Background: Toll-like receptor 4 (TLR4) mediates BAMBI down-regulation, which activates hepatic stellate cells (HSCs).
Results: LPS and TNF-α induced binding of NF-κBp50 to HDAC1, which suppressed BAMBI promoter activity and mRNA expression in HSCs.
Conclusion: TLR4-mediated HSC activation is regulated by promoter regulation of BAMBI.
Significance: Studying the regulation of BAMBI expression by TLR4 is important for understanding liver fibrosis.
Keywords: Hepatic Stellate Cells, Lipopolysaccharide (LPS), NF-κB, Toll-like Receptors (TLR), Transcription Promoter, Transforming Growth Factor β (TGF-β)
Abstract
TLR4 signaling induces down-regulation of the bone morphogenic protein (BMP) and activin membrane-bound inhibitor (BAMBI), which enhances TGF-β signaling during hepatic stellate cell (HSC) activation. We investigated the mechanism by which TLR4 signaling down-regulates BAMBI expression in HSCs and found that TLR4- and TNF-α-mediated BAMBI down-regulation is dependent on regulation of BAMBI promoter activity through the interaction with NF-κBp50 and HDAC1 in HSCs. Bambi was predominantly expressed in HSCs, at high levels in quiescent HSCs but at low levels in in vivo-activated and LPS-stimulated HSCs. In human HSCs, BAMBI expression was down-regulated in response to LPS and TNF-α. A BAMBI reporter assay demonstrated that the regulatory element to repress BAMBI transcription is located between 3384 and 1560 bp upstream from the transcription start site. LPS stimulation down-regulated BAMBI expression in cells with NF-κBp65 knockdown. However, it failed to down-regulate BAMBI in cells with inactivation of NF-κB or NF-κBp50 silencing, indicating that NF-κBp50 is a factor for BAMBI down-regulation. ChIP analysis revealed that LPS and TNF-α induced binding of the NF-κBp50/p50 homodimer to the BAMBI promoter region. We also found that HDAC1 is bound to this region as part of the NF-κBp50-HDAC1 complex, repressing transcriptional activity of the BAMBI promoter. Finally, we confirmed that LPS does not repress BAMBI reporter activity using a BAMBI reporter construct with a mutation at 3166 bp upstream of the coding region. In summary, our study demonstrates that LPS- and TNF-α-induced NF-κBp50-HDAC1 interaction represses BAMBI transcriptional activity, which contributes to TLR4-mediated enhancement of TGF-β signaling in HSCs during liver fibrosis.
Introduction
Liver fibrosis and its end stage, cirrhosis, are major public health problems worldwide. Cirrhosis causes life-threatening complications such as portal hypertension and liver failure and increases the risk of hepatocellular carcinoma (1–3). Cirrhosis resulting from chronic liver inflammation, and fibrosis is characterized by the deposition of excessive extracellular matrix proteins, including collagen fibers and regenerative nodules (4). In the past decade, notable advances have been made toward a deeper understanding of the molecular pathogenesis of liver fibrosis, moving closer to the treatment of liver fibrosis. Activation of hepatic stellate cells (HSCs)2 is a crucial step in the development of liver fibrosis. Therefore, the HSC is an attractive target for the development of new antifibrotic drugs (5).
We have reported previously that TLR4 enhances TGF-β signaling in HSCs, HSC activation, and hepatic fibrosis (6). TLR4-mutant mice had a significant reduction in liver fibrosis upon bile duct ligation and chronic carbon tetrachloride treatment compared with their respective wild-type controls. We showed that HSCs express high levels of TLR4 in the quiescent and activated states and that LPS, a TLR4 ligand, directly activates TLR4-dependent signaling pathways. LPS stimulation alone was insufficient to activate HSCs, but pretreatment with LPS enhanced the response of HSCs to profibrogenic cytokine TGF-β. We identified that the TGF-β pseudoreceptor, BAMBI, is the only TGF-β-related gene changed among 121 LPS-regulated genes in HSCs by gene profiling analysis. TGF-β is the best characterized fibrogenic mediator, and BAMBI is a functional inhibitor for the TGF-β receptor. The loss of BAMBI expression could amplify fibrogenic signaling. Thus, BAMBI plays a role in liver inflammation and fibrogenesis (7, 8). However, the mechanism by which BAMBI is down-regulated in HSCs is unclear.
This study demonstrates that BAMBI mRNA decreases in response to LPS and TNF-α in the human HSC cell line and primary HSCs. We found that the LPS and TNF-α-induced NF-κBp50 homodimer, in association with HDAC1, contributes to transcriptional repression of BAMBI expression in HSCs.
EXPERIMENTAL PROCEDURES
Cell Culture
The human HSC cell line LX-2 was provided by Dr. Scott Friedman (Mount Sinai Medical School, New York, NY) and cultured in DMEM with 2% FBS and 1% penicillin-streptomycin antibiotics.
Primary human HSCs (hHSCs) were isolated from wedge sections of normal human liver tissue unsuitable for transplantation by collagenase-Pronase digestion and centrifugation on Nicodenz gradients. The procedures used for cell isolation and characterization have been described extensively elsewhere (9). hHSCs were cultured in DMEM supplemented with 10% FBS, subcultured when confluent at a 1:3 split ratio, and used between passages 4 and 7. The procedures involving human materials were approved by the Investigational Review Board of the University of California San Diego.
Reagents
LPS (Sigma; Escherichia coli serotype 055:B5), recombinant human TNF-α (R&D Systems), and trichostatin A (TSA) (catalog no. T8552, Sigma) were used in this study. The antibodies used for the ChIP assay, Western blot analysis, and coimmunoprecipitation are p50 (catalog no. sc-8414x), HDAC1 (catalog no. sc-7872x), BAMBI (catalog no. sc-100681), and P-Smad2/3 (catalog no. sc-11769), all purchased from Santa Cruz Biotechnology, Inc. siRNA p50 (catalog no. sc-29407), siRNA HDAC1 (catalog no. sc-29343), siRNA p65, siRNA BAMBI (catalog no. sc-60243), and control siRNA (catalog no. sc-36869) were also purchased from Santa Cruz Biotechnology, Inc. Lipofectamine 2000 transfection regent was purchased from Invitrogen. The Dual-Luciferase reporter assay system (catalog no. E1910) was purchased from Promega.
Generation of the Human BAMBI Reporter Construct and Luciferase Reporter Assays
The BAMBI reporter constructs −3384/+82-luc, −1560/+82-luc, −1016/+82-luc, −586/+82-luc, −255/+82-luc, and −176/+82-luc were provided by Dr. Akiyama (Tokyo University, Tokyo, Japan) (10). The mutation was introduced to the κB binding site of the BAMBI promoter. GGG in the BAMBI promoter were converted to CTC using the QuikChange site-directed mutagenesis kit (Agilent). The adenovirus NF-κB supersuppressor expression construct has been described previously (6).
BAMBI Reporter Analysis
For luciferase assays, the human HSC cell line LX-2 was transfected with the BAMBI reporter-driven firefly luciferase plasmid and control reporter-driven Renilla luciferase by Lipofectamine 2000. LX2 cells were plated on 24-well plates 18 h prior to transfection. Transfection was performed with Lipofectamine 2000 according to the protocol of the manufacturer. Six hours after transfection of reporter plasmids, LX-2 cells were stimulated with or without LPS (100 ng/ml) and TNF-α (10 ng/ml) for 18 h. The cells were washed twice in PBS and then lysed in passive lysis buffer for 15 min at 4 °C with gentle shaking. A luciferase assay was performed using the Dual-Luciferase reporter assay system following the protocol of the manufacturer and measured in a microplate luminometer (Veritas, Turner Biosystems). Firefly luciferase activity was normalized to Renilla luciferase activity.
Bioinformatics Approaches
The BAMBI promoter sequences were obtained from Ensembl. We used rVista2.0. to analyze the BAMBI promoter sequence for their predicted transcription factor binding sites.
siRNA Transfection
siRNA transfection was done according to the protocol of the manufacturer. The control (scrambled) siRNA and NF-κBp50 siRNA, p65 siRNA, HDAC1 siRNA, and BAMBI siRNA were mixed with Lipofectamine 2000 at a final concentration of 80 pmol/liter in medium. mRNA and protein expression was determined by real-time PCR and Western blot analysis using a specific antibody to p50, p65, HDAC1, and BAMBI to confirm the significant reduction of p50, p65, HDAC1, and BAMBI 72 h after transfection.
Real-time PCR Analysis
Total RNA was extracted from LX2 and primary hHSCs using TRIzol reagent (Invitrogen). cDNA was synthesized using a high-capacity cDNA reverse transcription kit (Applied Biosystems). PCR amplification was conducted in 10 μl of solution containing 3 μl of cDNA, 5 μl of SYBR mixture, 1.6 μl of H2O, and 0.4 μl of primer (10 μm). The primers used were as follows: hBAMBI, 5′-GGCAGCATCACAGTAGCATC-3′ (forward) and 5′-GATCGCCACTCCAGCTACAT-3′ (reverse); 18 S rRNA, 5′-AGTCCCTGCCCTTTGTACAC-3′ (forward) and 5′-CGATCCGAGGGCCTCACTA-3′ (reverse). Amplification steps consisted of 40 cycles of denaturation at 94 °C for 40 s, annealing at 55 °C for 40 s, and extension at 72 °C for 40 s using a DNA cycler (Bio Rad) CFX96 real-time system.
Western Blot Analysis
Cell samples were collected in ice-cold radioimmune precipitation assay buffer. Samples were centrifuged for 10 min at 10,000 rpm. The supernatant was collected, and the protein concentration was measured using a BCA commercial kit (Thermo). Protein lysates were separated by SDS-PAGE and subsequently transferred onto nitrocellulose membranes. Membranes (GE Healthcare Life Science) were blocked with 5% nonfat dry milk buffer and incubated with antibodies against human pSmad2/3. Secondary antibody was used for chemiluminescent detection. The loading accuracy was evaluated by monoclonal antibodies against β-actin.
ChIP Assay
Cross-linking was performed by adding 1% formaldehyde directly into plates of cultured cells, followed by incubation at 37 °C for 10 min. Cells were washed twice with ice-cold PBS, collected, and pelleted by centrifugation at 2000 rpm for 5 min. The pellets were resuspended in sonication buffer (50 mm Tris-HCl (pH 8.1), 10 mm EDTA, 1% SDS, and protease inhibitors) and incubated on ice for 10 min to lyse the nuclei. Then they were further sonicated to obtain 200- to 500-bp fragments of chromatin. Immunoprecipitation was carried out according to the protocol provided by Upstate Biotechnology. Briefly, chromatin was diluted 10-fold in ChIP dilution buffer. A small amount of chromatin was kept aside at this step to be used as an input control in subsequent PCR reactions. Antibodies p50 (catalog no. sc-8414x) and HDAC1 (catalog no. sc-7872x) were incubated with diluted chromatin at 4 °C overnight. Immunoprecipitation was also carried out with normal IgG Ab. Protein A-Sepharose (Amersham Biosciences) blocked with sheared salmon sperm DNA was used to collect Ab-chromatin complexes. Immune complexes were then washed once with low-salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 8.1), and 150 mm NaCl), once with high-salt immune complex wash buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 8), and 150 mm NaCl), once with LiCl immune complex wash buffer (0.25 m LiCl, 1% deoxycholic acid, 1 mm EDTA, and 10 mm Tris-HCl (pH 8.1)), and twice with sterile Tris-EDTA buffer. The chromatin was eluted with freshly prepared elution buffer (1% SDS and 0.1 m NaHCO3), followed by reverse cross-linking with 0.3 m NaCl at 65 °C overnight. DNA was then recovered by phenol chloroform extraction and ethanol precipitation, followed by PCR amplification. The PCR conditions used were initial denaturation at 95 °C for 5 min, followed by 40 cycles of 95 °C (30 s), 58 °C (30 s), and 72 °C (30 s). The primers used to amplify the BAMBI promoter for the −3166 κB binding site were 5′-gtttctttgggttgcaagga-3′ and 5′-cacctgtctgcagaggaaga-3′.
Coimmunoprecipitation
For immunoprecipitation of p50 and HDAC1, LX2 cells were grown in 15-cm dishes to 85% confluence. The cells were washed twice in ice-cold PBS and collected. The samples were suspended in lysis buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 10% glycerol, 0.5% Nonidet P-40, 1 mm EDTA, and protease inhibitors). Then the samples were shaken for 40 min at 4 °C to lyse the protein completely. After lysis, the samples were centrifuged at 4 °C and 14,000 rpm for 10 min. The extracts were precleared with a 50% Sepharose A slurry (Millipore) and then incubated overnight at 4 °C with 2 μg of either the p50 polyclonal antibody (catalog no. sc-8414x, Santa Cruz Biotechnology, Inc.) or HDAC1 (catalog no. sc-7872x, Santa Cruz Biotechnology, Inc.) antibody. Antibody-protein complexes were collected with the 50% Sepharose A slurry, washed, and then boiled in sample buffer to remove the antibody-protein complex from the protein A slurry. Samples were then subjected to SDS-PAGE and immunoblotted.
Statistical Analysis
Differences between two groups were compared using Mann-Whitney U test or two-tailed unpaired Student's t test. Differences between multiple groups were compared using one-way analysis of variance using SPSS software (SPSS Inc., Chicago, IL). p <0.05 was considered significant.
RESULTS
BAMBI mRNA Decreases in Response to LPS or TNF-a Stimulation in HSCs
We initially investigated the responsible cell types expressing Bambi in mouse liver. As shown in Fig. 1A, Bambi mRNA was expressed at higher levels in HSCs than in Kupffer cells and hepatocytes. Among different activation states of HSCs, Bambi mRNA decreased to a low level in in vivo-activated HSCs isolated from mice after bile duct ligation and chronic CCl4 treatment and in LPS-treated quiescent HSCs but not in culture-activated HSCs (Fig. 1B), suggesting that TLR4-mediated Bambi down-regulation is crucial for HSC activation in vivo.
FIGURE 1.
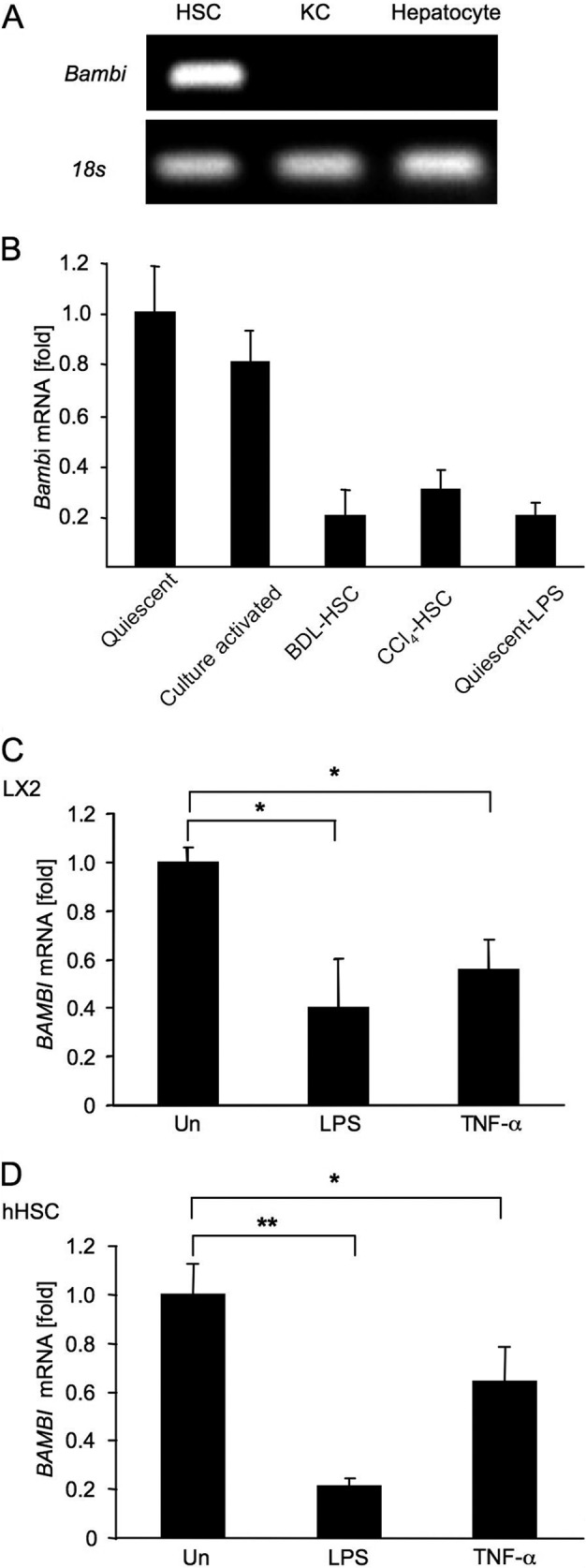
BAMBI regulation by TLR4 signaling. A, Bambi mRNA expression in primary mouse HSCs, Kupffer cells (KC), and hepatocytes was determined by RT-PCR. B, Bambi mRNA levels in quiescent and 5-day culture-activated HSCs, HSCs isolated from bile duct ligation (BDL) and CCl4-treated mice, and quiescent HSCs stimulated with LPS (100 ng/ml) for 4 h were determined by qPCR. C and D, BAMBI mRNA levels in LX2 cells (C) and hHSCs (D) 4 h after LPS (100 ng/ml) or TNF-α (10 ng/ml) stimulation were determined by qPCR. Un, untreated. Data represent mean ± S.D. *, p < 0.05; **, p < 0.01.
Next, we investigated whether BAMBI mRNA expression can be down-regulated in a human HSC cell line and in primary human HSCs in response to LPS or TNF-α. BAMBI mRNA was down-regulated 0.4- to 0.5-fold in LX2 cells in response to LPS and TNF-α compared with untreated cells (Fig. 1C). More dramatically, BAMBI mRNA decreased 0.21-fold in hHSCs in response to LPS compared with untreated hHSCs (Fig. 1D).
Regulatory Elements for Repression of BAMBI Promoter Activity after LPS and TNF-α Stimulation in HSCs
Quiescent HSCs express BAMBI at high levels, and LPS stimulation down-regulates BAMBI mRNA expression. To characterize the crucial elements for repression in the BAMBI promoter region after LPS and TNF-α stimulation, LX2 cells were transfected with hBAMBI promoter-luciferase reporter plasmids containing the hBAMBI transcriptional start site, the complete 82-bp untranslated region, and the 3384 region upstream of the coding region. The promoter activity analysis showed that the relative luciferase activity in the −3384 + 82-luc construct decreased significantly (p < 0.001) with LPS or TNF-α treatment (Fig. 2, A and B). In contrast, the relative luciferase activity in shorter deletion constructs was not reduced by either LPS or TNF-α stimulation. Luciferase activity in the −176 + 82-luc construct was significantly lower than that of longer constructs in untreated cells (Fig. 2, A and B). These results suggest that the regulatory element for repression of BAMBI transcription is located between −3384 and −1560 upstream from the BAMBI coding region and that the BAMBI promoter region between −255 and −176 is responsible for constitutive expression of BAMBI in hHSCs.
FIGURE 2.
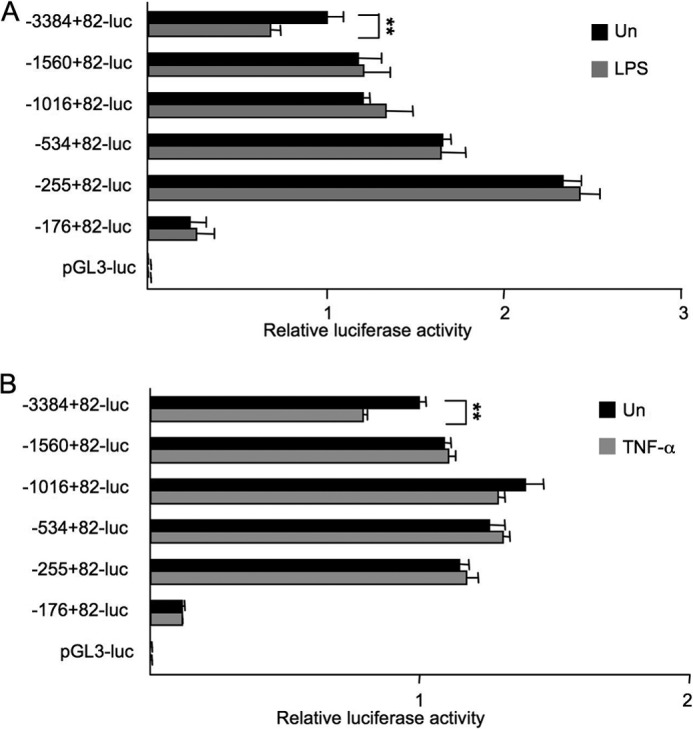
Identification of the element in the promoter region responsible for repression of BAMBI by LPS and TNF-α. Nucleotide sequence positions are indicated relative to the transcriptional start site. LX2 cells were transfected with the indicated hBAMBI reporter constructs. 6 h after transfection, cells were treated with LPS (100 ng/ml) (A) or TNF-α (10 ng/ml) (B) for 16 h, followed by measurement for luciferase activities. The Renilla luciferase reporter was cotransfected to normalize transfection efficiency. Un, untreated. Experiments were performed three times and are shown as mean ± S.D. **, p < 0.01.
NF-κBp50 Participates in BAMBI mRNA Repression in Response to LPS and TNF-α
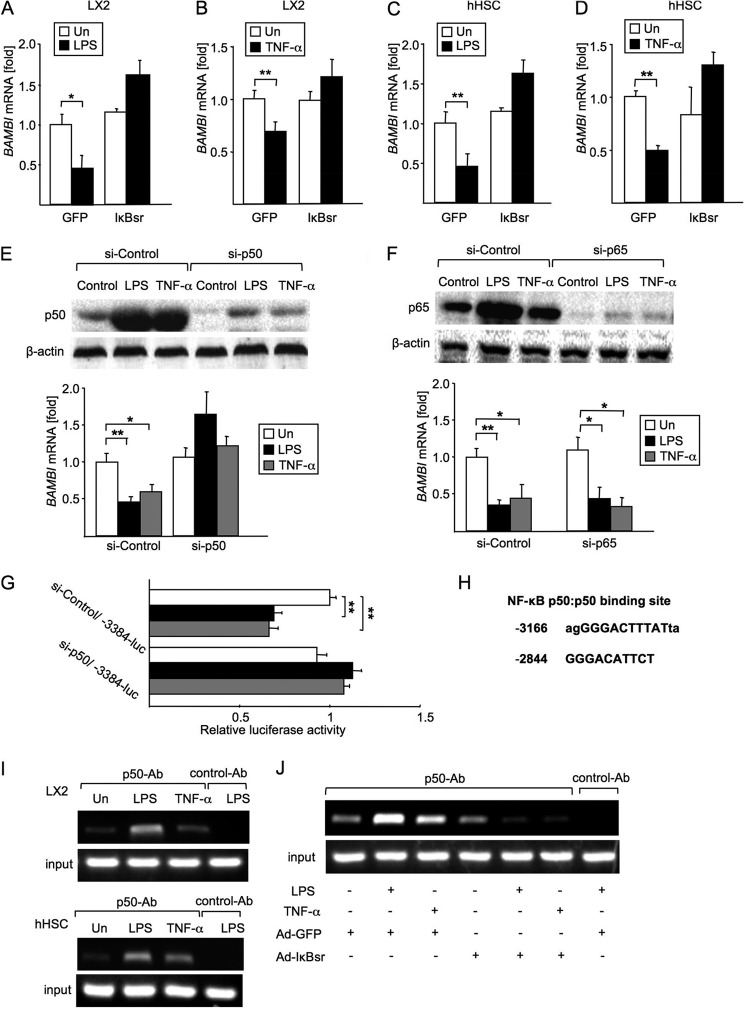
Because both TNF-α and LPS are strong activators of NF-κB, we hypothesized that NF-κB binds to a specific element on the BAMBI promoter region that is associated with the suppression of BAMBI gene transcription. We assessed the contribution of NF-κB activation to BAMBI repression by using an adenovirus that expresses an IκB superrepressor (IκBsr), a potent and specific inhibitor of NF-κB activation (6). In the presence of the NF-κB inhibitor adenovirus (Ad)-IκBsr, we observed the BAMBI mRNA levels after LPS and TNF-α treatment to be approximately equivalent to that of the untreated levels in both LX2 cells and hHSCs (Fig. 3, A–D), suggesting that NF-κB activation participates in BAMBI gene down-regulation.
FIGURE 3.
NF-κBp50 mediates BAMBI repression by LPS and TNF-α. A and B, BAMBI mRNA levels in LX2 cells after 4 h of stimulation with LPS (100 ng/ml) (A) or TNF-α (10 ng/ml) (B) were determined by qPCR. Before treatment, LX2 cells were transfected with the Ad-GFP or Ad-IκB superrepressor overnight. Un, untreated. Data represent mean ± S.D. *, p < 0.05; **, p < 0.01. C and D, BAMBI mRNA levels in hHSCs after 4 h of LPS (100 ng/ml) (C) or TNF-α (10 ng/ml) (D) treatment were determined by qPCR. Before treatment, hHSCs were transfected with control Ad-GFP or the Ad-IκB superrepressor overnight. Un, untreated. Data represent mean ± S.D. **, p < 0.01. E, after transfection with siRNA for p50 or a control for 72 h, LX2 cells were stimulated with LPS (100 ng/ml) or TNF-α (10 ng/ml) for 4 h. The knockdown efficiency of siRNA p50 was evaluated by Western blot analysis for p50. BAMBI mRNA was measured by qPCR. Un, untreated. Data represent mean ± S.D. *, p < 0.05; **, p < 0.01. F, LX2 cells were transfected with siRNA for p65 or a control for 72 h, followed by stimulation with LPS (100 ng/ml) or TNF-α (10 ng/ml) for 4 h. p65 expression was evaluated by Western blot analysis. BAMBI mRNA was measured by qPCR. Un, untreated. Data represent mean ± S.D. *, p < 0.05; **, p < 0.01. G, BAMBI luciferase reporter activity was measured. LX2 cells were cotransfected with −3384/+82-luc and siRNA p50. The Renilla luciferase reporter was used to normalize transfection efficiency. Data represent mean ± S.D. **, p < 0.01. H, two κB binding sites within −3384 to −1560 of the hBAMBI promoter region were identified using bioinformatics analysis. I, effects of LPS and TNF-α on the recruitment of p50 to the κB binding site in the hBAMBI promoter were assessed by ChIP analysis. LX2 cells (top panel) and hHSCs (bottom panel) were treated with LPS (100 ng/ml) or TNF-α for 2 h. After fixation, soluble chromatin was immunoprecipitated using anti-p50 or control Ab (normal mouse IgG). BAMBI promoter fragments containing the −3166 binding site were amplified by PCR. J, effects of Ad-IκBsr on the recruitments of p50 to the κB binding site in the hBAMBI promoter were assessed. LX2 cells were treated with control Ad-GFP or Ad-IκBsr overnight and then treated with LPS (100 ng/ml) or TNF-α for 2 h. A ChIP analysis using anti-p50 or control Ab (normal mouse IgG) was performed. BAMBI promoter fragments containing the −3166 binding site were amplified by PCR. Un, untreated.
To determine which subunit of NF-κB is associated with BAMBI down-regulation in HSCs, we selectively inhibited p50 and p65 translation using p50- and p65-specific siRNA (Fig. 3, E and F). We stimulated p50 and p65 knockdown LX2 cells or control siRNA-transfected LX2 cells with LPS and TNF-α and measured BAMBI mRNA (Fig. 3, E and F). In LX2 cells transfected with control siRNAs, LPS and TNF-α stimulation induced a 50–60% reduction of BAMBI mRNA expression compared with untreated LX2 cells (Fig. 3, E and F). The LPS-treated, p50-silenced LX2 cells showed similar levels of BAMBI mRNA as untreated cells (Fig. 3E). Of note, BAMBI mRNA was still reduced by LPS and TNF- α in p65-silenced LX2 cells (Fig. 3F). These results indicate that NF-κBp50, but not the p65 subunit, is required for the suppression of LPS- and TNF-α-induced down-regulation of BAMBI mRNA expression in LX2 cells.
Given that the NF-κBp50 subunit participates in BAMBI mRNA down-regulation in response to LPS and TNF-α, we investigated whether p50 is directly involved in BAMBI promoter activity using siRNA for p50 and BAMBI −3384/+82-luc. The relative luciferase activity was decreased in control siRNA-transfected LX2 cells in response to LPS and TNF-α (Fig. 3G). However, the relative luciferase activity did not decrease in LX2 cells transfected with siRNA for p50 in response to LPS and TNF-α treatment compared with cells transfected with p50 siRNA without LPS or TNF-α treatment.
It has been reported that the p50 homodimer negatively regulates gene expression (11–13). Our data strongly support the capability of the NF-κBp50 homodimer to suppress the BAMBI gene transcription upon LPS and TNF-α treatment. Sequence analysis of the human BAMBI promoter (-3384 to −1560) predicted two regions as p50-binding sites (Fig. 3H). ChIP analysis of the promoter using primers for these two regions showed that NF-κBp50 binds to the −3166 region (Fig. 3I). In LX2 cells and hHSCs, LPS and TNF-α stimulation increased p50 binding to the −3166 binding site (Fig. 3I) compared with untreated cells. However, p50 recruitment was decreased in Ad-IκBsr-infected cells in response to LPS and TNF-α stimulation (Fig. 3J), indicating that LPS and TNF-α induce p50 binding to the BAMBI promoter region.
Binding of HDAC1 to the BAMBI Promoter after LPS and TNF-α Stimulation
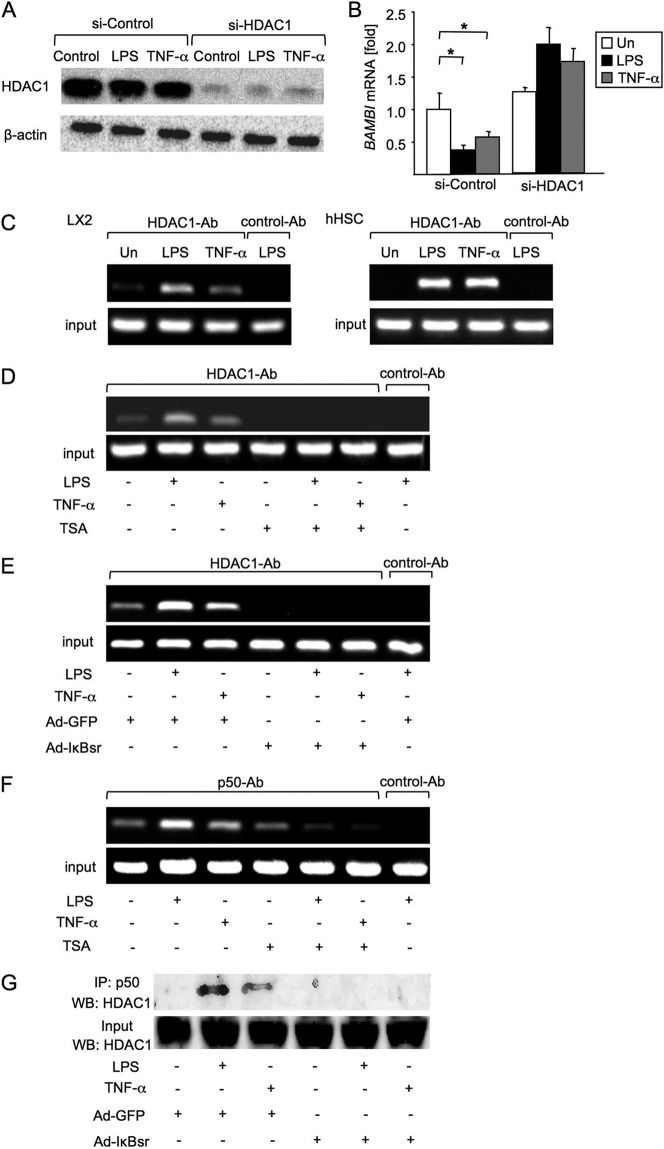
It has been reported previously that NF-κBp50 associates with the histone deacetylase (HDAC) that inhibits gene expression (14–16). Therefore, we examined whether HDAC1 participates in BAMBI mRNA regulation. When we knocked down HDAC1 in LX2 cells, the expression of BAMBI mRNA did not decrease in response to LPS and TNF-α stimulation (Fig. 4, A and B), suggesting that HDAC1 is associated with BAMBI down-regulation in response to LPS and TNF-α stimulation.
FIGURE 4.
Mechanism of BAMBI repression by HDAC1. A, the knockdown efficiencies of siRNA HDAC1 were estimated by Western blot analysis. LX2 cells were transected with siRNA HDAC1 or a control for 72 h, followed by treatment with LPS (100 ng/ml) or TNF-α (10 ng/ml) for 4 h. B, LX2 cells were transfected with siRNA HDAC1 for 72 h. Then, hBAMBI mRNA was measured by qPCR. 18 S was used as an internal control for normalization. Un, untreated. Data represent mean ± S.D. *, p < 0.05. C, effects of LPS and TNF-α on the recruitment of HDAC1 to the κB binding site in the hBAMBI promoter were assessed. LX2 (left panel) and hHSCs (right panel) were treated with LPS (100 ng/ml) or TNF-α (10 ng/ml) for 2 h. A ChIP analysis using anti-HDAC1 or control Ab (normal rabbit polyclonal IgG) was performed. BAMBI promoter fragments containing the −3166 binding site were amplified by PCR. D, effects of TSA on the recruitment of HDAC1 to the κB binding site in the hBAMBI promoter were analyzed. LX2 cells were pretreated with TSA (300 nm/ml) for 8 h and then treated with LPS or TNF-α for 2 h. A ChIP analysis using anti-HDAC1 or control Ab (normal rabbit polyclonal IgG) was employed. BAMBI promoter fragments containing the −3166 binding site were amplified by PCR. E, effects of NF-κB inhibition on the binding of HDAC1 to the κB binding site in the hBAMBI promoter. LX2 cells were treated with control Ad-GFP or Ad-IκBsr overnight and then treated with LPS or TNF-α for 2 h. A ChIP analysis on the BAMBI promoter using anti-HDAC1 or control Ab (normal rabbit polyclonal IgG) was performed. F, ChIP analysis for the assessment of the effects of TSA on the binding of p50 to the κB binding site in the hBAMBI promoter. G, direct interaction of p50 with HDAC1 after LPS or TNF-α stimulation was assessed. LX2 cells were treated with control Ad-GFP or Ad-IκBsr overnight and then treated with LPS and TNF-α for 2 h. After immunoprecipitation (IP) with anti-p50 antibody, Western blotting (WB) for HDAC1 was performed.
To identify whether HDAC1 binding to p50 contributes to BAMBI gene regulation, we carried out a ChIP analysis using anti-HDAC1 antibody directed at the −3166 κB-binding site. We observed that LPS and TNF-α stimulation led to higher HDAC1 binding to this κB-binding site of the BAMBI promoter in LX2 cells and hHSCs (Fig. 4C). Treatment with the HDAC1 inhibitor TSA inhibited LPS- or TNF-α-induced HDAC1 binding to the BAMBI promoter (Fig. 4D). As expected, LPS- or TNF-α-induced HDAC1 binding to the BAMBI promoter was also inhibited when NF-κB activation was inhibited in LX2 cells infected with Ad-IκBsr (Fig. 4E), indicating that NF-κB activation is required for binding of HDAC1 to the BAMBI promoter. Interestingly, TSA treatment prevented LPS- or TNF-α-induced binding of NF-κBp50 to the BAMBI promoter (Fig. 4F), suggesting that the HDAC1 activity is also required for NF-κBp50 binding to the BAMBI promoter. Finally, we tested the direct interaction between HDAC1 and NF-κBp50 by coimmunoprecipitation. This confirmed the interaction of HDAC1 with p50 in LX2 cells after LPS or TNF-α stimulation (Fig. 4G).
The −3166 Site of BAMBI Promoter Is Crucial for Repression of BAMBI Expression after LPS and TNF-α Stimulation
To further validate NF-κBp50 and BAMBI promoter interaction at the −3166 κB site, we created luciferase constructs of BAMBI with a mutated −3166 κB site (Fig. 5A). A luciferase reporter assay revealed that LPS and TNF-α-induced suppression of BAMBI reporter activity was not observed in cells transfected with BAMBI reporter constructs with a mutation at the −3166 site, indicating the importance of the −3166 binding κB site for the down-regulation of gene expression.
FIGURE 5.
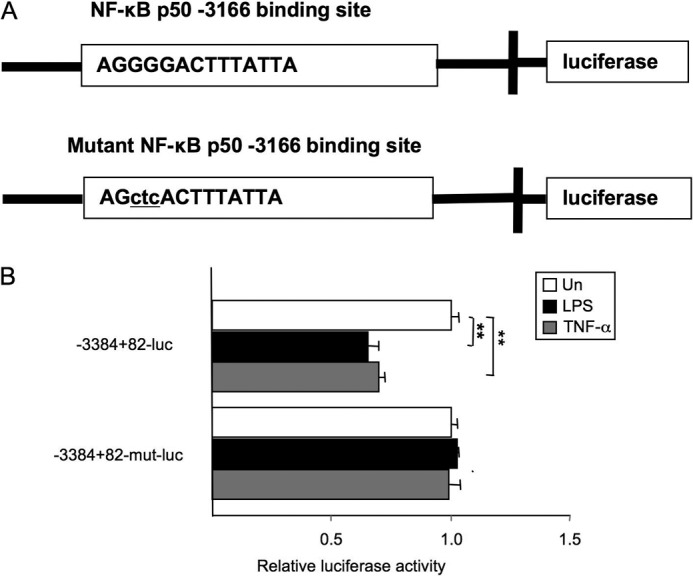
Effect of mutation of the −3166 κB-binding site in the BAMBI promoter. A, the predicted p50 binding sequence (top panel) and designed mutant sequence (bottom panel). B, hBAMBI p-3384+82-luc, which contains an NF-κB binding site, was used to assess the functional significance of the −3166 binding site. The p-3384+82-luc construct was responsive to an LPS or TNF-α stimulus, whereas the promoter constructs harboring a mutation of the predicted −3166 κB binding site (p-3384+82-mutant-luc) abrogated the LPS- or TNF-α-induced suppression of luciferase expression. Un, untreated. Data represent mean ± S.D. **, p < 0.01.
Assessment of the Role of BAMBI Down-regulation in Human LX-2 HSCs
As expected, TGF-β treatment increased phosphorylation of Smad2/3 in LX2 cells (Fig. 6A). In the context of down-regulation of BAMBI, LPS and TNF-α treatment further increased TGF-β-mediated phosphorylation of Smad2/3 in LX2 cells (Fig. 6A). Also, TGF-β-induced COL1A1 expression was further enhanced by pretreatment with LPS or TNF-α (Fig. 6B) in control adenovirus-infected LX2 cells. Consistently, inhibition of NF-κB decreased the enhancement of TGF-β-induced COL1A1 expression by pretreatment with LPS and TNF-α (Fig. 6B), suggesting that NF-κB-mediated BAMBI down-regulation is associated with LPS or TNF-α-mediated enhancement of TGF-β-induced COL1A1 expression. Lastly, we investigated the biological function of BAMBI in TGF-β signaling in human LX-2 HSC. TGF-β increased COL1A1 mRNA expression in control siRNA-transfected LX2 cells (Fig. 6C). COL1A1 mRNA was further increased after TGF-β stimulation in cells transfected with siRNA for BAMBI, confirming that BAMBI is a negative regulator of TGF-β signaling in human HSCs.
FIGURE 6.
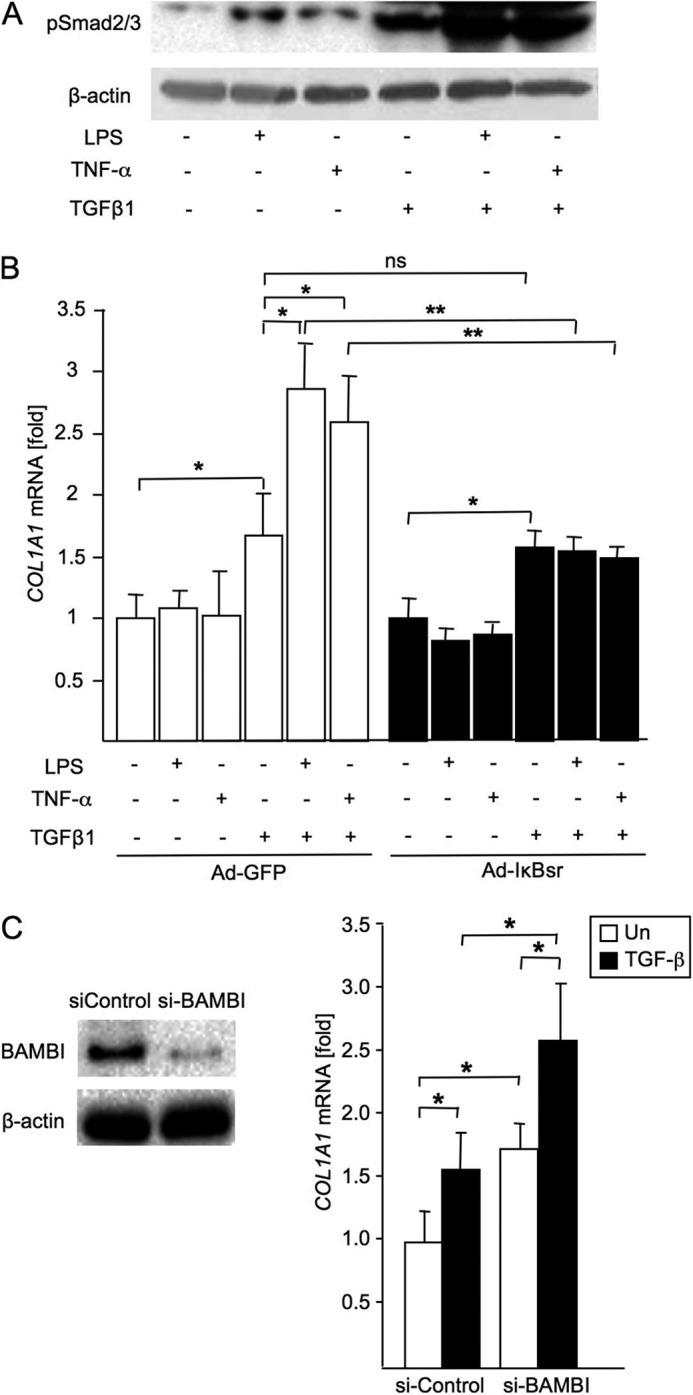
LPS enhances TGF-β signaling in hHSCs through down-regulation of BAMBI. A, LX2 cells were pretreated with LPS (100 ng/ml) or TNF-α (10 ng/ml) for 12 h and subsequently treated with TGF-β1 (5 ng/ml) for 30 min. A Western blot analysis for phospho-Smad2/3 was performed. B, LX2 cells were infected with Ad-IκBsr or Ad-GFP overnight, followed by treatment with LPS or TNF-α for 12 h. Subsequently, cells were treated with TGF-β1 (5 ng/ml) for 24 h. COL1A1 mRNA was measured by qPCR, and 18 S was used as an internal control. Data represent mean ± S.D. *, p < 0.05; **, p < 0.01. n.s., not significant. C, LX2 cells were transfected with siRNA BAMBI for 48 h and then treated with TGF-β1 (5 ng/ml) for an additional 24 h. BAMBI protein expression was analyzed by immunoblotting (left panel), and COL1A1 mRNA expression was measured by qPCR (right panel). Data represent mean ± S.D. *, p < 0.05.
DISCUSSION
Liver fibrosis is a complex pathological entity in which multiple components, including various proinflammatory cytokines and fibrotic matrix proteins, participate actively (17). There is overwhelming evidence that activated HSCs are the major producers of the fibrotic matrix (1–3). TGF-β is the key cytokine that mediates HSC activation, but the current model of TGF-β-dependent HSC activation does not account for the contribution of inflammatory mediators to hepatic fibrogenesis (6), and extensive studies are aimed to establish a mechanistic link between inflammation and fibrosis in chronic liver disease (6, 18). We have shown previously that BAMBI links inflammation and liver fibrosis via TLR4-dependent modification of TGF-β signaling in HSC. BAMBI plays an important role in HSC activation. However, the mechanism of the regulation of BAMBI gene expression was unknown.
In this study, we determined the molecular mechanism of BAMBI down-regulation in response to LPS and TNF-α stimulation in human HSCs. We demonstrated that BAMBI is mainly expressed in HSCs but not Kupffer cells and hepatocytes. TLR4 and TNF receptor signaling induce NF-κBp50 homodimer/HDAC1 binding to the −3166 site in the BAMBI promoter to down-regulate gene transcription.
BAMBI is a transmembrane glycoprotein structurally related to the TGF-β type I receptor, but it lacks the intracellular kinase domain (19). Several investigators have recognized the important role of BAMBI in recent years. BAMBI protects the murine heart from pressure overload biomechanical stress by restraining TGF-β signaling (20). BAMBI has also been reported as a negative regulator of adipogenesis and modulator of the anti- and proadipogenic effects of TGF-β (21). BAMBI blocks the differentiation of human bone marrow mesenchymal stem cells to carcinoma-associated fibroblasts via inhibition of TGF-β signaling (22).
BAMBI is coexpressed with TGF-β receptor family members during development and in cancer. It has been proposed that BAMBI may play a role in embryonic development (23) and in tumor growth and metastasis (24). Elevation of BAMBI expression may attenuate TGF-β-mediated growth arrest in colorectal and hepatocellular carcinomas (25) and induction of cell growth and invasion of human gastric cancer (26). Although BAMBI plays an important physiological and pathological role in the liver, heart, and cancer, its transcriptional regulation has not been studied intensively. Previous reports showed that BAMBI transcription is regulated by TGF-β signaling through direct binding of SMAD3 and SMAD4 to the BAMBI promoter in HepG2 cells (10). However, our results showed that BAMBI is expressed in HSCs but not in hepatocytes. NF-κB is a critical transcription factor induced by inflammatory mediators, including TNF-α and LPS (27). We found that inhibition of NF-κB prevented LPS and TNF-α from inhibiting BAMBI expression, suggesting that NF-κB participates in BAMBI down-regulation in human HSCs. In fact, BAMBI mRNA levels, in response to LPS and TNF-α, were slightly higher in Ad-IκBsr-treated cells compared with untreated cells, suggesting that BAMBI might be constitutively transcribed independently of NF-κB or be regulated at the posttranscriptional level (28). The stability of mRNA is regulated by degradation and, in part, by specific proteins binding to defined adenylate-uridylate-rich base sequences in the 3′ UTR. Our luciferase activity analysis showed that the relative luciferase activity decreased after LPS stimulation with transfection of a pBAMBI-3′ UTR reporter plasmid,3 suggesting that other mechanisms, such as miRNA, also participate in the regulation of BAMBI expression.
Different forms of NF-κB are known to have distinct functions in the inflammatory response (29, 30). The most abundant form of NF-κB in cells is a heterodimer of the p50 and p65 subunits. Our results showed that BAMBI mRNA still decreased after knocking down NF-κBp65, indicating that the NF-κBp65/p50 heterodimer is not important for BAMBI down-regulation in human HSCs in response to LPS and TNF-α treatment. This was also demonstrated by ChIP analysis. In contrast, when knocking down p50, BAMBI mRNA levels were protected from LPS or TNF-α stimulation, suggesting that the p50 homodimer, but not the p65/p50 heterodimer, participates in BAMBI repression by LPS or TNF-α. Generally, low levels of NF-κB, particularly the p50 homodimer, can be detected in the nuclei of most unstimulated cells. Following stimulation, the p50/p65 heterodimer containing phosphorylated p65 enters the nucleus and displaces the DNA-bound p50 homodimer (14). Intriguingly, our ChIP analysis showed that LPS and TNF-α increased p50 binding to the BAMBI promoter region. It is probable that the different κB sites exhibit a preference for specific subsets of NF-κB complexes (31). For instance, the regulatory region of the IL-8 gene contains a κB element that binds p65, c-Rel, and the p52 homodimer but not the p50 homodimer or p50/p65 heterodimer (32). Similarly, induction of the ICAM-1 gene occurs in response to inflammatory signals, such as TNF-α, that act through an NF-κB site in the proximal promoter region. This site binds only the p65 homodimer and p50/p65 heterodimer (33). p65 homodimer appears to be the transcriptionally active NF-κB complex for the ICAM-1 promoter, suggesting that p50/p65 may be excluded from binding (33). The mechanism by which the p50 homodimer inhibits gene expression is unknown, although it is unlikely that p50 subunits are intrinsically incapable to drive transcription. One possible mechanism is that nuclear p50 may interact with inhibitory proteins that recruit corepressor complexes containing HDACs to gene promoters (15).
Our experiments depleting HDAC1 and using TSA strongly suggest that the activity of HDAC1 maintains the transcriptional repression through a p50-HDAC1 complex because inhibition of HDAC activity by TSA blocks the repression of BAMBI mRNA expression. This is also supported by our ChIP analysis demonstrating that inhibition of HDAC1 activity by TSA treatment blocks HDAC1 binding to the promoter region induced by LPS and TNF-α. Furthermore, the inhibition of NF-κB activation or silencing of p50 by siRNA inhibited HDAC1 binding to p50 and the BAMBI promoter region, indicating that NF-κB activation is required for the formation of the p50-HDAC1 complex (15). Furthermore, inhibition of HDAC1 activity by TSA treatment decreased p50 binding to the BAMBI promoter region, suggesting that HDAC1 activity is required for the formation of the p50-HDAC1 complex and binding of this complex to the BAMBI promoter region.
In summary, BAMBI is predominantly expressed in HSCs in the liver. TNF-α or LPS stimulation generates an NF-κBp50 homodimer that interacts with HDAC1 to repress BAMBI transcription in human HSCs, which, in turn, increases the sensitivity to TGF-β signaling.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DK085252 and R01AA020172 and NIEHS, National Institutes of Health Grant P42ES010337 (Project 5) (to E. S.); National Institutes of Health/National Center for Advancing Translational Sciences/Clinical and Translational Research Institute Grant KL2TR000099 (to T. K.); and National Institutes of Health Grant R01 GM041804 (to D. A. B.).
C. Liu, D. A. Brenner, and E. Seki, unpublished observations.
- HSC
- hepatic stellate cell
- BAMBI
- BMP and activin membrane-bound inhibitor
- BMP
- bone morphogenic protein
- hHSC
- human hepatic stellate cell
- luc
- luciferase
- Ab
- antibody
- Ad
- adenovirus
- IκBsr
- IκB superrepressor
- HDAC
- histone deacetylase
- qPCR
- quantitative PCR
- TSA
- trichostatin A.
REFERENCES
- 1. Lee U. E., Friedman S. L. (2011) Mechanisms of hepatic fibrogenesis. Best. Pract. Res. Clin. Gastroenterol. 25, 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bataller R., Brenner D. A. (2005) Liver fibrosis. J. Clin. Invest. 115, 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aoyama T., Inokuchi S., Brenner D. A., Seki E. (2010) CX3CL1-CX3CR1 interaction prevents carbon tetrachloride-induced liver inflammation and fibrosis in mice. Hepatology 52, 1390–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. De Minicis S., Rychlicki C., Agostinelli L., Saccomanno S., Trozzi L., Candelaresi C., Bataller R., Millán C., Brenner D. A., Vivarelli M., Mocchegiani F., Marzioni M., Benedetti A., Svegliati-Baroni G. (2013) Semaphorin 7A contributes to TGF-β-mediated liver fibrogenesis. Am. J. Pathol. 183, 820–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Friedman S. L. (2004) Stellate cells. A moving target in hepatic fibrogenesis. Hepatology 40, 1041–1043 [DOI] [PubMed] [Google Scholar]
- 6. Seki E., De, Minicis S., Osterreicher C. H., Kluwe J., Osawa Y., Brenner D. A., Schwabe R. F. (2007) TLR4 enhances TGF-β signaling and hepatic fibrosis. Nat. Med. 13, 1324–1332 [DOI] [PubMed] [Google Scholar]
- 7. Luedde T., Trautwein C. (2008) A molecular link between inflammation and fibrogenesis. The bacterial microflora influences hepatic fibrosis via Toll-like receptor 4-dependent modification of transforming growth factor-β signaling in hepatic stellate cells. Hepatology 47, 1089–1091 [DOI] [PubMed] [Google Scholar]
- 8. Friedman S. L. (2007) A deer in the headlights. BAMBI meets liver fibrosis. Nat. Med. 13, 1281–1282 [DOI] [PubMed] [Google Scholar]
- 9. Schwabe R. F., Bataller R., Brenner D. A. (2003) Human hepatic stellate cells express CCR5 and RANTES to induce proliferation and migration. Am. J. Physiol. Gastrointest. Liver. Physiol. 285, G949-G958 [DOI] [PubMed] [Google Scholar]
- 10. Sekiya T., Oda T., Matsuura K., Akiyama T. (2004) Transcriptional regulation of the TGF-β pseudoreceptor BAMBI by TGF-β signaling. Biochem. Biophys. Res. Commun. 320, 680–684 [DOI] [PubMed] [Google Scholar]
- 11. Cao S., Zhang X., Edwards J. P., Mosser D. M. (2006) NF-κB1 (p50) homodimers differentially regulate pro- and anti-inflammatory cytokines in macrophages. J. Biol. Chem. 281, 26041–26050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grundström S., Anderson P., Scheipers P., Sundstedt A. (2004) Bcl-3 and NFκB p50-p50 homodimers act as transcriptional repressors in tolerant CD4+ T cells. J. Biol. Chem. 279, 8460–8468 [DOI] [PubMed] [Google Scholar]
- 13. Elsharkawy A. M., Oakley F., Lin F., Packham G., Mann D. A., Mann J. (2010) The NF-κB p50:p50:HDAC-1 repressor complex orchestrates transcriptional inhibition of multiple pro-inflammatory genes. J. Hepatol. 53, 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Williams S. A., Chen L. F., Kwon H., Ruiz-Jarabo C. M., Verdin E., Greene W. C. (2006) NF-κB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 25, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhong H., May M. J., Jimi E., Ghosh S. (2002) The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell. 9, 625–636 [DOI] [PubMed] [Google Scholar]
- 16. Ashburner B. P, Westerheide S. D., Baldwin A. S., Jr. (2001) The p65 (RelA) subunit of NF-κB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 21, 7065–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wynn T. A., Ramalingam T. R. (2012) Mechanisms of fibrosis. Therapeutic translation for fibrotic disease. Nat. Med. 18, 1028–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu C., Tao Q., Sun M, Wu J. Z., Yang W., Jian P., Peng J., Hu Y., Liu C., Liu P. (2010) Kupffer cells are associated with apoptosis, inflammation and fibrotic effects in hepatic fibrosis in rats. Lab. Invest. 90, 1805–1816 [DOI] [PubMed] [Google Scholar]
- 19. Onichtchouk D., Chen Y. G., Dosch R., Gawantka V., Delius H., Massagué J., Niehrs C. (1999) Silencing of TGF-β signalling by the pseudoreceptor BAMBI. Nature 401, 480–485 [DOI] [PubMed] [Google Scholar]
- 20. Villar A. V., García R., Llano M., Cobo M., Merino D., Lantero A., Tramullas M., Hurlé J. M., Hurlé M. A., Nistal J. F. (2013) BAMBI (BMP and activin membrane-bound inhibitor) protects the murine heart from pressure-overload biomechanical stress by restraining TGF-β signaling. Biochim. Biophys. Acta 1832, 323–335 [DOI] [PubMed] [Google Scholar]
- 21. Luo X., Hutley L. J., Webster J. A., Kim Y. H., Liu D. F., Newell F. S., Widberg C. H., Bachmann A., Turner N., Schmitz-Peiffer C., Prins J. B., Yang G. S., Whitehead J. P. (2012) Identification of BMP and activin membrane-bound inhibitor (BAMBI) as a potent negative regulator of adipogenesis and modulator of autocrine/paracrine adipogenic factors. Diabetes 61, 124–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shangguan L., Ti X., Krause U., Hai B., Zhao Y., Yang Z., Liu F. (2012) Inhibition of TGF-β/Smad signaling by BAMBI blocks differentiation of human mesenchymal stem cells to carcinoma-associated fibroblasts and abolishes their protumor effects. Stem Cells 30, 2810–2819 [DOI] [PubMed] [Google Scholar]
- 23. Grotewold L., Plum M., Dildrop R., Peters T., Rüther U. (2001) Bambi is co-expressed with Bmp-4 during mouse embryogenesis. Mech. Dev. 100, 327–330 [DOI] [PubMed] [Google Scholar]
- 24. Sekiya T., Adachi S., Kohu K., Yamada T., Higuchi O., Furukawa Y., Nakamura Y., Nakamura T., Tashiro K., Kuhara S., Ohwada S., Akiyama T. (2004) Identification of BMP and activin membrane-bound inhibitor (BAMBI), an inhibitor of transforming growth factor-β signaling, as a target of the β-catenin pathway in colorectal tumor cells. J. Biol. Chem. 279, 6840–6846 [DOI] [PubMed] [Google Scholar]
- 25. Yan X., Lin Z., Chen F., Zhao X., Chen H., Ning Y., Chen Y. G. (2009) Human BAMBI cooperates with Smad7 to inhibit transforming growth factor-β signaling. J. Biol. Chem. 284, 30097–30104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fritzmann J., Morkel M., Besser D., Budczies J., Kosel F., Brembeck F. H., Stein U., Fichtner I., Schlag P. M., Birchmeier W. (2009) A colorectal cancer expression profile that includes transforming growth factor β inhibitor BAMBI predicts metastatic potential. Gastroenterology 137, 165–175 [DOI] [PubMed] [Google Scholar]
- 27. Hatano E., Bennett B. L., Manning A. M., Qian T., Lemasters J. J., Brenner D. A. (2001) NF-κB stimulates inducible nitric oxide synthase to protect mouse hepatocytes from TNF-α- and Fas-mediated apoptosis. Gastroenterology 120, 1251–1262 [DOI] [PubMed] [Google Scholar]
- 28. Xavier S., Gilbert V., Rastaldi M. P., Krick S., Kollins D., Reddy A., Bottinger E., Cohen C. D., Schlondorff D. (2010) BAMBI is expressed in endothelial cells and is regulated by lysosomal/autolysosomal degradation. PLoS ONE 5, e12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Hara S. P., Splinter P. L., Gajdos G. B., Trussoni C. E., Fernandez-Zapico M. E., Chen X. M., LaRusso N. F. (2010) NF-κB p50-CCAAT/enhancer-binding protein β (C/EBPβ)-mediated transcriptional repression of microRNA let-7i following microbial infection. J. Biol. Chem. 285, 216–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao Z., Yin J., Zhang J., He Q., McGuinness O. P., Ye J. (2009) Inactivation of NF-κB p50 leads to insulin sensitization in liver through post-translational inhibition of p70S6K. J. Biol. Chem. 284, 18368–18376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baer M., Dillner A., Schwartz R. C., Sedon C., Nedospasov S., Johnson P. F. (1998) Tumor necrosis factor α transcription in macrophages is attenuated by an autocrine factor that preferentially induces NF-κB p50. Mol. Cell. Biol. 18, 5678–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kunsch C., Rosen C. A. (1993) NF-κ B subunit-specific regulation of the interleukin-8 promoter. Mol. Cell. Biol. 13, 6137–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ledebur H. C., Parks T. P. (1995) Transcriptional regulation of the intercellular adhesion molecule-1 gene by inflammatory cytokines in human endothelial cells. Essential roles of a variant NF-κ B site and p65 homodimers. J. Biol. Chem. 270, 933–943 [DOI] [PubMed] [Google Scholar]