Background: The amyloid precursor protein (APP) is strongly implicated in the pathogenesis of Alzheimer disease.
Results: The loss of Fbxo2, a brain-enriched substrate adaptor for ubiquitin ligases, leads to increased neuronal APP levels and processing.
Conclusion: Fbxo2 regulates APP levels in a brain region-specific manner.
Significance: These findings suggest a novel mechanism for ubiquitin-mediated regulation of amyloid-β production.
Keywords: Amyloid Precursor Protein, Neurobiology, Protein Targeting, Synapses, Ubiquitination
Abstract
The amyloid precursor protein (APP) is an integral membrane glycoprotein whose cleavage products, particularly amyloid-β, accumulate in Alzheimer disease (AD). APP is present at synapses and is thought to play a role in both the formation and plasticity of these critical neuronal structures. Despite the central role suggested for APP in AD pathogenesis, the mechanisms regulating APP in neurons and its processing into cleavage products remain incompletely understood. F-box only protein 2 (Fbxo2), a neuron-enriched ubiquitin ligase substrate adaptor that preferentially binds high-mannose glycans on glycoproteins, was previously implicated in APP processing by facilitating the degradation of the APP-cleaving β-secretase, β-site APP-cleaving enzyme. Here, we sought to determine whether Fbxo2 plays a similar role for other glycoproteins in the amyloid processing pathway. We present in vitro and in vivo evidence that APP is itself a substrate for Fbxo2. APP levels were decreased in the presence of Fbxo2 in non-neuronal cells, and increased in both cultured hippocampal neurons and brain tissue from Fbxo2 knock-out mice. The processing of APP into its cleavage products was also increased in hippocampi and cultured hippocampal neurons lacking Fbxo2. In hippocampal slices, this increase in cleavage products was accompanied by a significant reduction in APP at the cell surface. Taken together, these results suggest that Fbxo2 regulates APP levels and processing in the brain and may play a role in modulating AD pathogenesis.
Introduction
Alzheimer disease (AD)3 is the most common neurodegenerative disorder. Its progressive brain pathology robs patients of memory and cognitive abilities through complex mechanisms that remain unclear. Abnormalities in the processing of amyloid precursor protein (APP) are thought to play a central role in AD pathogenesis (1), largely based on the fact that mutations in APP and the APP cleavage enzyme γ secretase cause early-onset familial AD and replicate key disease features in animal models, including the accumulation of amyloid-β (Aβ) peptide (2). Generated from proteolytic processing of APP (3), Aβ is the major component of the characteristic amyloid plaques that accumulate in AD. Aβ has also been shown to alter synaptic function and decrease synapse numbers (4). Indeed, the loss of synaptic connections more closely correlates with cognitive deficits observed in AD patients than does the loss of neurons and cortical thinning observed later in AD (5).
The regulation of APP expression and processing is critically important to the development and progression of AD. APP is sequentially cleaved by a series of secretase enzymes through a complex process linked to neuronal metabolism and activity (6). Factors that increase the amount of APP available for cleavage inversely correlate with the age of onset of dementia and AD-like pathology (7). For example, patients with trisomy 21, a triplication of the chromosome harboring the APP gene, develop early onset dementia with characteristic AD pathology, which is not observed in patients with partial trisomy, excluding the APP gene (8). Genetic studies in animal models further support the notion that increased APP expression or processing, either through gene duplication or mutation, can lead to early onset dementia with AD-like pathologies (9).
The correlation of APP levels and APP processing to AD pathogenesis underscores the importance of understanding how neurons regulate APP levels and subcellular localization. The importance of APP, moreover, extends beyond its disease link: APP also plays numerous roles in neuronal function and development. Synapse formation and localization, maintenance of dendritic spines, neurite outgrowth, synaptic plasticity, and neuronal survival are all influenced by APP (10–14). Cleavage of APP is regulated by neuronal activity, and its cleavage products in turn seem to regulate neuronal activity, although the precise function of this feedback remains uncertain (15).
The current study seeks to define pathways that regulate APP levels in neurons, focusing on Fbxo2, a brain-enriched, ubiquitin ligase adaptor subunit that specifically recognizes glycoproteins containing high mannose glycans (16). As a type 1 transmembrane glycoprotein, APP is synthesized in the cytoplasm and translocated into the endoplasmic reticulum (ER) where it achieves native folding and becomes glycosylated. Properly folded and glycosylated APP then transits from the ER to the Golgi apparatus for further modification and is eventually delivered to the plasma membrane. APP is known to undergo rapid and constant bulk turnover (17); the extensive, newly synthesized APP that is continually produced by neurons must be appropriately governed for protein quality control purposes. ER-associated-degradation (ERAD) is a normal cellular process by which uncomplexed or improperly modified proteins are removed from the ER and degraded by the ubiquitin proteasome system. The ERAD ubiquitin ligase HRD1 has already been shown to play a role in clearing mutant or misfolded APP from the ER (18). Knocking down this ligase increases APP levels and promotes Aβ generation. Whether HRD1 is the only ubiquitin ligase that mediates APP clearance from the ER remains uncertain.
For several reasons, Fbxo2 is an attractive candidate to contribute to APP clearance. First, it is a neuron-enriched ubiquitin ligase substrate adaptor protein that binds glycoproteins containing high mannose N-linked glycans and facilitates their degradation through ERAD (19). Second, Fbxo2 was recently shown to decrease levels of the β secretase enzyme, BACE1, which is essential for Aβ generation, in an N-linked glycan-dependent manner (20). Overexpression of Fbxo2 resulted in decreased BACE1 levels, in turn diminishing Aβ production. And third, levels of Fbxo2 are decreased in AD patient brains, raising the intriguing possibility that age-related reduction in Fbxo2 levels might accelerate the amyloid process (20).
Although substrate specificity is conferred by ubiquitin ligases and their adaptors, an individual ligase can have numerous substrates. Here, using cell based studies and Fbxo2 knock-out (Fbxo2−/−) mice, we show that Fbxo2 also regulates additional glycoproteins beyond BACE1 in the amyloid precursor pathway, specifically APP itself and the α secretase enzyme, ADAM10. Our results suggest that the loss of Fbxo2 results in dysregulation of glycoprotein homeostasis in neurons with implications for APP processing.
EXPERIMENTAL PROCEDURES
Animals
Fbxo2−/− mice were previously generated through targeted deletion of the first five of six exons encoding Fbxo2. These mice were backcrossed to a C57BL/6J background and exhibited no abnormalities in brain size, weight, development, or adult gross brain structure, although they developed cochlear degeneration (21).
DNA Constructs, HEK Cell Culture, and Lysate Preparation
HEK-293 cells were cultured and maintained as described previously (22). For expression in HEK-293 cells, constructs for full-length APP (produced by D. Selkoe, Addgene plasmid 30154), Myc-Adam10 (produced by R. Derynck, Addgene plasmid 31717), BACE1 (Origene, clone SC115547) or Fbxo2 (gift of K. Glenn, University of Iowa) were transfected with Lipofectamine 2000 (Invitrogen) as per the manufacturer's directions. 48 h after transfection, cells were collected in hot denaturing lysis buffer containing 2% SDS and 100 mm DTT. Lysates were boiled for 5 min, centrifuged, and loaded onto 4–15 or 4–20% gradient SDS-PAGE gels (Bio-Rad).
Brain Extraction/Lysis
For Western blot experiments, animals were anesthetized with ketamine/xylazine and cardiac perfused with prewarmed PBS. Mice were then decapitated, and their brains were removed. When required, hippocampi were rapidly dissected under a dissecting microscope. Tissues were then lysed in hot SDS (2%) lysis buffer with 100 mm DTT in a dounce homogenizer, centrifuged, and boiled for 5 min. Protein concentrations were determined using a quantification kit (Macerey-Nagel, Duren, Germany), and equal amounts were loaded and run on 4–15 or 4–20% SDS-PAGE gels. For ELISA experiments, brains or hippocampi were lysed in cold radioimmune precipitation assay buffer with protease and phosphatase inhibitors (Roche Applied Science) in a dounce homogenizer, centrifuged, and kept on ice for immediate analysis.
Western Blotting
Proteins were immunoblotted using antibodies against APP (22C11, Millipore) or its C-terminal fragments (polyclonal anti-APP cytoplasmic domain G369 antibody, a gift from S. Gandy), Fbxo2 (a gift from K. Glenn, University of Iowa, directed against an amino-terminal domain of Fbxo2), Myc (Santa Cruz Biotechnology), BACE1 (gift from R. Vassar), transferrin receptor (Invitrogen), caspase-3 (Cell Signaling), or GAPDH (Millipore).
ELISAs
Radioimmune precipitation assay buffer-homogenized lysates and collected media were analyzed using ELISA kits according to the manufacturer's instruction. For brain tissue, Aβ42 (high sensitivity, Wako) and sAPPα (IBL) kits were used. For collected media from neurons, sAPPα (IBL), Aβ40 (Invitrogen), and Aβ42 (Novex/Invitrogen) were used.
Immunohistochemistry of Frozen Brain Sections
Briefly, mice were processed as for brain extraction but were perfused with 4% paraformaldehyde in PBS following the PBS flush. Upon removal, brains were post-fixed, rinsed, and cryopreserved. Once frozen, they were cut into 12-micron sections and preserved at −80 °C until use. Alexa Fluor 488 (Invitrogen) was used to visualize 22C11 staining and sections were imaged on a Nikon A1 confocal microscope. All lower intensity images were collected at the same laser, offset, and detection intensity regardless of brain region. Higher intensity images were generated by increasing the brightness equally for all pixels in all images in Fiji Image J software. Z-stack images were collected through equal stack dimensions. Images were cropped using Photoshop CS3 (Adobe). Heat maps were generated in Fiji Image J with the Rainbow RGB lookup table. The CA1 region of hippocampus and the portion of visual cortex (V1/V2) directly lateral and superior to CA1 were imaged. Three mice at the 6 month time point were used for imaging.
Acute Slice Biotinylation
Surface levels of proteins were compared with total amounts as described previously (23). Briefly, mice were anesthetized, and their brains were removed. Hippocampi were rapidly dissected in ice cold, oxygenated artificial cerebrospinal fluid. 350-μm slices were then cut using a MacIlwain Tissue Chopper, and alternating sections from both hippocampi were placed into cold, oxygenated artificial cerebrospinal fluid with or without EZ-Link Sulfo-NHS-LC-biotin (Pierce). After 45 min, slices were washed in artificial cerebrospinal fluid, and any unbound biotin was quenched by incubating the slices in lysine. Following additional washes, the non-biotinylated slices were homogenized in SDS (2%) lysis buffer with 100 mm DTT using a dounce homogenizer. Lysates were then boiled and centrifuged and retained at −80 °C as the “total” fraction. Slices with biotin were lysed in buffer containing 1% Triton X-100, 0.1% SDS, 1 mm EDTA, 50 mm NaCl, 20 mm Tris, pH 7.5, with protease inhibitors (Roche Applied Science) in a glass homogenizer. Biotinylated proteins were then precipitated by the addition of streptavidin resin (Pierce) and overnight incubation at 4 °C on a rotating platform. Precipitates were then collected by first centrifuging to separate supernatant from resin and then boiling the resin in SDS (2%) lysis buffer with 100 mm DTT. The resulting “surface” fraction was retained at −80 °C until being analyzed. 30 μg of protein from each total fraction was run alongside 2 μg of enriched surface protein.
Hippocampal Neuron Culture and Immunofluorescence
Hippocampal neurons were obtained and cultured from pups 3 days postnatal, maintained, and immunostained as described previously (24). All neurons were analyzed at 14 days in vitro. Antibodies against APP (22C11, Millipore), vesicular glutamate transporter (Millipore), spinophilin (Millipore), PSD-95 (Abcam), and binding protein (Abcam) were visualized using Alexa Fluor 488, 568, or 647 secondary antibodies (Invitrogen). Confocal z-stack images were collected using an A-1 confocal microscope (Nikon) at the University of Michigan Microscopy and Image Analysis Laboratory.
Quantification and Statistical Analysis
Immunoblot results were scanned into Adobe Photoshop and measured using ImageJ software. Prism software (version 6, GraphPad) was used for statistical analysis and to generate graphs. Immunofluorescence data were analyzed using ImageJ as described previously (25).
RESULTS
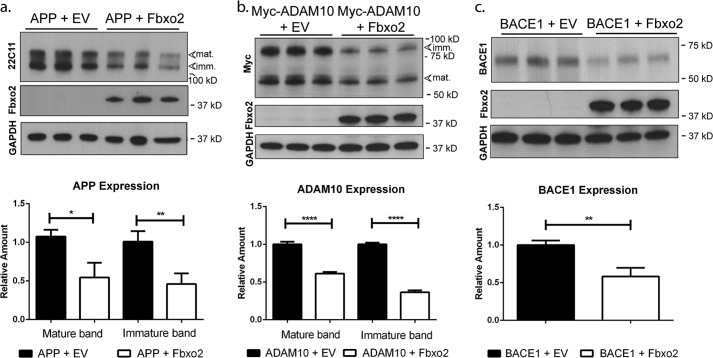
Fbxo2 has been implicated in the clearance of BACE1 (20). To determine whether Fbxo2 facilitates the degradation of other key glycoproteins in the amyloid processing pathway, we expressed constructs encoding full-length APP or a Myc epitope-tagged form of the α-secretase ADAM10 in HEK cells together with empty vector or FLAG-tagged Fbxo2 (Fig. 1). BACE1, already established as an in vitro substrate for Fbxo2, was included as a positive control (Fig. 1c). Co-expression of Fbxo2 resulted in a marked decrease in steady-state levels of each co-expressed substrate as measured by Western blot. The antibodies for APP and ADAM10 detect bands for both immature, high mannose glycan-bearing and mature, complex glycan-bearing forms of the protein on Western blot. Levels of APP are significantly decreased when co-expressed with Fbxo2 (Fig. 1a). A similar reduction is seen when Fbxo2 is co-expressed with ADAM10, with the immature form of ADAM10 being further reduced (Fig. 1b) (26). These ADAM10 results are consistent with the canonical view of the role of Fbxo2 in ERAD, as immature forms of these proteins still carry high mannose glycans (the major ligand for Fbxo2), which are later processed to complex glycans in the Golgi apparatus.
FIGURE 1.
Fbxo2 expression leads to decreased levels of key glycoproteins in the amyloid pathway. a, APP levels are decreased in the presence of Fbxo2. HEK293 cells expressing full-length APP together with empty vector (EV) or vector encoding Fbxo2 were collected 48 h after transfection. Lysates were examined by Western blot with antibodies recognizing APP (22C11), Fbxo2, or GAPDH (upper panel). The mature, complex glycan-bearing form is denoted at ∼130 kDa, and the immature, high mannose glycan-bearing form at ∼110 kDa. Results of triplicate experiments were quantified (lower panel). b, ADAM10 levels are decreased when Fbxo2 is co-expressed. Fbxo2 and ADAM10 were co-expressed as in a, and levels of ADAM10 were measured by anti-Myc antibody (upper panel) and quantified (lower panel). The mature form of ADAM10 is observed at ∼60 kDa and the immature form at ∼85 kDa. c, Fbxo2 expression leads to decreased BACE1. As in a, BACE1 and Fbxo2 were coexpressed, and BACE1 levels were assessed by Western blot using an anti-BACE1 antibody (upper panel) and quantified (lower panel). This antibody detects a single band of ∼60 kDa corresponding to mature BACE1. Shown in lower panels are mean values from triplicate experiments. Error bars, S.E. *, p < 0.05; **, p < 0.01; ****, p < 0.0001; (unpaired t test).
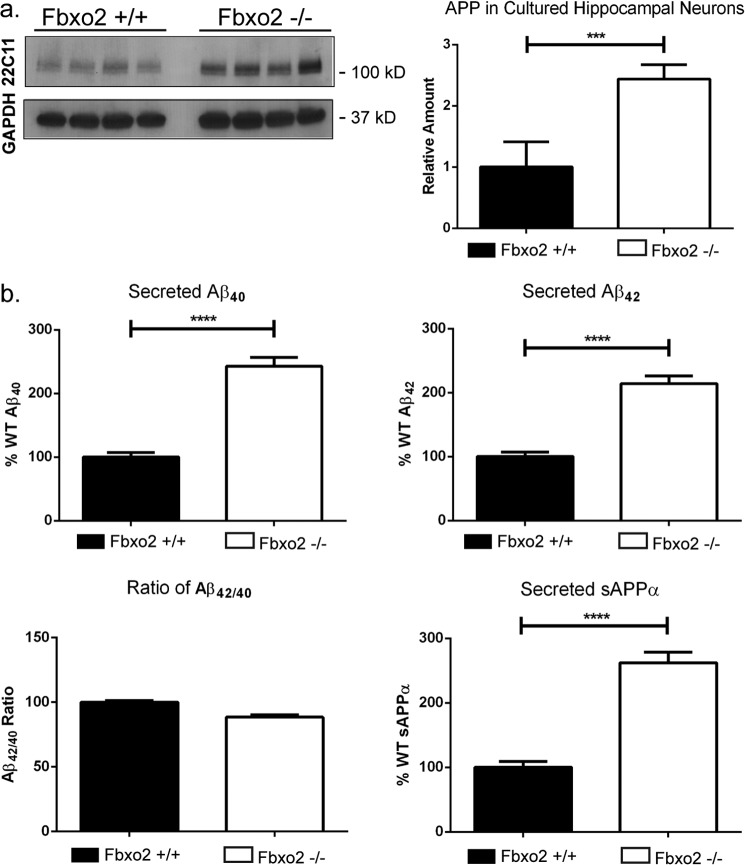
As Fbxo2 is a brain-enriched protein (27), we next sought to determine whether these APP pathway components, identified here as Fbxo2 substrates in heterologous, non-neuronal cell systems, are also physiologically relevant in neuronal cells. Cultured neurons isolated from wild-type and Fbxo2 knock-out mice generated previously in our laboratory offer three distinct advantages for addressing this question. First, we are able to measure endogenous proteins without the confounding factor of transient overexpression; second, media collected from cultures provide a feasible and practical way to assess the processing of APP into its secreted cleavage products; and third, neurons are more easily visualized in culture than in vivo, allowing for a better assessment of any potential differences in the localization of APP. Primary cultures of hippocampal neurons were prepared from mice at postnatal day 3. Cultured neurons lacking Fbxo2 differed markedly from wild-type neurons in several respects. Compared with levels in wild-type cultured neurons, total APP levels were elevated in Fbxo2−/− neurons as revealed by Western blot (Fig. 2a). ELISA assessment of media collected from Fbxo2−/− neuronal cultures revealed more than double the levels of secreted Aβ40, Aβ42, and sAPPα (Fig. 2b). The ratio of Aβ40 to Aβ42 remained unchanged, with both metabolites increasing proportionately (Fig. 2b). Neither ADAM10 nor BACE1 levels differed between wild-type and Fbxo2 knock-out neurons (data not shown).
FIGURE 2.
Dysregulated APP processing and levels in Fbxo2 −/− neurons. a, increased APP in cultured Fbxo2 null neurons. Cultured hippocampal neurons from p3 wild type and knock-out pups, 14 days in vitro, were lysed, and APP levels were measured by Western blot. Representative results from four dishes per genotype are presented (left panel). A significant increase in APP was observed and quantified (right panel); three replicates from different culture preparations were measured to control for potential prep-to-prep differences. b, loss of Fbxo2 results in a doubling of secreted APP cleavage products. Neuronal culture medium was exchanged and then collected 4 days later and assessed by ELISA for secreted APP products. Each measured product was approximately doubled in the knock-out neurons, with the ratio of Aβ42 to Aβ40 remaining unchanged (lower left panel). As in a, representative quantification of quadruplicate samples from a single preparation are presented; three different preparations showed similar results. Error bars, S.E. * and ***, p < 0.001; ****, p < 0.0001 (unpaired t test).
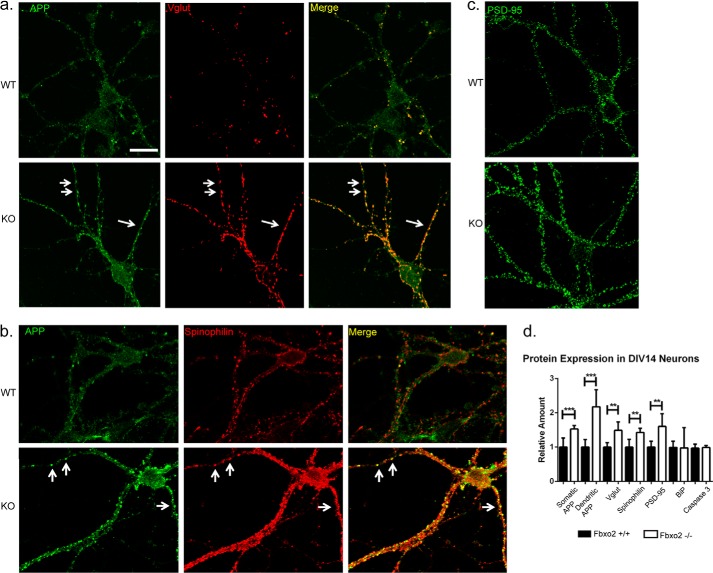
We next examined the distribution of the increased APP observed in Fbxo2 null neurons (Fig. 3). Quantitative immunofluorescence showed that although APP levels were higher throughout Fbxo2−/− neurons, APP accumulated to a greater degree in dendrites than in soma (Fig. 3, a and d). Immunoreactive APP puncta were observed along dendrites with greater frequency and size than in wild-type controls. Consistent with previous studies describing APP trafficking to synapses (28), APP puncta in dendrites colocalized with the pre- and post-synaptic markers vesicular glutamate transporter and spinophilin (Fig. 3, a and b). Remarkably, the amount of synaptic marker-positive puncta, additionally measured with the postsynaptic scaffolding protein PSD95, was significantly increased in neurons lacking Fbxo2 (Fig. 3, c and d). BiP, an ER-resident protein whose accumulation is indicative of ER stress, was not increased in knock-out neurons (Fig. 3d). Caspase-3, a cytosolic protein whose cleavage is believed to play a role in apoptosis, was also assessed by Western blot (Fig. 3d). No difference in the level of uncleaved caspase-3 was observed between wild-type and knock-out neurons, and no cleaved caspase-3 was detected for either genotype. These results suggest that the loss of Fbxo2 does not cause pathological consequences for neuronal health and does not universally affect protein expression.
FIGURE 3.
Loss of Fbxo2 results in increased APP, altered APP localization, and increased synaptic markers in cultured hippocampal neurons. a, APP is increased and distributed differently in Fbxo2 null neurons. 14 Days in vitro hippocampal neurons were fixed, permeabilized, and immunostained for APP and synaptic markers. Representative confocal images of wild-type and knock-out neurons reveal increased overall APP compared with wild type controls, with a greater increase in dendritic than somatic immunoreactivity. APP-immunoreactive puncta (arrows in a and b) were observed to co-localize with the presynaptic marker vesicular glutamate transporter (Vglut) (a) and with the postsynaptic marker spinophilin (b). Levels of vesicular glutamate transporter, spinophilin, binding protein (BiP), and PSD-95 (c) were also assessed and quantified (shown in d). Caspase-3 (d) was measured by Western blot as in Fig. 2. Quantification of immunofluorescence was performed in six to eight neurons per genotype. For somatic and dendritic analysis, three to six dendritic segments of 100 μm per neuron and six neurons per genotype were measured. Error bars, S.E. **, p < 0.01; ***, p < 0.001; (unpaired t test). Scale bar, 25 μm.
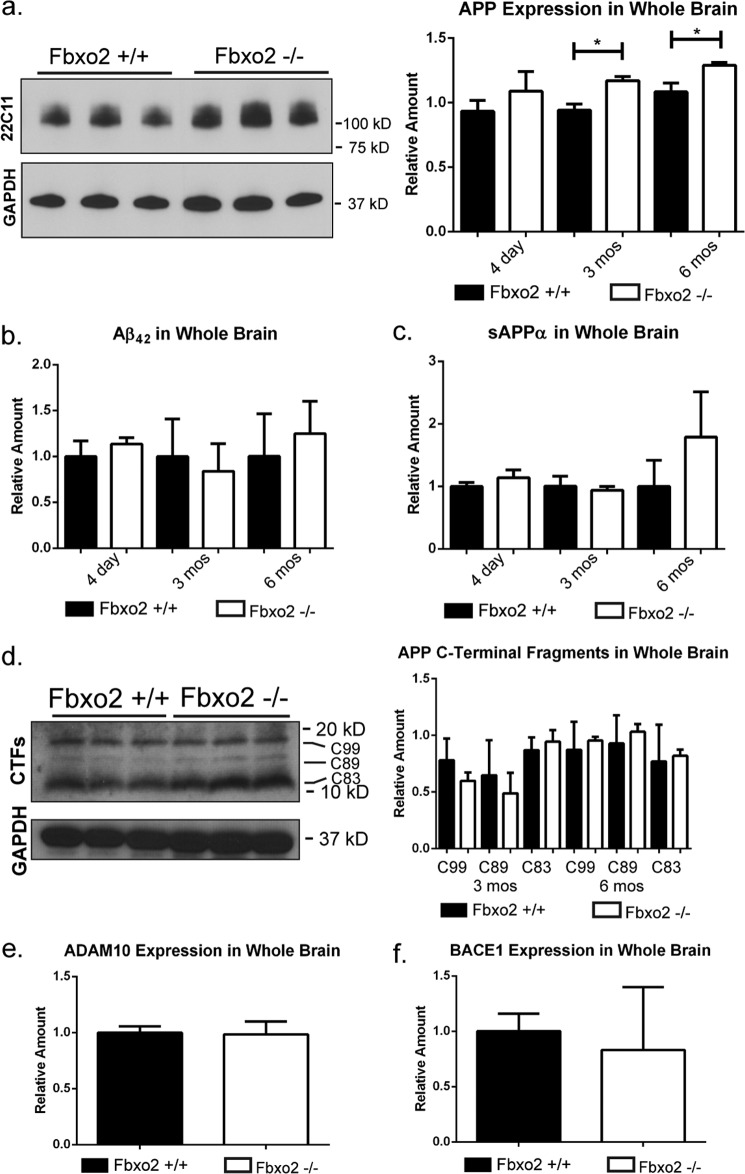
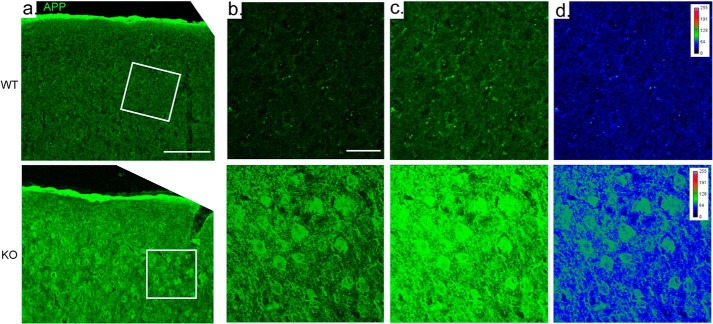
Having identified several changes in APP handling in cultured neurons, we next assessed these changes in the intact brain. Examination of brains from wild-type and Fbxo2 knock-out mice at several ages revealed age-dependent differences. We observed a significant increase in endogenous APP levels in Fbxo2 knock-out mouse brain at 3 and 6 months of age as measured by Western blot from whole brain lysates (Fig. 4a). The time course of this difference is consistent with the onset of murine Fbxo2 expression during the first month of age. We next sought to visualize the difference in APP expression between wild-type and knock-out mice. In cortex at 6 months, anti-APP immunofluorescence confocal imaging revealed that both the cell bodies of cortical neurons and the surrounding neuropils are more strongly labeled in knock-out cortex than in control cortex (Fig. 5a). Image brightness was equally increased (Fig. 5c) to help visualize the comparatively less bright staining in wild-type mice. Whereas in vitro, we observed a shift toward higher dendritic expression, in vivo, we observe increased expression throughout the cell bodies and neuronal projections of the cortex.
FIGURE 4.
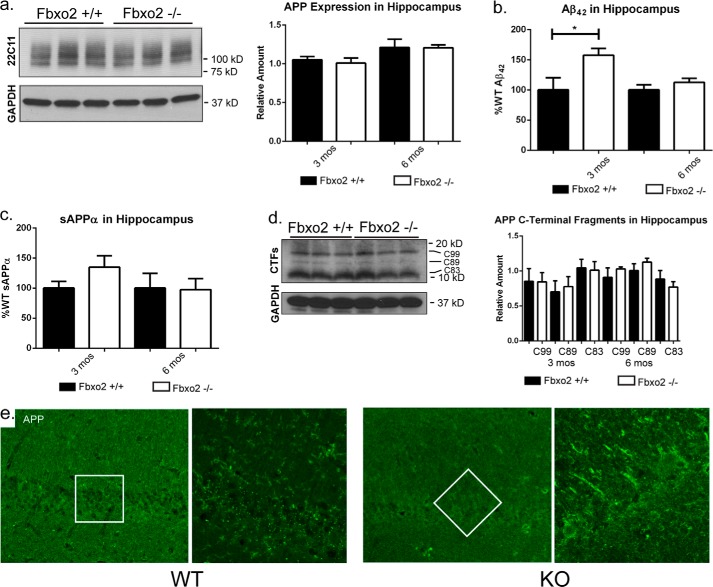
Increased levels of APP, but not amyloid-β, in Fbxo2−/− brain. a, the absence of Fbxo2 increases APP levels in whole brain. APP levels in brain lysates from wild-type and Fbxo2 null mice were assessed by Western blot. Representative immunoblot results from three mice of each genotype at six months are shown (left panel). APP levels were quantified and normalized at three time points, with an n of three animals per genotype at each time point (right panel). Error bars, S.E. *, p < 0.05 (unpaired t test). b and c, APP cleavage products are not increased in whole brain of Fbxo2 null mice. ELISA assays for Aβ42 (b) and sAPPα (c) were performed on protein homogenates from whole brains of three mice per genotype at the indicated time points. d, levels of APP C-terminal fragments were measured in triplicate at 3 and 6 months by Western blot. Results for the 3-month time point are presented (left panel), and results for both time points were quantified relative to loading control and total APP (right panel). e and f, ADAM10 (e) and BACE1 (f) expression are not changed in the absence of Fbxo2. Data shown represent quantification of three animals per genotype at 3 months. Additional time points were examined, and no differences were observed.
FIGURE 5.
Increased levels of APP in Fbxo2−/− brain. APP immunoreactivity is increased in the cortex of Fbxo2 null mice. a, confocal microscopy shows increased levels of APP in Fbxo2−/− mice (column a, lower row) compared with wild-type mice (upper row). From left to right, results from cortices are shown at 10× (a) and then in the denoted inset at 60× with a 2× optical zoom, at relatively lower (b) and higher brightness (c). d, a heat map at the far right illustrates differences in pixel intensity. Scale bar, 50 μm (a) and 20 μm (b).
ELISA experiments were used to assess whether the increased steady-state levels of APP seen in Fbxo2 knock-out mice corresponded to an increase in cleavage products. No change in the amyloid cleavage products Aβ42 and sAPPα was observed at the level of whole brain (Fig. 4, b and c). However, because amyloid cleavage products are rapidly removed from the brain through enzymatic degradation, glial uptake, and cerebrospinal fluid clearance (29), attempts to quantify secreted amyloid cleavage products by measuring brain tissue lysates can be confounded by these factors, making it difficult to gauge fully the extent of the role played by Fbxo2 in brain tissue. No differences were observed at three or six months of age in the levels of APP C-terminal fragments C99, C89, or C83 (Fig. 4d). It is possible that knock-out mice have no difficulty limiting these fragments to appropriate levels.
The previous study linking Fbxo2 to the regulation of BACE1 employed transient shifts in Fbox2 levels, either through heterologous cell expression or overexpression in mice (20). As shown earlier in Fig. 1, we similarly used transient expression to demonstrate facilitated degradation of BACE1 by Fbxo2 and to identify ADAM10 as an additional Fbxo2 substrate. When Fbxo2 is constitutively knocked out in the mouse, however, we did not detect any alteration in endogenous BACE1 or ADAM10 levels (Fig. 4, e and f).
Fbxo2 is expressed throughout the brain, but at higher levels in forebrain and midbrain than in hindbrain (27). AD preferentially affects specific brain regions, including the hippocampus (30). To test whether the effects of Fbxo2 knock-out on APP pathway components were more pronounced in this disease-relevant region, hippocampi were dissected and similarly assessed for APP levels. At three and six months of age, there was no statistical difference in total APP observed between Fbxo2+/+ and Fbxo2−/− in the hippocampus (Fig. 6a). Consistent with our Western blot data, anti-APP immunofluorescence of the hippocampus did not reveal a significant difference between wild-type and knock-out mice (Fig. 6e). There was, however, a significant increase in hippocampal levels of Aβ42 at 3 months of age in Fbxo2−/− mice and a non-statistically significant trend toward increased sAPPα (Fig. 6c). The observation that the whole brain of Fbxo2−/− mice carries more uncleaved APP (Fig. 4a), whereas the hippocampus carries more APP cleavage products may reflect regional differences in neuronal activity and amyloid processing, as described previously (15). In such a scenario, we might expect to see this change reflected in increased levels of C-terminal fragments of APP in Fbxo2 knock-out mouse hippocampus. We did not, however, observe differences in the levels of C-terminal fragments between wild-type and knock-out hippocampi at 3 or 6 months (Fig. 6d), despite a level of amyloid-β in knock-out hippocampus at 3 months, which is 50% greater than it is in wild-type hippocampus. Taken together, these data suggest that there are region-specific responses to the effects of eliminating Fbxo2.
FIGURE 6.
Unchanged APP levels, but increased amyloid-β, in Fbxo2−/− hippocampus. a, loss of Fbxo2 does not alter full-length APP levels in the hippocampus. Hippocampi from wild-type and Fbxo2 null mice were dissected, lysed, and examined by Western blot at 3 and 6 months of age. Representative immunoblots from 6 months are shown (left panel) and quantified in three animals per genotype at 3 and 6 months (right panel). b, APP cleavage products are increased in the hippocampus of Fbxo2 null mice. ELISAs show a significant increase in Aβ42 (b) at three months (3 mos) and suggest a nonstatistically significant increase in sAPPα (c). Results from three mice per genotype per time point were quantified. Error bars, S.E. *, p < 0.05 (unpaired t test). d, APP C-terminal fragment levels were also measured in triplicate at 3 and 6 months by Western blot. Results from the 3-month time point are shown (left panel) and quantified relative to loading control and total APP for both time points (right panel). e, APP immunoreactivity is unaltered in the hippocampus of Fbxo2 null mice. Confocal microscopy reveals no difference in the levels of APP in Fbxo2−/− mice (right panels) compared with wild-type mice (left panels). From left to right, results from the CA1 region of hippocampi are shown at 10× and then in the denoted inset at 60× with a 2× optical zoom.
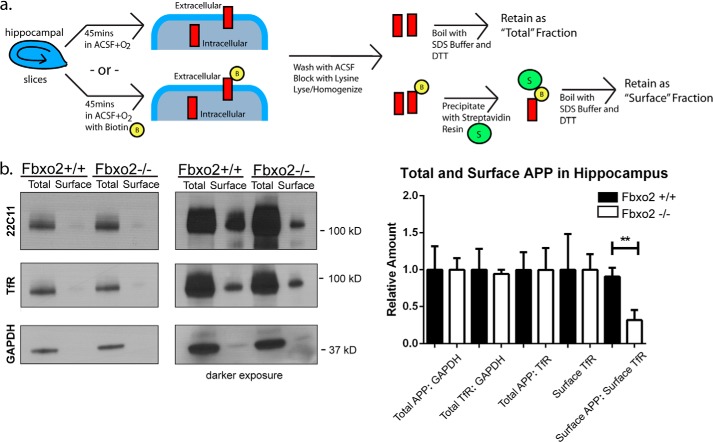
Aβ is primarily generated from APP that has been endocytosed from the cell surface and transported to the trans-Golgi network where it is cleaved by BACE1 (11, 31, 32). Alterations to surface APP levels have been shown to affect Aβ production (31, 33). To determine whether the increase in amyloid cleavage products observed in 3-month-old hippocampus corresponds to a decrease in surface APP, we performed in vivo biotinylation of acute hippocampal slices from three-month old mice, a technique that allows one to estimate the cell surface versus intracellular pools of APP or other protein of interest. In contrast to the lack of change in total hippocampal APP, the amount of surface APP in the hippocampus of Fbxo2−/− mice was indeed significantly decreased compared with wild-type controls (Fig. 7b). The transferrin receptor was used as a control glycoprotein known to have some surface expression, and no difference in the amount of surface transferrin receptor was observed between genotypes. These results suggest that although total levels of APP appear unaltered in the hippocampus of Fbxo2 knock-out mice, the handling of APP with respect to its surface localization is significantly different, which may reflect increased processing into Aβ.
FIGURE 7.
Decreased surface localization of APP in hippocampi of Fbxo2−/− mice. a, schematic of biotinylation procedure to assess surface APP levels in hippocampal slices. Acute hippocampal slices were incubated with oxygenated artificial cerebrospinal fluid with or without biotin for 45 min, washed, blocked with lysine, lysed, and homogenized, after which labeled surface proteins were affinity purified with streptavidin beads (see “Experimental Procedures”). b, surface levels of APP are reduced in the absence of Fbxo2. Acute slices were processed as above, and total and surface fractions were examined by Western blot for APP, GAPDH, or the transferrin receptor (TfR) as a control glycoprotein with surface expression. Representative results from a single animal of each genotype are shown (left panel). Three animals per genotype from 3 months of age were examined, and the results were quantified (right panel). Error bars, S.E. **, p < 0.01 (unpaired t test). ACSF, artificial cerebrospinal fluid.
DISCUSSION
Here, we describe a broader role for Fbxo2 than previously recognized in handling glycoprotein turnover relevant to APP processing. The constant demand for newly produced APP in neurons requires optimal protein quality control, and ERAD provides an effective means of clearing unwanted, immature glycoproteins still bearing high-mannose glycans. As a brain-enriched ubiquitin ligase adaptor subunit, Fbxo2 binds to such glycans on immature glycoproteins and facilitates their ubiquitin-dependent degradation. In the current study, we combined cell-based and animal models to show that Fbxo2 contributes to the regulation of both the steady-state levels of APP and its cleavage products.
Our results in cell-based, transient expression models support previous work (20) implicating Fbxo2 in handling BACE and extend Fbxo2 activity to two other critical amyloid pathway glycoproteins, APP and ADAM10. There are, however, important differences between our cell-based findings and our results in Fbxo2 knock-out mice that underscore the importance of combining in vitro and in vivo studies when seeking to define the role of a ubiquitin pathway protein such as Fbxo2. Overexpression in transient systems may elicit protein behaviors and protein-protein interactions not readily observed under normal, or more physiological, conditions. The fact that we do not see changes in BACE1 in Fbxo2 knock-out mice does not necessarily conflict with the previously findings by Gong et al. (20). Their studies employed transient changes to Fbxo2 levels in vivo and in vitro, and similarly, our transient overexpression studies in heterologous cell models strongly support the view that Fbxo2 facilitates the degradation of BACE1. The same is true for our transient expression studies showing Fbxo2 activity toward ADAM10. Under more physiologic conditions with endogenous ADAM10 and BACE1, however, no such effects are observed in Fbx02 knock-out mice. We suggest that the lifelong deprivation from Fbxo2 occurring in Fbxo2−/− mice expressing endogenous levels of BACE1 and ADAM10 may elicit redundant cellular mechanisms by which these proteins are regulated, resulting in only a partial effect of Fbxo2 absence on APP levels and no observed effect on BACE1 or ADAM10 levels in vivo. In the future, conditional Fbxo2 knock-out models could be utilized to address the discrepancy between previous in vitro data and the in vivo data presented here.
Our results suggest APP may be processed more extensively in Fbxo2−/− hippocampus than in wild-type hippocampus, as there is an increased amount of Aβ compared with cortex with a decreased amount of total APP. In opposition to this idea, we were unable to detect an increase in APP C-terminal fragments in cortex or hippocampus at 3 or 6 months. Although this seems counter-intuitive for the time point at which increased Aβ is observed, the processes by which these fragments are cleared are not entirely understood, although they may involve autophagic mechanisms. In Fbxo2 mice, presumably any excess in C-terminal fragments above the normal amount found in wild-type mice are cleared through a non-Fbx02 dependent manner as they no longer contain glycans.
There is brain region selectivity regarding the effect of Fbxo2 absence on the steady-state levels of APP as revealed by immunohistochemistry. In the cortex, imaging reveals strong staining for APP within neurons and throughout the neuropil. In the hippocampus, however, the signal is markedly less than that observed in the cortex, consistent with our finding that in Fbxo2−/− mice, full-length APP is increased in whole brain but not in the hippocampus per se. With no apparent difference in the level of Fbxo2 expression between cortex and hippocampus of wild-type mice, it remains to be determined why the loss of this protein would result in differing effects on a substrate in distinct brain regions.
Given the absence of changes to BACE1 and ADAM10 levels in Fbxo2 knock-out mice, the apparent increased cleavage of APP in the hippocampus of knock-out mice becomes more difficult to explain. If, in the absence of Fbxo2, APP bypasses ERAD and transits from the ER to the cell surface where it is subsequently endocytosed and cleaved, then we would anticipate that the ratio of surface to total APP might remain the same while seeing an increase in total cleavage products. Essentially both APP and its cleavage products would increase, as observed in our cultured hippocampal neurons. In Fbxo2 knock-out hippocampal neurons, however, the intracellular pool of APP and the cleavage products are both increased, presumably at the expense of the surface pool. What might cause the surface pool of APP to become more rapidly depleted in Fbxo2 knock-out mice remains to be determined. Conceivably, the absence of Fbxo2 drives excess APP to the neuronal cell surface, but into a different subregion of the plasma membrane or in an altered conformational state that allows it to be endocytosed and cleaved at a greater rate. Further analysis of Fbxo2−/− mice may reveal whether, for example, there is preferential distribution of surface APP to lipid rafts, allowing for more rapid processing into Aβ (34). Alternatively, the loss of Fbxo2 may permit the continued existence of a pool of improperly complexed or folded APP, which is not properly trafficked to the surface and is preferentially retained in cortical neurons over hippocampal neurons.
It is important to recognize that Fbxo2 may have functions beyond ERAD. It is expressed throughout neurons, including near synapses, far from the bulk of ER in neurons. The only known binding preference of Fbxo2 is for high mannose N-linked glycans on glycoproteins, which traditionally are modified during passage through the secretory pathway, losing their high mannose status in the process (35). There is, however, precedence for cell surface expression of high mannose forms of another known Fbxo2 substrate, the NMDA receptor subunit GRIN1 (36): all GRIN1 in neurons is susceptible to cleavage by endoglycosidase H, which cleaves high mannose N-linked glycans but not the complex glycans present on most glycoproteins after processing in the Golgi apparatus (37). Accordingly, even at the synaptic membrane GRIN1 remains a potential substrate for Fbxo2. The post-translational modifications to which APP is subjected are complex, and it remains uncertain whether immature (i.e. high mannose glycan-bearing) forms of APP exist in the cell beyond the ER. If APP retains high mannose forms of N-linked glycans on the cell surface, then Fbxo2 could play a role in regulating the endocytosis of APP through ubiquitination. An effect on differential trafficking of APP, namely, the retention of APP in specific cellular compartments, has been attributed to ubiquilin 1 (UBQLN1): polyubiquitination by UBQLN1 sequesters APP in the Golgi (38). Given this observation, we examined the relative distribution of APP in ER and Golgi of neurons in Fbxo2−/− and wild-type mice and found no differences in the pattern of expression (data not shown). Although these data suggest that altered sequestration of APP in the ER and Golgi does not occur, sequestration elsewhere in the secretory pathway or other vesicular bodies cannot be ruled out.
Our analysis of cultured hippocampal neurons suggests that neuronal connectivity is altered in the absence of Fbxo2. The marked changes in number and intensity of synaptic puncta in Fbxo2 knock-out neurons are especially intriguing. It is unlikely these data represent a universal increase in protein expression in the absence of Fbxo2, as the levels of two tested proteins that are not expected to interact with Fbxo2, binding protein, and caspase-3, are unchanged. However, whether these additional synapses function normally remains unknown. The presence of presynaptic terminals directly opposed to these postsynaptic markers suggests they are not “silent” synapses. But there may be other compensatory mechanisms by which these additional synapses are regulated. Future studies will be required to examine the composition and contribution of these synapses. If these additional synaptic connections are indeed active, their activity may feed back onto APP processing. Intriguingly, both Aβ and sAPPα possess dose-dependent neuromodulatory functions (14).
It is somewhat puzzling that cultured neurons from Fbxo2−/− mice maintain more synapses in the presence of elevated Aβ, which has been shown to eliminate synapses (4). It is not known how many substrates Fbxo2 has, but with regard to synaptic dynamics two may be of particular importance: β-integrin 1 (16) and GRIN1 (36). β-Integrin 1 is a membrane receptor that supports cell adhesion and responds to cues from surrounding cells. GRIN1 mediates the formation of NMDA receptors, which have profound effects on plasticity and structural change at the synapse. Both of these Fbxo2 substrates can significantly affect synapse formation, assembly, and maintenance. Therefore, we consider it unlikely that all or even most of the synaptic changes in Fbxo2 null neurons and brain can be attributed to altered APP alone.
In summary, our findings implicate Fbxo2 as a potentially important upstream regulator of key elements of neuronal health and function, APP processing and synaptic connectivity, that are also central to the pathogenesis of AD and perhaps other neurodegenerative diseases.
Acknowledgments
We thank Chris Valdez for assistance with experiments and Geoff Murphy, Shannon Moore, Biswa Ramani, and Yanzhuang Wang for helpful comments on the manuscript.
This work was supported by National Institutes of Health Grant RO1 AG034228, pilot research funds from the Michigan Alzheimer's Disease Center, and NIA, National Institutes of Health Grant T32-AG000114. This work was also supported by the University of Michigan Protein Folding Diseases Initiative.
- AD
- Alzheimer disease
- APP
- amyloid precursor protein
- Aβ
- amyloid-β
- ER
- endoplasmic reticulum
- ERAD
- ER-associated-degradation
- sAPP
- soluble APP
- BACE
- β-site APP-cleaving enzyme.
REFERENCES
- 1. De Strooper B., Vassar R., Golde T. (2010) The secretases: enzymes with therapeutic potential in Alzheimer disease. Nat. Rev. Neurol. 6, 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bertram L., Lill C. M., Tanzi R. E. (2010) The genetics of Alzheimer disease: back to the future. Neuron 68, 270–281 [DOI] [PubMed] [Google Scholar]
- 3. Thinakaran G., Koo E. H. (2008) Amyloid precursor protein trafficking, processing, and function. J. Biol. Chem. 283, 29615–29619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Purro S. A., Dickins E. M., Salinas P. C. (2012) The secreted Wnt antagonist Dickkopf-1 is required for amyloid β-mediated synaptic loss. J. Neurosci. 32, 3492–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Palop J. J., Chin J., Mucke L. (2006) A network dysfunction perspective on neurodegenerative diseases. Nature 443, 768–773 [DOI] [PubMed] [Google Scholar]
- 6. Citron M. (2010) Alzheimer's disease: strategies for disease modification. Nat. Rev. Drug Discov. 9, 387–398 [DOI] [PubMed] [Google Scholar]
- 7. Brouwers N., Sleegers K., Engelborghs S., Bogaerts V., Serneels S., Kamali K., Corsmit E., De Leenheir E., Martin J. J., De Deyn P. P., Van Broeckhoven C., Theuns J. (2006) Genetic risk and transcriptional variability of amyloid precursor protein in Alzheimer's disease. Brain 129, 2984–2991 [DOI] [PubMed] [Google Scholar]
- 8. Millan Sanchez M., Heyn S. N., Das D., Moghadam S., Martin K. J., Salehi A. (2012) Neurobiological elements of cognitive dysfunction in down syndrome: exploring the role of APP. Biol. Psychiatry 71, 403–409 [DOI] [PubMed] [Google Scholar]
- 9. Rovelet-Lecrux A., Hannequin D., Raux G., Le Meur N., Laquerrière A., Vital A., Dumanchin C., Feuillette S., Brice A., Vercelletto M., Dubas F., Frebourg T., Campion D. (2006) APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat. Genet. 38, 24–26 [DOI] [PubMed] [Google Scholar]
- 10. Tyan S. H., Shih A. Y., Walsh J. J., Maruyama H., Sarsoza F., Ku L., Eggert S., Hof P. R., Koo E. H., Dickstein D. L. (2012) Amyloid precursor protein (APP) regulates synaptic structure and function. Mol. Cell Neurosci. 51, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Strooper B., Annaert W. (2000) Proteolytic processing and cell biological functions of the amyloid precursor protein. J. Cell Sci. 113, 1857–1870 [DOI] [PubMed] [Google Scholar]
- 12. Mattson M. P. (1997) Cellular actions of β-amyloid precursor protein and its soluble and fibrillogenic derivatives. Physiol. Rev. 77, 1081–1132 [DOI] [PubMed] [Google Scholar]
- 13. Cousins S. L., Hoey S. E., Anne Stephenson F., Perkinton M. S. (2009) Amyloid precursor protein 695 associates with assembled NR2A- and NR2B-containing NMDA receptors to result in the enhancement of their cell surface delivery. J. Neurochem. 111, 1501–1513 [DOI] [PubMed] [Google Scholar]
- 14. Puzzo D., Privitera L., Fa' M., Staniszewski A., Hashimoto G., Aziz F., Sakurai M., Ribe E. M., Troy C. M., Mercken M., Jung S. S., Palmeri A., Arancio O. (2011) Endogenous amyloid-β is necessary for hippocampal synaptic plasticity and memory. Ann. Neurol. 69, 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bero A. W., Yan P., Roh J. H., Cirrito J. R., Stewart F. R., Raichle M. E., Lee J. M., Holtzman D. M. (2011) Neuronal activity regulates the regional vulnerability to amyloid-β deposition. Nat. Neurosci. 14, 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoshida Y., Chiba T., Tokunaga F., Kawasaki H., Iwai K., Suzuki T., Ito Y., Matsuoka K., Yoshida M., Tanaka K., Tai T. (2002) E3 ubiquitin ligase that recognizes sugar chains. Nature 418, 438–442 [DOI] [PubMed] [Google Scholar]
- 17. Morales-Corraliza J., Mazzella M. J., Berger J. D., Diaz N. S., Choi J. H., Levy E., Matsuoka Y., Planel E., Mathews P. M. (2009) In vivo turnover of tau and APP metabolites in the brains of wild-type and Tg2576 mice: greater stability of sAPP in the β-amyloid depositing mice. PLoS One 4, e7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaneko M., Koike H., Saito R., Kitamura Y., Okuma Y., Nomura Y. (2010) Loss of HRD1-mediated protein degradation causes amyloid precursor protein accumulation and amyloid-β generation. J. Neurosci. 30, 3924–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nelson R. F., Glenn K. A., Miller V. M., Wen H., Paulson H. L. (2006) A novel route for F-box protein-mediated ubiquitination links CHIP to glycoprotein quality control. J. Biol. Chem. 281, 20242–20251 [DOI] [PubMed] [Google Scholar]
- 20. Gong B., Chen F., Pan Y., Arrieta-Cruz I., Yoshida Y., Haroutunian V., Pasinetti G. M. (2010) SCFFbx2-E3-ligase-mediated degradation of BACE1 attenuates Alzheimer's disease amyloidosis and improves synaptic function. Aging Cell 9, 1018–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nelson R. F., Glenn K. A., Zhang Y., Wen H., Knutson T., Gouvion C. M., Robinson B. K., Zhou Z., Yang B., Smith R. J., Paulson H. L. (2007) Selective cochlear degeneration in mice lacking the F-box protein, Fbx2, a glycoprotein-specific ubiquitin ligase subunit. J. Neurosci. 27, 5163–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miller V. M., Nelson R. F., Gouvion C. M., Williams A., Rodriguez-Lebron E., Harper S. Q., Davidson B. L., Rebagliati M. R., Paulson H. L. (2005) CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. J. Neurosci. 25, 9152–9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thomas-Crusells J., Vieira A., Saarma M., Rivera C. (2003) A novel method for monitoring surface membrane trafficking on hippocampal acute slice preparation. J. Neurosci. Methods 125, 159–166 [DOI] [PubMed] [Google Scholar]
- 24. Aakalu G., Smith W. B., Nguyen N., Jiang C., Schuman E. M. (2001) Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 30, 489–502 [DOI] [PubMed] [Google Scholar]
- 25. Iliff A. J., Renoux A. J., Krans A., Usdin K., Sutton M. A., Todd P. K. (2013) Impaired activity-dependent FMRP translation and enhanced mGluR-dependent LTD in Fragile X premutation mice. Hum. Mol. Genet. 22, 1180–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Escrevente C., Morais V. A., Keller S., Soares C. M., Altevogt P., Costa J. (2008) Functional role of N-glycosylation from ADAM10 in processing, localization and activity of the enzyme. Biochim. Biophys. Acta 1780, 905–913 [DOI] [PubMed] [Google Scholar]
- 27. Glenn K. A., Nelson R. F., Wen H. M., Mallinger A. J., Paulson H. L. (2008) Diversity in tissue expression, substrate binding, and SCF complex formation for a lectin family of ubiquitin ligases. J. Biol. Chem. 283, 12717–12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kamenetz F., Tomita T., Hsieh H., Seabrook G., Borchelt D., Iwatsubo T., Sisodia S., Malinow R. (2003) APP processing and synaptic function. Neuron 37, 925–937 [DOI] [PubMed] [Google Scholar]
- 29. Wang Y. J., Zhou H. D., Zhou X. F. (2010) Modified immunotherapies against Alzheimer's disease: toward safer and effective amyloid clearance. J. Alzheimers Dis. 21, 1065–1075 [DOI] [PubMed] [Google Scholar]
- 30. Frisoni G. B., Ganzola R., Canu E., Rüb U., Pizzini F. B., Alessandrini F., Zoccatelli G., Beltramello A., Caltagirone C., Thompson P. M. (2008) Mapping local hippocampal changes in Alzheimer's disease and normal ageing with MRI at 3 Tesla. Brain 131, 3266–3276 [DOI] [PubMed] [Google Scholar]
- 31. Choy R. W., Cheng Z., Schekman R. (2012) Amyloid precursor protein (APP) traffics from the cell surface via endosomes for amyloid β (Aβ) production in the trans-Golgi network. Proc. Natl. Acad. Sci. U.S.A. 109, E2077–E2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Andersen O. M., Reiche J., Schmidt V., Gotthardt M., Spoelgen R., Behlke J., von Arnim C. A., Breiderhoff T., Jansen P., Wu X., Bales K. R., Cappai R., Masters C. L., Gliemann J., Mufson E. J., Hyman B. T., Paul S. M., Nykjaer A., Willnow T. E. (2005) Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. U.S.A. 102, 13461–13466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hoe H. S., Fu Z., Makarova A., Lee J. Y., Lu C., Feng L., Pajoohesh-Ganji A., Matsuoka Y., Hyman B. T., Ehlers M. D., Vicini S., Pak D. T., Rebeck G. W. (2009) The effects of amyloid precursor protein on postsynaptic composition and activity. J. Biol. Chem. 284, 8495–8506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rushworth J. V., Hooper N. M. (2010) Lipid Rafts: Linking Alzheimer's Amyloid-β production, aggregation, and toxicity at neuronal membranes. Int. J. Alzheimers Dis. 2011, 603052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Helenius A., Aebi M. (2004) Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 36. Kato A., Rouach N., Nicoll R. A., Bredt D. S. (2005) Activity-dependent NMDA receptor degradation mediated by retrotranslocation and ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 102, 5600–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huh K. H., Wenthold R. J. (1999) Turnover analysis of glutamate receptors identifies a rapidly degraded pool of the N-methyl-d-aspartate receptor subunit, NR1, in cultured cerebellar granule cells. J. Biol. Chem. 274, 151–157 [DOI] [PubMed] [Google Scholar]
- 38. El Ayadi A., Stieren E. S., Barral J. M., Boehning D. (2012) Ubiquilin-1 regulates amyloid precursor protein maturation and degradation by stimulating K63-linked polyubiquitination of lysine 688. Proc. Natl. Acad. Sci. U.S.A. 109, 13416–13421 [DOI] [PMC free article] [PubMed] [Google Scholar]