Background: Oocyte factors can reprogram the somatic nucleus efficiently, but these factors still need to be defined.
Results: Maternal vimentin acts as a genomic protector and results in p53 down-regulation during nuclear reprogramming.
Conclusion: Maternal vimentin is crucial for nuclear reprogramming.
Significance: We report the first evidence of vimentin as a reprogramming factor.
Keywords: Chromatin Remodeling, Cloning, DNA Damage, Oocyte, Reprogramming, Pig, Vimentin
Abstract
Nuclear reprogramming of somatic cells can be induced by oocyte factors. Despite numerous attempts, the factors responsible for successful nuclear reprogramming remain elusive. In the present study, we found that porcine oocytes with the first polar body collected at 42 h of in vitro maturation had a stronger ability to support early development of cloned embryos than porcine oocytes with the first polar body collected at 33 h of in vitro maturation. To explore the key reprogramming factors responsible for the difference, we compared proteome signatures of the two groups of oocytes. 18 differentially expressed proteins between these two groups of oocytes were discovered by mass spectrometry (MS). Among these proteins, we especially focused on vimentin (VIM). A certain amount of VIM protein was stored in oocytes and accumulated during oocyte maturation, and maternal VIM was specifically incorporated into transferred somatic nuclei during nuclear reprogramming. When maternal VIM function was inhibited by anti-VIM antibody, the rate of cloned embryos developing to blastocysts was significantly lower than that of IgG antibody-injected embryos and non-injected embryos (12.24 versus 22.57 and 21.10%; p < 0.05), but the development of in vitro fertilization and parthenogenetic activation embryos was not affected. Furthermore, we found that DNA double strand breaks dramatically increased and that the p53 pathway was activated in cloned embryos when VIM function was inhibited. This study demonstrates that maternal VIM, as a genomic protector, is crucial for nuclear reprogramming in porcine cloned embryos.
Introduction
Differentiated cell nuclei can be reprogrammed to a pluripotent state in several ways, including incubation with oocyte extracts, transfer into enucleated oocytes, and induced pluripotent stem cell technology (1–3). Nuclear transfer (NT)3-mediated reprogramming has been proven to be the most efficient method (4), and complex factors in oocytes are believed to be involved in the process. Therefore, identification and characterization of these oocyte factors will provide us important information on nuclear reprogramming.
During the past decades, several approaches analyzing the protein composition of oocytes have been conducted to obtain the proteome signatures of Xenopus, bovine, pig, and mouse oocytes (5–15). Calvert et al. (15) identified eight highly abundant heat shock proteins and related chaperones in the mature mouse egg by two-dimensional difference gel electrophoresis (DIGE). Vitale et al. (7) identified 12 proteins that appeared to be differentially expressed between germinal vesicle and metaphase II (MII) murine oocytes by two-dimensional DIGE and mass spectrometry (MS). And Miyamoto et al. (16) identified proteins that were incorporated into somatic nuclei after MII oocyte extract incubation by MS. However, only a few proteins have been identified as reprogramming factors, such as the imitation switch (ISWI) family, BRG1, nucleoplasmin, and PARK7 (16–19). Thus, exploration of reprogramming factors is still important.
During mammalian oogenesis, the oocyte nucleus undergoes germinal vesicle, germinal vesicle breakdown, metaphase I, and arrests at the MII stage. Accompanying the nuclear maturation process, many cytoplasmic changes, termed cytoplasmic maturation, occur (20, 21). Some proteins, regarded as reprogramming factors, are largely synthesized from stored mRNAs during the process of cytoplasmic maturation (9). Oocytes with full cytoplasmic maturation have been widely used to reprogram somatic cell nuclei to totipotency. By contrast, oocytes with incomplete cytoplasmic maturation have no or a very low reprogramming activity (5, 22). This information suggests that reprogramming factors can be explored by comparison of oocytes with different cytoplasmic qualities.
In general, porcine oocytes with the first polar body at 42 h of in vitro maturation (IVM) are used for in vitro fertilization (IVF), parthenogenetic activation (PA), and somatic cell nuclear transfer studies (23–26), but we found that the first polar body extrusion rate between the oocytes at 33 and 42 h of IVM had no significant difference. Therefore, in this study, we compared the proteome signatures of porcine oocytes with the first polar body collected at 33 h (33O) and 42 h (42O) of IVM by MS, and 18 differentially expressed proteins between 33O and 42O were discovered. The function of the identified proteins was then examined in cloned embryos, and we demonstrate that vimentin (VIM) is required for successful nuclear reprogramming in pig.
EXPERIMENTAL PROCEDURES
Porcine Oocyte IVM
Porcine ovaries were collected from a local slaughter house and kept in saline at 32–37 °C. Antral follicles (3–5 mm in diameter) were aspirated with an 18-gauge needle. Aspirated oocytes with an evenly granulated cytoplasm and at least three uniform layers of compact cumulus cells were selected and cultured in 4-well plates (Nunc, Naperville, IL) containing 500 μl of maturation medium, which was a TCM199 (Invitrogen)-based medium plus 0.05 μg/ml EGF and 0.5 μg/ml luteinizing hormone and FSH at 39 °C in 5% CO2 in air. The rates of the first polar body extrusion were calculated from 16 to 42 h of IVM. Porcine oocytes with the first polar body were obtained at 33 and 42 h for further experiments.
Oocyte Collection and Proteomic Analysis
Zonae pellucidae of more than 10,000 oocytes at 33 and 42 h of IVM were removed, and total proteins were extracted using ultrasonic waves and lysis buffer. The lysis buffer consisted of 7 m urea, 2 m thiourea, 4% (w/v) CHAPS, 65 mm DTT, 2% (v/v), and 1% (v/v) protease inhibitor mixture. The protein concentration was determined by the Bradford method, and pH was adjusted to 8.5 with 50 mm NaOH. In fluorescent two-dimensional DIGE, proteins from oocytes at 33 and 42 h of IVM were equally pooled together and labeled with Cy2 as internal standard, and the two samples were labeled with Cy3 or Cy5 separately. Labeled samples were mixed in the rehydration buffer before loading onto 24-cm pH 3–10 immobilized pH gradient strips (Bio-Rad) and run in a single two-dimensional gel. Then the gels were scanned using a Typhoon 9410 scanner with excitation/emission wavelengths of 488/520 nm for Cy2, 532/580 nm for Cy3, and 633/670 nm for Cy5. Image analyses were performed using DeCyder software suite 5.02 (GE Healthcare), which allows the comparison of the different combinations corresponding to the experimental conditions. An independent t test was used to determine the significance between the experimental groups. p values less than 0.05 and -fold changes greater than 1.5 were considered statistically significant.
For protein identification, the spots of interest were cut from the gels and washed with a solution of 25 mm NH4HCO3 and 50% acetonitrile, dehydrated with 100% acetonitrile sequentially, and dried by centrifugal lyophilization. The gels were digested with 15–20 μl of 0.01 μg/μl trypsin (Promega) in 25 mm ammonium bicarbonate for 15 h at 37 °C. The supernatants were collected, and the tryptic peptides were extracted from the gel sequentially with 5% TFA at 40 °C for 1 h and with 2.5% TFA and 50% acetonitrile at 30 °C for 1 h. The extracts were pooled and completely dried by centrifugal lyophilization. Digested peptides with matrix (50% acetonitrile and 0.1% TFA containing 3 mg/ml α-cyano-4-hydroxycinnamic acid matrix) were then spotted on the target plate. Samples were analyzed by MALDI-TOF-TOF/MS (4800 Proteomics Analyzer, Applied Biosystems) in positive reflectron mode at fixed laser fluency with a low mass gate and delayed extraction. Database searching was carried out using Mascot version 2.2 (Matrix Science, London, UK) via GPS explorer software (Applied Biosystems) version 3.6 combining MS and MS/MS interrogations on the NCBI pig protein database. For the searching parameters, modifications were set as carbamidomethylation and oxidation, and a maximum of one missed trypsin cleavage was permitted. Tolerances of precursor and fragment ions were both set to 0.2 Da. The reported proteins were always those with the highest number of peptide matches. All identified proteins were those with statistical significance (p ≤ 0.05) and the best ion score.
Embryo Manipulations
Before NT, IVF, and PA, 10 pl of VIM antibody (V6630, Sigma) or protein (ab84704, Abcam) solution was injected into matured oocytes. After injection, oocytes were kept for at least 2 h before manipulations, which allows the antibody to bind endogenous VIM. The procedure for porcine somatic cell nuclear transfer has been described previously (24). After 33 or 42 h of IVM, the oocytes were treated with 1 mg/ml hyaluronidase (H3506, Sigma-Aldrich) to remove the surrounding cumulus cells. Oocytes with a clearly extruded the first polar body were selected as recipient cytoplasts. Cumulus cell-free oocytes were enucleated by aspirating the first polar body and adjacent cytoplasm with a glass pipette 25 μm in diameter in TCM199-HEPES plus 0.3% bovine serum albumin (BSA) and 7.5 μg/ml cytochalasin B. Porcine fetal fibroblasts from ear serving as donor cells at passage 5 were injected into the perivitelline space of enucleated oocytes. Injected oocytes were placed in fusion/activation medium (0.3 m mannitol, 1.0 mm CaCl2, 0.1 mm MgCl2, and 0.5 mm HEPES). Fusion/activation was induced with two direct current pulses of 1.2 kV/cm for 30 ms on a BTX Electro Cell Manipulator 2001 (BTX, San Diego, CA). Cumulus cell-free oocytes were directly activated by the same parameters as for the somatic cell nuclear transfer procedure to produce PA embryos.
For IVF, freshly ejaculated sperm-rich fractions were collected from fertile boars, and following a short incubation at 39 °C, the semen was resuspended and washed three times in Dulbecco's phosphate-buffered saline (PBS) supplemented with 0.1% (w/v) BSA by centrifugation at 1500 × g for 4 min. The concentration of spermatozoa was measured using a hemocytometer, and the proportion of motile sperm was determined. The spermatozoa were diluted with modified Tris-buffered medium to an optimal concentration. Cumulus cell-free oocytes were washed three times in modified Tris-buffered medium. Approximately 30 oocytes were inseminated in 50-ml drops of modified Tris-buffered medium at a final sperm concentration of 3 × 105/ml for 6 h.
The embryos were cultured in porcine zygote medium-3 at 39 °C in 5% CO2 in air. The cleavage and blastocyst rates were assessed at 48 and 156 h after activation, and the number of blastocyst cells was examined by nuclear staining with 5 μg/ml Hoechst 33342.
Quantitative PCR Analysis
Total RNA was extracted using the PureLinkTM Micro-to-Midi System (Invitrogen) according to the manufacturer's instructions, and reverse transcription was used to generate cDNAs using the PrimeScriptTM RT Reagent kit (TaKaRa). Real time PCR was performed using SYBR Premix Ex TaqTM (TaKaRa) and the 7500 Real-Time PCR System (Applied Biosystems). The reaction parameters were 95 °C for 30 s followed by 40 two-step cycles of 95 °C for 5 s and 60 °C for 34 s. Primers for VIM were 5′-CTTCAGGAGGCGGAGGAGTGG-3′ (forward) and 5′-CTGCACGCGGCCAATAGTGTC-3′ (reverse), and primers for p53 were 5′-CTCTGACTGTACCACCATCCACTACAA-3′ (forward) and 5′-GGACAGGCACAAACACGCACCTC-3′ (reverse). 18 S rRNA was used as a reference gene, and the primer sequences were 5′-TCCAATGGATCCTCGCGGAA-3′ (forward) and 5′-GGCTACCACATCCAAGGAAG-3′ (reverse). The sizes of the amplification products were 249 bp for the VIM gene, 156 bp for the p53 gene, and 149 bp for the 18 S rRNA. Ct values were calculated using Sequence Detection System software (Applied Biosystems), and the amount of target sequence normalized to the reference sequence was calculated as 2−ΔΔCt.
Western Blot
Oocytes or embryos without zonae pellucidae (stored at −80 °C) were transferred to 10 μl of cold 40 mm sodium phosphate, pH 7.6 containing 50 mm NaCl, 50 μm sodium orthovanadate, 10 mm sodium fluoride, 20 μm MG132, 2 μm matrix metalloprotease inhibitor III (444264, Calbiochem), and 1% protease inhibitor mixture III (539134, Calbiochem). Homogenization was carried out at 4 °C with a Tekmar homogenizer by three 15-s bursts with a minute cooling in between. Homogenates were centrifuged at 4 °C for 1 h at 100,000 × g. The supernatant solutions are referred to as “soluble” fractions. The pellets were suspended in 0.2–0.25 ml of complete buffer containing 1% ASB-14 and were mixed every 15 min for 2 h with Radnoti glass pestles (Unitek, Monrovia, CA). After centrifugation at 4 °C for 1 h at 100,000 × g, the supernatants, referred to as “membrane extracts,” were removed, and the pellets were discarded. About 50 oocytes or embryos of each soluble and membrane extract for each gene tested were separated by lithium dodecyl sulfate polyacrylamide gel electrophoresis on 4–12% Bis-Tris NuPAGE gels and transferred to PVDF membranes (Invitrogen); nonspecific binding was blocked by overnight incubation in 1% casein in PBS at room temperature. Blots were then probed for 2–4 h at room temperature with antibodies against VIM (anti-VIM; V6630, Sigma) and p53 (anti-p53; sc-65226, Santa Cruz Biotechnology). Histone H2B (anti-H2B; ab40975, Abcam) served as a loading control. After a 2-h incubation at room temperature with secondary antibodies, protein bands were detected by enhanced chemiluminescence with the RPN2108 kit (Amersham Biosciences) and BioMax Light film (Eastman Kodak Co.).
Immunofluorescence Analysis
Oocytes and embryos without zonae pellucidae were washed twice in PBS, then fixed in freshly prepared 4% paraformaldehyde in PBS, permeabilized in 1% Triton X-100 in PBS, and left in blocking solution (1% BSA in PBS) for 1 h. For immunolabeling, the embryos were incubated overnight at 4 °C with anti-VIM subunit (V6630, Sigma), anti-p53 (sc-65226, Santa Cruz Biotechnology), or anti-γH2AX (ab26350, Abcam); washed three times; and incubated for 1 h with secondary antibody FITC-labeled donkey anti-mouse IgG (A21202, Invitrogen) diluted 1:1000 with blocking solution. Immunofluorescence of injected oocytes and one-cell cloned embryos without VIM primary antibody (only secondary antibody) was used to analyze VIM antibody injection and degradation. Samples were washed and counterstained with 5 μg/ml Hoechst 33342. Fluorescence was detected and imaged using a Nikon fluorescence microscope.
Statistical Analysis
Statistical analysis was performed using SPSS 13.0 for MicroSoftTM Windows. Data are shown as the mean ± S.D. One-way analysis of variance was used to assess any differences between groups. The Duncan method was used for pairwise comparisons followed by a Bonferroni correction. p < 0.05 (two-tailed) was considered statistically significant.
RESULTS
Effect of 33O and 42O on Development of Porcine Cloned Embryos
In the present study, we observed that there was no significant difference between the rates of the first polar body extrusion at 33 (76.72%) and 42 h (81.80%) of IVM (p > 0.05; Table 1), and most of 33O (79.71%) were arrested at the MII stage, showing no remarkable difference compared with 42O (83.86%; p > 0.05; Table 2). To confirm the effect of time of IVM on cloned embryo development, 33O and 42O were used for NT. The cleavage and blastocyst rates of cloned embryos from 42O were significantly higher than those from 33O (89.17 versus 67.60%; p < 0.05; 22.34 versus 13.54%; p < 0.05; Table 3), and the rates of enucleation of oocytes in both 33O and 42O were up to 90% (Table 4), excluding the influence of enucleation on cloned embryo development. The results suggest that the nuclear reprogramming ability of 42O is better than that of 33O.
TABLE 1.
The rates of first polar body extrusion of porcine oocytes at 33 and 42 h of IVM
| Culture period (h) | No. oocytes (repeats) | Rate of polar body extrusion (% ± S.E.) |
|---|---|---|
| 33 | 100 (3) | 77 (76.72 ± 2.29) |
| 42 | 101 (3) | 83 (81.80 ± 1.86) |
TABLE 2.
The rates of 33O and 42O arrested at the MII stage
| Culture period (h) | No. oocytes (repeats) | No. MII oocytes (% ± S.E.) |
|---|---|---|
| 33 | 119 (3) | 95 (79.71 ± 0.86) |
| 42 | 125 (3) | 105 (83.86 ± 0.76) |
TABLE 3.
The effect of 33O and 42O on cloned embryo development
| Culture period (h) | Repeats | No. oocytes | No. embryos fused (% ± S.E.) | No. embryos cleaved (% ± S.E.) | No. blastocysts (% ± S.E.) | Total cell no. blastocysts (mean ± S.E.) |
|---|---|---|---|---|---|---|
| 33 | 4 | 411 | 280 (67.56 ± 4.71) | 195 (67.60 ± 5.63)a | 36 (13.54 ± 2.58)a | 35 ± 2 (n = 33) |
| 42 | 4 | 750 | 467 (61.86 ± 4.68) | 233 (89.17 ± 2.15)b | 59 (22.34 ± 2.18)b | 37 ± 2 (n = 57) |
a Values with different superscripts in the same group differ significantly (p < 0.05).
b Values with different superscripts in the same group differ significantly (p < 0.05).
TABLE 4.
The enucleation rates of 33O and 42O
| Culture period (h) | Repeats | No. oocytes | No. oocytes enucleated (% ± S.E.) |
|---|---|---|---|
| 33 | 3 | 93 | 90 (99.12 ± 0.88)a |
| 42 | 3 | 79 | 73 (91.87 ± 2.46)b |
a Values with different superscripts within columns differ significantly (p < 0.05).
b Values with different superscripts within columns differ significantly (p < 0.05).
Identification of Differentially Expressed Proteins between 33O and 42O
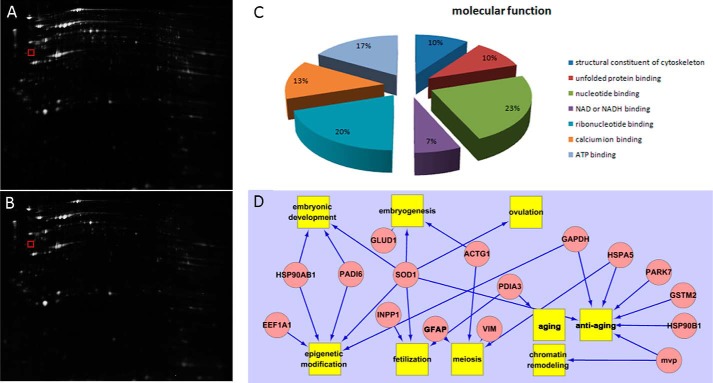
To uncover the different effects on nuclear reprogramming, global protein changes between 33O and 42O were examined by proteomic analysis. After oocyte collection and treatment, total proteins were separated by two-dimensional DIGE (Fig. 1A). Analysis of the gel images showed 994 paired protein spots, and we used an independent t test to calculate differentially expressed proteins. We considered only spots with -fold changes greater than 1.5 to exclude the possibility of false positives by multiple comparisons. Finally, 18 proteins were identified by MS. Based on MALDI-TOF/MS analysis, seven proteins were down-regulated, and 11 proteins were up-regulated in a comparison of 42O with 33O (supplemental Table 1). We categorized proteins identified in this study by searching gene ontology and performing a literature search. The differentially expressed proteins were classified into groups based on molecular function (Fig. 1B). DAVID and Agilent Literature Search were combined by Cytoscape to cluster the most affected molecular functions in oocyte maturation, namely embryonic development, embryogenesis, ovulation, epigenetic modification, fertilization, meiosis, aging, chromatin remodeling, and antiaging (Fig. 1C).
FIGURE 1.
Array data analysis. A, total proteins of 33O separated by two-dimensional DIGE. B, total proteins of 42O separated by two-dimensional DIGE. The gel marked by the red box contains the VIM protein spot. C, classification of the identified proteins was performed according to the gene ontology term “molecular function.” D, biological pathways deduced for the identified proteins using DAVID analysis and Agilent Literature Search. Proteins are indicated as pink ovals, and regulated processes are represented by yellow squares. Regulation events are displayed with arrows. GFAP, glial fibrillary acidic protein. MVP, major vault protein.
Expression Pattern of VIM
One unique protein spot expressed at high levels in 42O and expressed at low levels in 33O was detected (Figs. 1A and 2A). MALDI-TOF/MS analysis and database searches revealed that the spot matched VIM. The different expressions of VIM between 42O and 33O were confirmed by Western blot (Fig. 2B). VIM has been reported to interact strongly with genomic DNA (27–32), so we focused on its role in nuclear reprogramming. We first investigated the expression pattern of VIM. VIM expression was examined in porcine MII oocytes and NT and IVF embryos. A certain amount of VIM was stored in oocytes (Fig. 2, C–E). In IVF embryos, VIM was expressed at a low level at one- and two-cell stages, and expression increased at the four-cell stage (Fig. 2, C and D). In NT embryos, abundant VIM was observed in the one-cell embryos, but the amount of VIM rapidly decreased at the two-cell stage and increased at the four-cell stage (Fig. 2, C and E). The amount of VIM in cloned embryos was higher than in IVF embryos. VIM was dispersed in the MII oocyte cytoplasm (Fig. 3, A–A″) and was incorporated into transferred donor nuclei in cloned embryos at 2 h post-NT when the nuclei were condensed (Fig. 3, B–B″), and the signal was weaker in the pseudo-pronucleus at 6 h post-NT (Fig. 3, C–C″), indicating that VIM may function in the process of oocyte-mediated nuclear reprogramming. To further confirm oocyte VIM localization in donor nuclei, VIM protein with a His tag was injected into oocytes at least 2 h before NT, and the injected VIM incorporated into transferred donor nuclei in cloned embryos was detected by immunofluorescence of the His tag (Fig. 3, D–D″). In addition to the unique incorporation pattern of oocyte VIM protein into somatic nuclei, VIM appeared as a dense interconnected network surrounding or located near the donor nuclei similar to the pattern of donor cells (Fig. 3, E–E″), revealing that somatic cell-derived VIM may contribute to the formation of the network pattern. Taken together, oocyte VIM is likely incorporated into transferred somatic nuclei during nuclear reprogramming.
FIGURE 2.
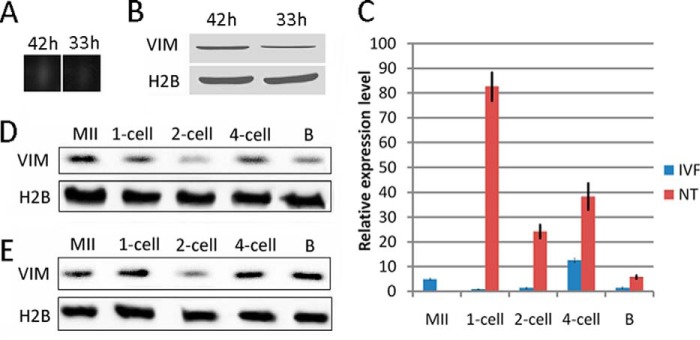
Identification of VIM protein in porcine oocytes. A, VIM spots in 33O and 42O. B, array data validation by Western blot. C, mRNA expression of VIM in cloned and IVF embryos. D, protein expression of VIM in porcine IVF embryos. E, protein expression of VIM in porcine cloned embryos. B, blastocyst. Error bars represent S.E.
FIGURE 3.
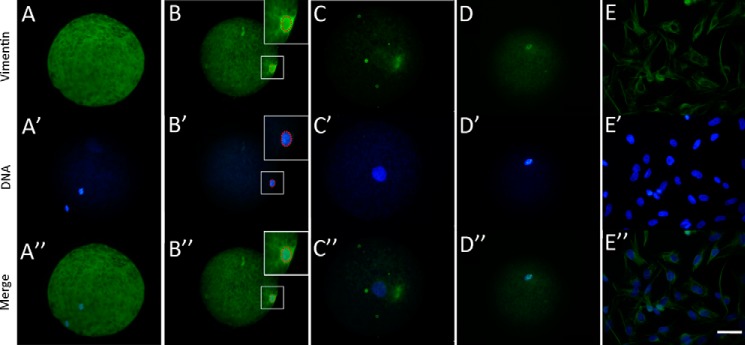
Location of VIM in cloned embryos by immunofluorescence analysis. A, A′, and A″, immunofluorescence analysis of oocyte VIM. B, B′, and B″, location of VIM in cloned embryos at 2 h post-NT. C, C′, and C″, location of VIM in cloned embryos at 6 h post-NT. D, D′, and D″, location of injected VIM in cloned embryos at 2 h post-NT. E, E′, and E″, expression pattern of VIM in porcine fibroblasts. Green, VIM; blue, DNA. Scale bar, 50 μm.
Inhibition of Maternal VIM Function in Cloned Porcine Embryos
To test the role of VIM in nuclear reprogramming, its function was inhibited by injection of anti-VIM antibody into MII oocytes at 42 h of IVM at least 2 h before NT, PA, and IVF. The successful injection of the antibody used here was verified by immunofluorescence analysis (Fig. 4, A A′, B, and B′), and the antibody is highly specific for recognizing the porcine VIM among whole oocyte proteins (Fig. 4E). Moreover, efficient knockdown of oocyte VIM protein by antibody injection was observed by Western blot (Fig. 4F). Next the effect of VIM on in vitro development of NT, IVF, and PA porcine embryos was examined. The rates of cleavage showed no significant difference among cloned embryos with no injection (Con-NT), anti-VIM antibody injection (anti-VIM-NT), and IgG (IgG-NT) injection, but the proportion of anti-VIM-NT embryos that developed to blastocysts was significantly lower than that of IgG-NT and Con-NT embryos (12.24 versus 22.57 and 21.10%, respectively; p < 0.05; Table 5). And more anti-VIM-NT embryos arrested at the two- or four-cell stage in comparison with IgG-NT and Con-NT embryos (49.68 versus 29.27 and 25.73%, respectively; p < 0.05; Table 5). In addition, detection by immunostaining of the anti-VIM antibody in injected oocytes at 2 h postinjection and in one-cell cloned embryos derived from injected oocytes showed no signal, indicating that the injected antibody had been degraded and would not affect somatic cell-derived VIM (Fig. 4, C, C′, D, and D′). In contrast to cloned embryos, inhibition of VIM did not disturb in vitro development of IVF and PA embryos (Table 5). Therefore, we suggest that maternal VIM is indispensible for successful nuclear reprogramming.
FIGURE 4.
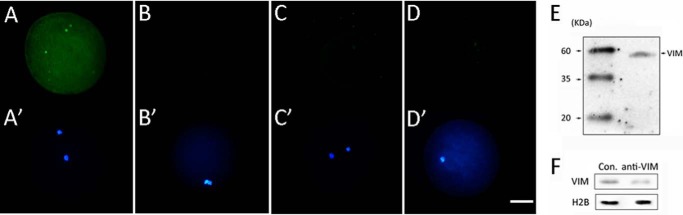
Efficient knockdown of oocyte VIM. A and A′, immunostaining of anti-VIM antibody-injected oocytes only by secondary antibody. B and B′, immunostaining of non-injected oocytes only by secondary antibody. C and C′, immunostaining of oocytes 2 h after anti-VIM antibody injection only by secondary antibody. D and D′, immunostaining of one-cell cloned embryos derived from anti-VIM antibody-injected oocytes only by secondary antibody. FITC-labeled donkey anti-mouse IgG was used as secondary antibody, which was used to immunostain anti-VIM antibody. E, specific match of anti-VIM antibody to the porcine VIM was verified by Western blot. F, efficient knockdown of oocyte VIM protein by antibody injection was confirmed by Western blot. Green, anti-VIM antibody; blue, DNA. Scale bar, 50 μm.
TABLE 5.
Effect of VIM on in vitro development of NT, IVF, and PA porcine embryos
| Groups | Repeats | Embryos | Cleavage (%) | Blastocysts (%) | No. blastocyst cells | Embryos arrested at two/four-cell stage (%) |
|---|---|---|---|---|---|---|
| NT | ||||||
| Con | 5 | 204 | 152 (73.48 ± 8.04) | 45 (21.10 ± 1.91)a | 36.43 ± 9.43 | 53 (25.73 ± 3.92)a |
| IgG | 5 | 191 | 135 (71.73 ± 10.43) | 41 (22.57 ± 3.61)a | 37.50 ± 12.72 | 54 (29.27 ± 4.27)a |
| Anti-VIM | 5 | 237 | 161 (71.31 ± 10.39) | 27 (12.24 ± 2.75)b | 31.67 ± 8.37 | 113 (49.68 ± 5.56)b |
| IVF | ||||||
| Con | 3 | 240 | 152 (62.50 ± 5.25) | 38 (14.96 ± 3.60) | 49.35 ± 5.43 | 49 (19.46 ± 5.72) |
| IgG | 3 | 240 | 158 (63.58 ± 3.62) | 35 (13.83 ± 3.60) | 47.60 ± 7.40 | 54 (21.27 ± 4.27) |
| Anti-VIM | 3 | 240 | 151 (60.63 ± 5.32) | 38 (14.83 ± 4.09) | 41.87 ± 4.87 | 41 (18.18 ± 7.99) |
| PA | ||||||
| Con | 3 | 180 | 147 (82.53 ± 10.43) | 58 (31.47 ± 3.22) | 43.50 ± 5.26 | 26 (13.56 ± 6.00) |
| IgG | 3 | 196 | 174 (87.75 ± 10.43) | 55 (29.06 ± 4.45) | 45.25 ± 7.12 | 54 (15.85 ± 5.74) |
| Anti-VIM | 3 | 211 | 180 (84.45 ± 10.39) | 63 (27.85 ± 5.05) | 42.53 ± 9.41 | 41 (17.31 ± 6.21) |
a Values with different superscripts within columns denote significant difference (p < 0.05).
b Values with different superscripts within columns denote significant difference (p < 0.05).
Maternal VIM Stabilizes Genomic DNA during Nuclear Reprogramming
We observed that maternal VIM was mainly incorporated into the condensed nuclei of transferred donor cells at 2 h post-NT, and it has been reported that nuclei tolerate severe remodeling at this time (33). During the process, DNA double strand breaks (DSBs) occur that cause serious genomic instability and increase the DNA damage response (34). Therefore, we checked the role of VIM in DSBs during NT by testing the presence of phosphorylated histone H2AX (γ-H2AX), which is rapidly formed following DSBs (35, 36). γ-H2AX-positive foci were detected in 37.29% of Con-NT embryos at 2 h post-NT, similar to the proportion of donor cells (45.71%), but the proportion increased to 64.86% when oocyte VIM was inhibited (Fig. 5F). Examples for one-cell cloned embryos and donor cells with or without γ-H2AX signals are shown (Fig. 5, A–E and A′–E′). γ-H2AX dramatically increased in two-cell anti-VIM-NT embryos compared with Con-NT embryos (Fig. 5, C, C′, D, and D′), and this was confirmed by Western blot (Fig. 5G). Next we investigated expression of p53, which can be activated by the DNA damage response and which acts as a barrier to nuclear reprogramming (37–39). Expression of p53 was significantly increased in anti-VIM-NT embryos compared with Con-NT embryos at the four-cell stage at both mRNA and protein levels (Fig. 5, H and I), suggesting that maternal VIM inhibition might induce activation of the p53 pathway in porcine cloned embryos. These results indicate that oocyte VIM improves nuclear reprogramming by protecting genomic DNA.
FIGURE 5.
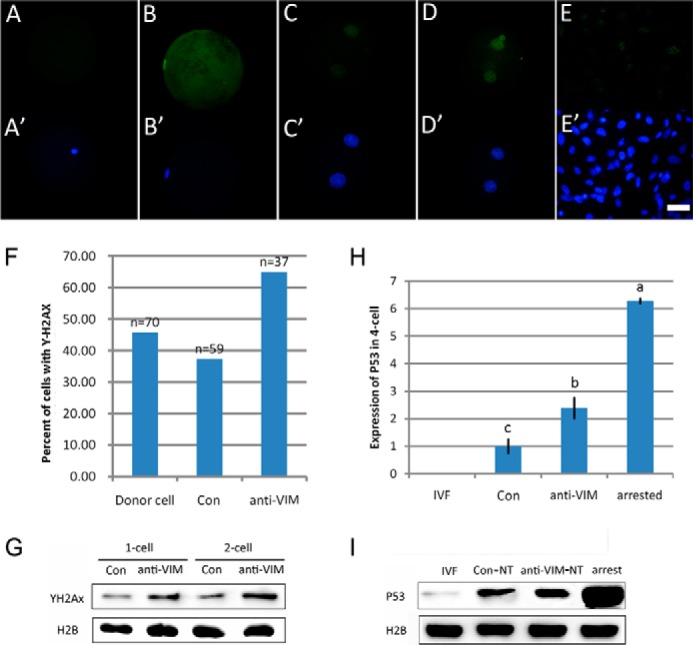
VIM improves genomic stability during nuclear reprogramming. A and A′, cloned embryo that showed negative γ-H2AX signal at 2 h post-NT. B and B′, cloned embryo that showed positive γ-H2AX signal at 2 h post-NT. C and C′, γ-H2AX signals in normal cloned embryos at the two-cell stage. D and D′, γ-H2AX signals in VIM-deficient cloned embryos at the two-cell stage. E and E′, γ-H2AX signals in porcine fibroblasts. Green, VIM; blue, DNA. Scale bar, 50 μm. F, γ-H2AX signals in cloned embryos. G, detection of γ-H2AX in cloned embryos by Western blot. H, mRNA expression of p53 in cloned embryos analyzed by quantitative PCR. Results are represented as mean ± S.E. (error bars). Data with different letters differ significantly (p < 0.05). I, protein expression of p53 in cloned embryos was analyzed by Western blot.
DISCUSSION
Factors essential for nuclear reprogramming in oocytes have been explored because of the production of cloned sheep; however, they remain largely unknown. Here, we compared the proteome signatures of 33O and 42O, which had different effects on nuclear reprogramming, by MS. An oocyte factor, VIM, was identified, and it is necessary for successful nuclear reprogramming in pig.
VIM is one type of intermediate filament, which is assembled by VIM subunit proteins (40–42). Intermediate filaments form an extensive and interconnected three-dimensional network that is distributed throughout the cytoplasm and functionally connected to the nuclear matrix (43–45). The major functions of intermediate filaments are maintenance of cellular integrity and acting as a scaffold that binds and regulates the activity of several effector proteins. The VIM network exists in endothelial and mesenchymal cells, such as fibroblasts, and appears in newly formed mesoderm in 7-day mouse embryos (41, 46). A previous report demonstrated that VIM network derived from donor cells can promote nuclear mechanical constraints during nuclear remodeling, inducing nuclear distortions and DNA damage (34). But nothing is known about the role of maternal VIM in nuclear reprogramming. In fact, oocytes and early embryos are devoid of a VIM network (46). However, we found that a certain amount of VIM subunit proteins was stored in oocytes, and it accumulated during IVM of oocytes. The VIM expression pattern showed that it is dispersed in MII oocyte cytoplasm and did not incorporate with the chromosomes. But interestingly, when somatic cells were transferred into the enucleated oocytes, VIM was incorporated into the transferred somatic nuclei, and inhibition of maternal VIM function resulted in failed development of cloned embryos. These data suggest that maternal VIM may play a crucial role in nuclear reprogramming.
During nuclear reprogramming, severe remodeling occurs in donor nuclei, which undergo nuclear envelop breakdown, premature chromosome condensation, decondensation, and pseudo-pronucleus formation (47–49). During the process of nuclear remodeling, for successful nuclear reprogramming, it is important that the somatic nuclei undergo premature chromosome condensation and form spermlike nuclei (50). In response to premature chromosome condensation, DSBs increase, leading to genomic instability (51, 52). Coincident with the DNA damage, we found that maternal VIM was mainly incorporated into the condensed somatic chromosomes during somatic nuclei remodeling. DSBs can induce phosphorylation of H2AX on serine 139 in the conserved C-terminal region, resulting in the formation of γ-H2AX, which is frequently used as a marker for DSBs (35). Therefore, we assayed γ-H2AX in cloned embryos. When oocyte VIM was inhibited, γ-H2AX-positive foci dramatically increased in cloned embryos, indicating that DSBs increase in VIM-deficient embryos. These results reveal that maternal VIM plays an important role as a protector of DNA from damage during reprogramming of somatic nuclei in oocytes.
In this study, a great number of VIM-deficient cloned embryos arrested at the two- or four-cell stage, and the p53 expression level significantly increased in the embryos. Because DSBs induce DNA damage response (53, 54) and the DNA damage response activates the p53 pathway (37), p53 activation in the VIM-deficient cloned embryos might be a result of DSBs. p53 responds to various cellular stresses and induces cell cycle arrest, DNA repair, and cell apoptosis (55–57) and finally affects embryo viability (58). Activation of p53 is one of the characteristics of in vitro cultured embryos with lower developmental competency (59). Thereby, the arrest of VIM-deficient cloned embryo development in this study could be the result of p53 activation that was induced by the stress of DSBs during nuclear remodeling. Knockdown or knock-out of p53 facilitates nuclear reprogramming during the induction of induced pluripotent stem cells, indicating that p53 acts as a barrier to nuclear reprogramming (38, 39). Thus, we can deduce that maternal VIM protects DNA from DSBs, resulting in p53 down-regulation, which is beneficial for successful nuclear reprogramming and promotes the survival of cloned embryos. In addition, we also suggest that VIM-promoted cell reprogramming mediated by oocytes may not target p53 directly and that down-regulation of p53 may be concomitant with the improvement of genomic stability.
Interestingly, we found that the function of VIM for development is only necessary for cloned embryos, not for IVF and PA embryos. It has been supposed that the transferred somatic nuclei tolerate more cellular stress during nuclear remodeling than sperm, which undergo a natural process (16). Cloned embryos also exhibit a higher rate of apoptosis and weaker stress coping functions than IVF embryos (60–62). Therefore, DNA damage occurs more frequently in cloned embryos. We have demonstrated that maternal VIM can protect DNA from damage during nuclear reprogramming. Thus, it can be assumed that the loss of VIM in cloned embryos is more harmful than in IVF embryos, meaning that VIM is more important for cloned embryos than IVF embryos.
In summary, we identified and characterized an oocyte factor, VIM, that was essential for successful nuclear reprogramming by comparing proteome signatures of oocytes with different effects on early development of cloned embryos. Maternal VIM acts as a genomic protector by inhibiting DSBs and leads to down-regulation of p53 during nuclear reprogramming. Our results show that the properties of VIM are crucial for nuclear reprogramming in porcine cloned embryos.
Supplementary Material
Acknowledgment
We gratefully acknowledge Dr. Jilong Liu in Huanan Agricultural University for proteomic analysis.
This work was supported by Major State Basic Research Development Program Grant 2011CB932004 and the Chang Jiang Scholar Candidates Programme for Provincial Universities in Heilongjiang.

This article contains supplemental Table 1.
- NT
- nuclear transfer
- PA
- parthenogenetic activation
- IVF
- in vitro fertilization
- IVM
- in vitro maturation
- 42O
- porcine oocytes with the first polar body collected at 42 h of in vitro maturation
- 33O
- porcine oocytes with the first polar body collected at 33 h of in vitro maturation
- MII
- metaphase II
- VIM
- vimentin
- DSB
- DNA double strand break
- γ-H2AX
- phosphorylated histone H2AX
- DIGE
- difference gel electrophoresis
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Con
- control.
REFERENCES
- 1. Miyamoto K., Tsukiyama T., Yang Y., Li N., Minami N., Yamada M., Imai H. (2009) Cell-free extracts from mammalian oocytes partially induce nuclear reprogramming in somatic cells. Biol. Reprod. 80, 935–943 [DOI] [PubMed] [Google Scholar]
- 2. Polejaeva I. A., Chen S. H., Vaught T. D., Page R. L., Mullins J., Ball S., Dai Y., Boone J., Walker S., Ayares D. L., Colman A., Campbell K. H. (2000) Cloned pigs produced by nuclear transfer from adult somatic cells. Nature 407, 86–90 [DOI] [PubMed] [Google Scholar]
- 3. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 4. Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M. J., Ji H., Ehrlich L. I., Yabuuchi A., Takeuchi A., Cunniff K. C., Hongguang H., McKinney-Freeman S., Naveiras O., Yoon T. J., Irizarry R. A., Jung N., Seita J., Hanna J., Murakami P., Jaenisch R., Weissleder R., Orkin S. H., Weissman I. L., Feinberg A. P., Daley G. Q. (2010) Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang S., Kou Z., Jing Z., Zhang Y., Guo X., Dong M., Wilmut I., Gao S. (2010) Proteome of mouse oocytes at different developmental stages. Proc. Natl. Acad. Sci. U.S.A. 107, 17639–17644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pfeiffer M. J., Siatkowski M., Paudel Y., Balbach S. T., Baeumer N., Crosetto N., Drexler H. C., Fuellen G., Boiani M. (2011) Proteomic analysis of mouse oocytes reveals 28 candidate factors of the “reprogrammome”. J. Proteome Res. 10, 2140–2153 [DOI] [PubMed] [Google Scholar]
- 7. Vitale A. M., Calvert M. E., Mallavarapu M., Yurttas P., Perlin J., Herr J., Coonrod S. (2007) Proteomic profiling of murine oocyte maturation. Mol. Reprod. Dev. 74, 608–616 [DOI] [PubMed] [Google Scholar]
- 8. Zhang P., Ni X., Guo Y., Guo X., Wang Y., Zhou Z., Huo R., Sha J. (2009) Proteomic-based identification of maternal proteins in mature mouse oocytes. BMC Genomics 10, 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Novak S., Paradis F., Savard C., Tremblay K., Sirard M.-A. (2004) Identification of porcine oocyte proteins that are associated with somatic cell nuclei after co-incubation. Biol. Reprod. 71, 1279–1289 [DOI] [PubMed] [Google Scholar]
- 10. Ellederova Z., Halada P., Man P., Kubelka M., Motlik J., Kovarova H. (2004) Protein patterns of pig oocytes during in vitro maturation. Biol. Reprod. 71, 1533–1539 [DOI] [PubMed] [Google Scholar]
- 11. Jiang G.-J., Wang K., Miao D.-Q., Guo L., Hou Y., Schatten H., Sun Q.-Y. (2011) Protein profile changes during porcine oocyte aging and effects of caffeine on protein expression patterns. PLoS One 6, e28996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. You J., Lee E., Bonilla L., Francis J., Koh J., Block J., Chen S., Hansen P. J. (2012) Treatment with the proteasome inhibitor MG132 during the end of oocyte maturation improves oocyte competence for development after fertilization in cattle. PLoS One 7, e48613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma M., Guo X., Wang F., Zhao C., Liu Z., Shi Z., Wang Y., Zhang P., Zhang K., Wang N., Lin M., Zhou Z., Liu J., Li Q., Wang L., Huo R., Sha J., Zhou Q. (2008) Protein expression profile of the mouse metaphase-II oocyte. J. Proteome Res. 7, 4821–4830 [DOI] [PubMed] [Google Scholar]
- 14. Cao S., Guo X., Zhou Z., Sha J. (2012) Comparative proteomic analysis of proteins involved in oocyte meiotic maturation in mice. Mol. Reprod. Dev. 79, 413–422 [DOI] [PubMed] [Google Scholar]
- 15. Calvert M. E., Digilio L. C., Herr J. C., Coonrod S. A. (2003) Oolemmal proteomics—identification of highly abundant heat shock proteins and molecular chaperones in the mature mouse egg and their localization on the plasma membrane. Reprod. Biol. Endocrinol. 1, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miyamoto K., Nagai K., Kitamura N., Nishikawa T., Ikegami H., Binh N. T., Tsukamoto S., Matsumoto M., Tsukiyama T., Minami N., Yamada M., Ariga H., Miyake M., Kawarasaki T., Matsumoto K., Imai H. (2011) Identification and characterization of an oocyte factor required for development of porcine nuclear transfer embryos. Proc. Natl. Acad. Sci. U.S.A. 108, 7040–7045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kikyo N., Wade P. A., Guschin D., Ge H., Wolffe A. P. (2000) Active remodeling of somatic nuclei in egg cytoplasm by the nucleosomal ATPase ISWI. Science 289, 2360–2362 [DOI] [PubMed] [Google Scholar]
- 18. Hansis C., Barreto G., Maltry N., Niehrs C. (2004) Nuclear reprogramming of human somatic cells by Xenopus egg extract requires BRG1. Curr. Biol. 14, 1475–1480 [DOI] [PubMed] [Google Scholar]
- 19. Tamada H., Van Thuan N., Reed P., Nelson D., Katoku-Kikyo N., Wudel J., Wakayama T., Kikyo N. (2006) Chromatin decondensation and nuclear reprogramming by nucleoplasmin. Mol. Cell. Biol. 26, 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Watson A. J. (2007) Oocyte cytoplasmic maturation: a key mediator of oocyte and embryo developmental competence. J. Anim. Sci. 85, (suppl.) E1–E3 [DOI] [PubMed] [Google Scholar]
- 21. Sun Q. Y., Lai L., Bonk A., Prather R. S., Schatten H. (2001) Cytoplasmic changes in relation to nuclear maturation and early embryo developmental potential of porcine oocytes: effects of gonadotropins, cumulus cells, follicular size, and protein synthesis inhibition. Mol. Reprod. Dev. 59, 192–198 [DOI] [PubMed] [Google Scholar]
- 22. Gao S., Gasparrini B., McGarry M., Ferrier T., Fletcher J., Harkness L., De Sousa P., Wilmut I. (2002) Germinal vesicle material is essential for nucleus remodeling after nuclear transfer. Biol. Reprod. 67, 928–934 [DOI] [PubMed] [Google Scholar]
- 23. Lai L., Kolber-Simonds D., Park K. W., Cheong H. T., Greenstein J. L., Im G. S., Samuel M., Bonk A., Rieke A., Day B. N., Murphy C. N., Carter D. B., Hawley R. J., Prather R. S. (2002) Production of α-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science 295, 1089–1092 [DOI] [PubMed] [Google Scholar]
- 24. Kong Q., Wu M., Huan Y., Zhang L., Liu H., Bou G., Luo Y., Mu Y., Liu Z. (2009) Transgene expression is associated with copy number and cytomegalovirus promoter methylation in transgenic pigs. PloS one 4, e6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Betthauser J., Forsberg E., Augenstein M., Childs L., Eilertsen K., Enos J., Forsythe T., Golueke P., Jurgella G., Koppang R., Lesmeister T., Mallon K., Mell G., Misica P., Pace M., Pfister-Genskow M., Strelchenko N., Voelker G., Watt S., Thompson S., Bishop M. (2000) Production of cloned pigs from in vitro systems. Nat. Biotechnol. 18, 1055–1059 [DOI] [PubMed] [Google Scholar]
- 26. Funahashi H., Day B. N. (1997) Advances in in vitro production of pig embryos. J. Reprod. Fertil. Suppl. 52, 271–283 [PubMed] [Google Scholar]
- 27. Tolstonog G. V., Li G., Shoeman R. L., Traub P. (2005) Interaction in vitro of type III intermediate filament proteins with higher order structures of single-stranded DNA, particularly with G-quadruplex DNA. DNA Cell Biol. 24, 85–110 [DOI] [PubMed] [Google Scholar]
- 28. Tolstonog G. V., Wang X., Shoeman R., Traub P. (2000) Intermediate filaments reconstituted from vimentin, desmin, and glial fibrillary acidic protein selectively bind repetitive and mobile DNA sequences from a mixture of mouse genomic DNA fragments. DNA Cell Biol. 19, 647–677 [DOI] [PubMed] [Google Scholar]
- 29. Hartig R., Shoeman R. L., Janetzko A., Tolstonog G., Traub P. (1998) DNA-mediated transport of the intermediate filament protein vimentin into the nucleus of cultured cells. J. Cell Sci. 111, 3573–3584 [DOI] [PubMed] [Google Scholar]
- 30. Wang X., Tolstonog G., Shoeman R. L., Traub P. (1996) Selective binding of specific mouse genomic DNA fragments by mouse vimentin filaments in vitro. DNA Cell Biol. 15, 209–225 [DOI] [PubMed] [Google Scholar]
- 31. Shoeman R. L., Traub P. (1990) The in vitro DNA-binding properties of purified nuclear lamin proteins and vimentin. J. Biol. Chem. 265, 9055–9061 [PubMed] [Google Scholar]
- 32. Shoeman R. L., Wadle S., Scherbarth A., Traub P. (1988) The binding in vitro of the intermediate filament protein vimentin to synthetic oligonucleotides containing telomere sequences. J. Biol. Chem. 263, 18744–18749 [PubMed] [Google Scholar]
- 33. Sutovsky P., Prather R. S. (2004) Nuclear remodeling after SCNT: a contractor's nightmare. Trends Biotechnol. 22, 205–208 [DOI] [PubMed] [Google Scholar]
- 34. Gall L., Brochard V., Ruffini S., Laffont L., Fleurot R., Lavin T. A., Jouneau A., Beaujean N. (2012) Intermediate filaments promote nuclear mechanical constraints during somatic cell nuclear transfer in the mouse. Cell. Reprogram. 14, 497–504 [DOI] [PubMed] [Google Scholar]
- 35. Paull T. T., Rogakou E. P., Yamazaki V., Kirchgessner C. U., Gellert M., Bonner W. M. (2000) A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10, 886–895 [DOI] [PubMed] [Google Scholar]
- 36. Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 [DOI] [PubMed] [Google Scholar]
- 37. Rashi-Elkeles S., Elkon R., Shavit S., Lerenthal Y., Linhart C., Kupershtein A., Amariglio N., Rechavi G., Shamir R., Shiloh Y. (2011) Transcriptional modulation induced by ionizing radiation: p53 remains a central player. Mol. Oncol. 5, 336–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li H., Collado M., Villasante A., Strati K., Ortega S., Cañamero M., Blasco M. A., Serrano M. (2009) The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460, 1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. (2009) Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 460, 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eriksson J. E., Opal P., Goldman R. D. (1992) Intermediate filament dynamics. Curr. Opin. Cell Biol. 4, 99–104 [DOI] [PubMed] [Google Scholar]
- 41. Fuchs E., Weber K. (1994) Intermediate filaments: structure, dynamics, function, and disease. Annu. Rev. Biochem. 63, 345–382 [DOI] [PubMed] [Google Scholar]
- 42. Steinert P. M., Roop D. R. (1988) Molecular and cellular biology of intermediate filaments. Annu. Rev. Biochem. 57, 593–625 [DOI] [PubMed] [Google Scholar]
- 43. Capco D. G., Wan K. M., Penman S. (1982) The nuclear matrix: three-dimensional architecture and protein composition. Cell 29, 847–858 [DOI] [PubMed] [Google Scholar]
- 44. Tolstonog G. V., Sabasch M., Traub P. (2002) Cytoplasmic intermediate filaments are stably associated with nuclear matrices and potentially modulate their DNA-binding function. DNA Cell Biol. 21, 213–239 [DOI] [PubMed] [Google Scholar]
- 45. Traub P., Shoeman R. L. (1994) Intermediate filament and related proteins: potential activators of nucleosomes during transcription initiation and elongation? BioEssays 16, 349–355 [DOI] [PubMed] [Google Scholar]
- 46. DePianto D., Coulombe P. A. (2004) Intermediate filaments and tissue repair. Exp. Cell Res. 301, 68–76 [DOI] [PubMed] [Google Scholar]
- 47. Campbell K. H., Alberio R. (2003) Reprogramming the genome: role of the cell cycle. Reprod. Suppl. 61, 477–494 [PubMed] [Google Scholar]
- 48. Peter M., Nakagawa J., Dorée M., Labbé J. C., Nigg E. A. (1990) In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell 61, 591–602 [DOI] [PubMed] [Google Scholar]
- 49. Wrenzycki C., Wells D., Herrmann D., Miller A., Oliver J., Tervit R., Niemann H. (2001) Nuclear transfer protocol affects messenger RNA expression patterns in cloned bovine blastocysts. Biol. Reprod. 65, 309–317 [DOI] [PubMed] [Google Scholar]
- 50. Whitworth K. M., Prather R. S. (2010) Somatic cell nuclear transfer efficiency: how can it be improved through nuclear remodeling and reprogramming? Mol. Reprod. Dev. 77, 1001–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Suzuki M., Watanabe M., Suzuki K., Nakano K., Matsui K. (1992) Heavy ion-induced chromosome breakage studied by premature chromosome condensation (PCC) in Syrian hamster embryo cells. Int. J. Radiat. Biol. 62, 581–586 [DOI] [PubMed] [Google Scholar]
- 52. Terzoudi G. I., Hatzi V. I., Donta-Bakoyianni C., Pantelias G. E. (2011) Chromatin dynamics during cell cycle mediate conversion of DNA damage into chromatid breaks and affect formation of chromosomal aberrations: biological and clinical significance. Mutat. Res. 711, 174–186 [DOI] [PubMed] [Google Scholar]
- 53. Podhorecka M., Skladanowski A., Bozko P. (2010) H2AX Phosphorylation: Its Role in DNA Damage Response and Cancer Therapy. J. Nucleic Acids 2010, 920161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Giunta S., Belotserkovskaya R., Jackson S. P. (2010) DNA damage signaling in response to double-strand breaks during mitosis. J. Cell Biol. 190, 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Toledo F., Wahl G. M. (2006) Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer 6, 909–923 [DOI] [PubMed] [Google Scholar]
- 56. Wade M., Li Y. C., Wahl G. M. (2013) MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 13, 83–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shiloh Y., Ziv Y. (2013) The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14, 197–210 [PubMed] [Google Scholar]
- 58. O'Neill C., Li Y., Jin X. L. (2012) Survival signaling in the preimplantation embryo. Theriogenology 77, 773–784 [DOI] [PubMed] [Google Scholar]
- 59. Fukuda A., Cao F., Morita S., Yamada K., Jincho Y., Tane S., Sotomaru Y., Kono T. (2010) Identification of inappropriately reprogrammed genes by large-scale transcriptome analysis of individual cloned mouse blastocysts. PLoS One 5, e11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hao Y., Lai L., Mao J., Im G. S., Bonk A., Prather R. S. (2003) Apoptosis and in vitro development of preimplantation porcine embryos derived in vitro or by nuclear transfer. Biol. Reprod. 69, 501–507 [DOI] [PubMed] [Google Scholar]
- 61. Boiani M., Gentile L., Gambles V. V., Cavaleri F., Redi C. A., Schöler H. R. (2005) Variable reprogramming of the pluripotent stem cell marker Oct4 in mouse clones: distinct developmental potentials in different culture environments. Stem Cells 23, 1089–1104 [DOI] [PubMed] [Google Scholar]
- 62. Rodriguez-Osorio N., Wang Z., Kasinathan P., Page G. P., Robl J. M., Memili E. (2009) Transcriptional reprogramming of gene expression in bovine somatic cell chromatin transfer embryos. BMC Genomics 10, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.