., Rha S. Y., Oh B. K. (2013) Increased alternative lengthening of telomere phenotypes of telomerase-negative immortal cells upon trichostatin: a treatment. Anticancer Res. 33, 821–829 [PubMed] [Google Scholar]
- 23. Lee T. H., Tun-Kyi A., Shi R., Lim J., Soohoo C., Finn G., Balastik M., Pastorino L., Wulf G., Zhou X. Z., Lu K. P. (2009) Essential role of Pin1 in the regulation of TRF1 stability and telomere maintenance. Nat. Cell Biol. 11, 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu Q., Meng L., Hsu J. K., Lin T., Teishima J., Tsai R. Y. (2009) GNL3L stabilizes the TRF1 complex and promotes mitotic transition. J. Cell Biol. 185, 827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen Y., Yang Y., van Overbeek M., Donigian J. R., Baciu P., de Lange T., Lei M. (2008) A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science 319, 1092–1096 [DOI] [PubMed] [Google Scholar]
- 26. Soohoo C. Y., Shi R., Lee T. H., Huang P., Lu K. P., Zhou X. Z. (2011) Telomerase inhibitor PinX1 provides a link between TRF1 and telomerase to prevent telomere elongation. J. Biol. Chem. 286, 3894–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. d'Adda di Fagagna F., Reaper P. M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N. P., Jackson S. P. (2003) A DNA damage checkpoint response in telomere-initiated senescence. Nature 426, 194–198 [DOI] [PubMed] [Google Scholar]
- 28. Takai H., Smogorzewska A., de Lange T. (2003) DNA damage foci at dysfunctional telomeres. Curr. Biol. 13, 1549–1556 [DOI] [PubMed] [Google Scholar]
- 29. Yonekawa T., Yang S., Counter C. M. (2012) PinX1 localizes to telomeres and stabilizes TRF1 at mitosis. Mol. Cell. Biol. 32, 1387–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martínez P., Thanasoula M., Muñoz P., Liao C., Tejera A., McNees C., Flores J. M., Fernández-Capetillo O., Tarsounas M., Blasco M. A. (2009) Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 23, 2060–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lackner D. H., Durocher D., Karlseder J. (2011) A siRNA-based screen for genes involved in chromosome end protection. PloS One 6, e21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Okamoto K., Iwano T., Tachibana M., Shinkai Y. (2008) Distinct roles of TRF1 in the regulation of telomere structure and lengthening. J. Biol. Chem. 283, 23981–23988 [DOI] [PMC free article] [PubMed] [Google Scholar]