Background: TDP-43 and hnRNPA1/A2 factors are implicated in neurodegeneration.
Results: The human and fruit fly TDP-43 and hnRNPA1/A2 orthologs show physical, genetic, and functional interplays.
Conclusion: The functional cooperation between TBPH/Hrp38 and TDP-43/hnRNP A/B is conserved throughout evolution.
Significance: TBPH/Hrp38 interplay can be critical for neurodegeneration, and Drosophila is a model suitable to study the impact of this interaction.
Keywords: Drosophila, Neuroscience, Retinal Degeneration, RNA Splicing, RNA-binding Proteins, Hrb98DE, Hrp38, TBPH, TDP-43, hnRNP A/B
Abstract
Human TDP-43 represents the main component of neuronal inclusions found in patients with neurodegenerative diseases, especially frontotemporal lobar degeneration and amyotrophic lateral sclerosis. In vitro and in vivo studies have shown that the TAR DNA-binding protein 43 (TDP-43) Drosophila ortholog (TBPH) can biochemically and functionally overlap the properties of the human factor. The recent direct implication of the human heterogeneous nuclear ribonucleoproteins (hnRNPs) A2B1 and A1, known TDP-43 partners, in the pathogenesis of multisystem proteinopathy and amyotrophic lateral sclerosis supports the hypothesis that the physical and functional interplay between TDP-43 and hnRNP A/B orthologs might play a crucial role in the pathogenesis of neurodegenerative diseases. To test this hypothesis and further validate the fly system as a useful model to study this type of diseases, we have now characterized human TDP-43 and Drosophila TBPH similarity in terms of protein-protein interaction pathways. In this work we show that TDP-43 and TBPH share the ability to associate in vitro with Hrp38/Hrb98DE/CG9983, the fruit fly ortholog of the human hnRNP A1/A2 factors. Interestingly, the protein regions of TDP-43 and Hrp38 responsible for reciprocal interactions are conserved through evolution. Functionally, experiments in HeLa cells demonstrate that TDP-43 is necessary for the inhibitory activity of Hrp38 on splicing. Finally, Drosophila in vivo studies show that Hrp38 deficiency produces locomotive defects and life span shortening in TDP-43 with and without animals. These results suggest that hnRNP protein levels can play a modulatory role on TDP-43 functions.
Introduction
TDP-432 (43-kDa TAR DNA-binding protein, TARDBP) is a nuclear factor involved in regulation of mRNA splicing, mRNA stability, and other cellular processes (1). Initially associated with the pathogenesis of monosymptomatic forms of cystic fibrosis transmembrane conductance regulator (CFTR) (2–4), TDP-43 has been found in the ubiquitin-positive cytosolic aggregates of neurons from patients with amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD-U) (5, 6). More recently, pathological TDP-43 inclusions have also been described in several additional neurodegenerative diseases such as Alzheimer, Huntington, and Parkinson diseases (7) and also MSP (formerly known as inclusion body myopathy with early-onset Paget disease and frontotemporal dementia (IBMPFD)/ALS) (8). Presently, however, the molecular mechanisms that link TDP-43 to neurodegeneration are not clear. Interestingly, the observation that in all patients carrying TDP-43 aggregates this region is cleaved and becomes aberrantly phosphorylated has focused particular attention on studying the C terminus of this protein in the pathogenesis of neurodegenerative disorders (9).
At the functional level recent studies have demonstrated that the C-terminal domain is important for TDP-43 splicing activity and is responsible for the interaction of this protein with many other cellular factors and bodies such as stress granules (10–12). In particular, the inhibitory role in pre-mRNA splicing played by TDP-43 relies on its ability to tether different members of the hnRNP A/B family in proximity of the inhibited exon through interactions mediated by the 342–366 Gln/Asn-rich region localized in the C-tail (13, 14). In keeping with this conclusion, mutants of human and Drosophila TDP-43 without this region are unable to interfere with exon inhibition (15). Most importantly, this Gln/Asn-rich sequence is also involved in modulating TDP-43 self-interaction and binding to polyglutamine aggregates, thus playing an important role in reducing TDP-43 natural tendency to aggregate (16, 17).
One open question that still carries considerable importance toward the development of animal models of TDP-43 pathology is to clarify how much the biological interactions and function have been conserved across different species, the reason being that a high degree of similarity will facilitate the use of these models for the development of effectors capable of modulating TDP-43 functional properties in humans. Multiple protein alignment across different species of sequence from TAR DNA-binding protein (TARDBP) have highlighted that this factor has been conserved throughout evolution (15, 18). This very high level of conservation can also be extended to all its basic functional properties. For example, it is clear that even TDP-43s from evolutionarily distant organisms share a binding specificity for similar (UG)-rich RNA sequences (15).
In particular, the functional overlap between human and Drosophila TDP-43 with regard to the splicing process was already well known after the observation that Drosophila TBPH could functionally complement the absence of human TDP-43 in HeLa cells (14).
The biological similarity between these two proteins was even more generally confirmed in a Drosophila melanogaster model of TDP-43 proteinopathy, where expression of the endogenous TBPH gene was abolished (19). The resulting flies showed locomotor dysfunctions and reduced life span that could be rescued by expression of the human TDP-43 factor (19). Presently, several different studies carried out in Drosophila strongly support the functional homology of the human and Drosophila TDP-43 orthologs, as recently reviewed by us (20). Altogether, therefore, most of the data collected so far promote the use of Drosophila models to investigate the molecular mechanisms underlying human neurodegenerative disorders derived from TDP-43 alterations.
In this work we have now further extended this connection by exploring the interaction of TDP-43 and TBPH with fruit fly hnRNP proteins. Using co-immunoprecipitation experiments we have found that the Drosophila ortholog of human hnRNP A1/A2 proteins (Hrb98DE/Hrp38) maintains the ability to bind both to TDP-43 and TBPH. Interestingly, we found that the regions involved in this interaction have been highly conserved in all these proteins, and there is a functional, genetic interaction between Hrp38 and TBPH.
EXPERIMENTAL PROCEDURES
GST Overlay (Far Western), HPLC Analysis, and Mass Spectrometry
Western blots containing 150 μg of HeLa nuclear (NE), HeLa cytoplasmic (S100), and Drosophila nuclear extract (L2NE) were incubated for 2 h with either GST-TDP-43 or GST-TBPH (10 μg of protein in 20 ml of PBS, 10% (w/v) of nonfat dried milk) for 1 h. The membrane was then washed 4 times with PBS plus 0.2% Tween 20 (polyoxyethylene sorbitan monolaurate) and was then incubated for another hour with a commercial anti-GST antibody (Sigma) at a dilution of 1:2000. The blots were then washed again 4 times using PBS plus 0.2% Tween 20 and incubated for another hour with anti-goat HRP antibody (Dako A/S) at a dilution of 1:2000. After four final washes, the Western blots were developed using ECL (Amersham Biosciences). Fractionation of nuclear extracts was performed on a HPLC Agilent 1100 Series using a Phenomenex C4 300A (250 × 4.6 mm) reverse-phase column. The proteins were eluted using a solution 95% acetonitrile + 0.1% TFA in a 20–70% linear gradient over 38 min. The different fractions from 35 to 43% mobile phase were first speed-dried for 2 h to reduce volume and then concentrated using YM-10 filters (Microcon). Each fraction was then divided in two and loaded on 10% SDS-PAGE gels, one to be stained with Coomassie and the other blotted on a Optitran Nitrocellulose BA-S 83 membrane (Schleicher & Schuell) and subjected to GST overlay as described above. Internal sequence analysis from the Coomassie Blue-stained bands excised from the SDS-PAGE gel was performed using an electrospray ionization mass spectrometer (LCQ DECA XP, Thermo Finnigan). The bands were digested by trypsin, and the resulting peptides were extracted with water and 60% acetonitrile, 1% trifluoroacetic acid. The fragments were then analyzed by mass spectrometry, and the proteins were identified by analysis of the peptide MS/MS data with Turbo SEQUEST (ThermoFinnigam) and MASCOT (Matrix Science).
Plasmid Construction
The generation of pFLAG TDP-43 and pFLAG TBPH constructs has been described previously (14, 15, 21). The apoAII (pTB-apoAIIm) splicing reporter minigenes have been described by Mercado et al. (22), respectively. The Hrp38 ORF was kindly provided by Prof. Joan Steitz. The Hrp38 ORF was amplified by PCR with the oligos HRP38_s (5′-tcaGAATTCaagcttATGGTGAACTCGAACCAGAACCAGAA-3′) and HRP38_as (5′-tcaGGTACCgcggccgcctcgagTCAATATCTGCGGTTGTTGCCACCGT-3′) and subsequently cloned in pFLAG-CMV2 vector for eukaryotic expression. The EGFP-TDP-43 (321–366) vector used in the co-immunoprecipitation experiments has already been described by Budini et al. (16).
The EGFP-Hrp38-(293–365) vector was generated by PCR amplification of the Hrp38 region spanning the residues 293-365 with the following oligos Hrp38 C-term XhoI_s (5′-AGATCTCGAGCGGCGGCAACAATTGGAACAATGGTG-3′) and Hrp38 C-term BamHI_as (5′-TCCGGTGGATCCTCAATATCTGCGGTTGTTGCCACCGTT-3′). All constructs were sequenced to confirm their identity and to exclude the presence of other mutations.
Transfections and RT-PCRs
The plasmid DNA used for transfections was purified with JetStar columns (Genomed). Liposome-mediated transfections of 2.5 × 105 human HeLa cells were performed using Effectene (Qiagen). The amount of constructs used for each transfection ranged between 0.5 and 1 μg. siRNA transfections were performed in HeLa cells by using the small interfering RNA (siRNA) oligonucleotide specific for TDP-43 (GCAAAGCCAAGAUGAGCCU) and the Oligofectamine reagent (Invitrogen) as previously described (14). At the end of transfections, the cells were harvested, and the RNA was extracted with Trifast reagent (EuroGold). The cDNA was prepared with M-MLV reverse transcriptase (Invitrogen) and poly-dT primer according to the manufacturer's instructions. PCRs were carried out for 35 amplification cycles (95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s), and PCR products were analyzed on 1.5% agarose gels.
Co-immunoprecipitations
For co-immunoprecipitation assays, HeLa cells (60% of confluence) were transfected with 2 μg of each plasmid set and the Effectene reagent. The cells were collected in PBS supplemented with protease inhibitors (Roche Applied Science) and lysed by sonication. The lysates were incubated with 0.6 μg of anti-GFP antibody (Santa Cruz Biotechnology) for 3 h at 4 °C, and then 30 μl of A/G Plus-agarose beads (Santa Cruz Biotechnology) were added. After overnight incubation at 4 °C, the beads were precipitated and washed 3 times with PBS. After SDS-PAGE, the immunoprecipitates were probed with an anti-FLAG antibody (F1804, Sigma) in Western blot analysis.
Fly Stocks
RNA interference (RNAi) flies against TBPH (ID38377) and Hrp38/Hrb98DE/CG9983 (#31303) were obtained from VDRC Vienna and Bloomington Stock Center, respectively.
Quantitative Real Time PCRs
Total RNA was extracted from the heads of wild type W1118, ElavG4, tbphΔ23/+; Hrp38RNAi and ElavG4, tbph+/+; Hrp38RNAi by using TRIzol reagent (Invitrogen) according to the manufacturer' s protocol. cDNA was synthesized with 1 μg of RNA sample by using SuperscriptTM-III (Invitrogen) reverse transcriptase and oligo(dT) primers. Specific primers were designed to amplify the Hrb98DE/Hrp38 gene (forward, GTCTAGAATATGCGCAAGCTGTTCATC; reverse, AGAATTCCGTTTCTTGCCAGTCTCCTT), and the gene expression levels were checked by real-time PCR using SYBR Green technology. Housekeeping gene Rpl-11 (forward, CCATCGGTATCTATGGTCTGGA; reverse, CATCGTATTTCTGCTGGAACCA) and Rpl-32 (forward, AAGCGGCGACGCACTCTGTT and reverse, GCCCAGCATACAGGCCCAAG) were amplified and used to normalize the results. All amplifications were performed on CFX96TM, real-time PCR detection system (Bio-Rad). The relative expression levels were calculated according to the following equation: ΔCT = CT(target) − CT(normalized). Four PCR reactions for each genotype (2 duplicates) were performed.
Climbing Assay
Newly eclosed flies were transferred in batches of 20–25 (1:1 male:female) to fresh vials and aged for 4 days. They were then transferred without anesthesia to an empty transparent Duran 50-ml glass cylinder. The cylinder was divided into three parts (bottom, middle, and top). The climbing ability was quantified as the percentage of flies that reached the top of the tube in 15 s. Three trials were performed, and the average value was calculated. A minimum of 160 flies for each genotype were tested.
Life Span
Adult flies were collected for 2 days and transferred to fresh tubes at a density of 20–25 per vial (1:1 male:female). Every third day, flies were transferred to new tubes containing fresh medium, and deaths were scored. Survival rate graph was plotted with percentage of survival flies against day. All the experiments were performed in a humidified, temperature-controlled incubator at 25 °C and 60% humidity on a 12-h light and 12-h dark cycle. Flies were fed with standard cornmeal (2.9%), sugar (4.2%), and yeast (6.3%) fly food.
Immunohistochemistry
Drosophila adult brains were dissected in 1× PB (100 mm Na2HPO4/NaH2PO4, pH 7.2) 0.3%Triton X-100 and fixed in 4% paraformaldehyde for 20 min. Brains were washed in PB 0.3% Triton (20 min for 3 times) and then were blocked in 5% normal goat serum 30 min. Primary antibodies (anti-Elav DSHB 1:250 and anti anti-Futsch 22C10s DSHB 1:50) were incubated overnight at 4 °C. Secondary antibodies were incubated at room temperature for 2 h (Alexa Fluor® 488 mouse, 1:500; Alexa Fluor® 555 rat 1:500). Images were captured on a Zeiss 510 Meta confocal microscope. Labeling Futsch intensities normalized to anti-Elav staining/area were calculated with ImageJ.
RESULTS
Human and Drosophila TDP-43 Orthologs Share the Same Pattern of Human and Fruit Fly Nuclear Interactors
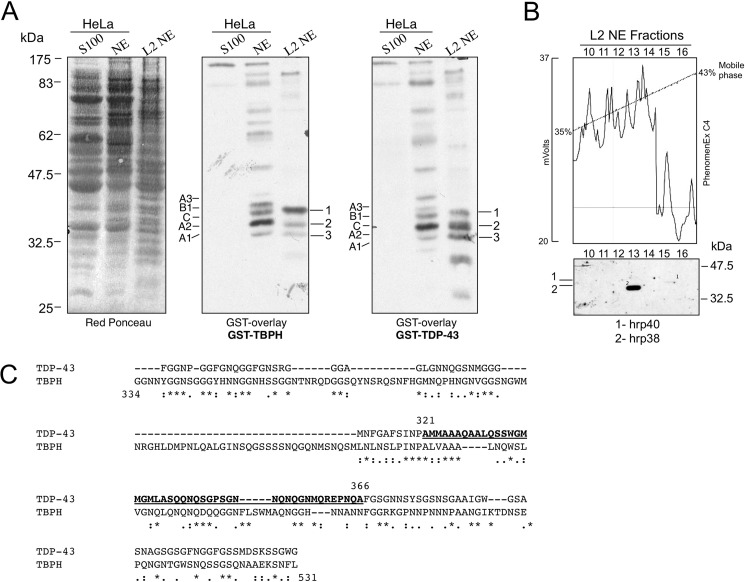
To compare the interactors of both human and Drosophila TDP-43, a GST overlay (Far Western) analysis was performed as previously described (13). In this experiment equal amounts of human HeLa nuclear (NE) and cytoplasmic (s100) extracts along with S2 Drosophila nuclear extract (L2NE) were blotted onto a Western blot membrane (Fig. 1A, left panel) and incubated with recombinant GST-TDP-43 (Fig. 1A, central panel) or GST-TBPH (Fig. 1A, right panel). Comparison of the signals in the NE and L2NE lanes confirmed that GST-TDP-43 and GST-TBPH shared a similar recognition pattern of interactors mostly localized in the 32.5–47.5-kDa molecular mass range. As control, both GST-TDP-43 and GST-TBPH yielded negligible signals in the cytoplasmic extract (S100 lane).
FIGURE 1.
Identification of TDP-43 and TBPH interactors by GST overlay and reverse-phase chromatography methods. A, staining (Red Ponceau) and GST-overlays (TBPH and TDP-43) of human HeLa cytoplasmic (S100), HeLa nuclear (NE), and Drosophila L2NE. Molecular weights (kDa) are shown on the left. The assays were carried out using recombinant GST-TDP-43 or GST-TBPH as probes. B, Reverse-phase chromatography (upper panel) and GST overlay (lower panel) of Drosophila L2NE-fractionated nuclear extract. Mass spectroscopic analysis of the Drosophila TDP-interactors has permitted identification of at least two factors, Squid/Hrp40 (1) and HRB98/Hrp38 (2). C, amino acid alignment of the C-terminal domains from TDP-43 and TBPH. The region of TDP-43 spanning residues 321–366 is underlined. Symbols below the alignment indicate: identity (asterisk), close similarity (colon), more distant similarity (period).
Previous characterization of human TDP-43 interacting factors using this kind of assay found that the strongest bands were hnRNP A1, hnRNP A2, hnRNP C1, hnRNP B1, and hnRNP A3 (13). Interestingly, GST-TBPH was also observed to bind specifically to these proteins in the HeLa nuclear extract (Fig. 1A, middle panel). This was also in keeping with previous reports from our laboratory that GST-TBPH could supershift human GST-hnRNP A2 in a band-shift experiment (14).
Most interestingly, a similar situation to the human extract recognition pattern could also be observed for the nuclear Drosophila proteins in the L2NE extract. In particular, three major bands that we named 1, 2, and 3 were specifically recognized both by GST-TDP-43 and GST-TBPH (Fig. 1A, middle and central panel). However, no further information was available with regard to the identity of these three bands.
Identification of the Drosophila Factors Interacting with TDP-43 and TBPH
To identify these factors, the L2NE nuclear extract was fractionated by HPLC with a C4 reverse-phase column (see Fig. 1B, upper panel, for the elution profile). After collection, equal amounts of each fraction were loaded onto two separate SDS-polyacrylamide gels. The first was stained with Coomassie Blue (data not shown), whereas the other was blotted and incubated with GST-TDP-43 in a Far Western assay (Fig. 1B, lower panel). By alignment of the Coomassie Blue-stained gel with the overlay signals we observed one faint and one strong band that migrated at the same height as the overlays shown in Fig. 1A, right panel. These bands were excised from the Coomassie Blue-stained gel and identified by mass spectroscopic analysis as Hrp38/Hrb98DE/CG9983 (P07909) (band 2) and as hnRNP Squid/Hrp40/CG16901 (Q08473) (band 1). Unfortunately, using this technique we could not discover the identity of band 3, probably because the reverse-phase fractionation of the Drosophila nuclear extract caused its denaturation. However, based on the nature and relative mobility of the two identified factors, it is highly probable that band 3 may represent HRB87F/hrp36/CG12749, another well known hnRNP A/B homologue in Drosophila.
Interestingly, both identified proteins belong to the Drosophila hnRNP A family and are orthologs of the human hnRNP A1 and hnRNP A2/B1 factors (23–25) that were previously shown to be the most efficient at interacting specifically with human TDP-43 (14). From these data we concluded that the Drosophila hnRNP A/B family is the most efficient interactor of Drosophila TBPH in the same way that their human orthologs, hnRNP A1 and hnRNP A2/B1 factors, are of human TDP-43 (14).
The observation that Drosophila TBPH and human hnRNP proteins can cross-interact is also consistent with the substantial conservation in TBPH of the 321–366 region located within the C-tail of TDP-43 (Fig. 1C). In fact, we have already established in previous studies that the 321–366 region of TDP-43 is important for TDP-43 hnRNP A/B protein-protein interactions (14, 16).
Mapping the Sequences Required for the Interaction of TDP-43 with Drosophila Hrp38
After the identification of Hrp38/Hrb98DE/CG9983 and Squid/Hrp40/CG16901 as two putative interactors of both human and Drosophila TDP-43, we focused the attention on Hrp38 because its signal in the GST overlay using human GST-TDP-43 was much stronger than that of Hrp40 both before (Fig. 1A, right panel) and after fractionation (Fig. 1B, lower panel). This is in keeping with the observation that there is an extremely high sequence similarity also between Drosophila Hrp38 and human hnRNP A1 and A2 at the level of both the RRM and C-terminal sequences that are known to be mostly involved in determining RNA-protein and protein-protein interactions, respectively.
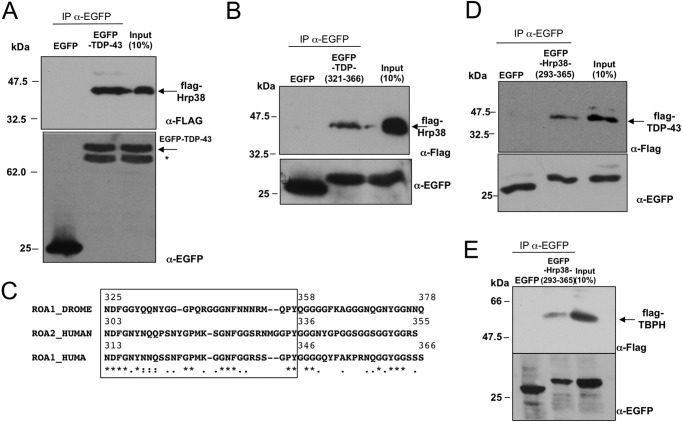
First of all, therefore, using co-immunoprecipitation, we sought to validate the interaction between TDP-43 and Hrp38 in vivo. To achieve this aim, we cloned the Hrp38 cDNA in the pFLAG eukaryotic expression vector (FLAG-Hrp38) and set up a co-immunoprecipitation assay by co-transfecting in HEK293 cells a GFP-TDP-43 wild type fusion protein (EGFP-TDP-43) in combination with the plasmid expressing FLAG-Hrp38. The results shown in Fig. 2A demonstrate that FLAG-Hrp38 can efficiently co-immunoprecipitate EGFP-TDP-43 but not EGFP alone. As a control, Western blot analysis showed that both EGFP and EGFP-TDP-43 were strongly expressed at the protein level (Fig. 2A, lower panel, α-EGFP).
FIGURE 2.
Hrp38 co-precipitates with TDP-43. A, co-immunoprecipitation of Hrp38 with TDP-43. HEK293 cells were cotransfected with FLAG-Hrp38 and EGFP-TDP43 or EGFP-empty constructs and subjected to immunoprecipitation with polyclonal anti-EGFP (IP α-EGFP) followed by immunoblotting with antibodies as indicated. A nonspecific band (*), probably derived from a cleavage in the TDP-43 portion, as it is not seen in EGFP alone, was also detected by anti-EGFP antibody. B, co-immunoprecipitation of Hrp38 with the region of TDP-43 spanning residues 321–366. HEK293 cells were cotransfected with FLAG-Hrp38 and EGFP-TDP43 (321–366) or EGFP-empty constructs and subjected to immunoprecipitation with polyclonal anti-EGFP (IP α-EGFP) followed by immunoblotting with antibodies as indicated. C, amino acid alignment of the Gly-rich tract of Hrp38 (ROA1_DROME) with the regions from the human hnRNP A1 (ROA1_HUMAN) hnRNP A2 (ROA2_HUMAN) critical for protein-protein interactions according to the mapping previously performed (26). Symbols below the alignment indicate: identity (asterisk), close similarity (colon), more distant similarity (period). The MUSCLE program was used for alignment (61). D, co-immunoprecipitation of TDP-43 with the region of Hrp38 spanning residues 293–365. HEK293 cells were cotransfected with FLAG-TDP-43 and EGFP-Hrp38 (293–365) or EGFP-empty constructs and subjected to immunoprecipitation with polyclonal anti-EGFP (IP α-EGFP) followed by immunoblotting with antibodies as indicated. E, co-immunoprecipitation of TBPH with the region of Hrp38 spanning residues 293–365. HEK293 cells were cotransfected with FLAG-TBPH and EGFP-Hrp38 (293–365) or EGFP-empty constructs and subjected to immunoprecipitation with polyclonal anti-EGFP (IP α-EGFP) followed by immunoblotting with antibodies as indicated.
To perform a more precise mapping of the interacting region, recent experiments aiming at mapping the TDP-43 domain responsible of binding to the various hnRNPs demonstrated that the region spanning residue 321 to 366 is responsible for the interaction with hnRNP A2 (14, 16).
In this respect, the alignment of the human and fly TDP-43 C-terminal domains highlights that this area of human TDP-43 shows a very high degree of similarity within the Gly-rich domain of TBPH spanning residues 428–481 (Fig. 1C). Therefore, we repeated the co-immunoprecipitation experiments of FLAG-Hrp38 (Fig. 2B, upper panel, FLAG-Hrp38) in combination with a fusion protein consisting of EGFP carrying only the human region 321–366 (Fig. 2B, upper panel, EGFP-TDP-(321–366)). As shown in Fig. 2B, FLAG-Hrp38 co-immunoprecipitated also with TDP-321–366 but not with EGFP alone.
Mapping the Hrp38 Sequence Required for the Interaction with Human TDP-43
With regard to identifying the Hrp38 region involved in this interaction, previous studies on hnRNP A1-mediated protein interactions have shown that they mostly rely on a short region within the Gly-rich domain of the human hnRNP A1 (26). In keeping with this original observation, we have also recently shown by immunoprecipitation analysis that the region of hnRNP A2 involved in the interaction between this protein and TDP-43 must be comprised within its C terminus region that spans residues 288–341 (16) and that the C terminus of TBPH is required to supershift hnRNP A2 in a band-shift assay (14).
Based on these considerations, the alignment of the C-terminal regions of human hnRNPA1 and hnRNP A2 with Hrp38 showed that within the hnRNP A1, hnRNP A2, and Hrp38 sequences there is a highly conserved motif that could be responsible for this cross-interaction (Fig. 2C).
Therefore, we prepared a fusion vector of EGFP with the region of Hrp38 spanning residues 293–365 (EGFP-Hrp38 (293–365)) and carried out co-transfection and co-immunoprecipitation experiments in combination with a flagged version of human TDP-43 (FLAG-TDP-43) or of fly TBPH. As shown in Fig. 2, D and E, the Hrp38 region spanning these amino acids was able to specifically co-immunoprecipitate both FLAG-TDP-43 and FLAG-TBPH, thus establishing this region as the one responsible for the interaction of Hrp38 with both human and fly TDP-43 orthologs.
Hrp38 Supports the Inhibitory Role of TDP-43 for ApoAII Exon 3 Splicing
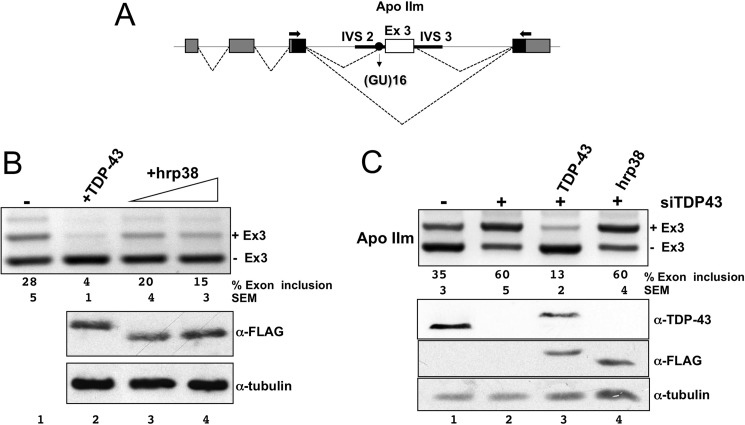
After confirming the in vivo interaction between TDP-43 and Hrp38 and mapping the interacting sequences, it was then interesting to test whether this interaction was also functionally significant. To achieve this, we used the Hrp38 cDNA eukaryotic expression vector to test whether Hrp38 could affect the splicing of TDP-43-related reporter minigenes similarly to what had been observed for the human hnRNP A2 ortholog (14). Initially, the effects of Hrp38 overexpression in HEK293 cells were monitored by using the apoAII splicing reporter assay (Fig. 3A) where the involvement of TDP-43 was previously well characterized (22, 27). In this system the independent overexpression of Hrp38 was able to decrease the amount of exon inclusion (Fig. 3B, lanes 3 and 4) although to a lesser extent compared with TDP-43 overexpression (Fig. 3B, lane 2).
FIGURE 3.
Hrp38 factor inhibits splicing of the apoAII exon 3 in a TDP-43-dependent manner. A, schematic representation of the minigene apoAII-ISEm used in the splicing assay. The human apoAII exon 3 and its flanking introns were cloned in the α-globin/fibronectin reporter system (pTB). α-Globin, fibronectin EDB exons, and human apoAII exon 3 are indicated in gray, black, and white boxes, respectively. Solid lines indicate apoAII IVS2 and IVS3. The black circle indicates the (GT)16 tract. Dashed lines indicate splicing options. B, Hrp38 overexpression has an inhibitory effect on splicing of apoAII-ISEm (lane 4). The RT-PCRs show the increasing splicing inhibitory activity of Hrp38 with respect to the controls (lane 1, −). The percentages of exon inclusion along with S.E. obtained in three independent transfection experiments are reported. Western blots against the FLAG peptide and tubulin are shown below to show equal transgene expression. The overexpression of TDP-43 was used as a control of splicing inhibition (lane 3). C, the inhibitory effect of Hrp38 on the splicing of apoAII-ISEm is lost after TDP-43 silencing. The percentages of exon inclusion along with S.E. obtained in three independent transfection experiments are reported. Western blots against TDP-43, tubulin, and FLAG peptide are shown below to show silencing efficiency (anti-TDP-43), transgene overexpression (anti-FLAG), and comparable protein load (anti-tubulin). The overexpression of TDP-43 was used as a control of splicing inhibition (lane 3).
Most importantly, to assess whether the activity of Hrp38 on splicing was direct or indirect (i.e. dependent on the presence of TDP-43), we repeated the experiment after endogenous TDP-43 silencing by siRNA treatment. The hypothesis behind this experiment is that if the effects of Hrp38 on apoAII splicing were mediated by TDP-43, then the silencing of endogenous TDP-43 should abrogate the Hrp38 splicing inhibition observed in Fig. 3B.
Under normal conditions, the apoAIIm minigene reporter displays ∼35 ± 5% of exon inclusion (Fig. 3C, lane 1). As expected, the RNAi with anti-TDP-43 resulted in a significant increase in the amount of the apoAII exon 3 inclusion (Fig. 3C, lane 2), whereas the overexpression of siRNA-resistant wild type TDP-43 promoted strong apoAII exon 3 skipping (Fig. 3C, lane 3). Unlike TDP-43, however, the overexpression of Hrp38 in the absence of endogenous TDP-43 did not modify the splicing pattern of both reporters (Fig. 3C, lane 4).
Similar results were obtained by using another well-characterized splicing reporter assay (cystic fibrosis transmembrane conductance regulator C155T; data not shown) (3, 28, 29). In conclusion, these results demonstrate that Drosophila Hrp38 can interact with human TDP-43 also at the functional level. It was, therefore, important to see whether Hrp38 could modulate the disease phenotype in our Drosophila models.
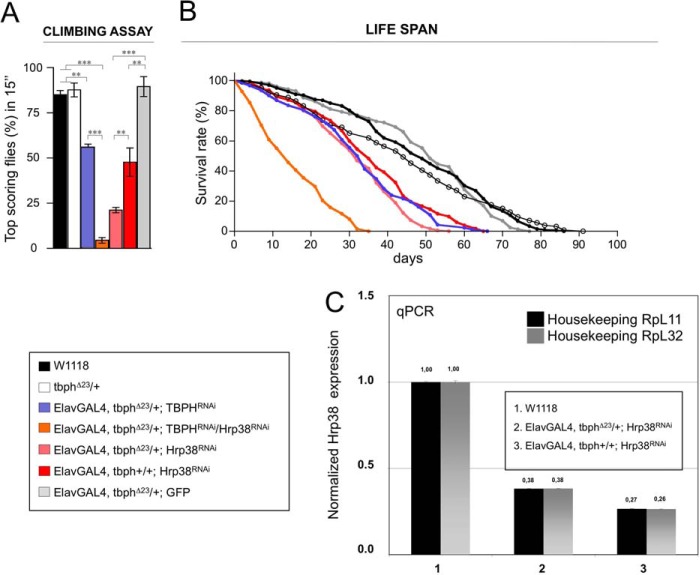
Hrp38 and TBPH Genetically Interact to Prevent Locomotor Defects and Reduced Life Span in a Drosophila Model of Amyotrophic Lateral Sclerosis
We have previously shown that the suppression of TBPH exclusively in neurons by RNAi closely mirrored the main symptoms observed in ALS, such as drastic alterations in locomotive behaviors and shortening of life span (19, 30). We now sought to investigate whether the suppression of Hrp38 in Drosophila neurons could induce similar ALS-related phenotypes in vivo or enhance the neurological alterations provoked by the reduction of TBPH expression. It should first of all be noted that the in vivo depletion of the two single genes are lethal when homozygous (19, 31). For these reasons, to test the presence of a genetic interaction between TBPH and Hrp38 genes, hypomorphic alleles and heterozygous backgrounds were used for both genes.
Therefore, efficient pan-neuronal RNAi-mediated knockdown of Hrp38 was obtained using the Elav-GAL4 driver. The reduction of Hrp38 expression in Drosophila brains (Fig. 4C) produced significant locomotor alterations, revealed by defects in climbing assay associated to a severe shortening of the life span, compared with controls (Fig. 4, A and B). These phenotypes closely resemble the defects observed in the TBPH-KO flies (19) and suggest that these proteins might be involved in the regulation of similar genetic traits.
FIGURE 4.
Hrp38 and TBPH genetically interact to regulate flies locomotion and life span. A, a climbing ability analysis of Hrp38-silenced flies demonstrates the existence of a genetic interaction between TBPH and Hrp38. The phenotype of Hrp38 neuronal silencing becomes significantly stronger if one copy of TBPH is removed as compared with the silencing in a wild type background (Elav-GAL4, tbphΔ23/+; Hrp38RNAi versus Elav-GAL4, tbph+/+; Hrp38RNAi, p < 0.01). The results of Hrp38 silencing in a TBPH sensible background further support this genetic interaction. In fact, Hrp38 silencing exacerbated the phenotype caused by TBPH silencing (Elav-GAL4, tbphΔ23/+; TBPHRNAi versus Elav-GAL4, tbphΔ23/+; TBPHRNAi/Hrp38RNAi, p < 0.001). n = 200 flies for genotype, error bars indicate S.E. Statistics were performed using GraphPad Prism. One-way analysis of variance was performed using Bonferroni's multiple comparison test to compare the genotypes (** indicates p < 0.01; ***, p < 0.001). B, percentage of flies survivors during aging. Median lifespan is: 49 days for wild type (n = 208), 44 days for tbphΔ23/+ (n = 126), 33 days for Elav-GAL4, tbphΔ23/+; TBPHRNAi (n = 150), 14 days for Elav-GAL4, tbphΔ23/+; TBPHRNAi/Hrp38RNAi (n = 138), 51 days for Elav-GAL4, tbphΔ23/+; GFP (n = 159), 32 days for Elav-GAL4, tbphΔ23/+; Hrp38RNAi (n = 312), and 36 days for Elav-GAL4, tbph+/+; Hrp38RNAi (n = 235). Log rank test and p value: wild type versus tbphΔ23/+ (p = not significant), wild type versus Elav-GAL4, tbphΔ23/+; TBPHRNAi (p < 0.0001), wild type versus Elav-GAL4, tbphΔ23/+; TBPHRNAi/Hrp38RNAi (p < 0.0001), wild type versus Elav-GAL4, tbphΔ23/+; GFP (p = not significant), wild type versus Elav-GAL4, tbphΔ23/+; Hrp38RNAi (p < 0.0001), wild type versus Elav-GAL4, tbph+/+; Hrp38RNAi (p < 0.0001), Elav-GAL4, tbphΔ23/+; TBPHRNAi versus Elav-GAL4, tbphΔ23/+; TBPHRNAi/Hrp38RNAi (p < 0.0001). and Elav-GAL4, tbph+/+; Hrp38RNAi versus Elav-GAL4, tbphΔ23/+; Hrp38RNAi (p < 0.0001). Statistics were performed using GraphPad Prism. A log rank test was performed to compare survival distribution of the genotypes (***, indicates p < 0.001). C, RNAi treatment against Hrp38 reduced mRNA expression levels in Drosophila heads, as compared with control untreated flies.
Indeed, on one hand, we found that TBPH gene dose modifications in Hrp38-RNAi backgrounds strongly enhanced the above described phenotypes. This is outlined by the remarkable differences observed in climbing activity between Elav-GAL4, tbph+/+; Hrp38RNAi flies and in flies where TBPH expression levels were reduced by either removing one copy of the gene (Elav-GAL4, tbphΔ23/+; Hrp38RNAi) or by expression of a double RNAi against these two proteins (Elav-GAL4, tbphΔ23/+; TBPHRNAi/Hrp38RNAi) (Fig. 4A). On the other hand, the observation that reduction in life span was directly proportional to the TBPH gene copy number in the Elav-GAL4/Hrp38RNAi flies (Fig. 4B) suggested that these genes are functionally related to common metabolic pathways.
To determine whether the differences in locomotive behaviors and life span described above were effectively due to increased degeneration of Drosophila neurons, we used the human homolog microtubule binding molecule MAP-1B, futsch in Drosophila, as the maker of neuronal damage, as demonstrated by our previous studies on TBPH-dependent synaptic microtubules organization (30).
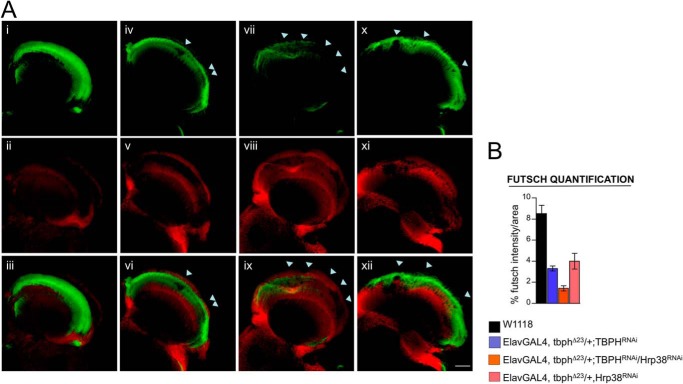
Therefore, we stained adult brains with a monoclonal antibody against futsch and analyzed the distribution of the axons in the lamina and medulla, a highly innervated cerebral area sited in the Drosophila optic lobes, immediately behind the retina. We observed that the neuropil of Elav-GAL4, tbphΔ23/+; Hrp38RNAi or Elav-GAL4, tbphΔ23/+; TBPHRNAi individually treated flies in the lamina appeared less stained and thinner compared with wild type controls with the presence of irregular holes in these structures due to the loss of innervating axons. Nevertheless, these phenotypes became drastically increased after the expression of a double RNAi against these two proteins (Elav-GAL4, tbphΔ23/+/Hrp38RNAi; TBPHRNAi) (Fig. 5A). Demonstrating that these proteins are required to maintain the innervation of complex cerebral areas and indicating that the neurological defects observed in TBPH and Hrp38 suppressed flies involved an enlarged neurodegenerative process.
FIGURE 5.
Increased neuropil degeneration in RNAi-treated adult flies. A, confocal images of optic lobe of adult brains stained with anti-Elav (in red) and anti-22C10 (futsch) (in green) antibodies in 1/2 days old flies reveals strong defects in the innervation of the lamina and medulla regions in control brains (i–iii) compared with Elav-GAL4,tbphΔ23/+; TBPHRNAi (iv–vi), Elav-GAL4,tbphΔ23/+; TBPHRNAi/Hrp38RNAi (vii–ix), and Elav-GAL4, tbphΔ23/+; Hrp38RNAi (x–xii). Scale bar, 30 μm. B, quantification of futsch intensity in lamina and medulla regions indicated that the density of axons in these regions became dramatically reduced after RNAi treatments. n = 5.
DISCUSSION
TDP-43 is a nuclear factor highly conserved through evolution (18). A high degree of homology exists among the TDP-43 orthologs from human, rodents, D. melanogaster, and Caenorhabditis elegans (15). From a structural point of view, TDP-43 belongs to the hnRNP family of nuclear proteins (32), and its functions are mostly involved in the regulation of all aspects of mRNA metabolism starting from transcriptional regulation, the control of pre-mRNA splicing, mRNA transport/stability, microRNA expression, and probably mRNA translation (1). The central importance played by this factor to maintain cellular viability comes from knock-out transgenic mice lines in which it was observed that embryonic development was severely impaired when in a homozygous state (33–35). From a human disease point of view, several studies have demonstrated that TDP-43 has a primary role in the origin and development of different neurodegenerative diseases, but the exact molecular mechanism(s) mediating such an involvement is still largely unknown (36).
To better understand its biological role in health and disease it is obviously of the utmost importance to characterize its molecular targets and cellular partners. In fact, as with almost all members of the hnRNP family, the biological functions mostly depend on the presence of complex interaction networks that form distinct functional modules (37–39). For example, it has been long established that within the nuclear compartment hnRNP proteins are usually assembled into ribonucleoprotein particles, (40), and binary interactions have been described between several of these members, such as polypyrimidine tract-binding protein (PTB) and hnRNP L (41), hnRNP K and hnRNP A1, and hnRNP A2/B1 and other nuclear proteins (42). In addition, it has been demonstrated that hnRNP A1, C1, E2, I, K, and L can form both homodimeric and heterodimeric interactions with each other (43). This behavior has also been observed for TDP-43. This protein, in fact, has been proposed to exist as a dimer or as part of ribonucleoprotein complexes within the nuclear environment (44–47).
Considering these similarities, therefore, it is expected that knowledge of TDP-43 protein binding network will help researchers to better understand the pathways underlying the pathogenesis of diseases, as has been previously shown to occur for several other factors involved in neurodegeneration (48–51).
In this direction, recent proteomic studies have shown that TDP-43 can associate with several factors involved in trafficking of biological membranes and transcription (52–54) as well as RNA metabolism both in the nucleus and in the cytoplasm (55, 56). At present, however, the number of verified partners of TDP-43 is still limited. Nonetheless, it is now well known that among the better characterized interactors, there are several hnRNP A/B family members such as hnRNP A2/B1, A1, A3, and C2 (13, 14, 57, 58).
In keeping with this situation, we show here that one Drosophila ortholog of the human hnRNP A/B family (Hrp38) can bind very efficiently to both TBPH and human TDP-43. Most importantly, our results show that the regions of the human and fly factors involved in this protein-protein interaction seem to be structurally conserved. In fact, TDP-43 residues 321–366 that were previously mapped as the minimal binding region required for interaction with hnRNP A2 (14) retain the ability to bind Hrp38. This conservations appears to be reciprocated by Hrp38 itself, as the cluster of amino acids implicated in this protein-protein interaction also show a very good level of evolutionary conservation between Hrp38 and hnRNPs A1/A2, with 36% identity and 33% similarity over 33 residues.
Most importantly, the high degree of conservation of these interactions can be carried over at the functional level, as Hrp38 can modulate the splicing of two model exons only in the presence of TDP-43. Even at a more general level, in vivo studies carried out in Drosophila also support this view. In particular, the locomotor deficit and reduction in life span as well as the level of neuropil degeneration caused by the simultaneous decrease in expression of both factors were higher than that expected by an additive effect of reduction in the level of the two single factors.
Taken together, these observations support the hypothesis that the functional cooperation between TBPH/Hrp38 and TDP-43/hnRNP A/B are fundamental also in vivo and are extremely well conserved throughout evolution despite the observation that the general structural conservation of the human and Drosophila TDP-43 C-terminal domains is low.
This is a particularly important conclusion, considering the recent criticism that research focused on neurodegeneration based on classic laboratory animal models can lead to potentially misleading results if sufficient care is not taken to establish whether the chosen model is sufficiently similar to the human disease under study (59). What these results mean, therefore, is that the Drosophila system should be considered quite adequate to study ways that modulate the interaction of TDP-43 and other hnRNP factors.
Most importantly, it has recently been found that mutations in hnRNPA2/B1 and hnRNPA1 can also lead to multisystem proteinopathy and ALS (60). Interestingly, the position of these mutations D290V/D302V in hnRNP A2B1 and D262V/D314V are in the region that is involved with the binding to TDP-43. The expression of these mutant forms of human hnRNPA2 and hnRNPA1 as well as of mutant forms of the fly homologue Hrp38 in transgenic Drosophila led to severe muscle degeneration and enhanced formation of fibrils by the mutated proteins due to the fact that these missense substitutions are localized in a prion-like domain (60). For these reasons, therefore, a better characterization of this interaction could be quite important to provide us with additional insights on disease mechanisms and the factors that can potentially affect its origin and severity.
Acknowledgment
The Hrp-38 ORF was kindly provided by Prof. Joan Steitz.
This work was supported by AriSLA (TARMA; to F. B.) and (ALSMNDTDP-43; to F. F.), Thierry Latran Foundation (REHNPALS; to E. B. and F. F.), and University of Trieste-Finanziamento per Ricercatori di Ateneo.
- TDP-43
- TAR DNA-binding protein
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- ALS
- amyotrophic lateral sclerosis
- NE
- nuclear extract
- L2NE
- second instar larva nuclear extracts
- TBPH
- TAR DNA-binding protein-43 homolog.
REFERENCES
- 1. Buratti E., Baralle F. E. (2008) Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front. Biosci. 13, 867–878 [DOI] [PubMed] [Google Scholar]
- 2. Buratti E., Baralle F. E. (2001) Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 276, 36337–36343 [DOI] [PubMed] [Google Scholar]
- 3. Buratti E., Dörk T., Zuccato E., Pagani F., Romano M., Baralle F. E. (2001) Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 20, 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buratti E., Brindisi A., Pagani F., Baralle F. E. (2004) Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9. A functional link with disease penetrance. Am. J. Hum. Genet. 74, 1322–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 6. Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., Oda T. (2006) TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611 [DOI] [PubMed] [Google Scholar]
- 7. Baloh R. H. (2011) TDP-43. The relationship between protein aggregation and neurodegeneration in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. FEBS J. 278, 3539–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Greenberg S. A. (2011) Inclusion body myositis. Curr. Opin. Rheumatol. 23, 574–578 [DOI] [PubMed] [Google Scholar]
- 9. Banks G. T., Kuta A., Isaacs A. M., Fisher E. M. (2008) TDP-43 is a culprit in human neurodegeneration and not just an innocent bystander. Mamm. Genome 19, 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bentmann E., Neumann M., Tahirovic S., Rodde R., Dormann D., Haass C. (2012) Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43-kDa (TDP-43). J. Biol. Chem. 287, 23079–23094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Colombrita C., Zennaro E., Fallini C., Weber M., Sommacal A., Buratti E., Silani V., Ratti A. (2009) TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 111, 1051–1061 [DOI] [PubMed] [Google Scholar]
- 12. Kawahara Y., Mieda-Sato A. (2012) TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. U.S.A. 109, 3347–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buratti E., Brindisi A., Giombi M., Tisminetzky S., Ayala Y. M., Baralle F. E. (2005) TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail. An important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 280, 37572–37584 [DOI] [PubMed] [Google Scholar]
- 14. D'Ambrogio A., Buratti E., Stuani C., Guarnaccia C., Romano M., Ayala Y. M., Baralle F. E. (2009) Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 37, 4116–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ayala Y. M., Pantano S., D'Ambrogio A., Buratti E., Brindisi A., Marchetti C., Romano M., Baralle F. E. (2005) Human, Drosophila, and C. elegans TDP43. Nucleic acid binding properties and splicing regulatory function. J. Mol. Biol. 348, 575–588 [DOI] [PubMed] [Google Scholar]
- 16. Budini M., Buratti E., Stuani C., Guarnaccia C., Romano V., De Conti L., Baralle F. E. (2012) Cellular model of TAR DNA-binding protein 43 (TDP-43) aggregation based on its C-terminal Gln/Asn-rich region. J. Biol. Chem. 287, 7512–7525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuentealba R. A., Udan M., Bell S., Wegorzewska I., Shao J., Diamond M. I., Weihl C. C., Baloh R. H. (2010) Interaction with polyglutamine aggregates reveals a Q/N-rich domain in TDP-43. J. Biol. Chem. 285, 26304–26314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang H. Y., Wang I. F., Bose J., Shen C. K. (2004) Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics 83, 130–139 [DOI] [PubMed] [Google Scholar]
- 19. Feiguin F., Godena V. K., Romano G., D'Ambrogio A., Klima R., Baralle F. E. (2009) Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 583, 1586–1592 [DOI] [PubMed] [Google Scholar]
- 20. Romano M., Feiguin F., Buratti E. (2012) Drosophila Answers to TDP-43 Proteinopathies. J. amino acids 2012, 356081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ayala Y. M., Zago P., D'Ambrogio A., Xu Y. F., Petrucelli L., Buratti E., Baralle F. E. (2008) Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Sci. 121, 3778–3785 [DOI] [PubMed] [Google Scholar]
- 22. Mercado P. A., Ayala Y. M., Romano M., Buratti E., Baralle F. E. (2005) Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 33, 6000–6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matunis M. J., Matunis E. L., Dreyfuss G. (1992) Isolation of hnRNP complexes from Drosophila melanogaster. J. Cell Biol. 116, 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matunis E. L., Matunis M. J., Dreyfuss G. (1992) Characterization of the major hnRNP proteins from Drosophila melanogaster. J. Cell Biol. 116, 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zu K., Sikes M. L., Haynes S. R., Beyer A. L. (1996) Altered levels of the Drosophila HRB87F/hrp36 hnRNP protein have limited effects on alternative splicing in vivo. Mol. Biol. Cell 7, 1059–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cartegni L., Maconi M., Morandi E., Cobianchi F., Riva S., Biamonti G. (1996) hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J. Mol. Biol. 259, 337–348 [DOI] [PubMed] [Google Scholar]
- 27. Arrisi-Mercado P., Romano M., Muro A. F., Baralle F. E. (2004) An exonic splicing enhancer offsets the atypical GU-rich 3` splice site of human apolipoprotein A-II exon 3. J. Biol. Chem. 279, 39331–39339 [DOI] [PubMed] [Google Scholar]
- 28. Niksic M., Romano M., Buratti E., Pagani F., Baralle F. E. (1999) Functional analysis of cis-acting elements regulating the alternative splicing of human CFTR exon 9. Hum. Mol. Genet. 8, 2339–2349 [DOI] [PubMed] [Google Scholar]
- 29. Pagani F., Buratti E., Stuani C., Romano M., Zuccato E., Niksic M., Giglio L., Faraguna D., Baralle F. E. (2000) Splicing factors induce cystic fibrosis transmembrane regulator exon 9 skipping through a nonevolutionary conserved intronic element. J. Biol. Chem. 275, 21041–21047 [DOI] [PubMed] [Google Scholar]
- 30. Godena V. K., Romano G., Romano M., Appocher C., Klima R., Buratti E., Baralle F. E., Feiguin F. (2011) TDP-43 regulates Drosophila neuromuscular junctions growth by modulating Futsch/MAP1B levels and synaptic microtubules organization. PLoS One 6, e17808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ji Y., Tulin A. V. (2012) Poly(ADP-ribose) controls DE-cadherin-dependent stem cell maintenance and oocyte localization. Nat. Commun. 3, 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dreyfuss G., Kim V. N., Kataoka N. (2002) Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3, 195–205 [DOI] [PubMed] [Google Scholar]
- 33. Sephton C. F., Good S. K., Atkin S., Dewey C. M., Mayer P., 3rd, Herz J., Yu G. (2010) TDP-43 is a developmentally regulated protein essential for early embryonic development. J. Biol. Chem. 285, 6826–6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu L. S., Cheng W. C., Hou S. C., Yan Y. T., Jiang S. T., Shen C. K. (2010) TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis 48, 56–62 [DOI] [PubMed] [Google Scholar]
- 35. Kraemer B. C., Schuck T., Wheeler J. M., Robinson L. C., Trojanowski J. Q., Lee V. M., Schellenberg G. D. (2010) Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 119, 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buratti E., Baralle F. E. (2012) TDP-43. Gumming up neurons through protein-protein and protein-RNA interactions. Trends Biochem. Sci. 37, 237–247 [DOI] [PubMed] [Google Scholar]
- 37. Hartwell L. H., Hopfield J. J., Leibler S., Murray A. W. (1999) From molecular to modular cell biology. Nature 402, C47–C52 [DOI] [PubMed] [Google Scholar]
- 38. Oltvai Z. N., Barabási A. L. (2002) Systems biology. Life's complexity pyramid. Science 298, 763–764 [DOI] [PubMed] [Google Scholar]
- 39. Shen-Orr S. S., Milo R., Mangan S., Alon U. (2002) Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 31, 64–68 [DOI] [PubMed] [Google Scholar]
- 40. Piñol-Roma S., Choi Y. D., Matunis M. J., Dreyfuss G. (1988) Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 2, 215–227 [DOI] [PubMed] [Google Scholar]
- 41. Hahm B., Cho O. H., Kim J. E., Kim Y. K., Kim J. H., Oh Y. L., Jang S. K. (1998) Polypyrimidine tract-binding protein interacts with hnRNP L. FEBS Lett. 425, 401–406 [DOI] [PubMed] [Google Scholar]
- 42. Mikula M., Dzwonek A., Karczmarski J., Rubel T., Dadlez M., Wyrwicz L. S., Bomsztyk K., Ostrowski J. (2006) Landscape of the hnRNP K protein-protein interactome. Proteomics 6, 2395–2406 [DOI] [PubMed] [Google Scholar]
- 43. Kim J. H., Hahm B., Kim Y. K., Choi M., Jang S. K. (2000) Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J. Mol. Biol. 298, 395–405 [DOI] [PubMed] [Google Scholar]
- 44. Kuo P. H., Doudeva L. G., Wang Y. T., Shen C. K., Yuan H. S. (2009) Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 37, 1799–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shiina Y., Arima K., Tabunoki H., Satoh J. (2010) TDP-43 dimerizes in human cells in culture. Cell. Mol. Neurobiol. 30, 641–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sephton C. F., Cenik C., Kucukural A., Dammer E. B., Cenik B., Han Y., Dewey C. M., Roth F. P., Herz J., Peng J., Moore M. J., Yu G. (2011) Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J. Biol. Chem. 286, 1204–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tollervey J. R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., König J., Hortobágyi T., Nishimura A. L., Zupunski V., Patani R., Chandran S., Rot G., Zupan B., Shaw C. E., Ule J. (2011) Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 14, 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen H. K., Fernandez-Funez P., Acevedo S. F., Lam Y. C., Kaytor M. D., Fernandez M. H., Aitken A., Skoulakis E. M., Orr H. T., Botas J., Zoghbi H. Y. (2003) Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell 113, 457–468 [DOI] [PubMed] [Google Scholar]
- 49. Goehler H., Lalowski M., Stelzl U., Waelter S., Stroedicke M., Worm U., Droege A., Lindenberg K. S., Knoblich M., Haenig C., Herbst M., Suopanki J., Scherzinger E., Abraham C., Bauer B., Hasenbank R., Fritzsche A., Ludewig A. H., Büssow K., Buessow K., Coleman S. H., Gutekunst C. A., Landwehrmeyer B. G., Lehrach H., Wanker E. E. (2004) A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol. Cell 15, 853–865 [DOI] [PubMed] [Google Scholar]
- 50. Ravikumar B., Vacher C., Berger Z., Davies J. E., Luo S., Oroz L. G., Scaravilli F., Easton D. F., Duden R., O'Kane C. J., Rubinsztein D. C. (2004) Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 36, 585–595 [DOI] [PubMed] [Google Scholar]
- 51. Tsuda H., Jafar-Nejad H., Patel A. J., Sun Y., Chen H. K., Rose M. F., Venken K. J., Botas J., Orr H. T., Bellen H. J., Zoghbi H. Y. (2005) The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/Senseless proteins. Cell 122, 633–644 [DOI] [PubMed] [Google Scholar]
- 52. Lehner B., Sanderson C. M. (2004) A protein interaction framework for human mRNA degradation. Genome Res. 14, 1315–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sato S., Tomomori-Sato C., Parmely T. J., Florens L., Zybailov B., Swanson S. K., Banks C. A., Jin J., Cai Y., Washburn M. P., Conaway J. W., Conaway R. C. (2004) A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14, 685–691 [DOI] [PubMed] [Google Scholar]
- 54. Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F. H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Timm J., Mintzlaff S., Abraham C., Bock N., Kietzmann S., Goedde A., Toksöz E., Droege A., Krobitsch S., Korn B., Birchmeier W., Lehrach H., Wanker E. E. (2005) A human protein-protein interaction network. A resource for annotating the proteome. Cell 122, 957–968 [DOI] [PubMed] [Google Scholar]
- 55. Freibaum B. D., Chitta R. K., High A. A., Taylor J. P. (2010) Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 9, 1104–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ling S. C., Albuquerque C. P., Han J. S., Lagier-Tourenne C., Tokunaga S., Zhou H., Cleveland D. W. (2010) ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc. Natl. Acad. Sci. U.S.A. 107, 13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang Y. J., Xu Y. F., Cook C., Gendron T. F., Roettges P., Link C. D., Lin W. L., Tong J., Castanedes-Casey M., Ash P., Gass J., Rangachari V., Buratti E., Baralle F., Golde T. E., Dickson D. W., Petrucelli L. (2009) Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Budini M., Romano V., Avendaño-Vázquez S. E., Bembich S., Buratti E., Baralle F. E. (2012) Role of selected mutations in the Q/N rich region of TDP-43 in EGFP-12xQ/N-induced aggregate formation. Brain Res. 1462, 139–150 [DOI] [PubMed] [Google Scholar]
- 59. Bolker J. (2012) Model organisms. There's more to life than rats and flies. Nature 491, 31–33 [DOI] [PubMed] [Google Scholar]
- 60. Kim H. J., Kim N. C., Wang Y. D., Scarborough E. A., Moore J., Diaz Z., MacLea K. S., Freibaum B., Li S., Molliex A., Kanagaraj A. P., Carter R., Boylan K. B., Wojtas A. M., Rademakers R., Pinkus J. L., Greenberg S. A., Trojanowski J. Q., Traynor B. J., Smith B. N., Topp S., Gkazi A. S., Miller J., Shaw C. E., Kottlors M., Kirschner J., Pestronk A., Li Y. R., Ford A. F., Gitler A. D., Benatar M., King O. D., Kimonis V. E., Ross E. D., Weihl C. C., Shorter J., Taylor J. P. (2013) Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Edgar R. C. (2004) MUSCLE. Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]