Background: Monoubiquitination of FANCD2 and FANCI is critical in the FA pathway.
Results: Specific mutations in FANCE that disrupt FANCD2 binding impair the FA pathway.
Conclusion: Inhibition of the FANCE-FANCD2 interaction via FANCE point mutations in the C terminus disrupts the FA pathway.
Significance: Recruitment of FANCD2 and FANCI to the FA E3 ligase is a critical step in the FA pathway.
Keywords: DNA Damage, DNA Repair, E3 Ubiquitin Ligase, Protein Degradation, Protein Dynamics, Protein Folding, Protein Targeting, Protein-protein Interactions, Ubiquitin
Abstract
Fanconi anemia (FA) is a genome instability syndrome characterized by bone marrow failure and cellular hypersensitivity to DNA cross-linking agents. In response to DNA damage, the FA pathway is activated through the cooperation of 16 FA proteins. A central player in the pathway is a multisubunit E3 ubiquitin ligase complex or the FA core complex, which monoubiquitinates its substrates FANCD2 and FANCI. FANCE, a subunit of the FA core complex, plays an essential role by promoting the integrity of the complex and by directly recognizing FANCD2. To delineate its role in substrate ubiquitination from the core complex assembly, we analyzed a series of mutations within FANCE. We report that a phenylalanine located at the highly conserved extreme C terminus, referred to as Phe-522, is a critical residue for mediating the monoubiquitination of the FANCD2-FANCI complex. Using the FANCE mutant that specifically disrupts the FANCE-FANCD2 interaction as a tool, we found that the interaction-deficient mutant conferred cellular sensitivity in reconstituted FANCE-deficient cells to a similar degree as FANCE null cells, suggesting the significance of the FANCE-FANCD2 interaction in promoting cisplatin resistance. Intriguingly, ectopic expression of the FANCE C terminus fragment alone in FA normal cells disrupts DNA repair, consolidating the importance of the FANCE-FANCD2 interaction in the DNA cross-link repair.
Introduction
Fanconi anemia is an autosomal recessive disorder characterized by bone marrow failure, chromosomal instability, and increased incidence of cancers, including acute myeloid leukemia (1). There are 16 Fanconi anemia (FA)2 genes identified so far (A, B, C, D1, D2, E, F, G, I, J, L, M, N, O, P, and Q), the protein products of which cooperate in a cellular pathway that regulates the cellular resistance to DNA interstrand cross-links (1–3). Most of the mutations in the FA pathway inactivate a multisubunit nuclear complex, the FA core complex, which consists of at least eight FA proteins (A, B, C, E, F, G, L, and M) and four FAAPs (Fanconi-associated proteins; FAAP16, FAAP20, FAAP24, and FAAP100). The main function of the FA core complex is to monoubiquitinate its two substrates, FANCD2 and FANCI (4–7), which form a heterodimer (8). Inactivation of the FA core complex results in failure in the FANCD2-FANCI monoubiquitination, leading to a defect in downstream DNA repair signaling. The modified FANCD2 colocalizes with factors that mediate DNA homologous recombination such as RAD51 and BRCA1 and FANCD2 may physically interact with BRCA2 at the chromatin (1, 9, 10). Monoubiquitinated FANCD2 also recruits ubiquitin zinc finger domain-containing DNA repair proteins such as FAN1, FANCP (SLX4), and translesion synthesis polymerases η (11–16). Therefore, monoubiquitination of FANCD2 by the FA E3 ligase complex is a critical regulatory step in the response to DNA damaging agents. Defective monoubiquitination of FANCD2 and FANCI constitutes >90% of all FA cases, further suggesting the importance of this enzymatic reaction in the biology of the FA pathway and disease progression.
The hierarchy of the FA core complex is highly modular with the FANCL subunit possessing a RING domain, which provides E3 ligase activity, and the FANCE subunit serving as a probable adaptor that directly recognizes the substrate FANCD2 (1). FANCE forms a subcomplex with FANCC and FANCF and is essential for nuclear accumulation and assembly of the FA core complex (17–19). FANCE is also phosphorylated by Chk1 upon DNA damage, and this phosphorylation is required for cellular resistance to mitomycin C (MMC) (20). FANCE may therefore mediate cross-talk between the FA pathway and upstream DNA checkpoint machinery.
To delineate the role of FANCE in inducing substrate ubiquitination from its general role in the FA core complex assembly, we analyzed a series of mutations of FANCE for their ability to support monoubiquitination of FANCD2 and FANCI, and resistance to the DNA cross-linker MMC. We mapped the FANCD2-interacting region to be within the highly conserved extreme C terminus of the protein and showed that the phenylalanine 522 residue is among the critical determinants for physical interaction with FANCD2, accounting for the above phenotypes. Mutation of the FANCE Phe-522 failed to support the cellular resistance to MMC due to the abrogation of both FANCD2 and FANCI monoubiquitination. Importantly, the complemented FANCE F522D system provides a useful tool for elucidating the mechanism of recruitment of the FANCD2-FANCI heterodimer to the FA core complex.
EXPERIMENTAL PROCEDURES
Cell Lines, Plasmids, and Antibodies
DF1179 FA-E fibroblasts were maintained in Chang medium (Irvine Scientific). EUFA130 FA-E lymphoblasts were maintained in RPMI 1640 medium. 293T and HeLa cells were maintained in DMEM. All media were supplemented with 10% bovine calf serum and l-glutamine. For generation of the FANCE mutant cell lines, viruses were produced by co-transfecting 293T cells with pMMP vectors and helper plasmids encoding retroviral gag, pol, and env genes. DF1179 and EUFA130 cells were infected with the viruses with 4 μg/ml polybrene. 2 days later, infected cells were selected by media containing 1 μg/ml puromycin. Site-directed mutagenesis was performed using pMMP-puro-FLAG-FANCE construct as a template, and procedures from the manufacturer's instructions were followed. For generating stable cell lines that express FANCE C-terminal truncates, pMSCV-puro-FLAG-FANCE (amino acids 273–536) was used. Antibodies used in this study are as follows: anti-FANCD2, anti-PCNA, (Santa Cruz Biotechnology), anti-FANCI (Bethyl Laboratory), anti-phospho-CHK1(Ser-317) (Cell Signaling Technology), anti-γ-H2AX (Upstate), anti-γ-Tubulin, anti-FLAG antibodies (Sigma). Anti-FANCA and FANCG antibodies were produced from rabbit and described previously (21).
Yeast Two-hybrid Analysis
cDNA for human FANCD2 and FANCE were cloned into MatchmakerTM GAL4 two-hybrid system 3 vector pGBKT7, and those for FANCC, FANCF, FANCE, FANCD2, and FANCI were cloned into vector pGADT7 (Clontech). Site-directed mutagenesis of FANCE was performed using the pGBKT7-FANCE template. Yeast strain AH109 was sequentially transformed with pGBKT7 and pGADT7 constructs using the small-scale transformation method described in the Yeast Protocols Handbook (Clontech). 10-Fold serial dilutions of the transformants were spotted in triplicate on selection media lacking tryptophan, leucine, and histidine, and containing 3 mm 3-amino-1,2,4-triazole and 20 μg/ml X-α-gal.
Cell Survival Assays
FA-E subtype EUFA130 lymphoblasts were seeded in triplicate in 96-well microplates at a density of 500 cells/well. MMC was added at a final concentration of 0 to 200 μm. Cells were then incubated for 5 days, and survival of the cells was then determined by staining nucleic acids with a proprietary dye (CyQUANT; Molecular Probes) and analyzed with a fluorescence microplate reader according to the manufacturer's instructions. HeLa cells stably selected for expression of the C terminus of FANCE (pMSCV-puro-FLAG-FANCE(273–536) WT and F522D were seeded in triplicate in 12-well plates, similarly as described above. Cisplatin was added at a final concentration of 0 to 2 μm. Cells were incubated for 5 days, and survival of the cells was determined as described above.
Immunofluorescence Assay
HeLa cells were pretreated with extraction buffer (0.25% Triton X-100 in PBS buffer) on ice for 3 min prior to fixation with 4% paraformaldehyde. For the FANCD2 foci in Fig. 3E, anti-FANCD2 (Santa Cruz Biotechnology) primary antibody and Alexa Fluor 488-conjugated donkey anti-mouse secondary antibody (Invitrogen) were used. Images were collected by a Zeiss Axiovert 200 microscope equipped with a Perkin Elmer ERS spinning disk confocal imager and a 63x/1.45NA oil objective using Volocity software (Perkin Elmer).
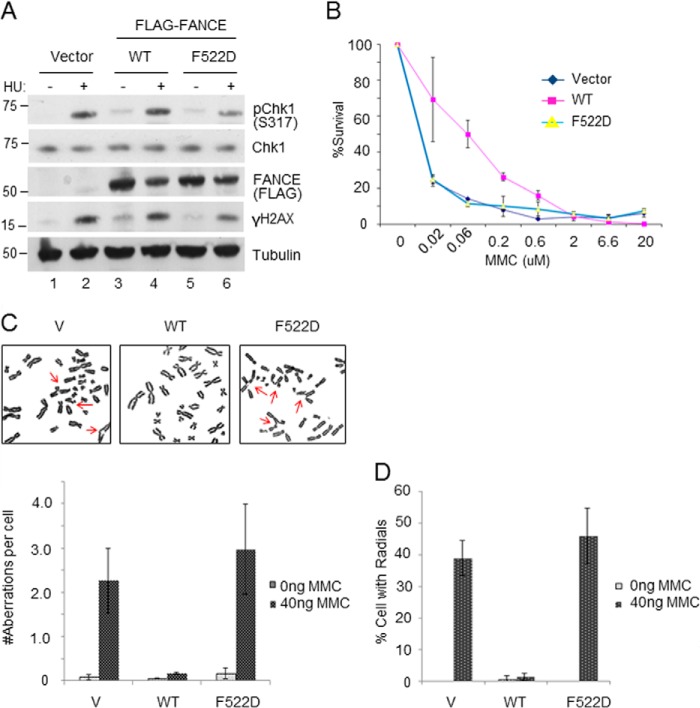
FIGURE 3.
FANCE F522D expressing cells are phenotypically similar to FANCE null cells. A, the F522D mutant EUFA130 cells elicit a normal DNA damage response. B, MMC survival assay. Chromosomal breakage analyses with representative images (C) and percentage of chromosomal aberrations per cell (D) in EUFA130 cells expressing either WT FANCE or the F522D mutant. These are based on three independent experiments. V, vector.
Cytogenetic Analysis
For each cell line, one of two plates was treated with 20 ng/ml MMC for 48 h. Following treatment, the cells were exposed to colcemid (final concentration of 100 ng/ml) for 2 h, treated with a hypotonic solution (0.075 m KCl) for 20 min and fixed with 3:1 methanol/acetic acid. Slides were stained with Wright's stain and when possible, 50 metaphase spreads were scored for aberrations. Metaphase spreads were observed using a Zeiss Axio Imager microscope and captured using CytoVision software (Applied Imaging).
Immunoprecipitation Assay
EUFA130 lymphoblasts were lysed with lysis buffer (50 mm Tris, pH 7.5, 0.5% Nonidet P-40, 100 mm NaCl supplemented with protease inhibitors). The whole cell lysates were centrifuged at 14,000 rpm for 15 min and diluted 2-fold with water. The lysates were precleared by incubating with Sepharose 6B (Sigma) for 1 h, followed by incubating with anti-FLAG M2 agarose (Sigma) overnight at 4 °C. The beads were washed three times with wash buffer (50 mm Tris, pH 7.5, 100 mm NaCl, 0.1% Nonidet P-40) before proteins were eluted by boiling in 1× SDS loading buffer for SDS-PAGE and Western blot analysis. HeLa cells stably expressing truncated FLAG-FANCE WT and F522D were treated with 2 mm hydroxylurea for 18 h, harvested, and lysed (50 mm Tris, pH 7.5, 0.1% Nonidet P-40, 100 mm NaCl supplemented with protease inhibitors). Whole cell extracts were centrifuged at 14,000 prm for 15 min, and the resultant supernatant was used for IP. The lysate was incubated overnight with anti-FLAG M2-agarose (Sigma). The beads were washed three times with lysis buffer, and proteins were eluted and analyzed as described above.
RESULTS
The FANCE C Terminus Is Essential in Monoubiquitination of FANCD2 and Resistance to MMC
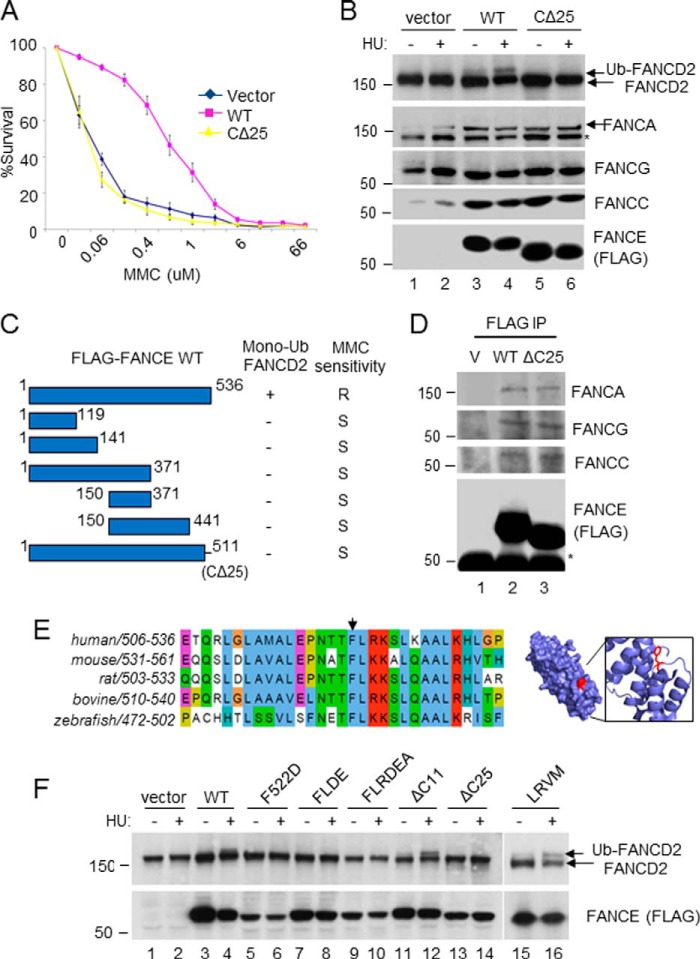
To identify a critical region of FANCE required for promoting the FANCD2 monoubiquitination and the FA pathway, we made serial truncations of FANCE and expressed them in a patient-derived FANCE null lymphoblast cell line, EUFA130. We summarized the results for the monoubiquitination of FANCD2 and the sensitivity to MMC, a hallmark of FA (1), in each FANCE mutant cell line (Fig. 1C). Notably, all of the mutants that lacked the extreme C terminus of FANCE failed to restore monoubiquitination of FANCD2 and resistance to MMC. Particularly, the mutant that minimally lacked the highly conserved C-terminal 25 amino acids (FANCE 1–511; CΔ25) did not support FANCD2 monoubiquitination and MMC sensitivity, suggesting that this region plays an important role. Interestingly, the CΔ25 mutant still supported the integrity of the FA core complex, judged by the expression level of other FA proteins, although it was still unable to induce the monoubiquitination of FANCD2 and resistance to MMC (Fig. 1, A and B). Co-immunoprecipitation confirmed that both wild type and the mutant (CΔ25) associate with similar amount of other FA core members (FANCA, FANCC, FANCG; Fig. 1D), further suggesting that the integrity of the FA core complex remains unaffected in the mutant cells. Consistently, the region required for interaction with other subunits of the FA core complex is mapped outside of the C-terminal region (19). These results suggest that the C-terminal region is not simply involved in providing a scaffolding role. The C-terminal region is highly conserved throughout vertebrate species, suggesting its important role in the FA pathway (Fig. 1E). We therefore further examined the functions of the C-terminal region of FANCE with the hypothesis that this region might be important for the FA core complex to recognize its substrates. To identify a critical residue within the C terminus that is required for FANCD2 monoubiquitination, we generated a set of point mutations within the region. Sequence alignment analysis revealed that the residues surrounding Phe-522 are highly conserved (Fig. 1F). When we mutated the residues surrounding Phe-522, we observed that all of the mutants with a mutation in the Phe-522 residue (FD, FL-DE, FLR-DEA) failed to restore the monoubiquitination of FANCD2, suggesting that the Phe-522 is a critical residue to mediate FANCD2 monoubiquitination (Fig. 1G). Consistently, a truncation mutant of FANCE (Δ11), which was missing the C-terminal 11 amino acids but retained the intact F522, allowed FANCD2 monoubiquitination. These results are highly consistent with the structural study, which suggested that the residues surrounding the Phe-522 residue are energetically favorable for protein-protein interaction and thus important for interaction with FANCD2 (22).
FIGURE 1.
The C terminus of FANCE is essential in promoting the FANCD2 monoubiquitination. MMC survival assays (A) and the FANCD2 monoubiquitination (Ub; B) for the CΔ25 mutant expressed in patient-derived FANCE-deficient fibroblasts (DF1179). An asterisk indicates a nonspecific band in the anti-FANCA blot. C, a summary of FANCE truncations and their ability to promote the FA pathway. D, anti-FLAG immunoprecipitation from the DF1179 lysate, followed by anti-FANCA, FANCC, FANCG Western blotting. E, left panel, sequence alignment of the FANCE C terminus. Right panel, solvent-accessible structure of the FANCE C terminus. The Protein Data Bank file was downloaded from RCSB Protein Data Bank code 2ILR. Residue Phe-522 is shown in red. F, analysis of FANCE point mutants within the C terminus for their ability to support FANCD2 monoubiquitination in the DF1179 FANCE-null fibroblasts. HU, hydroxylurea; R, resistant; S, sensitive; V, vector.
The FANCE Phe-522 Residue Is Required for Inducing Monoubiquitination of Both FANCD2 and FANCI
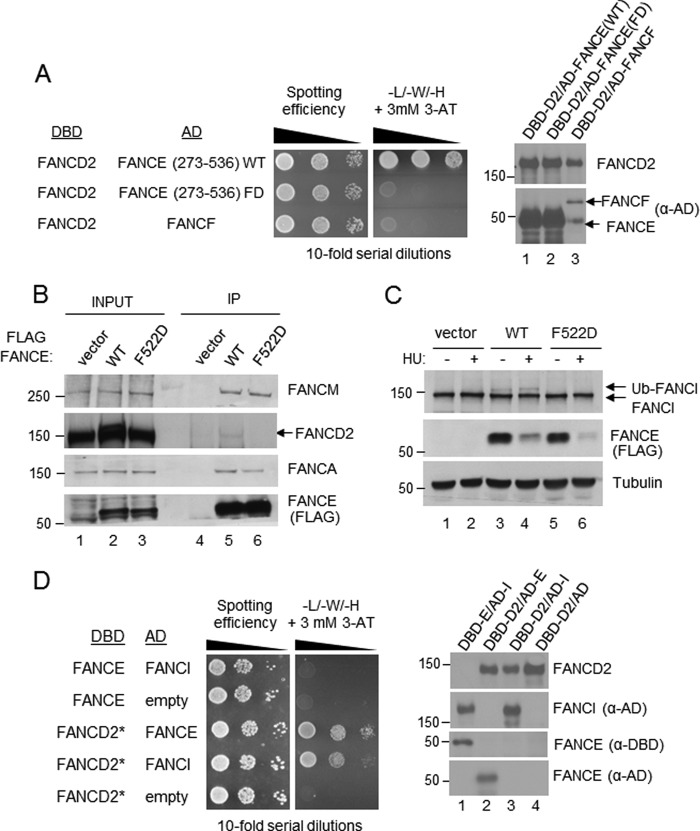
To test whether the deficiency of FANCE F522D mutant cells in FANCD2 monoubiquitination resulted from the inability of the mutant FANCE to interact with FANCD2, a yeast two-hybrid assay was performed (Fig. 2A). While being expressed at a similar level to that of wild type FANCE, the AD-FANCE C-terminal fragment harboring the F522D mutation failed to interact with DBD-FANCD2. To further test the ability of the FANCE mutant to physically interact with FANCD2, a co-immunoprecipitation assay was performed (Fig. 2B). A small fraction of FANCD2 was present only in the wild type FANCE IP, and not in the FANCE F522D mutant IP, confirming that the mutant was unable to interact with FANCD2. Similar amounts of the FANCA and FANCM subunits of the FA core complex were present in the wild type as well as mutant complexes, further supporting that the mutant is still supporting an intact core complex.
FIGURE 2.
The FANCE Phe-522 residue is required for its interaction with FANCD2. A, yeast two-hybrid analysis of FANCD2 paired with FANCE and FANCF (see “Experimental Procedures”). B, immunoprecipitation of FLAG-FANCE WT and F522D from EUFA130 cells show that the Phe-522 residue is essential for the FA core complex to recognize an unmodified form of FANCD2. C, monoubiquitination (Ub) of FANCI is impaired in EUFA130 cells reconstituted with FLAG-FANCE FD mutant. Expression of the recombinant FANCE proteins (both WT and FD) is reduced upon hydroxylurea (HU) treatment, for unclear reason. D, yeast two-hybrid analysis of FANCE paired with FANCD2 and FANCI. Because the full-length DBD-FANCD2 did not yield GAL4 activation when coexpressed with AD-FANCI, the C terminus truncation containing residues 1–1196 was expressed as a DBD hybrid, in this experiment. It has been shown that the C terminus of FANCD2 is dispensable for interaction with FANCE (not shown).
The F522D mutant also failed to induce monoubiquitination of FANCI (Fig. 2C), suggesting that this epitope in FANCE is involved in interacting with both FANCD2 and FANCI. Although the FANCE-FANCD2 interaction is readily detectable in the two-hybrid assay, we were not able to detect the FANCI-FANCE interaction in the same assay conditions (Fig. 2D). As FANCD2 and FANCI form a stable heterodimer (8), the FANCD2-FANCI interaction was used as a control for expression and folding of the AD-FANCI fusion protein in yeast cells.
FANCE F522D-expressing Cells Are Phenotypically Similar to FANCE Null Cells
When we compared the status of phospho-Chk1 and γ-H2AX in the EUFA130 FANCE-deficient lymphoblasts expressing wild type or mutant cells exposed to hydroxylurea, there were no major differences (Fig. 3A), suggesting that the F522D mutant cells are specifically defective in the FANCD2-FANCI monoubiquitination and not in general DNA damage responses. Having acquired the useful mutant cell line that specifically disrupts the FANCE-FANCD2 interaction, we sought to determine to what extent this interaction is critical for conferring cellular resistance to DNA cross-linking agents. Intriguingly, the FANCE-deficient cells expressing F522D FANCE mutant were as sensitive to MMC as FANCE null background, measured by cell survival assay (Fig. 3B). Chromosomal breakage analysis further confirmed this result, as the F522D-expressing cells showed a greater level of breakages and aberrations compared with the wild type, to a similar degree to that of the null background (Fig. 3, C and D). The expression level of FANCE wild type and the F522D mutant were similar (Fig. 3A), suggesting that the observed phenotypes are not due to overall disruption of the FANCE protein. These results underscore that the specific interaction between FANCE and FANCD2 is critical in the FA pathway and perhaps suggests that the primary function of the FA core complex during the DNA ICL repair is to promote the recruitment and activation of FANCD2 and FANCI.
Expression of a FANCE C Terminus Fragment in FA Wild Type Cells Disrupts FANCD2 Monoubiquitination and FA DNA Repair
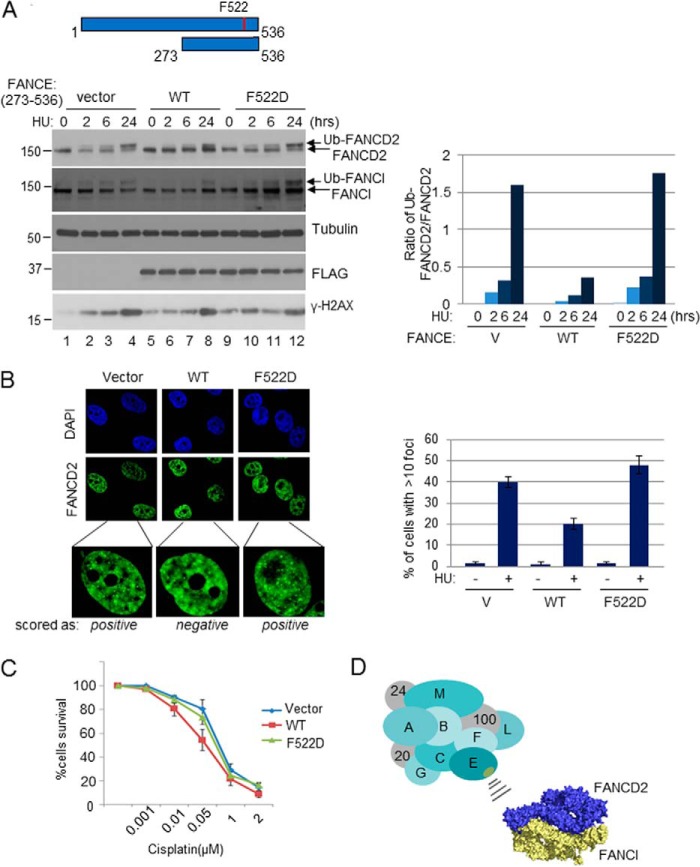
Having established the functional importance of the FANCE C terminus in promoting the FA pathway through FANCD2 recruitment, we sought to use the information to test the feasibility of disrupting the FANCE-FANCD2 interaction in FA normal cells. We stably expressed the C terminus of FANCE (residues 273–536) in HeLa and tested whether the fragment can behave as a dominant negative. Interestingly, the stable expression of the FLAG-tagged FANCE C-terminal fragment (WT) severely suppressed the damage-inducible monoubiquitination of FANCD2, whereas the same fragment harboring F522D (FD) mutation had no effect (Fig. 4A). Expression of this segment is not expected to disrupt the complex formation with FANCC, as the region of FANCE that binds to FANCC was mapped to the outside of 273–536 (23). Similarly, damage-inducible induction of FANCD2 foci formation was significantly inhibited by the expression of the FANCE fragment (Fig. 4B). Consistent with the role of FANCD2 monoubiquitination in DNA ICL repair, HeLa cells that expressed the FANCE WT fragment became sensitive to cisplatin (Fig. 4C). Expression of the FD fragment essentially had no effect on all of these phenotypes, suggesting that the effects are attributed to the specific disruption in the interaction between FANCD2 and FANCE. Altogether, these results consolidate the notion that the FANCE-FANCD2 interaction is critical in promoting the FA pathway.
FIGURE 4.
Expression of a C-terminal fragment of FANCE in FA-wild type cells disrupts the FA pathway. A, HeLa cells stably expressing pMSCV-FLAG-FANCE (273–536) WT, F522D, or empty vector (V) were treated with 2 mm hydroxylurea for 2, 6, or 24 h. Right panel, ImageJ software was used to quantify the monoubiquitination (Ub) status of FANCD2 as seen in the immunoblot. B, left panel, immunofluorescence of FANCD2 in the HeLa cell lines treated with 2 mm hydroxylurea (HU) for 24 h. Right panel, data obtained from counting ∼80 cells with at least 10 FANCD2 foci. C, cisplatin survival assay of HeLa cells stably expressing the corresponding FANCE fragments. D, a schematic model for the recruitment of the FANCD2-FANCI proteins to the FA core complex through the FANCE C terminus. The surface area of FANCE containing the Phe-522 residue is indicated as yellow oval. The determinant within the FANCD2-FANCI that interacts with FANCE is not well understood. PyMOL software was used for modeling the FANCD2-FANCI heterodimer (8).
DISCUSSION
Our report establishes the functional significance of the FANCE C terminus in promoting the FA pathway. Specifically, the Phe-522 within the region is a critical residue for the ability of FANCE to recruit FANCD2 to the FA core complex and to induce the subsequent monoubiquitination reaction. Although the monoubiquitination of FANCI is also disrupted by the Phe-522 mutation, it is not clear whether FANCI also directly interacts with the C terminus of FANCE. Our analysis using the yeast two-hybrid assay suggests that the FANCE-FANCI interaction may not occur directly (Fig. 2D). This raises a fundamental issue in the critical reaction in the FA pathway. How are FANCD2 and FANCI proteins recruited to the FA E3 ligase complex? FANCD2 and FANCI proteins form a stable complex as non-ubiquitinated form, demonstrated by the co-crystalization study by Joo et al. (8). Furthermore, the FANCD2-FANCI interaction is readily detectable in co-immunoprecipitation analysis (5, 6) and even in yeast cells (Fig. 2D). Therefore, it is possible that FANCD2 and FANCI proteins are recruited to the FA core complex simultaneously as a heterodimer through a specific interaction between FANCE and FANCD2. An alternative mechanism may exist, as it was shown that the monoubiquitination kinetics of FANCI can be different from FANCD2, depending on the type of DNA intermediates provided in the Xenopus extract system (24). The precise mechanism for the recruitment process remains to be determined.
Although FANCE has been suggested as the probable substrate adaptor subunit for the FA core complex by several studies (18, 22, 23), our work consolidates the functional significance of FANCE in promoting the interaction between FANCD2 and the FA core complex, FANCD2 and FANCI monoubiquitination, and overall DNA ICL repair. We show that the Phe-522 residue within the C terminus is a critical determinant in the monoubiquitination process by an unbiased mapping analysis. Our result corroborates the result obtained from the structural analysis of FANCE, which showed that the Phe-522 residue forms a surface pocket, which is likely to mediate protein interactions (22). Our results further demonstrate that mutation in the Phe-522 residue or the deletion of the C-terminal 25 amino acids specifically disrupts the FANCE-FANCD2 interaction, without significantly compromising the ability of FANCE to support the structural integrity of the FA core complex. The FA core complex has been suggested to have multiple functions independent of monoubiquitinating its substrates (25, 26). To our surprise, the FANCE FD point mutant cells were as hypersensitive to DNA crosslinking agents as FANCE-null cells (Fig. 3). Given that the integrity of the FA core complex is preserved in the mutant cells, these results demonstrate that the FANCE-FANCD2 interaction is a critical determinant in promoting the resistance to the DNA ICL agent. This result further implicates that, with regard to conferring cellular resistance to DNA ICL agents, the primary role of the FA core complex is to mediate the monoubiquitination of FANCD2 and FANCI. However, we could not rule out the possibility that the same surface of FANCE may mediate other interactions than that with FANCD2. Furthermore, this conclusion assumes that the E3 ubiquitin ligase activity of the FA core complex remains normal in the F522D mutant. Further analysis of the relevant FANCE mutants within the C terminus will bolster this concept.
Finally, our successful attempt to inhibit the FA pathway by way of overexpressing the dominant negative FANCE fragment provides a proof of principle that the interface between FANCD2 and FANCE can be exploited as a strategy for developing a targeted chemosensitizer. It was suggested that hyper-activation or restoration of the FA pathway might be an underlying mechanism by which certain cancer cells acquire resistance to chemotherapeutic drugs (27). Furthermore, disruption of the FA pathway has been partly attributed as one mechanism for cellular sensitization to anti-cancer agents (28–31). Thus, inhibiting the FA pathway in drug-resistant cancers with a more specific manner is a potentially promising anti-cancer approach. The information provided here can be useful for designing epitope-based therapeutics to inhibit the FA pathway, directed toward the interface of FANCD2 and FANCE binding. Further studies directed at the precise understanding of this interaction will enhance our knowledge of the critical substrate-enzyme recognition in DNA repair.
Acknowledgments
We thank members of the Kee laboratory, Drs. Meera Nanjundan and Lucian Moldovan for helpful suggestions and critical reading of the manuscript. We thank Patricia Stuckert, Kailin Yang, and Anne Mainardi for technical assistance for cellular complementation and cell growth assays. We thank Robert Buzzeo in the Cell Biology, Microbiology, and Molecular Biology department core facility for technical assistance with confocal microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1HL52725 (to A. D. D.) and University of South Florida departmental startup funds (to Y. K.).
- FA
- Fanconi anemia
- MMC
- mitomycin C.
REFERENCES
- 1. Kee Y., D'Andrea A. D. (2012) Molecular pathogenesis and clinical management of Fanconi anemia. J. Clin. Investig. 122, 3799–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bogliolo M., Schuster B., Stoepker C., Derkunt B., Su Y., Raams A., Trujillo J. P., Minguillón J., Ramírez M. J., Pujol R., Casado J. A., Baños R., Rio P., Knies K., Zúñiga S., Benítez J., Bueren J. A., Jaspers N. G., Schärer O. D., de Winter J. P., Schindler D., Surrallés J. (2013) Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am. J. Hum. Genet. 92, 800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Osorio A., Bogliolo M., Fernandez V., Barroso A., de la Hoya M., Caldes T., Lasa A., Ramon Y. C. T., Santamarina M., Vega A., Quiles F., Lazaro C., Diez O., Fernandez D., Gonzalez-Sarmiento R., Duran M., Piqueras J. F., Marin M., Pujol R., Surralles J., Benitez J. (2013) Evaluation of Rare Variants in the New Fanconi Anemia Gene ERCC4 (FANCQ) as Familial Breast/Ovarian Cancer Susceptibility Alleles. Hum. Mutat. 34, 1615–1618 [DOI] [PubMed] [Google Scholar]
- 4. Kennedy R. D., D'Andrea A. D. (2005) The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 19, 2925–2940 [DOI] [PubMed] [Google Scholar]
- 5. Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E. R., 3rd, Hurov K. E., Luo J., Ballif B. A., Gygi S. P., Hofmann K., D'Andrea A. D., Elledge S. J. (2007) Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129, 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sims A. E., Spiteri E., Sims R. J., 3rd, Arita A. G., Lach F. P., Landers T., Wurm M., Freund M., Neveling K., Hanenberg H., Auerbach A. D., Huang T. T. (2007) FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 14, 564–567 [DOI] [PubMed] [Google Scholar]
- 7. Dorsman J. C., Levitus M., Rockx D., Rooimans M. A., Oostra A. B., Haitjema A., Bakker S. T., Steltenpool J., Schuler D., Mohan S., Schindler D., Arwert F., Pals G., Mathew C. G., Waisfisz Q., de Winter J. P., Joenje H. (2007) Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol. 29, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Joo W., Xu G., Persky N. S., Smogorzewska A., Rudge D. G., Buzovetsky O., Elledge S. J., Pavletich N. P. (2011) Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway. Science 333, 312–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang X., Andreassen P. R., D'Andrea A. D. (2004) Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol. Cell. Biol. 24, 5850–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taniguchi T., Garcia-Higuera I., Andreassen P. R., Gregory R. C., Grompe M., D'Andrea A. D. (2002) S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100, 2414–2420 [DOI] [PubMed] [Google Scholar]
- 11. Kim H., D'Andrea A. D. (2012) Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 26, 1393–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kratz K., Schöpf B., Kaden S., Sendoel A., Eberhard R., Lademann C., Cannavó E., Sartori A. A., Hengartner M. O., Jiricny J. (2010) Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell 142, 77–88 [DOI] [PubMed] [Google Scholar]
- 13. Smogorzewska A., Desetty R., Saito T. T., Schlabach M., Lach F. P., Sowa M. E., Clark A. B., Kunkel T. A., Harper J. W., Colaiácovo M. P., Elledge S. J. (2010) A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol. Cell 39, 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamamoto K. N., Kobayashi S., Tsuda M., Kurumizaka H., Takata M., Kono K., Jiricny J., Takeda S., Hirota K. (2011) Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc. Natl. Acad. Sci. U.S.A. 108, 6492–6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fu D., Dudimah F. D., Zhang J., Pickering A., Paneerselvam J., Palrasu M., Wang H., Fei P. (2013) Recruitment of DNA polymerase eta by FANCD2 in the early response to DNA damage. Cell Cycle 12, 803–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu T., Ghosal G., Yuan J., Chen J., Huang J. (2010) FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science 329, 693–696 [DOI] [PubMed] [Google Scholar]
- 17. Léveillé F., Ferrer M., Medhurst A. L., Laghmani el H., Rooimans M. A., Bier P., Steltenpool J., Titus T. A., Postlethwait J. H., Hoatlin M. E., Joenje H., de Winter J. P. (2006) The nuclear accumulation of the Fanconi anemia protein FANCE depends on FANCC. DNA Repair 5, 556–565 [DOI] [PubMed] [Google Scholar]
- 18. Pace P., Johnson M., Tan W. M., Mosedale G., Sng C., Hoatlin M., de Winter J., Joenje H., Gergely F., Patel K. J. (2002) FANCE: the link between Fanconi anaemia complex assembly and activity. EMBO J. 21, 3414–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gordon S. M., Alon N., Buchwald M. (2005) FANCC, FANCE, and FANCD2 form a ternary complex essential to the integrity of the Fanconi anemia DNA damage response pathway. J. Biol. Chem. 280, 36118–36125 [DOI] [PubMed] [Google Scholar]
- 20. Wang X., Kennedy R. D., Ray K., Stuckert P., Ellenberger T., D'Andrea A. D. (2007) Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol. Cell. Biol. 27, 3098–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garcia-Higuera I., Kuang Y., Denham J., D'Andrea A. D. (2000) The fanconi anemia proteins FANCA and FANCG stabilize each other and promote the nuclear accumulation of the Fanconi anemia complex. Blood 96, 3224–3230 [PubMed] [Google Scholar]
- 22. Nookala R. K., Hussain S., Pellegrini L. (2007) Insights into Fanconi Anaemia from the structure of human FANCE. Nucleic Acids Res. 35, 1638–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gordon S. M., Buchwald M. (2003) Fanconi anemia protein complex: mapping protein interactions in the yeast 2- and 3-hybrid systems. Blood 102, 136–141 [DOI] [PubMed] [Google Scholar]
- 24. Sareen A., Chaudhury I., Adams N., Sobeck A. (2012) Fanconi anemia proteins FANCD2 and FANCI exhibit different DNA damage responses during S-phase. Nucleic Acids Res. 40, 8425–8439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mirchandani K. D., McCaffrey R. M., D'Andrea A. D. (2008) The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair 7, 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsushita N., Kitao H., Ishiai M., Nagashima N., Hirano S., Okawa K., Ohta T., Yu D. S., McHugh P. J., Hickson I. D., Venkitaraman A. R., Kurumizaka H., Takata M. (2005) A FancD2-monoubiquitin fusion reveals hidden functions of Fanconi anemia core complex in DNA repair. Mol. Cell 19, 841–847 [DOI] [PubMed] [Google Scholar]
- 27. Taniguchi T., Tischkowitz M., Ameziane N., Hodgson S. V., Mathew C. G., Joenje H., Mok S. C., D'Andrea A. D. (2003) Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat. Med. 9, 568–574 [DOI] [PubMed] [Google Scholar]
- 28. Jacquemont C., Taniguchi T. (2007) Proteasome function is required for DNA damage response and fanconi anemia pathway activation. Cancer Res. 67, 7395–7405 [DOI] [PubMed] [Google Scholar]
- 29. Kee Y., Huang M., Chang S., Moreau L. A., Park E., Smith P. G., D'Andrea A. D. (2012) Inhibition of the Nedd8 system sensitizes cells to DNA interstrand cross-linking agents. Mol. Cancer Res. 10, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jacquemont C., Simon J. A., D'Andrea A. D., Taniguchi T. (2012) Non-specific chemical inhibition of the Fanconi anemia pathway sensitizes cancer cells to cisplatin. Mol. Cancer 11, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen C. C., Kennedy R. D., Sidi S., Look A. T., D'Andrea A. (2009) CHK1 inhibition as a strategy for targeting Fanconi Anemia (FA) DNA repair pathway deficient tumors. Mol. Cancer 8, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]