Background: Polyubiquitin chains are signaling polypeptides altering the fate of substrates.
Results: A Lys-48-ubiquitin chain of a length greater than four, but not its Lys-63 linkage counterparts, slowed the rate of additional ubiquitin conjugation.
Conclusion: The ubiquitin chain length and linkage may impact kinetic rates of chain elongation.
Significance: Our findings suggest a self-restraining mechanism that limits Lys-48-polyubiquitination.
Keywords: Catenin, E3 Ubiquitin Ligase, Enzymes, Post Translational Modification, Protein Degradation, Ubiquitination, Cdc34, E2 Conjugating Enzyme, SCF, UbcH5c
Abstract
We have explored the mechanisms of polyubiquitin chain assembly with reconstituted ubiquitination of IκBα and β-catenin by the Skp1-cullin 1-βTrCP F-box protein (SCFβTrCP) E3 ubiquitin (Ub) ligase complex. Competition experiments revealed that SCFβTrCP formed a complex with IκBα and that the Nedd8 modified E3-substrate platform engaged in dynamic interactions with the Cdc34 E2 Ub conjugating enzyme for chain elongation. Using “elongation intermediates” containing β-catenin linked with Ub chains of defined length, it was observed that a Lys-48-Ub chain of a length greater than four, but not its Lys-63 linkage counterparts, slowed the rate of additional Ub conjugation. Thus, the Ub chain length and linkage impact kinetic rates of chain elongation. Given that Lys-48-tetra-Ub is packed into compact conformations due to extensive intrachain interactions between Ub subunits, this topology may limit the accessibility of SCFβTrCP/Cdc34 to the distal Ub Lys-48 and result in slowed elongation.
Introduction
Eukaryotic cells require maintenance of proteins at optimal levels. The majority of unwanted and harmful proteins are eliminated through a process called the ubiquitin (Ub)4-proteasome system (1). Ub-proteasome system requires tagging a target protein with chains of Ub molecules, typically connected through the Lys-48–Gly-76 isopeptide linkage, which enable affinity interactions with the 26 S proteasome for the degradation of protein substrates. Defective regulation of Ub-proteasome system is manifested in human diseases including cancers.
Cullin-RING E3 Ub ligase complexes (CRL) are the largest E3 family that function to orchestrate ubiquitination by simultaneously binding to a protein substrate and anchoring an E2 Ub conjugating enzyme through the ROC1/Rbx1 RING domain (2, 3). CRLs, capable of directing numerous substrates for Ub-dependent degradation, build a cellular regulatory network of fundamental importance in controlling protein homeostasis and are critical for the function of a wide range of biological processes ranging from cell cycle regulation to signal transduction. Cullin 1 (CUL1)-based CRL1, more commonly known as SCF (Skp1-CUL1-F box protein), is historically the prototype of all CRLs.
Over the past decade abundant genetic, biochemical and high resolution structural evidence have provided considerable information on the framework of SCF-directed ubiquitination. SCF is organized as a modular complex with the C terminus of CUL1 binding to ROC1. The CUL1-ROC1 interactions establish an intermolecular α/β hydrophobic core containing the CUL1 α/β domain and the N-terminal S1 β-strand of ROC1 (4). SCF functions through a substrate receptor subunit known as the F-box protein, which is characterized by a 40-amino acid-long F-box domain and comprises a family of 69 members in humans (5, 6). Via the Skp1 adaptor that binds both to the CUL1 N terminus and to the F-box domain, most of the F-box proteins can be assembled into the SCF E3 complex. Studies with F-box proteins including β-TrCP, Skp2, and Fbw7 demonstrate an ability of one substrate recognition protein to bind multiple substrates, thereby expanding the SCF functional range.
SCF and most of the CRLs are regulated by Nedd8, the COP9 signalosome, CAND1 (cullin-associated and neddylation-dissociated-1), and glomulin. Covalent conjugation of Nedd8 to cullin, termed neddylation, activates the E3 ligase activity of CRLs by promoting substrate polyubiquitination (7). Subsequent studies have suggested conformation-based mechanisms that explain the activating role of neddylation. The results of in vitro mutagenesis experiments have uncovered autoinhibitory interactions between human ROC1 and CUL1 C-terminal tail in the unmodified state that render the SCF inactive (8). It was proposed that CUL1 neddylation induces conformational changes that disrupt this autoinhibitory interaction, thereby driving SCF into an active state. This hypothesis was supported by the structural studies of Schulman and co-workers (9) that revealed extensive conformational changes in CUL5 when conjugated with Nedd8. Schulman and co-workers (9) further suggest that the neddylation-mediated conformational changes in cullin enabled the repositioning of the RING-tethered E2-S∼Ub to a bound substrate for catalysis. In support of this model, in vitro cross-linking experiments have revealed that neddylation brought an SCF substrate into a close proximity with an E2 Ub-conjugating enzyme (10). Neddylation is reversed by the COP9 signalosome that enzymatically removes Nedd8 from a cullin molecule (11). It is believed that the CRL activities are dynamically controlled by the cullin neddylation-deneddylation cycles. Intriguingly, it was observed that an SCF complex bound to a substrate contained higher levels of neddylated CUL1, thereby suggesting that substrate-E3 interactions may trigger neddylation (12). CAND1 was recently shown to act as an exchange factor that controls the dynamics of CRL assembly (13–15). Recent studies have discovered a new SCF inhibitor called glomulin that binds to ROC1/Rbx1 and blocks its association with the Cdc34 E2 conjugating enzyme (16–17).
Biochemical reconstitution experiments have demonstrated that SCF-directed ubiquitination is a multistep reaction composed of preinitiation, initiation, and elongation. Typically, the reaction begins with activation of a degron by post-translational modification such as phosphorylation, which is often triggered in response to a diversified array of cellular signaling pathways (5, 6). The subsequent step concerns substrate-SCF E3 complex formation, driven by interactions between the substrate phospho-degron and the E3 F-box protein subunit. One example is the complex formed between the β-TrCP F-box protein and the DpSGφXpS degron motif present on IκBα and β-catenin (18). The SCF-substrate association may trigger neddylation, converting the E3 into an active state (8–10, 12). Last, SCF, bound substrate, and Ub-charged E2 conjugating enzyme(s) are engaged in multifaceted interactions that produce Lys-48-linked polyubiquitin chains. In this context, emerging studies have revealed that the joint action of the UbcH5 and Cdc34 E2 enzymes ensures efficient ubiquitination on IκBα (19). It was proposed that in humans, SCF reactions require UbcH5, which acts as an initiator E2 and attaches a substrate with a single Ub molecule, upon which Cdc34 builds the Ub chain. The current study explores mechanistic insights into the SCF-directed polyubiquitination concerning the assembly of a substrate, linked with Ub chains and E3 and E2 complexes. By analyzing ubiquitination of a substrate-linked with Ub chains of defined length, this work provides a first glimpse into the actions of SCF/Cdc34 toward a growing Lys-48-Ub chain.
EXPERIMENTAL PROCEDURES
Protein Preparations
Substrates
IκBα-(1–54) was derived from GST-IκBα-(1–54) that contains an engineered cAMP phosphorylation consensus sequence and a thrombin cleavage site (20). 32P-Labeled IκBα-(1–54) was prepared by using a multistep procedure as previously published (19), including phosphorylation of GST-IκBα-(1–54) at IκBα Ser-32 and Ser-36 by FLAG-IKKβS177E/S181E, adsorption to glutathione-Sepharose (GE Healthcare), radioactive labeling by cAMP kinase, digestion with biotinylated thrombin, and removal of thrombin with streptavidin-Sepharose adsorption. 32P-IκBα-(1–54)-Ub, used in Fig. 3, referred to as I20-Ub-I23 (19) and prepared as described in the same paper. The concentrations of the resulting 32P-labeled substrates were estimated as 1 pmol/μl.
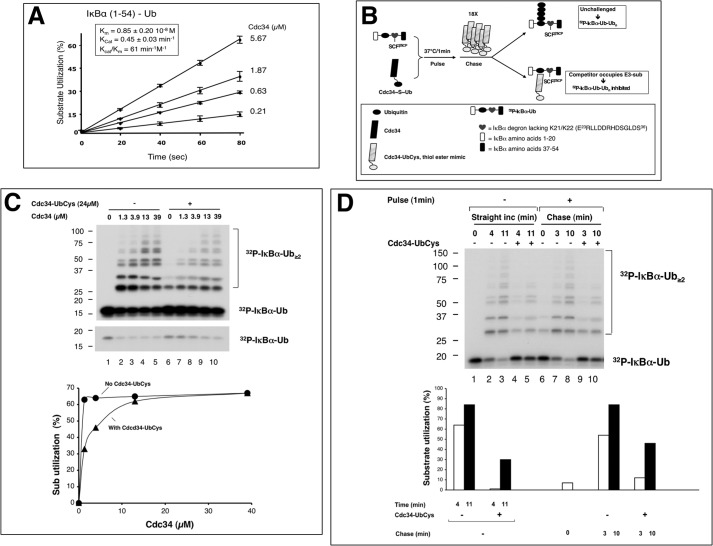
FIGURE 3.
Dynamic association between Cdc34 and the SCFβTrCP1-IκBα-Ub complex. A, kinetic parameters of polyubiquitination of IκBα-(1–54)-Ub by SCFβTrCP2 and Cdc34. 32P-Labeled IκBα-(1–54)-Ub (2pmol) was incubated with SCFβTrCP2 (∼2 pmol) for 10 min. Separate mixtures of Cdc34∼Ub at differing concentrations (5.67, 1.87, 0.63, and 0.21 μm) of Cdc34 were incubated at 37 °C for 5 min. The two mixtures were combined, and reactions were stopped by the addition of SDS-loading buffer after 20, 40, 60, or 80 s. Substrate utilization of three independent data sets was calculated and shown graphically. Km and Kcat were estimated by fitting to Michaelis-Menten kinetics and shown in the inset box. B, the competition scheme. The detailed procedure is described under “Experimental Procedures.” C, the inhibition of Cdc34-UbCys on Cdc34-catalyzed elongation of 32P-IκBα-Ub. The inhibitory effects were reversed by high concentrations of Cdc34. A short exposure of the input substrate is shown to better reveal its reduction with reaction time. D, the competition reaction. The effects of Cdc34-UbCys, which was added after the reaction had been initiated, are shown. The substrate conversion was quantified and is shown graphically.
The β-catenin peptide substrate was prepared as previously described (10), including an engineered cAMP phosphorylation consensus sequence. This substrate contains chemically incorporated phosphoserines at residues 33 and 37. The β-catenin peptide substrate was 32P-labeled by cAMP kinase.
SCFβTrCP
Two preparations of SCFβTrCP were used in this study. Experiments shown in Figs. 1, 2 (A–C), 3A, 5–7, and 8A, employed SCFβTrCP2, which was isolated from FCHT293 cells, a HEK293-based cell line generated previously to constitutively co-express HA-βTrCP2 and FLAG-CUL1 (8). SCFβTrCP2 was affinity-purified as described in Wu et al. (19) and typically contained CUL1, of which 30–40% was in the neddylated form. The concentrated SCFβTrCP2 was ∼1–2 pmol/μl.
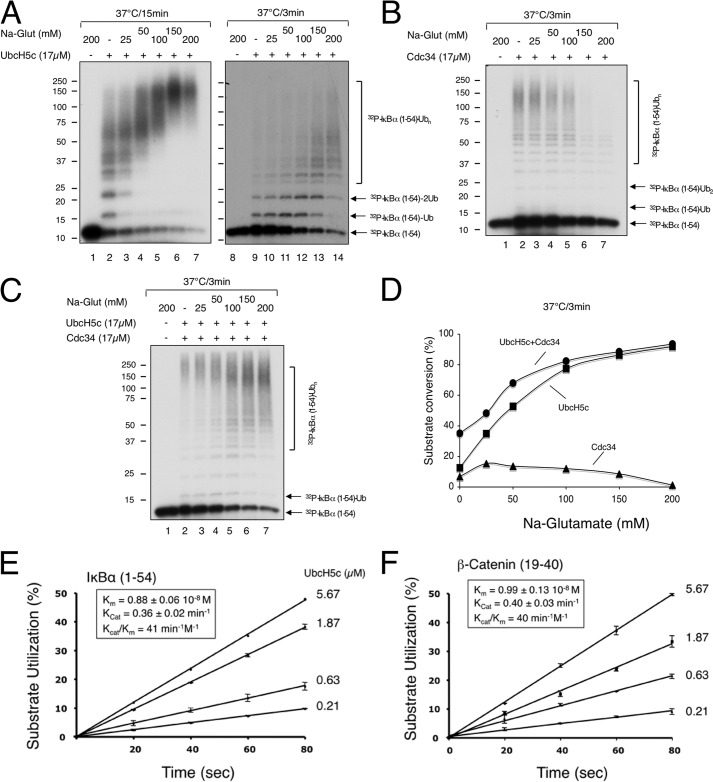
FIGURE 1.
Characterization of polyubiquitination of IκBα-(1–54) and β-catenin by SCFβTrCP2, UbcH5c, and Cdc34. A–D, sodium glutamate activates UbcH5c but inhibits Cdc34 activity. The effects of increasing concentrations of sodium glutamate (as indicated) on polyubiquitination of IκBα-(1–54) by SCFβTrCP2 in the presence of UbcH5c (panel A), Cdc34 (panel B), or both UbcH5c and Cdc34 (panel C). Note that sodium glutamate (Na-Glut), at 150 and 200 mm, affects gel migration such that the input substrate bands appear wider (panel B, lanes 1, 6, and 7). Phosphorimaging revealed no substrate utilization under these conditions. The detailed procedure is described under “Experimental Procedures” with incubation times as indicated. The reactions were quantified and shown graphically (panel D). E and F, kinetic parameters of polyubiquitination of IκBα-(1–54) and β-catenin by SCFβTrCP2 and UbcH5c. 32P-Labeled IκBα-(1–54) (2pmol) or β-catenin (2pmol) were incubated with SCFβTrCP2 (∼2pmol) for 10 min. Separate mixtures of UbcH5c∼Ub at differing concentrations (5.67, 1.87, 0.63, and 0.21 μm) of UbcH5c were incubated at 37 °C for 5 min. The two mixtures were combined, and reactions were stopped by the addition of SDS-loading buffer after 20, 40, 60, or 80 s. Substrate utilization of three independent data sets was calculated and shown graphically for the reaction of IκBα-(1–54) (panel E) or β-catenin (panel F). Km and Kcat were estimated by fitting to Michaelis-Menten kinetics and shown in the inset box.
FIGURE 2.
In vitro ubiquitination of IκBα and β-Catenin by SCFβTrCP2, UbcH5c, and Cdc34. A, kinetic analysis of polyubiquitination of IκBα-(1–54) by SCFβTrCP2 in the presence of UbcH5c, Cdc34, or both UbcH5c and Cdc34. Ubiquitination was carried out as described under “Experimental Procedures” with substrate (0.1 μm), E3 (0.1 μm), UbcH5c (17 μm), and Cdc34 (17 μm). B, effect of Cdc34 concentration on polyubiquitination of IκBα-(1–54) by SCFβTrCP2 in the presence of UbcH5c. Ubiquitination was carried out as in panel A at 37 °C for 3 min. Reaction products are marked on the gel. Although Ub1 denotes mono-ubiquitinated IκBα-(1–54) at either Lys-21 or Lys-22, 2Ub represents the di-Ub products with Ub conjugated at both Lys-21 and Lys-22. * refers to IκBα-(1–54) attached with a di-Ub chain formed at either Lys-21 or Lys-22. C, combined actions of UbcH5c and Cdc34 drive polyubiquitination of β-catenin by SCFβTrCP2. Ubiquitination was carried out as in panel A with the wild type Ub or variants as indicated at 37 °C for 15 min. Both short and long exposures are shown. Substrate and reaction products are marked on the gel, with numbers denoting the number of Ub moieties. D, analysis of polyubiquitination of β-Catenin by SCFβTrCP1 with UbcH5c or Cdc34 in a range of concentrations of substrate or E3. Varying concentrations of β-catenin or SCFβTrCP1, as indicated, were used for the ubiquitination assay. The results were quantified and are shown graphically. E, immunoblot analysis of cellular concentrations of UbcH5 and Cdc34. Protein extracts from the indicated cell lines were subject to immunoblot analysis along with purified UbcH5c and Cdc34 in amounts as indicated. For the immunoblots of UbcH5 or Cdc34, protein lysates derived from 300,000 or 150,000 cells, respectively, were used. The box indicates the estimated number of molecules for UbcH5 or Cdc34 in the indicated cell line.
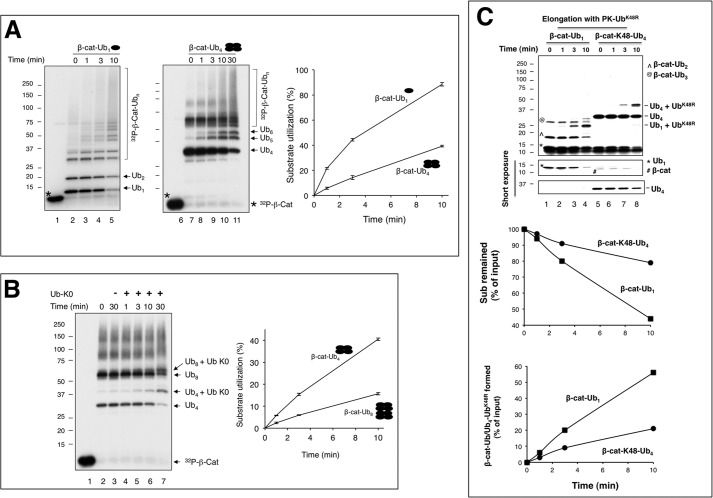
FIGURE 5.
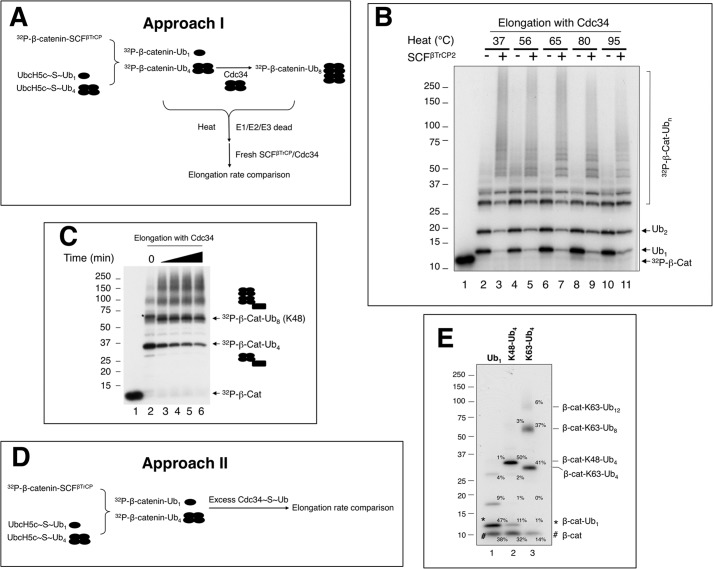
Building substrate-Ub chain intermediates. A, Approach I, a scheme for the preparation of β-cat-Ub chains using heat denaturation. The detailed procedure is described under “Experimental Procedures.” B, SCFβTrCP2/Cdc34 is capable of elongating heat-treated β-cat-Ub. To prepare β-cat-Ub, SCFβTrCP2-β-cat and UbcH5c∼Ub were mixed and incubated at 37 °C for 10 min. Note that under this condition, all of the substrate was utilized, yielding β-cat-Ub as the predominant product (lane 2). The β-cat-Ub-containing reaction mixture was then heat-treated at various temperatures as indicated (lanes 4–11) or not (lanes 2 and 3). The heat or mock-treated mixture was mixed with the preformed Cdc34∼Ub charging mix. In reactions shown in lanes 3, 5, 7, 9, and 11, fresh SCFβTrCP2 was added. The final incubation was at 37 °C for 10 min. C, the synthesis of β-cat-Ub8. The time of incubation with Cdc34 is 30, 60, 90, and 120 min. Note that in lane 5, the ratio of β-cat-Ub8 to β-cat-Ub4 is 2.5:1. This reaction mixture was used for experiments shown in Fig. 6B. D, Approach II, a scheme for the preparation of β-cat-Ub chains without heat denaturation. The detailed procedure is described under “Experimental Procedures.” E, the distribution of the β-cat-Ub1, β-cat-Lys-48-Ub4, and β-cat-Lys-63 Ub8 preparations. The relative percentages of the input and products are indicated.
FIGURE 6.
Increasing Lys-48-Ub chain length on a substrate results in decreased conjugation rates. A) Comparison of Cdc34-catalyzed elongation of β-cat-Ub1 with β-cat-Ub4, ∼0.1 μm each, using approach I (Fig. 5A) with heat denaturation. The quantification of β-cat-Ub1 or β-cat-Ub4 was based on phosphorimaging analysis of experiment shown in Fig. 5E. B, same as in A, but comparing β-cat-Ub4 with β-cat-Ub8. β-cat-Ub8 was prepared as described for reaction shown in Fig. 5C, lane 5. Note that the graph shown in panels A and B integrates the results of three independent experiments, with error bars for the calculated S.D. The experiment shown in panel C was carried out using approach II without heat denaturation (Fig. 5D).
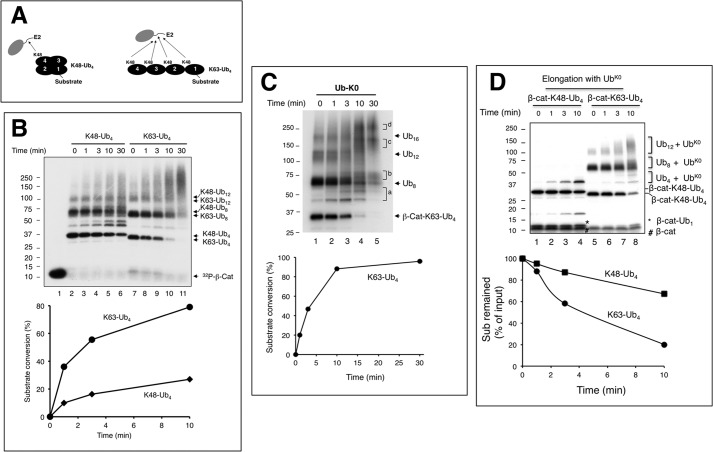
FIGURE 7.
SCFβTrCP2/Cdc34 elongates β-catenin-Lys-63-Ub4 more efficiently than β-catenin-Lys-48-Ub4. Panel A shows the reaction scheme comparing elongation of β-cat-Lys-48-Ub4 with β-cat-Lys-63-Ub4, ∼0.1 μm each. Note that there are four Lys-48 residues within β-cat-Lys-63-Ub4 that can act as a receptor for elongation by Cdc34. The detailed procedure is described under “Experimental Procedures”. Panel B shows a comparison of Cdc34-catalyzed elongation of β-cat-Lys-48-Ub4 with β-cat-Lys-63-Ub4 in the presence of wild type Ub. Panel C shows Cdc34-catalyzed elongation of β-cat-Lys-63-Ub4–16 in the presence of Ub-K0. Letters (a–d) on the gel mark the reaction products in the regions as indicated. Note that the heterogeneity of β-cat-Lys-63-Ub8–12 precludes accurate assessment of their conversion into products. Although experiments in panels B and C were carried out using Approach I (Fig. 5A) with heat denaturation, the experiment in panel D was performed with approach II (Fig. 5D) without heat denaturation.
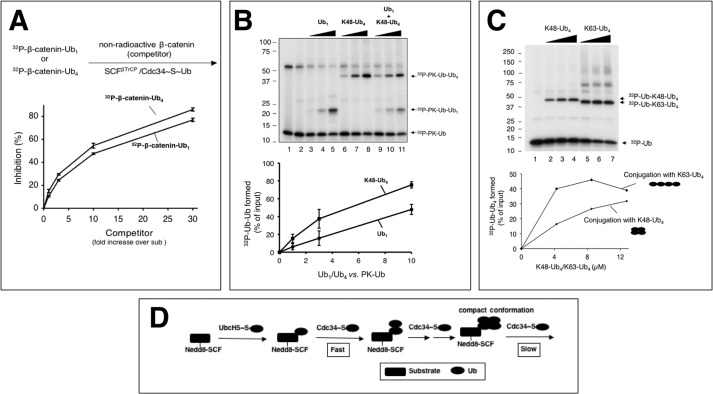
FIGURE 8.
A, competition with β-catenin-Ub1 and β-catenin-Ub4 as substrates. β-cat-Ub1 or β-cat-Ub4 were prepared as described under “Experimental Procedures.” These preformed β-cat-Ub1 (∼2 pmol) or β-cat-Ub4 (∼2 pmol) were mixed with increasing amounts of β-catenin peptide (1-, 3-, 10-, and 30-fold excess) in the presence of SCFβTrCP2, and the resulting mixture was incubated for 10 min at 25 °C. A separate mixture containing Cdc34∼Ub (17 μm E2) was added, and the reaction was incubated at 37 °C for 10 min. For quantification, the difference in substrate utilization between the reaction with or without competitor was calculated. The calculated difference was expressed as % inhibition. This graph integrates the results of three independent experiments, with error bars for the calculated S.D. B, substrate-independent di-Ub synthesis by Cdc34 with Ub or Lys-48-Ub4 as the receptor. The detailed procedure is described under “Experimental Procedures.” The concentrations of Ub or Lys-48-Ub4 are 0.85, 2.55, and 8.5 μm. This graph integrates the results of three independent experiments, with error bars for the calculated standard deviation. C, comparison of Cdc34-catalyzed di-Ub synthesis with Lys-48-Ub4 or Lys-63-Ub4 as the receptor. The concentrations of Lys-48-Ub4 or Lys-63-Ub4 are 4.25, 8.5, and 12.75 μm. D, an overall model for SCF-directed ubiquitination. See “Discussion” for detail.
Experiments shown in Figs. 2D and 3, C--D, employed SCFβTrCP1, which was assembled by combining ROC1-CUL1 and βTrCP1-Skp1 subcomplexes, each purified using baculovirus/SF9 overexpression system according to the previously published protocol (4). To assemble neddylated SCFβTrCP1, ROC1-CUL1 (1.5pmol) was incubated in a mixture (3 μl) that contained 50 mm Tris-HCl, pH7.4, 5 mm MgCl2, 2 mm NaF, 10 nm okadaic acid, 2 mm ATP, 0.5 mm DTT, 0.1 mg/ml BSA, Nedd8 (20 μm), APP-BP1/Uba3 (83 nm), and Ubc12 (15 μm). The reaction was incubated for 10 min at room temperature. βTrCP1-Skp1 (3pmol) was added and incubation continued for 10 min at room temperature. The resulting SCFβTrCP1 contained CUL1, of which ∼50% was in the neddylated form.
Other Proteins
Other enzymes and proteins were prepared as described: the FLAG-IKKβS177E/S181E kinase (20), 32P-PK-Ub (21), ROC1/Rbx1-CUL1 (4), ROC1/Rbx1-CUL1-(411–776), Cdc34-UbCys (32), Nedd8, Ubc12, UbcH5c, and Cdc34 (23). E1, APP-BP1/Uba3, Ub-K0, and Ub chains were purchased from Boston Biochem (Cambridge, MA).
Protocols for Ubiquitination
Substrate Ubiquitination
In all ubiquitination experiments, the reaction was initiated by combining two preformed mixtures that contained E2∼S∼Ub and E3-substrate. The E2 charging reaction was assembled typically in a mixture (5 μl) that contained 50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 2 mm NaF, 10 nm okadaic acid, 2 mm ATP, 0.5 mm DTT, 0.1 mg/ml BSA, Ub (50 μm), E1 (0.1 μm), and UbcH5c or Cdc34 (concentrations as specified). The reaction was incubated for 5 min at 37 °C. To assemble the E3-substrate complex, a mixture (5 μl) containing SCFβTrCP and 32P-substrate at the concentrations specified was incubated in the presence of 0.1 m sodium glutamate for 10 min at room temperature. Finally, the above two reaction mixtures were combined (in a final volume of 10 μl) and incubated at 37 °C for times as indicated. The reaction products were visualized by autoradiography after separation by 4–20% SDS-PAGE, and the levels of input substrate and products were quantified by phosphorimaging.
Competition
32P-IκBα-(1–54)-Ub (0.5pmol) and SCFβTrCP1 (1.5pmol) were used to prepare the E3-substrate-Ub complex. Cdc34 (1.3 μm) and Ub (2 μm) were used to prepare E2∼Ub. After combining the two mixtures, the reaction was incubated for 1 min at 37 °C before the addition of Cdc34-UbCys (22) (0.5 μl; 18× excess over Cdc34) or H2O (0.5 μl). The reaction was chased at 37 °C for times as indicated.
Elongation of β-Catenin Linked with Ub Chains of Defined Length
Approach I
We devised a two-step assay to prepare β-catenin linked with Ub chains of defined length. In the first step, 32P-labeled β-catenin (2 pmol) and SCFβTrCP2 (∼2 pmol) were used to prepare the E3-substrate complex. UbcH5c (4 μm) and Ub (30 μm), Lys-48-Ub4 (5 μm), or Lys-63-Ub4 (5 μm) were used for the E2 charging reaction. After mixing the two mixtures (in a final volume of 10 μl), incubation was at 37 °C for 5, 30, or 90 min to prepare β-cat-Ub1, β-Lys-63-cat-Ub4, or β-Lys-48-cat-Ub4, respectively. The resulting mixture was heated at 65 °C for 5 min to inactivate the enzymes.
In the second step, the heat-treated mixture was combined with preformed Cdc34∼Ub charging mix (17 μm E2 and 20 μm Ub) and SCFβTrCP2 (∼2pmol) to a final volume of 15 μl. The incubation was at 37 °C for times as indicated. β-cat-Ub8 was prepared similarly as β-cat-Ub4 except that an additional mixture (5 μl) containing Cdc34 (6 μm) charged with Lys-48-Ub4 (5 μm) was added, and the entire reaction was incubated at 37 °C for 90 min.
Approach II
32P-Labeled β-catenin (2 pmol) and Nedd8-SCFβTrCP (∼2 pmol) were used to prepare the E3-substrate complex. UbcH5c (1.2 μm) and Ub (10 μm), Lys-48-Ub4 (10 μm), or Lys-63-Ub4 (10 μm) were used for the E2 charging reaction (5 μl). After mixing the two mixtures (in a final volume of 10 μl), incubation was at 37 °C for 5, 30, or 60 min to prepare β-cat-Ub1, β-Lys-63-cat-Ub4, or β-Lys-48-cat-Ub4, respectively.
One-fifth of the above reaction mixture (2 μl) was mixed with Nedd8-SCFβTrCP (∼2 pmol; in 3 μl) and Cdc34∼Ub K0 (20 μm Cdc34, 40 μm Ub K0; in 5 μl) to a final volume of 10 μl. Incubation was at 37 °C for times as indicated. Note that the concentration of Ub-K0 was 20× greater than Ub, Lys-48-Ub4, or Lys-63-Ub4, and the concentration of Cdc34 was 83× more than UbcH5c.
Di-Ub Synthesis
Multiple Rounds
For the experiment shown in Fig. 4A, Cdc34∼S∼32P-Ub was prepared as described above except with 32P-PK-Ub (1.7 μm 32P-PK-Ub, 0.5 μm Cdc34, in 5 μl). The resulting mixture was then chased (in a final volume of 10 μl) with human native Ub (43 μm in 10 μl) and ROC1-CUL1-(411–776) in the amount indicated in the presence or absence of Nedd8 modification. The incubation was at 37 °C for 10 min.
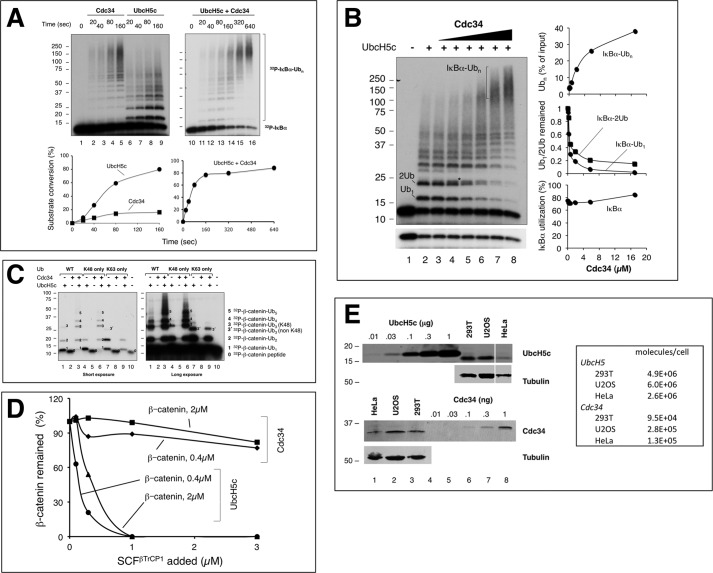
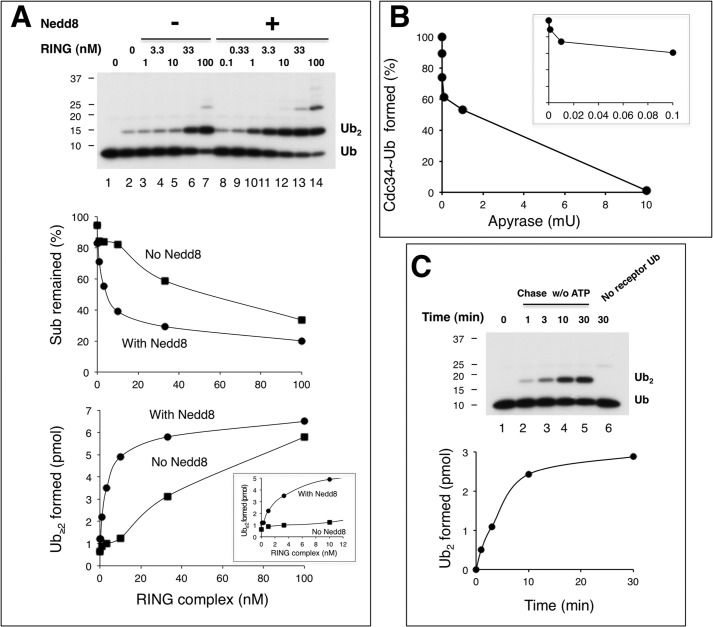
FIGURE 4.
ROC1-CUL1-Nedd8 acts catalytically, converting Cdc34∼Ub to di-Ub. Multiple (panel A) and single (panel C) round di-Ub synthesis reactions were performed as described under “Experimental Procedures.” RING complex refers to ROC1-CUL1-(411–776). The inset graph in panel A reveals the effects of Nedd8 at low concentrations of the RING complex. Panel B shows that apyrase blocks the formation of E1∼Ub and Cdc34∼Ub. The Cdc34∼S∼32P-Ub (5 μl) was prepared with 1.7 μm 32P-PK-Ub, 0.05 μm E1, and 0.5 μm Cdc34. Increasing amounts of apyrase, as indicated, was added to the mix before 32P-PK-Ub, and the incubation was at 37 °C for 1min. The inset graph in panel B reveals the effects of apyrase at low concentrations. mU, milliunits.
For the experiment as described in Fig. 8, B and C, Cdc34∼S∼32P-Ub was prepared as described above except with 32P-PK-Ub (1.7 μm in 5 μl). The resulting mixture was then chased (in a final volume of 10 μl) with ROC1/Rbx1-CUL1 (0.4 μm) and human Ub, or Lys-48-Ub4, or Lys-63-Ub4 in concentrations as specified. The incubation was at 37 °C for 10 min.
Single Round
For the experiment as described in Fig. 4C, Nedd8-ROC1-CUL1-(411–776) was prepared (5 μl; 10 nm RING complex). Cdc34∼S∼32P-Ub was prepared (5 μl; 1.7 μm 32P-PK-Ub, 0.5 μm Cdc34). Each mixture was treated with apyrase (30 milliunits) at 37 °C for 1 min. The two resulting mixes were then combined for further incubation at 37 °C for times as indicated.
RESULTS
Optimization of in Vitro Ubiquitination of IκBα and β-Catenin by SCFβTrCP, UbcH5c, and Cdc34
Previously, we developed an in vitro ubiquitination system employing IκBα 1–54 as the substrate and SCFβTrCP2 as the E3 and using UbcH5c as the initiator E2, which predominantly catalyzes mono-ubiquitination and then hands off the reaction to Cdc34 for polyubiquitination (19). We recently observed that sodium glutamate stimulated the ubiquitination of IκBα 1–54 (Fig. 1, A and D). Although the exact mechanism remains to be elucidated, sodium glutamate likely reduces nonspecific interactions between SCFβTrCP, UbcH5c, and substrate in a manner that inhibits catalysis. Sodium glutamate at high concentrations, however, inhibited the Cdc34 activity (Fig. 1, B and D). The optimal concentration of sodium glutamate was found at 0.1 m, a condition in which a majority of the IκBα 1–54 input substrate was converted to high molecular weight products in the presence of both UbcH5c and Cdc34 (Fig. 1, C and D).
This study also employed the β-catenin peptide substrate, which contains a phospho-degron nearly identical to IκBα, with chemically incorporated phosphoserine residues required for binding to βTrCP and a single lysine residue that acts as the receptor for Ub conjugation (10, 18). We determined the catalytic parameters of the ubiquitination of IκBα 1–54 and β-catenin by SCFβTrCP2 and UbcH5c, with Kcat/Km values of 41 min−1/m−1 (Fig. 1E) and 40 min−1/m−1 (Fig. 1F), respectively.
Properties of the in Vitro Ubiquitination of IκBα and β-Catenin by SCFβTrCP, UbcH5c, and Cdc34
With the optimized ubiquitination system we examined the reaction properties of SCFβTrCP, UbcH5c, and Cdc34 with IκBα or β-catenin as substrate. Time course experiments revealed that 1) UbcH5c displayed efficient substrate conversion with a rate that was at least 4-fold greater than that observed in reactions with Cdc34 (Fig. 2A), 2) whereas UbcH5c produced Ub conjugates of small sizes (Fig. 2A, lanes 6–9), Cdc34 yielded high molecular weight products centered around 150 kDa (Fig. 2A, lanes 1–5), and 3) UbcH5c and Cdc34 in combination catalyzed ubiquitination with characteristics of both E2s, converting the substrate rapidly in a rate similar to the UbcH5c reaction and producing Cdc34-patterned Ub conjugates of extensive length (Fig. 2A, lanes 10–16). Further E2 titration experiments showed that Cdc34 efficiently transformed UbcH5c-produced, mono-ubiquitinated substrate into high molecular weight species in a dose-dependent fashion while leaving the input substrate levels unchanged (Fig. 2B).
The results of reactions using the β-catenin peptide substrate were similar to those with IκBα 1–54. UbcH5c converted the β-catenin substrate into mono-ubiquitinated species quantitatively (Fig. 2C, left, lane 1). Cdc34, alone inactive (Fig. 2C, left, lane 2), drove nearly all mono-ubiquitinated species into chains (Fig. 2C, left, lane 3), which were of Lys-48-linkage specificity exclusively (Fig. 2C, left, compare lanes 3, 6, and 9). Finally, the differential roles for UbcH5c in mono-ubiquitination and for Cdc34 in Ub chain elongation were confirmed with reactions employing a range of concentrations of SCFβTrCP1 and β-catenin (Fig. 2D).
Taken together these results have substantiated the dual E2 model for efficiently driving the polyubiquitination of SCF substrates. The consecutive actions by UbcH5c and Cdc34 ensure rapid substrate mono-ubiquitination and processive Ub chain elongation.
Immunoblot analysis revealed that UbcH5 is more abundant than Cdc34 in cells by a factor of 27 on average (Fig. 2E). It should also be noted that UbcH5c is capable of polyubiquitination of IκBα at higher concentrations of E2 with longer periods of incubation (Figs. 1A and 2A). However, as demonstrated by previous work (19), the UbcH5c-generated polyubiquitin chains are of mixed linkages, in sharp contrast to those made by Cdc34 that has exclusive Lys-48 specificity. These studies have raised intriguing possibilities that unrecognized cellular mechanisms might exist to restrain the abundant UbcH5 activities in the elongation phase of the ubiquitination reaction, thereby preventing non-Lys-48-linked polyubiquitination of proteasomal substrates.
SCF-Substrate Engages in Dynamic Interactions with Cdc34 for Ub Chain Elongation
Previous work has revealed electrostatic interactions between the Cdc34 acidic C terminus and CUL1 basic canyon that enable rapid assembly and disassembly of the E2-SCF complex (24). However, the dynamics between SCF-substrate and Cdc34∼Ub have yet to be determined in the context of the ubiquitination reaction directly. To this end we employed a competition-based strategy using a previously established Ub chain elongation system with the IκBα-Ub fusion substrate (19). It was determined that the elongation reaction by Cdc34 with IκBα-Ub displayed Kcat/Km of 61min−1/m−1 (Fig. 3A), a value greater than that for the reaction with IκBα and UbcH5c (Fig. 1E). These findings underscore a capability by IκBα-Ub to function as a receptor for directing SCFβTrCP–dependent Ub chain assembly reactions by Cdc34.
To evaluate the stability of the SCFβTrCP-IκBα-Ub/Cdc34∼Ub complex, we developed a scheme using Cdc34-UbCys in which an Ub variant containing a cysteine substitution at position 76 has been disulfide-bonded to cysteine 93 in human Cdc34 (32). Cdc34-UbCys is a structural mimic of Cdc34∼Ub but cannot be used for ubiquitination (22) (Fig. 3B). In this protocol excess Cdc34-UbCys was added after the elongation reaction was initiated for 1 min. If the E3-substrate-Ub-Cdc34 complex were unstable, the excess E2 competitor would out-compete Cdc34 for binding to the SCFβTrCP-substrate-Ub complex, thereby inhibiting the ubiquitination of radioactive IκBα-Ub. Alternatively, a stable E3-substrate-Ub-Cdc34 complex would be resistant to challenge by the E2 competitor, allowing the E3-substrate-bound radioactive IκBα-Ub to be converted to elongation products. The results revealed that Cdc34-UbCys effectively inhibited ubiquitination (Fig. 3C; Fig. 3D, compare lanes 2 and 4). Even when this competitor was added 1 min post reaction initiation, Cdc34-UbCys still inhibited ubiquitination by 90% (Fig. 3D, compare lanes 7 and 9). These findings demonstrated that Cdc34-UbCys was able to compete with Cdc34∼Ub for interactions with the SCFβTrCP-IκBα-Ub complex regardless of whether the E3-substrate-Ub complex was allowed to encounter E2∼Ub first. Therefore, Cdc34∼Ub is associated with the SCFβTrCP-IκBα-Ub complex in a dynamic fashion.
The observed dynamic association between SCF-substrate-Ub and Cdc34∼Ub suggests that the SCF ROC1 RING-based Ub ligase acts to facilitate Ub-Ub conjugation in a catalytic fashion. To address this possibility, we measured di-Ub synthesis by Cdc34 with the ROC1-CUL1 complex in the presence or absence of modification by Nedd8. The results revealed that neddylated ROC1-CUL1 in 0.01 or 0.1 pmol yielded 2.2 or 4.9 pmol of di-Ub product within 10 min of incubation (Fig. 4A), suggesting that ROC1-CUL1-Nedd8 acts catalytically. In addition, it was observed that in the low range of the ROC1-CUL1 complex used, Nedd8 modification stimulated di-Ub synthesis by >10-fold (Fig. 4A, bottom graph). These findings underscore a critical role for Nedd8 in the rapid turnover of Cdc34∼Ub to ubiquitinated products. To confirm these observations in single turnover conditions, we employed apyrase that potently hydrolyzes ATP. Using apyrase at a concentration that eliminated Ub conjugation by E1 and Cdc34 (Fig. 4B), it was observed that 0.1pmol of ROC1-CUL1-Nedd8 yielded 2.5 pmol of di-Ub product within 10 min of incubation (Fig. 4C), reaffirming its ability to act catalytically. Note that in this single turnover assay, the yield of di-Ub was low in comparison to multiple-round reactions (Fig. 4A), presumably due to the treatment of apyrase (1 min at 37 °C) that likely caused substantial dissociation of Cdc34∼Ub.
Altogether the above results suggest that upon receiving a Ub moiety by the action of UbcH5c, the neddylated SCFβTrCP-substrate core complex is engaged in dynamic interactions with Cdc34 leading to repeated cycles of chain elongation, ultimately forming Ub polymers of suitable length to trigger proteasomal degradation.
Strategies to Investigate Ub Conjugation on a Growing Chain
Little is known whether and how a growing Ub chain might impact the rate of Ub conjugation by E3/E2 interactions. To this end we have developed Ub chain intermediates, having β-catenin linked with Ub chains of defined length, including one or four Ub moieties of Lys-48 or other linkage by SCFβTrCP and UbcH5c (Figs. 5A and Fig. 7A; for details see “Experimental Procedures”). To examine the effects of these intermediates on Ub conjugation, we have employed two protocols. Approach I employed a heat denaturation step to inactivate all enzymes (Fig. 5A). Given that the β-catenin peptide is unstructured (18) and that Ub has been shown to be heat-stable (25) and remains folded up to 85 °C (26), the newly synthesized β-cat linked with one or four Ub moieties is expected to refold properly and can serve as a substrate for elongation catalyzed by a set of freshly added enzymes (E1, Cdc34, and SCFβTrCP2). Fig. 5B validated this procedure by testing elongation of β-cat-Ub1 after heat treatment ranging from 56 to 95 °C. In all cases the addition of fresh E1, Cdc34, Ub, and SCFβTrCP2 resulted in conversion of UbcH5c-generated, mono-ubiquitinated species into high molecular weight products (Fig. 5B, lanes 4–11). Importantly, the elongation efficiency with these heat-denatured-then-re-natured substrates was similar to those prepared in the absence of heat treatment (Fig. 5B, compare lanes 2 and 3 with lanes 4–11), demonstrating that the re-natured Ub-linked β-catenin was fully functional for elongation by Cdc34. To generate β-cat-Ub8, β-cat-Ub4 was first formed by SCFβTrCP2 and UbcH5c∼Ub4 then heat-treated. Subsequently, fresh SCFβTrCP2 and Cdc34∼Ub4 were added, resulting in formation of β-cat-Ub8 (Fig. 5C, lanes 3–6).
Approach II avoids the heat denaturation step. Instead, a dilution protocol was developed to allow excess Cdc34∼Ub to react with β-cat conjugated with Ub or Ub chain (Fig. 5D; for details see “Experimental Procedures”). Fig. 5E shows the distribution of the reaction products as well as input substrate after initial UbcH5c reaction. Importantly, of the reaction mixtures, β-cat-Ub1,β-cat-Lys-48-Ub4, and β-cat-Lys-63-Ub4 were all major species in approximately equal abundance (∼50%), thus allowing comparison of conjugation rates with these as substrates. In addition, β-cat-Lys-63-Ub8 and β-cat-Lys-63-Ub12 were visible (Fig. 5E, lane 3), which lead to elongation products as analyzed in Fig. 7.
Increasing Lys-48-Ub Chain Length on a Substrate Results in Decreased Conjugation Rates
We first compared Ub conjugation rates by performing time course experiments with preformed β-cat-Ub1 or β-cat-Lys-48-Ub4 as the substrate using the heat denaturation protocol (Approach I, Fig. 5A). As shown, the protein levels of β-cat-Ub1 decreased with time and declined by as much as 90% at the 10-min mark, with concomitant accumulation of high molecular weight conjugates (Fig. 6A, lanes 2–5). In contrast, the reaction with β-cat-Lys-48-Ub4 proceeded in a pace that was ∼3× slower than β-cat-Ub1 (Fig. 6A, lanes 7–11; graph).
We next compared the elongation of β-cat-Lys-48-Ub4 and β-cat-Lys-48-Ub8 (Fig. 6B). As shown in Fig. 5C (lane 6), β-cat-Lys-48-Ub4 and β-cat-Lys-48-Ub8 migrated distinctly. We reasoned that elongation with Lys-less Ub (Ub-K0) in place of the wild type would allow for the assessment of substrate reduction as well as the accumulation of ubiquitination product differing from the input substrate in gel mobility. The results revealed that both β-cat-Lys-48-Ub4 and β-cat-Lys-48-Ub8 decreased as a function of time, coinciding with the appearance of Ub K0-conjugated products (Fig. 6B). Analysis of the quantitative differences indicates that β-cat-Lys-48-Ub8 exhibited a rate ∼2–3 times slower than β-cat-Lys-48-Ub4 (Fig. 6B, graph). Note that experiments with Approach II, which does not involve the heat denaturation step (Fig. 5D), confirmed a significantly slowed reaction rate with β-cat-Lys-48-Ub4 in comparison to β-cat-Ub1 (Fig. 6C), thereby excluding the possibility that heat denaturation rendered β-cat-Lys-48-Ub4 less active for Ub conjugation.
The above findings have led to a conclusion that increasing Lys-48-Ub chain length on a substrate slowed the rate of additional Ub conjugation by Cdc34. Thus, Lys-48-Ub chains of a length greater than four appear to restrict the actions by SCFβTrCP2/Cdc34 for further elongation.
SCFβTrCP/Cdc34 Elongates β-Catenin-Lys-63-Ub4 More Efficiently Than β-Catenin-Lys-48-Ub4
Several possibilities could have accounted for the decreased rate of ubiquitination of β-catenin linked with Lys-48-Ub chains by Cdc34. The covalent linkage of Lys-48-tetra-Ub to β-catenin might 1) significantly weaken the interaction of substrate with the E3 complex, 2) approach an inhibitory threshold determined by chain length, or 3) restrict the actions by SCFβTrCP2/Cdc34 due to the conformation of the Lys-48-Ub4 being inhibitory. To address these possibilities, we sought to compare elongation reactions with β-cat-Lys-48-Ub4 or β-cat-Lys-63-Ub4 as the substrate. Lys-63-Ub4 possesses the same number of Ub moieties as Lys-48-Ub4 but differs substantially from Lys-48-Ub4 in conformation. It has been well established that Lys-48-Ub4 exhibits compact conformations, whereas Lys-63-Ub4 adopts an open, extended state (27–29).
Unlike β-cat-Lys-48-Ub4, which contains a single receptor Lys-48, β-cat-Lys-63-Ub4 has four Lys-48 residues that could potentially act as a receptor lysine (Fig. 7A). The results of time course experiments using Approach I (heat denaturation, Fig. 5A), revealed that Cdc34 converted β-cat-Lys-63-Ub4 about 3× faster than β-cat-Lys-48-Ub4 to ubiquitinated products (Fig. 7B, compare lanes 2–6 and lanes 7–11; graph). In keeping with its rapid turnover, β-cat-Lys-63-Ub4 accumulated Ub chains at levels and lengths that were far greater than that of β-cat-Lys-48-Ub4 (Fig. 7B). It was also noted that input substrate preparations contained very large conjugates including β-cat-Lys-48-Ub8 and β-cat-Lys-48-Ub12 (Fig. 7B, lane 2) as well as β-cat-Lys-63-Ub8 and β-cat-Lys-63-Ub12 (Fig. 7B, lane 7). It should also be reminded that β-cat contains a single N-terminally located lysine, thereby providing only one receptor for a polyubiquitin chain. Evidently, reactions with both β-cat-Lys-63-Ub8 and β-cat-Lys-63-Ub12 showed significantly faster turnover rates and lengthier products than those with β-cat-Lys-48-Ub8 and β-cat-Lys-48-Ub12 (Fig. 7B, compare lanes 2–6 and lanes 7–11).
We tested the ability of Cdc34 to use multiple Ub Lys-48 residues within β-cat-Lys-63-Ub4 by performing reactions with Ub-K0 in place of the wild type. The results showed rapid reduction of the input β-cat-Lys-63-Ub4 and accumulation of distinct products corresponding to β-cat-Lys-63-Ub4-Ub K01, β-cat-Lys-63-Ub4-Ub K02, and β-cat-Lys-63-Ub4-Ub K03 (Fig. 7C). Quantification showed that Cdc34 elongated the β-cat-Lys-63-Ub4 in a rate similarly to that observed with the β-cat-Ub1 substrate (compare Fig. 7C, graph, to Fig. 6A, graph). In addition, in contrast to β-cat-Lys-48-Ub8–16 that showed a slowed reaction rate with Ub K0 (Fig. 6B), β-cat-Lys-63-Ub8–16 were used very effectively (Fig. 7C). Altogether, these results demonstrate that SCFβTrCP/Cdc34 elongates β-catenin linked with Lys-63-polyubiquitin chains more efficiently than those of the Lys-48 linkage.
Additional Properties Revealed by Elongation Reactions with Ub Chains as Substrate
The Ub-K0 conjugation analysis revealed additional intriguing reaction properties with β-catenin linked with Lys-63-Ub chains of substantial length up to 16 Ub moieties (Fig. 7C). First, β-cat-Lys-63-Ub8, β-cat-Lys-63-Ub12, and β-cat-Lys-63-Ub16, which were synthesized by SCFβTrCP and UbcH5c, displayed significant heterogeneity in gel mobility (Fig. 7C, lane 1), presumably reflecting a diversity in linkage and in chain branching due to the UbcH5c ability to use a variety of Ub lysine residues for conjugation (19). Second, it was evident that all these substrates reacted with SCFβTrCP/Cdc34 effectively to form distinct conjugates (Fig. 7C). Considering that the product species “d” is about 60 kDa larger than the input substrate β-cat-Lys-63-Ub16 (Fig. 7C, lanes 4 and 5), it suggests that SCFβTrCP/Cdc34 is able to conjugate ∼10 Ub-K0 to 16 possible Lys-48 receptor sites on this substrate.
The results of experiments with Approach II, which does not involve the heat denaturation step (Fig. 5D), confirmed a significantly slowed reaction rate with β-cat-Lys-48-Ub4 in comparison to β-cat-Lys-63-Ub4 (Fig. 7D). In addition, β-cat-Lys-63-Ub4 supported the formation of lengthy products (Fig. 7D). These findings exclude a possibility that the slowed elongation on β-cat-Lys-48-Ub chains results from improper folding of the Lys-48-Ub chain-linked substrates after heat denaturation (Fig. 7, B and C). In all, the above experiments underscored a remarkable flexibility of SCFβTrCP/Cdc34 for locating a receptor lysine.
Given the SCFβTrCP/Cdc34 ability to interact with β-catenin linked with Lys-63 chains ranging from 4 to 16 Ub moieties for elongation, it is unlikely that Ub chain length alone acts as a determining factor that sets an inhibitory threshold to inhibit further chain assembly. These findings are also at odds with the competition hypothesis, suggesting that Ub chains attached to a substrate would weaken its interaction with the E3. In further support of this note, competition experiments revealed comparable affinity between β-cat-Ub1 and β-cat-Lys-48-Ub4 to SCFβTrCP2 (Fig. 8A).
The above findings raised a possibility that Cdc34 might recognize Lys-48-Ub4 poorly, thereby diminishing the elongation rate. To address this question, we compared the conjugation of radioactive Ub (as a donor) to a single Ub or to a Lys-48-Ub4 receptor. The results showed that Cdc34 along with the ROC1-CUL1 complex catalyzed the conjugation reaction more efficiently with the Lys-48-Ub4 receptor than with the single Ub (Fig. 8B). These findings demonstrate an ability of the ROC1 RING-activated Cdc34 to interact with Lys-48-Ub4 with increased efficiency as compared with that observed with single Ub, presumably because Lys-48-Ub4 provides multiple interaction surfaces. Additionally, it was observed that Cdc34 (activated by CUL1-ROC1) catalyzed the ligation of 32P-PK-Ub to Lys-63-Ub4 about 3× more efficiently than to Lys-48-Ub4 (Fig. 8C), in keeping with observations that Cdc34 was able to utilize multiple Lys-48 sites on Lys-63-Ub4 (Fig. 7C). Thus, Lys-48-Ub4 is inhibitory to elongation by Cdc34 only when conjugated to a substrate. Taken together, these results suggest that Lys-48-Ub4 on the SCFβTrCP-bound β-catenin may be in a constrained state that is less optimal for interactions with Cdc34∼Ub for further elongation.
DISCUSSION
Polyubiquitin chains act as signaling polypeptides that upon attachment to a target protein can either commit a substrate to proteolytic destruction or, alternatively, change the substrate protein function. Both of these outcomes can cause profound impact to a variety of intracellular pathways at both the cellular and organismal levels. Using the reconstituted SCFβTrCP/UbcH5c/Cdc34 ubiquitination system, our current study has provided mechanistic insights into polyubiquitination concerning the assembly of substrate, E3 and E2 complexes, and has revealed the impact of Ub chain length and linkage on the kinetic rates of chain elongation.
Establishing an SCF-Substrate Core Complex to Engage in Dynamic Interactions with Cdc34
During elongation the SCFβTrCP-substrate core complex must interact with Cdc34∼Ub to drive Ub conjugation. Our present work suggests that the interactions between SCFβTrCP-IκBα-Ub and Cdc34∼Ub are dynamic. The results of competition experiments showed that a Cdc34∼Ub-mimicking inhibitor effectively outcompeted Cdc34 to block ubiquitination despite that the inhibitor was added after sufficient time was allowed for Cdc34 to encounter the SCFβTrCP-IκBα-Ub complex (Fig. 3). Furthermore, we observed that ROC1-CUL1-Nedd8 acts catalytically in the synthesis of di-Ub products (Fig. 4), suggesting a capability of the Nedd8-linked SCFβTrCP-substrate core complex to rapidly turnover Cdc34∼Ub for chain elongation. It is worth pointing out that the potent stimulatory effect by the Nedd8 in the catalysis of Ub-Ub conjugation (Fig. 4) strongly argues for a direct role of Nedd8-dependent open conformation of the SCF in productive interactions with Cdc34∼Ub in addition to closing the gap between the E3-bound substrate and ROC1 RING protein as proposed previously (9).
Due to recent biochemical and structural studies, details concerning the SCF/Cdc34/Ub interactions have emerged. Quench-flow experiments have revealed that SCF/Cdc34 catalyzes ubiquitination in a progressive manner, with the addition of one Ub at a time (30). SCF appears to engage Cdc34 during the elongation reaction via a two-step mechanism. In the first step, the SCF CUL1 subunit uses its basic canyon to form electrostatic interactions with the acidic C terminus of Cdc34 (24). Upon thiolester formation of Cdc34, the proximity of the SCF Rbx1/ROC1 subunit favors an interaction with the Cdc34∼Ub conjugate (22), which likely promotes Ub transfer to a substrate. In this context, Cdc34 may utilize both the core domain (22) and C terminus (31, 32) for interactions with Ub. A recent study has provided structural evidence suggesting that the RING/E2/Ub interactions position the Ub carboxyl tail in the E2 active site groove for catalysis (33). Our observation on the dynamic association between the Nedd8-linked SCFβTrCP-IκBα-Ub and Cdc34∼Ub suggests that once the Ub transfer is completed, Cdc34 is released from SCF, in turn resetting the system to allow for repeated cycles of Ub transfer.
Restraining Chain Elongation by Lys-48-Ub4
In SCF/Cdc34-catalyzed chain elongation, conjugation of an Ub to the distal Ub Lys-48 requires the assembly of a productive substrate-E3-E2 complex that properly aligns Lys-48 of the receptor Ub to Gly-76 of the E2-linked donor Ub. Before this work, how E3/E2 interacts with growing Ub chains during elongation remained largely unexplored. Our study for the first time probed an SCF/Cdc34-catalyzed elongation reaction by employing preformed β-catenin linked with Lys-48-Ub chains of defined length. The results revealed an inverse correlation between conjugation rates by Cdc34 and chain length. β-cat-Lys-48-Ub4 exhibits a 3-fold decrease of conjugation rate in comparison to β-cat-Ub1 (Fig. 6A). Increasing Ub chain length from four to eight caused a further drop in conjugation rate by 2–3-fold (Fig. 6B). Taken together, these findings suggest that a Lys-48-Ub chain of a length greater than four restricted the actions by SCFβTrCP/Cdc34 for further elongation.
It may appear paradoxical that the proposed Lys-48-Ub4-slowing-elongation mechanism would predict accumulation of tetra-ubiquitinated species, which was not observed in reactions using monomeric Ub (Fig. 2). However, it should be noted that in such reactions the level of an ubiquitinated product, designated as substrate-Ubn, depends on the transfer rates of [substrate-Ubn−1] → [substrate-Un] as well as [substrate-Ubn] → [substrate-Ubn+1]. In fact, Deshaies and co-workers (30) have employed quench flow to show that β-catenin-Ub4 exhibits a 2-fold drop of transfer rate in comparison to β-catenin-Ub1. Taken together, we believe that β-catenin linked with Lys-48-Ub4 is more slowly elongated than β-catenin-Ub1.
Perhaps the most striking observation of this work is the SCFβTrCP/Cdc34 ability to interact with β-catenin linked with Lys-63 chains ranging from 4 to 16 Ub moieties for elongation (Fig. 7, C and D). Given their heterogeneous gel mobilities (Fig. 7C, lane 1), β-catenin-Lys-63-Ub4–16 are likely the mixtures of branched Ub chains connected through different linkages. The fact that SCFβTrCP/Cdc34 readily finds Lys-48 residues within these complex structures points to a remarkable flexibility of this E3/E2 pair for locating a receptor Ub. It thus appears that SCF, Cdc34∼Ub, and/or substrate-linked Lys-63-Ub chains have considerable rotational freedom and the proper positioning of these components can be readily established to promote catalysis. However, such plasticity is at odds with the severely compromised ability exhibited by SCFβTrCP/Cdc34 to elongate β-cat-Lys-48-Ub4 (Fig. 6).
A possible explanation for the above paradox lies in the topological difference between β-cat-Lys-48-Ub4 and β-cat-Lys-63-Ub4. Previous studies have reported three high resolution structures of Lys-48 tetra-Ub, which are formed at different pH values (27–29). In one such structural model built with both crystallography and NMR data, the Lys-48 tetra-Ub chain at near physiological pH (pH 6.7) adopts a conformation consisting of a dimer of di-Ubs, with each di-Ub subunit in the closed conformation (28). In this structure, the hydrophobic patch of each Ub, comprising residues Leu-8, Ile-44, and Val-70, is located at the di-Ub interface. Another crystal structure of the Lys-48-tetra-Ub at acidic pH (5.0) has revealed an extremely compact conformation that is stabilized by extensive intrachain, hydrogen bonding, and dipolar interactions between Ub subunits (27). This conformation is supported by a recent crystallography and NMR study on a Lys-48-di-Ub structure (34). Furthermore, using single-molecule fluorescence resonance energy transfer, it was observed that Lys-48-linked di-Ub adopts predominantly compact conformations (35). However, additional conformations of Lys-48-tetra-Ub may also exist (36, 37). Thus, although Lys-48-tetra-Ub overall is packed in a compact state, its topology is dynamic. Lys-48-tetra-Ub can adopt closed or open conformations that are stabilized by hydrophobic or polar intrachain interactions between Ub subunits, respectively. In contrast, Lys-63-tetra-Ub exists in an open and extended conformation (29).
To account for how Lys-48-tetra-Ub might restrain chain elongation, we have considered two possibilities. One possibility is that the distal Ub hydrophobic patch, which is buried in the di-Ub interface within the Lys-48-Ub4 in closed conformation, is required for elongation. It is also conceivable that Ub contains an unidentified Lys-48 recognition motif critical for conjugation. In the compact form of Lys-48-Ub4, either intrachain hydrophobic or polar interactions between Ub subunits may render the distal Ub Lys-48 recognition motif inaccessible. In either scenario, the elongation of Lys-48-Ub4 may require relaxation of such structural constraints that could be rate-limiting. In this context, Lys-48-Ub4 on the SCFβTrCP-bound β-catenin could be more populated in a locked compact conformation. As a consequence, the distal Ub hydrophobic patch and/or Lys-48 recognition motif are inaccessible for interactions with Cdc34∼Ub, resulting in slowed conjugation as observed in Fig. 6. In contrast, the open and extended conformation of Lys-63-Ub4 presents no restriction to SCFβTrCP/Cdc34 for finding Lys-48, thereby explaining the robust ubiquitination seen with Lys-63 chains in this study (Fig. 7). Intriguingly, the restrained elongation by Lys-48-tetra-Ub appears context-dependent, as substrate-free Lys-48-Ub4 was elongated at a rate similar to that observed with monomeric Ub receptor (Fig. 8, B and C). It is possible that substrate-free Lys-48-Ub4 may be more readily “opened” for interactions with Cdc34∼Ub and/or SCF RING-CUL1 complex.
It should be noted that the current work does not impose a “rule” of Lys-48 poly-ubiquitination about n = 4 being a cutoff. The possible role of Lys-48-Ub4 is discussed extensively because of its well documented high resolution structures and its correlation with slowed ubiquitination. In addition, conclusive evidence for a role of conformation in regulating Lys-48-Ub chain elongation requires studies employing mutant forms of Ub polymers with altered conformation. This task would likely involve mutational disruption of the Ub hydrophobic patch, which is required to establish the closed conformation of Lys-48-tetra-Ub (28). However, such alteration would also diminish the Ub capability to interact with E1 and E2, thereby perturbing its function as a donor in conjugation (38) and limiting the incorporation of the mutant Ub into the tetrameric form using the currently widely accepted, enzyme-based procedure (39). Future work is required to overcome such technical difficulty.
An Overall Model for SCF-directed Polyubiquitination
Altogether, these studies suggest a model for SCF-directed substrate polyubiquitination (Fig. 8D). It follows that the SCF E3 forms a complex with a substrate such as β-catenin. Upon mono-ubiquitination by UbcH5, the Nedd8-linked, E3-substrate-Ub complex is engaged in dynamic interactions with Cdc34∼Ub leading to repeated cycles of chain elongation. The elongation begins with a fast phase in which Cdc34 rapidly catalyzes conjugation of an Ub to SCF-substrate-Ub. As the chain is extended to four Ub molecules, hydrophobic or polar interactions between Ub subunits drive the packing of the chain into a compact conformation. Such topology may severely limit the accessibility of E3/E2 to the distal Ub Lys-48 and result in slowed elongation. This in-elongation-Lys-48-chain-packing may offer a self-restraining mechanism to inhibit conjugation, leading to eventual chain termination.
Acknowledgments
We are grateful to R. Deshaies, B. Schulman, and N. Zheng for reagents. We thank J. Hurwitz for constructive suggestions during the course of this work. We thank I. Tappin and F. Du for assistance in the preparation of baculoviruses-infected insect cells expressing SCFβTrCP1 components.
This work was supported, in whole or in part, by National Institutes of Health Grants GM61051 and CA095634 (Public Health Service; to Z.-Q. P.). This work was also supported by Canadian Institutes of Health Research Grant MOP-14606 (to G. S. S.).
- Ub
- CRL, Cullin-RING E3 Ub ligase
- CUL1
- Cullin 1
- SCF
- Skp1-CUL1-F box protein
- CAND1
- cullin-associated and neddylation-dissociated-1.
REFERENCES
- 1. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2. Petroski M. D., Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 3. Sarikas A., Hartmann T., Pan Z. Q. (2011) The cullin protein family. Genome Biol. 12, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zheng N., Schulman B. A., Song L., Miller J. J., Jeffrey P. D., Wang P., Chu C., Koepp D. M., Elledge S. J., Pagano M., Conaway R. C., Conaway J. W., Harper J. W., Pavletich N. P. (2002) Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 [DOI] [PubMed] [Google Scholar]
- 5. Skaar J. R., Pagan J. K., Pagano M. (2009a) SnapShot. F box proteins I. Cell 137, 1160–1160.e1 [DOI] [PubMed] [Google Scholar]
- 6. Skaar J. R., D'Angiolella V., Pagan J. K., Pagano M. (2009b) SnapShot. F Box Proteins II. Cell 137, 1358–1358 [DOI] [PubMed] [Google Scholar]
- 7. Pan Z. Q., Kentsis A., Dias D. C., Yamoah K., Wu K. (2004) Nedd8 on cullin. Building an expressway to protein destruction. Oncogene 23, 1985–1997 [DOI] [PubMed] [Google Scholar]
- 8. Yamoah K., Oashi T., Sarikas A., Gazdoiu S., Osman R., Pan Z. Q. (2008) Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1s C-terminal tail. Proc. Natl. Acad. Sci. U.S.A. 105, 12230–12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duda D. M., Borg L. A., Scott D. C., Hunt H. W., Hammel M., Schulman B. A. (2008) Structural insights into NEDD8 activation of cullin-RING ligases. Conformational control of conjugation. Cell 134, 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saha A., Deshaies R. J. (2008) Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cope G. A., Suh G. S., Aravind L., Schwarz S. E., Zipursky S. L., Koonin E. V., Deshaies R. J. (2002) Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298, 608–611 [DOI] [PubMed] [Google Scholar]
- 12. Read M. A., Brownell J. E., Gladysheva T. B., Hottelet M., Parent L. A., Coggins M. B., Pierce J. W., Podust V. N., Luo R. S., Chau V., Palombella V. J. (2000) Nedd8 modification of cul-1 activates SCF(β(TrCP))-dependent ubiquitination of IκBα. Mol. Cell. Biol. 20, 2326–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pierce N. W., Lee J. E., Liu X., Sweredoski M. J., Graham R. L., Larimore E. A., Rome M., Zheng N., Clurman B. E., Hess S., Shan S. O., Deshaies R. J. (2013) Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell 153, 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu S., Zhu W., Nhan T., Toth J. I., Petroski M. D., Wolf D. A. (2013) CAND1 controls in vivo dynamics of the cullin 1-RING ubiquitin ligase repertoire. Nat. Commun. 4, 1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zemla A., Thomas Y., Kedziora S., Knebel A., Wood N. T., Rabut G., Kurz T. (2013) CSN- and CAND1-dependent remodelling of the budding yeast SCF complex. Nat. Commun. 4, 1641. [DOI] [PubMed] [Google Scholar]
- 16. Tron A. E., Arai T., Duda D. M., Kuwabara H., Olszewski J. L., Fujiwara Y., Bahamon B. N., Signoretti S., Schulman B. A., DeCaprio J. A. (2012) The glomuvenous malformation protein Glomulin binds Rbx1 and regulates cullin RING ligase-mediated turnover of Fbw7. Mol. Cell 46, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duda D. M., Olszewski J. L., Tron A. E., Hammel M., Lambert L. J., Waddell M. B., Mittag T., DeCaprio J. A., Schulman B. A. (2012) Structure of a glomulin-RBX1-CUL1 complex. Inhibition of a RING E3 ligase through masking of its E2-binding surface. Mol. Cell 47, 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu G., Xu G., Schulman B. A., Jeffrey P. D., Harper J. W., Pavletich N. P. (2003) Structure of a β-TrCP1-Skp1-β-catenin complex. Destruction motif binding and lysine specificity of the SCF(β-TrCP1) ubiquitin ligase. Mol. Cell 11, 1445–1456 [DOI] [PubMed] [Google Scholar]
- 19. Wu K., Kovacev J., Pan Z. Q. (2010) Priming and extending. An UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol. Cell 37, 784–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gazdoiu S., Yamoah K., Wu K., Pan Z. Q. (2007) Human Cdc34 employs distinct sites to coordinate attachment of ubiquitin to a substrate and assembly of polyubiquitin chains. Mol. Cell. Biol. 27, 7041–7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan P., Fuchs S. Y., Chen A., Wu K., Gomez C., Ronai Z., Pan Z. Q. (1999) Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol. Cell 3, 527–533 [DOI] [PubMed] [Google Scholar]
- 22. Spratt D. E., Wu K., Kovacev J., Pan Z. Q., Shaw G. S. (2012) Selective recruitment of an E2∼ubiquitin complex by an E3 ubiquitin ligase. J. Biol. Chem. 287, 17374–17385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu K., Chen A., Tan P., Pan Z. Q. (2002) The Nedd8-conjugated ROC1-CUL1 core ubiquitin ligase utilizes Nedd8 charged surface residues for efficient polyubiquitin chain assembly catalyzed by Cdc34. J. Biol. Chem. 277, 516–527 [DOI] [PubMed] [Google Scholar]
- 24. Kleiger G., Saha A., Lewis S., Kuhlman B., Deshaies R. J. (2009) Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell 139, 957–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ciehanover A., Hod Y., Hershko A. (1978) A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem. Biophys. Res. Commun. 81, 1100–1105 [DOI] [PubMed] [Google Scholar]
- 26. Safadi S. S., Shaw G. S. (2007) A disease state mutation unfolds the parkin ubiquitin-like domain. Biochemistry 46, 14162–14169 [DOI] [PubMed] [Google Scholar]
- 27. Cook W. J., Jeffrey L. C., Kasperek E., Pickart C. M. (1994) Structure of tetraubiquitin shows how multiubiquitin chains can be formed. J. Mol. Biol. 236, 601–609 [DOI] [PubMed] [Google Scholar]
- 28. Eddins M. J., Varadan R., Fushman D., Pickart C. M., Wolberger C. (2007) Crystal structure and solution NMR studies of Lys-48-linked tetraubiquitin at neutral pH. J. Mol. Biol. 367, 204–211 [DOI] [PubMed] [Google Scholar]
- 29. Datta A. B., Hura G. L., Wolberger C. (2009) The structure and conformation of Lys-63-linked tetraubiquitin. J. Mol. Biol. 392, 1117–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pierce N. W., Kleiger G., Shan S. O., Deshaies R. J. (2009) Detection of sequential polyubiquitylation on a millisecond timescale. Nature 462, 615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Choi Y. S., Wu K., Jeong K., Lee D., Jeon Y. H., Choi B. S., Pan Z. Q., Ryu K. S., Cheong C. (2010) The human Cdc34 carboxyl terminus contains a non-covalent ubiquitin binding activity that contributes to SCF-dependent ubiquitination. J. Biol. Chem. 285, 17754–17762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Spratt D. E., Shaw G. S. (2011) Association of the disordered C terminus of CDC34 with a catalytically bound ubiquitin. J. Mol. Biol. 407, 425–438 [DOI] [PubMed] [Google Scholar]
- 33. Plechanovová A., Jaffray E. G., Tatham M. H., Naismith J. H., Hay R. T. (2012) Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lai M. Y., Zhang D., Laronde-Leblanc N., Fushman D. (2012) Structural and biochemical studies of the open state of Lys-48-linked diubiquitin. Biochim. Biophys. Acta 1823, 2046–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ye Y., Blaser G., Horrocks M. H., Ruedas-Rama M. J., Ibrahim S., Zhukov A. A., Orte A., Klenerman D., Jackson S. E., Komander D. (2012) Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature 492, 266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Phillips C. L., Thrower J., Pickart C. M., Hill C. P. (2001) Structure of a new crystal form of tetraubiquitin. Acta Crystallogr. D. Biol. Crystallogr. 57, 341–344 [DOI] [PubMed] [Google Scholar]
- 37. Hirano T., Serve O., Yagi-Utsumi M., Takemoto E., Hiromoto T., Satoh T., Mizushima T., Kato K. (2011) Conformational dynamics of wild-type Lys-48-linked diubiquitin in solution. J. Biol. Chem. 286, 37496–37502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wickliffe K. E., Lorenz S., Wemmer D. E., Kuriyan J., Rape M. (2011) The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Piotrowski J., Beal R., Hoffman L., Wilkinson K. D., Cohen R. E., Pickart C. M. (1997) Inhibition of the 26 S proteasome by polyubiquitin chains synthesized to have defined lengths. J. Biol. Chem. 272, 23712–23721 [DOI] [PubMed] [Google Scholar]