Background: Histone acetyltransferase MORF4L1 forms a homodimer to perform its epigenetic function, but the molecular mechanisms for its homodimerization are unknown.
Results: Histone deacetylase HDAC2 deacetylates MORF4L1 at Lys-148 to enhance MORF4L1 homodimerization.
Conclusion: HDAC2-dependent deacetylation of MORF4L1 enhances MORF4L1 homodimerization, which facilitates complex formation to repress cellular proliferation.
Significance: The molecular control of MORF4L1 homodimerization may impact fundamental cellular processes such as proliferation.
Keywords: Epigenetics, Histone Acetylase, Histone Deacetylase, Posttranslational Modification, Proliferation, Protein Complexes, Protein Structure, Site-directed Mutagenesis
Abstract
Histone acetyltransferase mortality factor 4-like 1 (MORF4L1) is a relatively new histone acetyltransferase component that exists as a homodimer to exert its epigenetic function. The mechanism of MORF4L1 self-assembly is unknown. Here we report that Lys-148 deacetylation is indispensable for facilitating MORF4L1 self-assembly into a homodimeric unit. Among a stretch of ∼10 amino acids in the NH2 terminus between the chromodomain and MORF4-related gene (MRG) domain within MORF4L1, Lys-148 is normally acetylated. Substitution of Lys-148 with arginine augments MORF4L1 self-assembly. However, acetylation mimics of MORF4L1, including K148L and K148Q, abolished its self-assembly of the histone acetyltransferase component. HDAC2, a deacetylase, interacts with and keeps MORF4L1 in a deacetylation status at Lys148 that triggers MORF4L1 self-assembly. Knockdown of HDAC2 reduces MORF4L1 self-assembly. HDAC2-dependent deacetylation of MORF4L1 enhances MORF4L1 homodimerization, thus facilitating the functionality of complex formation to repress cell proliferation.
Introduction
Histone posttranslational modification has emerged as a major epigenetic mechanism that regulates a variety of life processes (1–3). Mortality factor 4-like 1 (MORF4L1) is one member of a subgroup of histone acetyltransferases belonging to the mortality factor on chromosome 4 (MORF4) class of proteins (4, 5). Most histone acetyltransferases activate gene transcription and promote cell proliferation. In contrast, MORF4 was initially cloned and characterized as a senescence gene because cellular expression of MORF4 produces massive cell death and senescence (5, 6). MORF4L1 was cloned from chromosome 15 as a paralogue of MORF4, and it is also termed mortality factor-related gene on chromosome 15 (MRG15) (7). Sequence analysis indicates that the primary amino acid composition is highly identical (94%) between MORF4 and MORF4L1. However, MORF4 lacks a chromodomain located within the NH2 terminus of MORF4L1 (8). MORF4L1 is highly conserved from single-cell eukaryotic yeast to vertebrates and mammals, indicating a fundamental role for this gene. MORF4L1 is also ubiquitously expressed. Deletion of MORF4L1 gene in knockout mice leads to embryonic lethality and developmental delay (9–11). MORF4L1 has been reported to be potentially important in neuronal development (7, 12), and it may be proapoptotic when expressed in cells (13). Further, MORF4L1 null and heterozygotic fibroblast cells exhibit DNA repair defects (14). MORF4L1 may also participate in the maintenance of the interphase of the cell cycle (15). Genome-wide association studies implicate MORF4L1 in coronary artery disease (16) and breast cancer (17).
Like other histone acetyltransferases, MORF4L1 forms a complex to execute its epigenetic function. MORF4L1 has been reported to interact with retinoblastoma protein (Rb) and Pam14 (18, 19), with histone acetyltransferase hMOF (human males absent on the first) (20), and with two histone acetyltransferases, Sin3A and TLE (transducing-like enhancer of split) (5). MORF4L1 forms a corepressor complex with PF1, HDAC1/2, and Sin3A (21, 22). These molecular interactions endow MORF4L1 with dual roles in both transcriptional activation and repression. Crystallographic studies indicate that MORF4L1 forms a homodimer (23, 24). Several functional domains within MORF4L1 have been studied. A chromodomain (CD)3 at its NH2 terminus is believed to bind to methylated histone lysine residues (19). The CD specifically binds to trimethylated histone H3K36 and bimethylated H2BK4 (23, 25). An MRG domain resides within the carboxyl terminus of MORF4L1 (5, 24). This domain contains the acetyltransferase activity that modifies lysine residues of histone substrates at a lower affinity and a leucine zipper domain that provides a platform to mediate interaction with many transcriptional regulators (23). Between the CD and MRG domain, a stretch of amino acids of about 20 residues may act as a nuclear localization signal to shift MORF4L1 from the cytoplasmic compartment into the nucleus to exert its epigenetic function.
Homodimeric formation of MORF4L1 is important in regulating its acetyltransferase activity (22). Interestingly, the 20-amino acid span between the CD and MRG domain contains several lysine residues typical of a nuclear localization signal. We observed that several residues within this region are necessary for MORF4L1 homodimerization. In particular, a lysine residue (Lys-148) within this region serves as a major acetylation site of MORF4L1 and is critical for its homodimerization. Importantly, reversible acetylation at this acceptor site serves as a molecular signature in the maintenance of the MORF4L1 homodimeric formation and function.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
Murine lung epithelial (MLE-12) cells were maintained with HITES medium (500 ml of DMEM/F12, 2.5 mg of insulin, transferrin, sodim selenite, 2.5 mg of transferrin, 10 μm hydrocortisone, 10 μm β-estradiol, 10 mm Hepes, and 2 mm l-glutamine) supplemented with 10% FBS in a 37 °C incubator and 5% CO2 as described previously (26). MORF4L1 antibody and Myc antibody were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). V5 antibody was from Invitrogen. Acetyl-lysine antibody, HDAC2 antibody, and PF1 antibody were from Sigma. HDAC2 shRNA retroviral plasmid was from Origene (Rockville, MD).
Plasmid Expression and shRNA Knockdown
Cells were nucleofected with plasmids as described previously (16). Briefly, 1 million MLE cells in their exponential growth stage were suspended in 100 μl of nucleofection buffer (20 mm of Hepes in PBS buffer) and well mixed with 3 μg of plasmid DNA in an electroporation cuvette. Electroporation was performed with preset program T-013 in the NucleofectionTM II system (Amaxa Biosystems, Gaithersburg, MD), and the cells were cultured in 2 ml of complete HITES medium for 48 h. For gene silencing, a retroviral shRNA plasmid was also delivered into cells by using nucleofection with the same protocol, and 4 μg of retroviral shRNA plasmid was used in each nucleofection. In some experiments, cells were first nucleofected with shRNA plasmid for 24 h and followed by coexpression of V5- or Myc-tagged MORF4L1 plasmids for an extra 24 h. After 48 h of incubation, cells were subjected to immunoprecipitation and immunoblotting analysis.
Immunoblot Analysis
Immunoblotting was performed as described previously (26). Cell lysates were prepared by brief sonication of harvested cells in buffer A (150 mm NaCl, 50 mm Tris-HCl, 1 mm EDTA, 2 mm DTT, 0.025% sodium azide, 1 mm phenylmethylsulfonyl fluoride, 1% Triton X-100, and appropriate amounts of protease inhibitor mixture (pH 7.4)). The cell lysates were separated by SDS-PAGE and subjected to immunoblotting analysis. Images were obtained with a Carestream Kodak in vivo Image F Pro System. Antibodies were diluted 1:1000 except when specifically indicated.
Size Exclusion Spin Column Fractionation
Cells expressed with wild-type or the mutant MORF4L1 plasmid were harvested and lysed with buffer (150 mm NaCl, 50 mm Tris-HCl, 1 mm EDTA, 2 mm DTT, 0.1% Triton X-100 (v/v), and 1:1000 protease inhibitor mixture). Cell lysates were subjected to size exclusion spin column fractionation (Millipore, Billerica, MA) following the protocol of the manufacturer. Fractions of less than 50 kDa, 50–100 kDa, and more than 100 kDa were collected for immunoblot analysis.
Protein Expression and Purification
Recombinant NH2 terminus-truncated MORF4L1 (amino acids 130–362) was expressed and purified following the instructions of the manufacturer (Invitrogen). Briefly, MORF4L1 recombinant protein expression was conducted by introducing the pcDNA3.1/MOF4L1 (amino acids 130–362) plasmid into Escherichia coli strain BL21. Recombinant protein was purified with NI60 his-resin. The elution was separated by electrophoresis and visualized by Coomassie Blue stain.
Pulldown Assay
Recombinant proteins of V5-tagged or Myc-tagged wild-type MORF4L1 and the cysteine mutants C19S, C30S, and C19S/C30S were generated by using a TnT® coupled reticulocyte lysate transcription/translation system (Promega, Madison, WI) following the instructions of the manufacturer. The recombinant proteins were mixed together in pulldown buffer (150 mm NaCl, 50 mm Tris-HCl, 1 mm EDTA, 0.3% Triton X-100 (v/v), and 1:1000 protease inhibitor mixture) and rotated in 4 °C for 2 h. V5 antibody conjugated with agarose beads (30 μl) was added to the mixture and incubated overnight. Beads were spun down and washed with pulldown buffer three times. The precipitates were subjected to immunoblot analysis.
Immunoprecipitation
MLE cells during exponential growth were lysed with lysis buffer (150 mm NaCl, 50 mm Tris-HCl, 1 mm EDTA, 2 mm DTT, 0.3% Triton X-100 (v/v), and 1:1000 protease inhibitor mixture). Cells were subjected to sonication, and samples were spun at 13,000 rpm for 10 min. Cell lysates (containing 1 mg of protein) were incubated and rotated with 2 μg of V5 antibody at 4 °C overnight. After incubation with 30 μl of protein A/G-agarose beads for another 3 h, beads were spun down and washed with lysis buffer three times. The immunoprecipitates were mixed with 50 μl of SDS loading dye (containing 1% SDS and 10 mm DTT) and heated at 95 °C for 5 min. The samples were subjected to SDS-PAGE and immunoblot analysis.
Deletion Mutation and Site-directed Mutagenesis
Human MORF4L1 cDNA was purchased from Origene, and the coding region of the gene was cloned into pcDNA 3.1 by PCR using the forward primer 5′-atggcgccgaagcaggacccg-3′ and reverse primer 5′-cacagctttccgatggtactcagg-3′. NH2-terminal truncations of MORF4L1 were generated by PCR using the following primers: 5′-atgggggctgccccaggaaagaag-3′ (forward) and 5′-cacagctttccgatggtactcagg-3′ (reverse), 5′-atgcaacagaaaaatgttgaagtgaaaacg-3′ (forward) and 5′-cacagctttccgatggtactcagg-3′ (reverse), and 5′-atgaagaacaaacagaaaacacctggaaatgg-3′ (forward) and 5′-cacagctttccgatggtactcagg-3′ (reverse), respectively. MORF4L1 site-directed mutants of K135R, K136R, K143R, K148R, K150R, K148L, K148Q, C19S, C30S, and C19S/C30S were generated by site-directed mutagenesis (Stratagene, La Jolla, CA) with the following primers: K135R, 5′-gggctgccccaggaaggaagacatctggtctgcaacag-3′ (forward) and 5′-ctgttgcagaccagatgtcttccttcctggggcagccc-3′ (reverse); K136R, 5′-Gggctgccccaggaaagaggacatctggtctgcaacag-3′ (forward) and 5′-ctgttgcagaccagatgtcctctttcctggggcagccc-3′ (reverse); K143R, 5′-ctggtctgcaacagagaaatgttgaagtgaaaacg-3′ (forward) and 5′-cgttttcacttcaacatttctctgttgcagaccag-3′ (reverse); K148R, 5′-gaagtgaaaacgaaaaggaacaaacagaaaacacctg-3′ (forward) and 5′-caggtgttttctgtttgttccttttcgttttcacttc-3′ (reverse); K148L, 5′-gaagtgaaaacgaaactgaacaaacagaaaacacctg-3′ (forward) and 5′-caggtgttttctgtttgttcagtttcgttttcacttc-3′ (reverse); K148Q, 5′-gaagtgaaaacgaaacagaacaaacagaaaacacctg-3′ (forward) and 5′-caggtgttttctgtttgttctgtttcgttttcacttc-3′ (reverse); K150R forward (5′-gtgaaaacgaaaaagaacagacagaaaacacctgg-3′) and 5′-cctggtgttttctgtctgttctttttcgttttcac-3′ (reverse); C19S, 5′-ggtgagcgagtgctgtcctttcatgggcctcttc-3′ (forward) and 5′-gaagaggcccatgaaaggacagcactcgctcacc-3′ (reverse); and C30S, 5′-ctttatgaagcaaagtctgtaaaggttgccataaag-3′ (forward) and 5′-ctttatggcaacctttacagactttgcttcataaag-3′ (reverse).
Cell Growth Analysis
Cell proliferation was assayed as described previously (27). MLE cells were transduced to overexpress wild-type MORF4L1, K135R, K148R, and K150R, respectively. The cells were seeded at 3 × 104 cells/ml in 6-well plates and allowed to grow in a standard cell culture incubator. For each cell line, three independent wells were harvested after 48 h post-seeding. The cells were counted using a T10 automated cell counter (Bio-Rad).
Statistics
All data were statistically analyzed by Student's t test and presented as mean ± S.E.
RESULTS
MORF4L1 Is a Homodimer
Gel filtration and size exclusion chromatographic studies reported that histone acetyltransferase MORF4L1 is located in two fractions: one is around 70 kDa and another approximately 700 kDa (20, 28). The primary sequence of MORF4L1 is comprised of 362 amino acids. Crystallographic studies show that MORF4L1 is a homodimer and that MORF4L1 forms a complex with other transcription factors to exert its epigenetic function (22). To confirm these observations, we fractioned the cell lysates with a size exclusion spin column followed by immunoblot analysis. Low levels of MORF4L1 were detected in a monomeric form. MORF4L1 was predominately detected in the fractions of 50–100 kDa and a more than 100-kDa-size fraction (Fig. 1A). These data are consistent with published observations, indicating that MORF4L1 may be a homodimer, as reflected by its size (50–100 kDa), and a component of the large corepressor complex (20, 28). To further test this hypothesis, we constructed V5-tagged and Myc-tagged MORF4L1 expression plasmids, coexpressed the plasmids in MLE cells, and subjected the cell lysates to coimmunoprecipitation analysis. Results from V5 immunoprecipitation showed that Myc-tagged MORF4L1 interacts with V5-tagged MORF4L1 (Fig. 1B). We then synthesized V5-tagged and Myc-tagged recombinant MORF4L1 proteins for in vitro binding assays. Pulldown assays proved that V5-tagged recombinant MORF4L1 interacts with Myc-tagged MORF4L1 protein (Fig. 1C). These data indicate that MORF4L1 exists as a dimer and a component in the high molecular complex under normal culture conditions.
FIGURE 1.
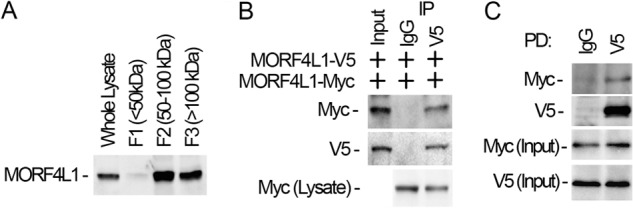
MORF4L1 is a homodimer. A, MLE cells (6 × 100 mm dish) were collected, lysed, and then cellular proteins were applied to a size exclusion spin column. The fractions were collected and separated by SDS-PAGE followed by immunoblotting analysis. B, V5- and Myc-tagged MORF4L1 were coexpressed in cells. Cell lysates were subjected to V5 immunoprecipitation (IP). The precipitates were analyzed by immunoblotting as indicated. C, V5- and Myc-tagged MORF4L1 were synthesized in vitro. The recombinant proteins were mixed with V5 antibody-conjugated agarose beads overnight. The washed beads were analyzed by immunoblotting as indicated. Input was 10% of the recombinant protein used in the pulldown (PD) assays. These data are representative of three separate experiments.
Deletion of the MORF4L1 NH2 Terminus Ablates Its Homodimerization
Because MORF4L1 appears to form a homodimer, we examined the mechanism of its homodimeric formation. In general, cysteine residues within a protein play a crucial role in dimerization. In the NH2 terminus of MORF4L1, there are two cysteine residues, Cys-19 and Cys-30. To assess the contribution of these molecular sites in MORF4L1 homodimerization, we generated MORF4L1-V5 cysteine point mutant plasmids and expressed the plasmids in cells. The immunoprecipitation results indicate that MORF4L1 interacts with each cysteine variant (Fig. 2A). Interestingly, the C19S mutant, a C30S mutant, and a C19S/C30S double mutant each bind to wild-type MORF4L1 at lower but comparable levels as wild-type MORF4L1. These data suggest that two cysteine residues within MORF4L1 do not play essential roles in MORF4L1 homodimerization. To explore further molecular elements that may impact MORF4L1 homodimerization, we generated MORF4L1 deletion mutant plasmids, expressed them in cells, and performed coimmunoprecipitation to better define the self-interaction region within MORF4L1 (Fig. 2B). The results showed that deletion of the first 130 amino acids of the NH2-terminal chromodomain does not affect its homodimerization (Fig. 2C). We then deleted an additional 10 amino acids within the NH2 terminus of MORF4L1 and found that deletion of 140 amino acids slightly affected its homodimerization. Notably, deletion of 150 amino acids in its NH2 terminus completely abolished MORF4L1 homodimeric formation (Fig. 2C). In these studies, cotransfection of V5-truncated MORF4L1 mutants with Myc-tagged, wild-type MORF4L1 plasmids resulted in a lower level of expression of the truncated variant plasmids. However, the absence of any V5 signal despite modest expression of the 150–362 MORF4L1 variant (Fig. 2C, top row, right lane versus bottom row) strongly suggests that a region harbors molecular signatures important in protein dimerization. This region between the chromodomain and the MRG domain contains a stretch of amino acids of ∼20 amino acids in length. In addition, we expressed and purified a NH2-terminal truncated MORF4L1 (amino acids 130–362). The recombinant protein was separated by electrophoresis and visualized by Coomassie Blue stain (Fig. 2D). The results show that the NH2 terminus-truncated MORF4L1 efficiently forms a dimer. These results suggest that the amino acids from 140–150 are necessary for MORF4L1 homodimerization.
FIGURE 2.
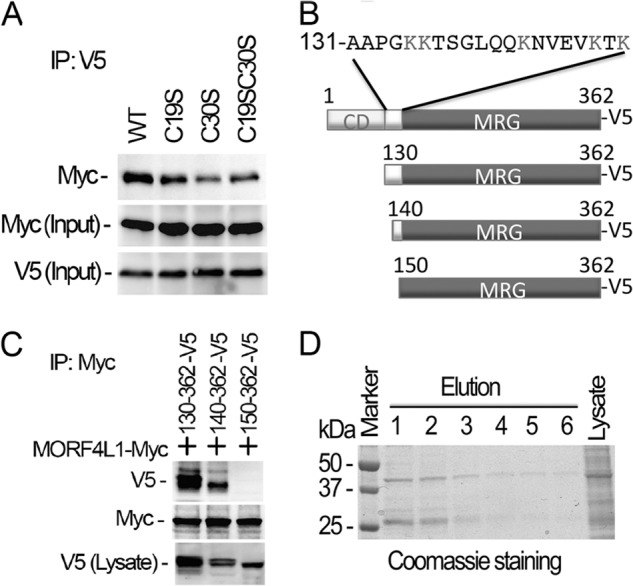
Deletion of the NH2 terminus of MORF4L1 ablates its homodimerization. A, MORF4L1 cysteine residue mutants tagged with V5 were coexpressed with Myc-tagged, wild-type MORF4L1 in the cells. The cell lysates were used for immunoprecipitation (IP) studies as indicated. Input was 10% of the cell lysate used in the studies. B, schematic presentation of the deletion mutations. C, NH2-terminal deletion mutants were expressed in MLE cells. The cell lysates were subjected to V5 immunoprecipitation, and the immunoprecipitates were analyzed by immunoblotting as indicated. NH2-terminal deletional mutant expression in the cells was checked by V5 immunoblotting. D, purified NH2-terminal truncated MORF4L1 forms a dimer efficiently. NH2-terminal truncated MORF4L1 was expressed and purified. Elution products were separated by electrophoresis, and the proteins were visualized by Coomassie Blue stain. These data are representative of three separate experiments.
Replacement of Lys-148 with Arginine Enhances Its Dimerization, whereas Acetylation Mimics Block MORF4L1 Self-dimerization
The 20 amino acids between the chromodomain and MRG domain are enriched with five lysine residues (Fig. 2B). Lysine residues are highly prone to a variety of modifications, particularly acetylation, methylation, and ubiquitination. We hypothesized that posttranslational modification of these lysine residues may affect MORF4L1 homodimerization. Thus, we carried out site-directed mutagenesis by substitution of the lysine residue with a similarly charged arginine. We expressed the point mutant MORF4L1 plasmids in cells and processed cells as above to evaluate MORF4L1 homodimerization. Interestingly, among the mutants, we found that mutant K148R remarkably enhanced MORF4L1 homodimerization ∼3.7-fold (Fig. 3, A and B). Because arginine substitution at this molecular site would likely result in an unmodified residue, the data suggest that Lys-148 deacetylation may be required for MORF4L1 homodimerization. Following this clue, we constructed acetylation mimics of MORF4L1 (K148Q and K148L plasmid mutants (myc-tagged)) and coexpressed these acetylation mimic plasmids in cells together with wild-type V5-MORF4L1 to assess homodimer formation ability. Strikingly, none of the acetylation mimic mutants bind to wild-type MORF4L1 (Fig. 3C). We also treated the cells with the acetyltransferase inhibitor anacardic acid (50 mm) and observed that MORF4L1 homodimerization was augmented (Fig. 3D). Conversely, treatment of the cells with the deacetylase inhibitor trichastatin A, a pan-HDAC deacetylase inhibitor, effectively abrogated MORF4L1 homodimerization. These data demonstrate that deacetylation of MORF4L1 at Lys-148 is crucial for its homodimerization.
FIGURE 3.
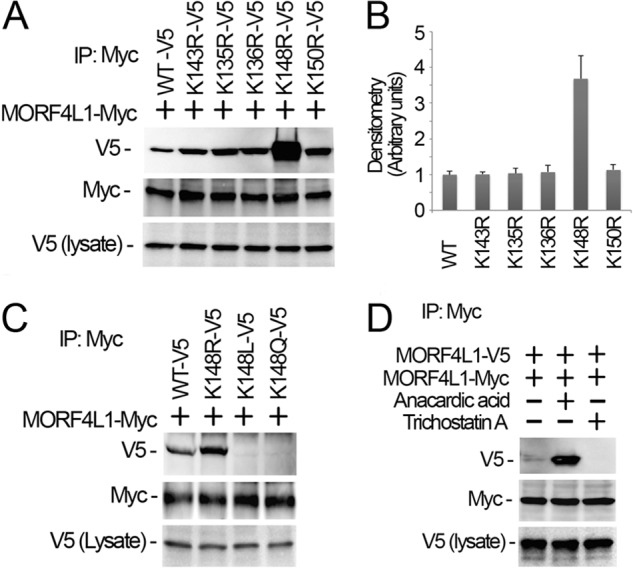
Reversible MORF4L1 acetylation at Lys-148 modulates its dimerization. A and B, lysine mutants and myc-tagged, wild-type MORF4L1 were coexpressed in cells. Cell lysates were subjected to myc immunoprecipitation (IP), and the precipitates were analyzed by V5 and myc immunoblot analysis. The cell lysates were analyzed with V5 immunoblotting to verify MORF4L1 mutant expression. The densitometry results of A were plotted in B. C, K148R and the acetylation mimic K148L and K148Q mutants were coexpressed with myc-tagged, wild-type MORF4L1 in the cells. The cell lysates were used for myc immunoprecipitation, followed by immunoblotting analysis as indicated. MORF4L1 mutant expression levels were analyzed by V5 immunoblotting. D, wild-type of MORF4L1 tagged with V5 or myc were expressed in cells for 40 h, and cells were treated with anacardic acid (final concentration of 50 μm) or trichastatin A (final concentration of 100 nm) for 6 h. Cell lysates were subjected to myc immunoprecipitation and followed by immunoblot analysis as indicated. Tagged MORF4L1 expression was verified by immunoblotting. These data are representative of three separate experiments.
HDAC2 Interacts with MORF4L1 and Is Critical for MORF4L1 Homodimerization
Histone deacetylase HDAC2 is a critical enzyme that interacts with and modifies a variety of substrates to regulate gene transcriptional activity. To evaluate whether MORF4L1 is associated with HDACs, we conducted MORF4L1 immunoprecipitation followed by HDAC2 immunoblotting. The results showed that MORF4L1 coprecipitated with HDAC2 (Fig. 4A). In reciprocal studies, we performed HDAC2 immunoprecipitation. Analyzing the HDAC2 precipitates, we detected MORF4L1, indicating that MORF4L1 interacts with HDAC2 (Fig. 4B). Because MORF4L1 interacts with HDAC2, we next tested whether HDAC2 is necessary for MORF4L1 homodimerization. We knocked down HDAC2 by introduction of a retroviral HDAC2 shRNA plasmid into MLE cells. Immunoblot analysis indicated that HDAC2 is effectively silenced at the protein level (Fig. 5A). We then checked MORF4L1 homodimerization and found that MORF4L1 homodimerization is abrogated in HDAC2-silenced cells (Fig. 5B). To rule out the possibility that HDAC2 may act as a scaffold to recruit MORF4L1 binding, we conducted V5- and Myc-tagged MORF4L1 in vitro binding assays in the absence or presence of HDAC2 protein. The addition of HDAC2 protein does not affect MORF4L1 self-binding, indicating that HDAC2 does not act as a scaffold to recruit MORF4L1 (Fig. 5C). These results indicate that HDAC2 is associated with and necessary for MORF4L1 homodimerization.
FIGURE 4.
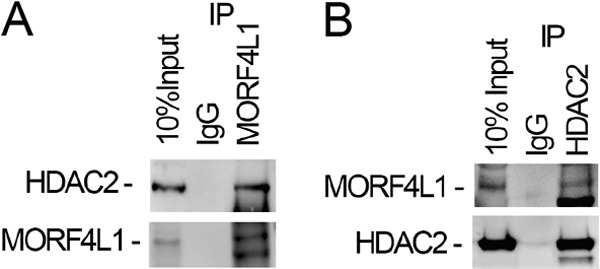
HDAC2 interacts with MORF4L1. A, cell lysates were used for MORF4L1 immunoprecipitation (IP). The precipitates were analyzed by HDAC2 immunoblotting. B, reciprocally, cell lysates were immunoprecipitated with HDAC2 antibody, and the precipitates were immunoblotted using MORF4L1 antibody. These data are representative of three separate experiments.
FIGURE 5.
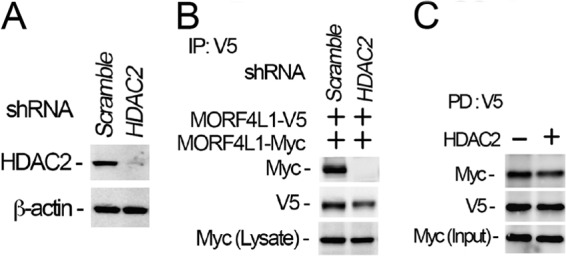
HDAC2 is necessary for MORF4L1 self-assembly. A, HDAC2 was depleted in cells by introducing shRNA retroviral constructs for 48 h. Cell lysates were analyzed with HDAC2 immunoblotting. A scrambled retroviral plasmid was used as a shRNA specificity control. β-actin was used as a loading control. B, HDAC2 was first knocked down by shRNA for 24 h in the cells, and then V5- or Myc-tagged MORF4L1 were expressed for another 24 h. Cell lysates were analyzed by V5 immunoprecipitation (IP), followed by immunoblotting. C, V5- and Myc-tagged MORF4L1 and HDAC2 were synthesized in vitro and the MORF4L1 recombinant proteins were mixed with V5 antibody-conjugated agarose beads in the presence or absence of HDAC2 overnight. The washed beads were analyzed by immunoblotting as indicated. Input was 10% of the recombinant protein used in the pulldown (PD) assays. These data are representative of three separate experiments.
Lys-148 within MORF4L1 Is the HDAC2-dependent Deacetylation Site
Because HDAC2 mediated deacetylation is critical to maintain MORF4L1 homodimerization, we next determined whether Lys-148 is a HDAC2-dependent deacetylation site. Under normal culture conditions, MORF4L1 is highly acetylated. To determine the major acetylation site(s) within the 20-amino acid region, we expressed the lysine mutants and analyzed the acetylation levels of the mutants by V5 tag immunoprecipitation, followed by acetyl-lysine immunoblot analysis. Results showed that both the K143R and K148R mutants exhibited reduced acetylation levels, indicating that Lys-148 is one of the acetylation sites (Fig. 6A). In addition, knockdown of HDAC2 increased wild-type MORF4L1 but not K148R mutant acetylation levels (Fig. 6B). These data suggest that Lys-148 is an HDAC2-dependent deacetylation molecular site within MORF4L1. We then examined whether the Lys-148 acetylation and deacetylation statuses may impair MORF4L1 homodimerization. Wild-type MORF4L1 and the K148R, K148Q, and K148L mutants were expressed in cells. Thereafter, the cells were harvested and lysed, and cellular proteins were subjected to size exclusion spin columns. The individual fractions were then analyzed by immunoblotting. The results showed, remarkably, that the acetylation mimic mutants displayed reduced MORF4L1 homodimerization, whereas deacetylation enhanced MORF4L1 homodimerization (Fig. 6C).
FIGURE 6.
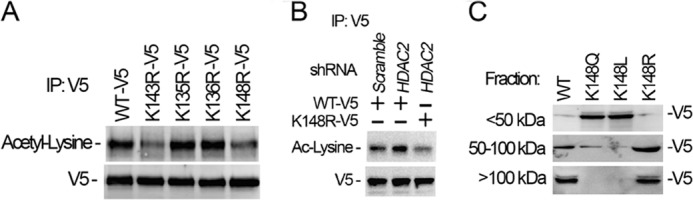
HDAC2-dependent deacetylation of MORF4L1 at Lys-148. A, wild-type or a variety of lysine-mutant MORF4L1 plasmids were expressed in the cells. Cell lysates were subjected to V5 immunoprecipitation (IP), and the precipitates were analyzed by acetyl lysine immunoblotting. B, HDAC2 was depleted by introducing shRNA retroviral constructs into the cells for 24 h. Wild-type or the K148R mutant MORF4L1 plasmid was expressed in cells for another 24 h. Cell lysates were analyzed with V5 immunoprecipitation, followed by immunoblot analysis. A scrambled retroviral plasmid was used as a shRNA specificity control. C, wild-type, K148R, or acetylation mimic mutant MORF4L1 plasmids were expressed in cells. The cell lysates were fractionated by size exclusion spin columns, and all fractions were analyzed by immunoblotting. These data are representative of three separate experiments.
Dimerization Augments MORF4L1 Corepressor Complex Formation and Regulates Cell Proliferation
We next studied whether MORF4L1 homodimerization would affect MORF4L1 corepressor complex formation. MORF4L1 forms a corepressor complex with PF1. Thus we checked whether knockdown of HDAC2 would affect complex formation. As expected, knockdown of HDAC2 abolished MORF4L1 and PF1 interaction and reduced Sin3A association (Fig. 7A). Although HDAC2 knockdown may directly impair complex formation, one possibility is via its deacetylation of Lys-148 at MORF4L1. To test this possibility, we expressed the K148R mutant plasmid in cells and observed that this deacetylated mutant enhanced MORF4L1 and PF1-Sin3A interactions compared with the wild type of MORF4L1 (Fig. 7B). These data indicate augmented complex formation by deacetylation of Lys-148 that directs MORF4L1 homodimerization. Further, to evaluate a physiologic role of these constructs, we expressed the K148R mutant plasmid and assayed cell proliferation. The cell proliferation rate in K136R- and K150R-transfected cells were at comparable levels with wild-type MORF4L1. However, expression of the K148R mutant resulted in significantly reduced cell proliferative rates (Fig. 7C). As a control, protein levels of wild-type MORF4L1 and mutants were equally expressed (Fig. 7C, top panel). These data indicate that HDAC2-dependent deacetylation of MORF4L1 at Lys-148 enhances corepressor complex assembly and that deacetylation at this molecular site is important in modulating cell proliferation.
FIGURE 7.
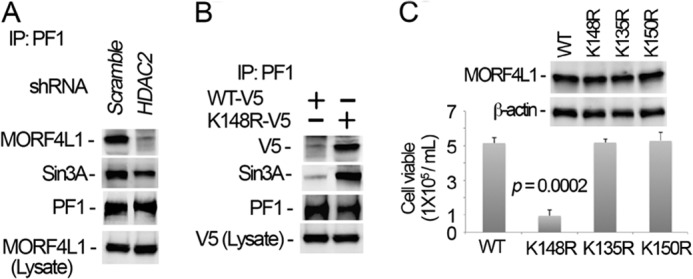
Dimerization augments MORF4L1 complex formation and cellular viability. A, HDAC2 was depleted in cells by introducing shRNA retroviral constructs for 48 h. Cell lysates were immunoprecipitated (IP) with PF1 antibody, and the precipitates were analyzed by immunoblotting. A scrambled retroviral plasmid was used as a shRNA specificity control. B, wild-type and K148R mutant MORF4L1 plasmids were coexpressed in cells for 48 h. Cell lysates were used for PF1 immunoprecipitation, followed by immunoblot analysis. Wild-type and K148R mutant MORF4L1 plasmids in the cells were validated by V5 immunoblotting. C, wild-type and K135R, K148R, and K150R mutant MORF4L1 plasmids were expressed in the cells for 48 h. Viable cells were counted in each transfected cell line. The expression levels of the proteins were analyzed by immunoblotting. The data are representative of three separate experiments.
DISCUSSION
The major finding in this study is that HDAC2-dependent deacetylation of MORF4L1 at Lys-148 regulates MORF4L1 homodimerization and that this dimeric assembly augments MORF4L1 corepressor complex formation to regulate cell proliferation. Gel filtration and size exclusion chromatographic studies demonstrate that MORF4L1 exists in the cells mainly as two complexes (20, 28). MORF4L1 exists as a component of a large complex, such as the corepressor Sin3A-PF1-HDAC1/2-MORF4L1 complex, at a molecular weight of ∼670 kDa. MORF4L1 also appears to be the component in a smaller complex at a molecular weight of around 70 kDa as a homodimer (20, 28). MORF4L1 is 362 amino acids in length. Thus, the molecular mass of a homodimer of MORF4L1 is ∼70 kDa. Crystallographic studies identified that two copies of MORF4L1 are detected in the corepressor complex (22). Consistent with these reports, we demonstrate that MORF4L1 interacts with itself to form a homodimer, as shown in our studies of coimmunoprecipitation, pulldown assays, size exclusion fractionation, and bands from Coomassie staining of gels using purified recombinant protein electrophoresis. The biological function of MORF4L1 homodimers is not clear. One possibility is that MORF4L1 homodimers probably exist as an intermediate product for further assembly into larger functional complexes, such as a corepressor complex.
In general, cysteine residues play an indispensable role in dimerization by forming a disulfide bond between two subunits. Interestingly, two cysteine residues in the NH2 terminus within MORF4L1 are not essential in MORF4L1 homodimeric assembly because replacement of these two residues does not totally impair MORF4L1 homodimerization. We speculate that these residues may contribute to further assembly with other components in corepressor formation. Instead, our site-directed mutagenesis and deletional studies indicate that Lys-148 plays an important role in homodimerization. Lysine residues are prone to many posttranslational modifications, such as acetylation, methylation, and ubiquitination. As key posttranslational modifications, acetylation or deacetylation of lysine residues contribute to protein dimerization of other signaling proteins. For example, dimerization of signaling transducer and activator of transcription 3 (STAT3) is triggered by histone acetyltransferase p300-mediated acetylation (29). Deacetylation by HDAC1 abolishes STAT3 dimerization. Conversely, acetylation mimic mutants of Lys-148 impair MORF4L1 homodimerization, whereas deacetylation of the protein enhances MORF4L1 homodimerization. Silencing of HDAC2 by shRNA or treatment of the cells with the deacetylation inhibitor trichastatin A completely inhibits MORF4L1 dimerization. Notably, both MORF4L1 and histone deacetylase HDAC2 proteins are components associated with a corepressor complex. These observations underscore a unique model for deacetylation in MORF4L1 assembly. Furthermore, HDAC1/2 also exist as heterodimers or homodimers (30), and dimerization of HDAC1/2 is tightly regulated as well. A recent study indicates that phosphorylation controls HDAC1/2 heterodimer or homodimer formation during mitosis (30). HDAC1/2 are highly phosphorylated, and protein kinase CK2 phosphorylates HDAC1/2. Hyperphosphorylation of HDAC1/2 impairs heterodimeric but not homodimeric formation. Interestingly, the phosphorylation status did not affect the integrity of the corepressor complex.
It is believed that the role of HDAC1/2 as a component in the corepressor complex may deacetylate acetylated lysine residues in the histone tail. Our results show HDAC2-dependent deacetylation of MORF4L1, suggesting that one of the functions of HDAC1/2 in the corepressor complex is to keep MORF4L1 in a deacetylated status to maintain integrity of the corepressor complex. It will be interesting to determine when the deacetylation occurs, either before complex formation or while the complex is being assembled in vivo. One possibility is that MORF4L1 may be deacetylated before complex formation because a homodimer is identified as an intermediate product, as shown in size exclusion chromatographic studies. How the corepressor complex is assembled has yet to be characterized. The order of the members in the assembly, the modification status of each subunit in the complex, and the overall pathophysiological settings that trigger complex formation are yet to be studied.
Given the fact that MORF4L1 has been reported to be involved in many disorders, further investigations on the molecular assembly and functionality of MORF4L1 may shed light onto diverse processes, including neoplasia and inflammation. For example, given the role of histone acetyltransferases in cell proliferation, the status of MORF4L1 complex formation in human tumors may provide insight into chemotherapeutic targeting of MORF4L1 in specific pathological settings. This may involve specific modulation of subunits expressed with MORF4L1 or against broader targets (e.g. HDACs). The Lys-148 acetylation/deacetylation status may also provide a unique platform to regulate corepressor complex integrity, thus modulating the function of the acetyltransferase. Further investigation of the modification of molecular acceptor sites between the CD and MRG domain of MORF4L1 are of interest, as is their physiologic role in governing MORF4L1 localization and acetyltransferase activity. In this regard, there are five lysine residues within this bridging region. The studies of posttranslational modification of these lysine residues and their purported roles in MORF4L1 behavior are ongoing.
This work was supported, in whole or in part, National Institutes of Health Grants HL096376, HL097376, HL098174, and P01 HL114453 (to R. K. M.). This work was also supported by American Heart Association Award 12SDG12040330 (to C. Z.); by the United States Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development; and by a merit review award from the United States Department of Veterans Affairs.
- CD
- chromodomain
- HDAC
- histone deacetylase
- MLE
- murine lung epithelial
- MRG
- MORF4-related gene.
REFERENCES
- 1. Qi H. H., Sarkissian M., Hu G. Q., Wang Z., Bhattacharjee A., Gordon D. B., Gonzales M., Lan F., Ongusaha P. P., Huarte M., Yaghi N. K., Lim H., Garcia B. A., Brizuela L., Zhao K., Roberts T. M., Shi Y. (2010) Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466, 503–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Price B. D., D'Andrea A. D. (2013) Chromatin remodeling at DNA double-strand breaks. Cell 152, 1344–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Falvo J. V., Jasenosky L. D., Kruidenier L., Goldfeld A. E. (2013) Epigenetic control of cytokine gene expression. Regulation of the TNF/LT locus and T helper cell differentiation. Adv. Immunol. 118, 37–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bertram M. J., Pereira-Smith O. M. (2001) Conservation of the MORF4 related gene family. Identification of a new chromo domain subfamily and novel protein motif. Gene 266, 111–121 [DOI] [PubMed] [Google Scholar]
- 5. Yochum G. S., Ayer D. E. (2002) Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and transducin-like enhancer of Split. Mol. Cell. Biol. 22, 7868–7876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bryce S. D., Forsyth N. R., Fitzsimmons S. A., Clark L. J., Bertram M. J., Cuthbert A. P., Newbold R. F., Pereira-Smith O. M., Parkinson E. K. (1999) Genetic and functional analyses exclude mortality factor 4 (MORF4) as a keratinocyte senescence gene. Cancer Res. 59, 2038–2040 [PubMed] [Google Scholar]
- 7. Matsuoka Y., Matsuoka Y., Shibata S., Ban T., Toratani N., Shigekawa M., Ishida H., Yoneda Y. (2002) A chromodomain-containing nuclear protein, MRG15 is expressed as a novel type of dendritic mRNA in neurons. Neurosci. Res. 42, 299–308 [DOI] [PubMed] [Google Scholar]
- 8. Tominaga K., Pereira-Smith O. M. (2002) The genomic organization, promoter position and expression profile of the mouse MRG15 gene. Gene 294, 215–224 [DOI] [PubMed] [Google Scholar]
- 9. Tominaga K., Kirtane B., Jackson J. G., Ikeno Y., Ikeda T., Hawks C., Smith J. R., Matzuk M. M., Pereira-Smith O. M. (2005) MRG15 regulates embryonic development and cell proliferation. Mol. Cell. Biol. 25, 2924–2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen M., Takano-Maruyama M., Pereira-Smith O. M., Gaufo G. O., Tominaga K. (2009) MRG15, a component of HAT and HDAC complexes, is essential for proliferation and differentiation of neural precursor cells. J. Neurosci. Res. 87, 1522–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang H., Li Y., Yang J., Tominaga K., Pereira-Smith O. M., Tower J. (2010) Conditional inactivation of MRG15 gene function limits survival during larval and adult stages of Drosophila melanogaster. Exp. Gerontol. 45, 825–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boije H., Ring H., Shirazi Fard S., Grundberg I., Nilsson M., Hallbook F. (2013) Alternative splicing of the chromodomain protein Morf4l1 pre-mRNA has implications on cell differentiation in the developing chicken retina. J. Mol. Neurosci. 51, 615–628 [DOI] [PubMed] [Google Scholar]
- 13. Liang Y., Lin J. C., Wang K., Chen Y. J., Liu H. H., Luan R., Jiang S., Che T., Zhao Y., Li de F., Wang da C., Guo L., Sun H. (2010) A nuclear ligand MRG15 involved in the proapoptotic activity of medicinal fungal galectin AAL (Agrocybe aegerita lectin). Biochim. Biophys. Acta 1800, 474–480 [DOI] [PubMed] [Google Scholar]
- 14. Garcia S. N., Kirtane B. M., Podlutsky A. J., Pereira-Smith O. M., Tominaga K. (2007) Mrg15 null and heterozygous mouse embryonic fibroblasts exhibit DNA-repair defects post exposure to γ ionizing radiation. FEBS Lett. 581, 5275–5281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith H. F., Roberts M. A., Nguyen H. Q., Peterson M., Hartl T. A., Wang X. J., Klebba J. E., Rogers G. C., Bosco G. (2013) Maintenance of interphase chromosome compaction and homolog pairing in Drosophila is regulated by the condensin cap-h2 and its partner mrg15. Genetics 195, 127–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coronary Artery Disease (C4D) Genetics Consortium (2011) A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 43, 339–344 [DOI] [PubMed] [Google Scholar]
- 17. Martrat G., Maxwell C. M., Tominaga E., Porta-de-la-Riva M., Bonifaci N., Gómez-Baldó L., Bogliolo M., Lázaro C., Blanco I., Brunet J., Aguilar H., Fernández-Rodríguez J., Seal S., Renwick A., Rahman N., Kühl J., Neveling K., Schindler D., Ramírez M. J., Castellà M., Hernández G., EMBRACE, Easton D. F., Peock S., Cook M., Oliver C. T., Frost D., Platte R., Evans D. G., Lalloo F., Eeles R., Izatt L., Chu C., Davidson R., Ong K. R., Cook J., Douglas F., Hodgson S., Brewer C., Morrison P. J., Porteous M., Peterlongo P., Manoukian S., Peissel B., Zaffaroni D., Roversi G., Barile M., Viel A., Pasini B., Ottini L., Putignano A. L., Savarese A., Bernard L., Radice P., Healey S., Spurdle A., Chen X., Beesley J., kConFab, Rookus M. A., Verhoef S., Tilanus-Linthorst M. A., Vreeswijk M. P., Asperen C. J., Bodmer D., Ausems M. G., van Os T. A., Blok M. J., Meijers-Heijboer H. E., Hogervorst F. B., Hebon, Goldgar D. E., Buys S., John E. M., Miron A., Southey M., Daly M. B., Bcfr, Swe B., Harbst K., Borg A., Rantala J., Barbany-Bustinza G., Ehrencrona H., Stenmark-Askmalm M., Kaufman B., Laitman Y., Milgrom R., Friedman E., Domchek S. M., Nathanson K. L., Rebbeck T. R., Johannsson O. T., Couch F. J., Wang X., Fredericksen Z., Cuadras D., Moreno V., Pientka F. K., Depping R., Caldes T., Osorio A., Benitez J., Bueren J., Heikkinen T., Nevanlinna H., Hamann U., Torres D., Caligo M. A., Godwin A. K., Imyanitov E. N., Janavicius R., Collaborators G. S., Sinilnikova O. M., Stoppa-Lyonnet D., Mazoyer S., Verny-Pierre C., Castera L., de Pauw A., Bignon Y. J., Uhrhammer N., Peyrat J. P., Vennin P., Ferrer S. F., Collonge-Rame M. A., Mortemousque I., McGuffog L., Chenevix-Trench G., Pereira-Smith O. M., Antoniou A. C., Ceron J., Tominaga K., Surralles J., Pujana M. A. (2011) Exploring the link between MORF4L1 and risk of breast cancer. Breast Cancer Res. 13, R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leung J. K., Berube N., Venable S., Ahmed S., Timchenko N., Pereira-Smith O. M. (2001) MRG15 activates the B-myb promoter through formation of a nuclear complex with the retinoblastoma protein and the novel protein PAM14. J. Biol. Chem. 276, 39171–39178 [DOI] [PubMed] [Google Scholar]
- 19. Pardo P. S., Leung J. K., Lucchesi J. C., Pereira-Smith O. M. (2002) MRG15, a novel chromodomain protein, is present in two distinct multiprotein complexes involved in transcriptional activation. J. Biol. Chem. 277, 50860–50866 [DOI] [PubMed] [Google Scholar]
- 20. Hayakawa T., Ohtani Y., Hayakawa N., Shinmyozu K., Saito M., Ishikawa F., Nakayama J. (2007) RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes Cells 12, 811–826 [DOI] [PubMed] [Google Scholar]
- 21. Pena A. N., Pereira-Smith O. M. (2007) The role of the MORF/MRG family of genes in cell growth, differentiation, DNA repair, and thereby aging. Ann. N.Y. Acad. Sci. 1100, 299–305 [DOI] [PubMed] [Google Scholar]
- 22. Kumar G. S., Xie T., Zhang Y., Radhakrishnan I. (2011) Solution structure of the mSin3A PAH2-Pf1 SID1 complex. A Mad1/Mxd1-like interaction disrupted by MRG15 in the Rpd3S/Sin3S complex. J. Mol. Biol. 408, 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang P., Du J., Sun B., Dong X., Xu G., Zhou J., Huang Q., Liu Q., Hao Q., Ding J. (2006) Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 34, 6621–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang P., Zhao J., Wang B., Du J., Lu Y., Chen J., Ding J. (2006) The MRG domain of human MRG15 uses a shallow hydrophobic pocket to interact with the N-terminal region of PAM14. Protein Sci. 15, 2423–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blus B. J., Wiggins K., Khorasanizadeh S. (2011) Epigenetic virtues of chromodomains. Crit. Rev. Biochem. Mol. Biol. 46, 507–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zou C., Butler P. L., Coon T. A., Smith R. M., Hammen G., Zhao Y., Chen B. B., Mallampalli R. K. (2011) LPS impairs phospholipid synthesis by triggering β-transducin repeat-containing protein (β-TrCP)-mediated polyubiquitination and degradation of the surfactant enzyme acyl-CoA:lysophosphatidylcholine acyltransferase I (LPCAT1). J. Biol. Chem. 286, 2719–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zou C., Chen Y., Smith R. M., Snavely C., Li J., Coon T. A., Chen B. B., Zhao Y., Mallampalli R. K. (2013) SCFFbxw15 mediates histone acetyltransferase binding to origin recognition complex (HBO1) ubiquitin-proteasomal degradation to regulate cell proliferation. J. Biol. Chem. 288, 6306–6316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kirkwood K. J., Ahmad Y., Larance M., Lamond A. I. (2013) Characterisation of native protein complexes and protein isoform variation using size-fractionation based quantitative proteomics. Mol. Cell. Proteomics 12, 3851–3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yuan Z. L., Guan Y. J., Chatterjee D., Chin Y. E. (2005) Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science 307, 269–273 [DOI] [PubMed] [Google Scholar]
- 30. Khan D. H., He S., Yu J., Winter S., Cao W., Seiser C., Davie J. R. (2013) Protein kinase CK2 regulates the dimerization of histone deacetylase 1 (HDAC1) and HDAC2 during mitosis. J. Biol. Chem. 288, 16518–16528 [DOI] [PMC free article] [PubMed] [Google Scholar]


