Background: Schistosome tetraspanin Sm-TSP-2 is a vaccine antigen.
Results: We describe the structure of the large extracellular domain of Sm-TSP-2, develop a model of its interactions with Tetraspanin-enriched-microdomain proteins and plasma membrane, and identify TEM constituents.
Conclusion: Structural conservation of the domain means this model is likely applicable to TSPs in general.
Significance: Tetraspanin-enriched-microdomain proteins provide further targets for multiplex vaccines and/or novel drug targets.
Keywords: Membrane Proteins, Protein Structure, Proteomics, Tetraspanins, Vaccines, Schistosoma mansoni, Schistosomiasis
Abstract
The tetraspanins (TSPs) are a family of integral membrane proteins that are ubiquitously expressed at the surface of eukaryotic cells. TSPs mediate a range of processes at the surface of the plasma membrane by providing a scaffold for the assembly of protein complexes known as tetraspanin-enriched microdomains (TEMs). We report here the structure of the surface-exposed EC2 domain from Sm-TSP-2, a TSP from Schistosoma mansoni and one of the better prospects for the development of a vaccine against schistosomiasis. This is the first solution structure of this domain, and our investigations of its interactions with lipid micelles provide a general model for interactions between TSPs, membranes, and other proteins. Using chemical cross-linking, eight potential protein constituents of Sm-TSP-2-mediated TEMs were also identified. These include proteins important for membrane maintenance and repair, providing further evidence for the functional role of Sm-TSP-2- and Sm-TSP-2-mediated TEMs. The identification of calpain, Sm29, and fructose-bisphosphate aldolase, themselves potential vaccine antigens, suggests that the Sm-TSP-2-mediated TEMs could be disrupted via multiple targets. The identification of further Sm-TSP-2-mediated TEM proteins increases the available candidates for multiplex vaccines and/or novel drugs targeting TEMs in the schistosome tegument.
Introduction
Tetraspanins (TSPs)5 are a family of integral membrane proteins that are ubiquitously expressed on the plasma membranes of eukaryotic organisms. In recent years TSPs have emerged as essential participants in a range of processes that occur on cell surfaces. TSPs act as “scaffold” proteins providing a framework of homo- and heterodimers that form complexes with many other proteins. These complexes are clustered into tetraspanin-enriched microdomains (TEMs) that possess the capacity to facilitate vesicular fusion and/or fission (1). By modulating, inhibiting, or stabilizing associated proteins, TSPs facilitate functionality at the surface of the plasma membrane, and they are now thought to be essential in events such as fertilization, parasite and viral infection, cellular development, proliferation, immune response induction, metastasis suppression, and tumor progression (for review, see Hemler (1, 2)). Moreover, large quantities of TSPs are released from the cell in small vesicles called exosomes (3), leading to the hypothesis that TSPs orchestrate changes in the plasma membrane essential for the formation and budding of the exosomes (4).
Recently, TSPs have been shown to be promising vaccine targets for schistosomiasis, a debilitating parasitic disease predominantly caused by one of the three main species of human blood flukes; Schistosoma mansoni, Schistosoma japonicum, and Schistosoma haematobium (5). Schistosomiasis is a disease of poverty and affects ∼210 million people in developing countries of Asia, Africa, and South America (6). Although the anthelmintic drug praziquantel provides effective treatment, concerns over rapid reinfection and drug resistance have prompted a great deal of effort to be invested in the development of anti-schistosome vaccines and novel anthelmintics. One of the most promising candidate antigens for a schistosomiasis vaccine is a TSP located on the surface of the parasite, Sm-TSP-2. This protein was initially discovered because of its unique recognition by antibodies from individuals with acquired resistance to schistosomiasis and protection induced by vaccination of mice in a challenge model of schistosomiasis using a recombinant form of the EC2 domain (7). Large scale phase I clinical trials in humans are currently under way (8), and Sm-TSP-2 has been manufactured under cGMP conditions. In the parasite, Sm-TSP-2 appears to play a critical structural role in maintaining the integrity of the tegument, the syncytial outer covering of the parasite. Knockdown of the Sm-tsp-2 gene in the immature human stage of the parasite, the schistosomula, resulted in an 83% reduction in adult survival rates and caused large-scale degradation of the tegument (9), probably indicating a specific role for Sm-TSP-2 in both tegument formation and in the plasticity of the membrane.
Although an individual organism will normally express multiple TSP proteins, e.g. humans express 33 different TSPs, they are all characterized by a conserved topology comprising four trans-membrane helices, a short intracellular N- and C-terminal loop, and two extracellular domains; one short domain (EC1) and one longer domain (EC2). The EC2 plays a major role in mediating protein-protein interactions (2) and further defines the TSP structures by the presence of at least four cysteine residues, two of which are incorporated into a highly conserved CCG motif, involved in stabilizing disulfide bonds. Despite the importance of the TSPs as ubiquitous eukaryotic mediators of cell surface functionality, remarkably little is known about their structure. Accordingly, there is no clear understanding of how they assemble into protein complexes or how they interact with each other, with other proteins, and with the plasma membrane.
The only structural information available about TSPs is the crystal structure of the human CD81 EC2 domain (hCD81-LEL; Ref. 10). The EC2 domain of CD81 comprises five α-helices packed into a “mushroom”-like domain in which three helices form the stem that supports a head region. Extensive structural modeling of TSPs suggests that the stem and head regions are, respectively, conserved and variable (11) domains. Based on homodimeric crystal forms observed in the x-ray structure, it has been suggested that the EC2 domain mediates the formation of TSP homodimers in vitro. However, the physiological relevance of the crystal homodimer has been questioned on the basis of both topological concerns (2) and mutational studies showing no disruption of CD81 homodimers after disruption of the putative dimerization interface (12). Accordingly, to better understand the immune protection provided by the Sm-TSP-2 vaccine and to delineate the structure/function relationship of TSPs in general, we have structurally characterized the EC2 domain of Sm-TSP-2 and present the first solution structure of this domain. Furthermore, we have addressed the functional role of Sm-TSP-2 by identifying proteins potentially involved in TEMs at the surface of the schistosome tegument. Based on these findings we are able to propose a refined model of the interactions of TSPs with the plasma membrane, other TSPs, and the proteins comprising TEMs in S. mansoni.
EXPERIMENTAL PROCEDURES
Recombinant Protein Expression and Purification
TSP topology was predicted using TMHMM (13), and the region of Sm-TSP-2 corresponding to the Sm-TSP-2-EC2 was PCR-amplified from a pET-41a-derived construct and cloned into the pLic-MBP vector via ligation-independent cloning for bacterial expression (14) using the forward primer (TACTTCCAATCCAATGAAAAGCCCAAGGTCAAAAAACAC) and reverse primer (TTATCCACTTCCAATGTTAGTGCGCTTTGCTTAGATCGC). The vector was constructed by Stephen Shouldice and was a kind gift from Prof. Jenny Martin (University of Queensland, Brisbane, Australia).6 This vector encoded a MalE signal sequence for periplasmic export, a His6 affinity tag inserted in two residues after MalE, a maltose-binding protein (MBP) fusion tag to aid the folding and solubility, and a tobacco etch virus (TEV) protease recognition site. The TEV was mutated to the thrombin site by a single step mutagenesis (15) using the forward primer (GTGCCGCGTGGCTCCAATGAAAAGCCCAAGGTCAAAAAACACATCAC) and reverse primer (GGAGCCACGCGGCACCAGGTTCTCGGTACCTGGGATATC). The plasmid encoding His6-MBP-Throm-Sm-TSP-2-EC2 was transformed into Escherichia coli BL21(DE3) cells and, 13C,15N/-labeled or 15N-labeled Sm-TSP-2-EC2 was expressed using established methods (16). The cells were harvested by centrifugation (6000 × g, 10 min, 4 °C), and the His6-MBP-TEV-Sm-TSP-2-EC2 was extracted from the bacterial periplasmic space by osmotic shock using 30 mm sodium phosphate, pH 7.4, 20% sucrose, 1 mm EDTA, and 5 mm MgSO4 in ice-cold water (80 ml for each gram of cells wet weight). The protein was desalted to PBS buffer, pH 7.4, with a PD-10 column (GE healthcare). The MBP tag was removed by incubating the eluted protein with thrombin protease (Sigma) in PBS at room temperature overnight. After thrombin cleavage, Sm-TSP-2-EC2 was recovered using a nickel-nitrilotriacetic acid column and desalted to PBS buffer, pH 7.4, in the presence of 3 mm glutathione (GSH) and 0.3 mm glutathione disulfide (GSSG) for 24 h. Sm-TSP-2-EC2 was then purified by high performance liquid chromatography (HPLC) using a linear gradient of 20–60% Buffer B (10% H2O in CH3CN) over 80 min (Phenomenex Jupiter C18 column of 5 μm; 150 mm × 150 μm inner diameter). After the expected mass of Sm-TSP-2-EC2 was confirmed using mass spectroscopy, the protein was lyophilized and dissolved in pure water at a concentration of 1.4 mm, and the pH was adjusted to 5.2. For structure determination, CHAPS was added to a concentration of 5 mm.
Protein Structure Determination Using NMR Spectroscopy
Data were acquired at 298 K using a 900-MHz Avance II NMR spectrometer (Bruker BioSpin GmbH, Rheinstetten, Germany) equipped with a cryogenically cooled probe. Data for backbone resonance assignments, including HNCA, HNCACB, CBCA(CO)NH, HNCO, HNCACO, HBHACONH, were acquired using nonuniform sampling. Sampling schedules that approximated the signal decay in each indirect dimension were generated using sched3D (17). Nonuniform sampling data were processed using the Rowland NMR toolkit with the automatically generated maximum entropy processing scripts (17). A nonuniform sampling 4D HCC(CO)NH-TOCSY (18) was used for rapid side-chain assignments, which were confirmed by 13C-edited NOESYSHQC (aliphatic side chain). (H)CB(CGCC-TOCSY)Har (19), Tyr-selective (H)CB(CGCC-TOCSY)Har, and Phe-selective (H)CB(CGCC-TOCSY)Har were also acquired to assist the assignment of the aromatic spin system. The complete chemical shift assignments are available from BioMagResBank (accession number 19217). A 15N-edited NOESYHSQC and two 13C-edited NOESYHSQC (aliphatic and aromatic side chains, respectively) with a mixing time of 120 ms were used for NOE derived distance restraints. φ and ψ dihedral angles restraints were derived from TALOS chemical shift analysis (20), with the boundaries of the restraint used for structural calculations set to twice the estimated standard deviation. NOESY spectra were manually peak picked and integrated using CCPNMR (21), peak lists were automatically assigned, distance restraints were extracted, and an ensemble of structures was calculated using CANDID within the torsion angle dynamics package CYANA (22). The tolerances used for CYANA were 0.03 ppm in the direct 1H dimension, 0.040 ppm in the indirect 1H dimension, and 0.45 ppm for the heteronuclei (13C,15N). Disulfide topology was initially left undefined, but initial structure calculations showed only one disulfide topology was possible based on the observed NOEs, and in the final structure calculation the disulfide topology was defined (Cys-42–Cys-70 and Cys-43–Cys-59). Using CYANA (22), 100 random conformers were annealed in 8000 steps using torsion-angle dynamics, and 20 conformers with the lowest residual restraint violations were selected as the representative structures. Coordinates of the final ensembles were deposited into the Protein Data Bank with code 2m7z. Structure quality was assessed using MolProbity (23).
Pulse Field Gradient NMR Diffusion Measurements
Sm-TSP-2-EC2 was dissolved in H2O (1.1 mm, pH 5.2) containing 10% D2O, and 20 μl of 1% dioxane in D2O was added as an internal radius standard (2.12 Å) (24). A translational self-diffusion coefficient of 1.1 mm 15N-labeled Sm-TSP-2-EC2 was measured at 298 K on the aforementioned NMR spectrometer. Pulse field gradient NMR diffusion measurements were performed with a stimulated echo sequence using bipolar gradients and water suppression using a 3-9-19 pulse sequence with gradients (24–26). The lengths of the diffusion gradient and the stimulated echo were optimized to 2 and 100 ms to give a total decay in the protein signal of ∼93%. A series of 80 or 128 diffusion-weighted one-dimensional spectra were recorded in a pseudo two-dimensional manner with the strength of the diffusion gradient varying between 2 and 95% that of its maximum value. Diffusion coefficients of 1.17 ± 0.037 × 10−10 m2/s (D) were obtained by fitting intensities of several peaks (7.659, 7.559, 7.356, 6.989, 6.891, 6.845, 6.723 ppm) in the aromatic region to the following equation with the program Simfit in Topspin 3.2 (Bruker), I = I0e(−Dγ2g2δ2(Δ−δ/3−τ/2)), where I is the observed intensity, I0 the reference intensity (unattenuated signal intensity), D the diffusion coefficient, γ is the gyromagnetic ratio of 1H, g is the gradient strength, δ the length of the gradient, Δ the diffusion time, and τ the time between bipolar gradients.
D2O Exchange and Detergent Titration
500 μl of 0.1 mm 15N-labeled Sm-TSP-2-EC2 in H2O, pH 5.2, was lyophilized and redissolved in 500 μl of D2O in the presence of 5 mm CHAPS. A 15N HSQC was recorded at 0, 2, 3, 6, 7.5, 10, 13, and 14 h after the addition of D2O. Amide protons involved in hydrogen bonds were identified by the presence of their NH resonances in 15N HSQC at least 6 h after dissolving in D2O. The interactions of Sm-TSP-2-EC2 with CHAPS was determined by titration of a 0.1 mm solution of 15N-labeled Sm-TSP-2-EC2 with a 10 mm solution of CHAPS to a final protein:CHAPS ratio of 1:0.5, 1:1, 1:2, 1:5, 1:10, 1:20, 1:30, 1:40, 1:50, 1:60 and monitoring of the chemical shift changes and intensity changes of the protein peaks by 15N HSQC spectra. The binding affinity was obtained by the least squares fitting of the chemical shift changes to a binding isotherm assuming a single binding site of CHAPS on Sm-TSP-2-EC2. To characterize the interaction of Sm-TSP-2-EC2 with tegument membrane, 0.53 mm 15N-Sm-TSP-2-EC2 was titrated with a 10 or 100 mm stock of the 1:0.8 mixture of dodecylphosphocholine (DPC) and CHAPS to approximate the cholesterol-rich schistosome Tegument (27, 28). The titration was performed at a [protein]:[DPC 1:CHAPS 0.8] ratio of 1:0.5, 1:1, 1:4, 1:8, 1:16, and 1:24 to reach the critical micelle concentration (CMC) of DPC 1:CHAPS 0.8. Chemical shift and intensity changes of the cross-peaks were monitored by 15N HSQC spectra.
Animal Ethics and Parasite Lifestyle Maintenance
Research involving animals in this study was approved by the Animal Ethics Committee of the Queensland Institute of Medical Research. The study was conducted according to guidelines of the National Health and Medical Research Council of Australia as published in the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes, 7th edition, 2004. The Puerto Rican strain of S. mansoni was maintained in ARC Swiss mice and Biomphalaria glabrata snails at QIMR from stocks originating from the National Institute of Allergy and Infectious Diseases Schistosomiasis Resource Centre, Biomedical Research Institute (Rockville, MD). Worm pairs were perfused from mice 6 weeks post-infection.
Tegumental Protein Cross-linking, Purification, and SDS-PAGE
Approximately 57 worm pairs were perfused from infected mice and washed thoroughly in perfusion buffer (145 mm NaCl, 60 mm sodium citrate). For control samples the perfusion buffer was removed, and worms were snap-frozen on dry ice and stored in liquid nitrogen for later processing. For cross-linking of surface proteins with bis(sulfosuccinimidyl)suberate (BS3), worms were perfused and washed thoroughly in Hanks' balanced salt solution (HBSS; Sigma) and then incubated in 5 mm BS3 in HBSS for 30 min at room temperature with occasional agitation. The reaction was quenched by the removal of BS3 and the addition of 10 mm glycine in HBSS for 15 min at room temperature. Worms were washed 3 times with HBSS, and all liquid was removed before freezing on dry ice. The tegument was removed from both cross-linked and uncross-linked worms by the freeze/thaw/vortex method (29) with slight modifications. Briefly, the worms were thawed, initially at room temperature and then on ice, and washed quickly in ice cold TBS (10 mm Tris/HCl, 0.84% NaCl, pH 7.4). Worms were incubated on ice for 5 min in 10 mm Tris/HCl, pH 7.4, vortexed 5 times for 1 s each, and the supernatant was transferred to a new tube. The supernatant was centrifuged at 12,000 × g for 30 min at 4 °C, and the resulting pellet was resuspended in 21 μl of 10 mm Tris/HCl, pH 7.4. 4 × Laemmli sample buffer (62.5 mm, 1 m Tris, pH 6.8, 2% SDS, 0.05% bromphenol blue, 20% glycerol, 0.71 mm β-mercaptoethanol) was added to a final concentration of 1× to each sample before boiling for 5 min at 95 °C. The samples were then applied to 1-mm-thick 4% stacking, 10% resolving gel for SDS-PAGE. Electrophoresis was carried out at 100 V for 20 min and then 200 V for 50 min. The gels were stained using Coomassie Brilliant Blue and destained in 25:10:65 methanol/acetic acid/water (v/v/v).
Blue Native PAGE
Approximately 60 freshly perfused worm pairs were washed thoroughly in perfusion buffer. The tegument was isolated according to the standard freeze/thaw/vortex protocol (above), except the tegument was resuspended in a final concentration of 1 × NativePAGE sample buffer, 1 × protease inhibitor (Roche Applied Science), and 1% n-dodecyl-β-d-maltoside using the NativePAGE Sample Prep kit (Invitrogen). Samples were incubated on ice for 15 min before centrifugation at 12,000 × g at 4 °C for 30 min. NativePAGE G-250 Sample Additive (Invitrogen) was added to isolated proteins to a final concentration of 0.25%. Fractionation was carried out on 1.0-mm NativePAGE 3–12% Bis-Tris Gel (Invitrogen) using 1× NativePAGE Anode Running Buffer and 1× Native PAGE Dark Cathode Buffer (Invitrogen). The sample was fractionated under native conditions at 4 °C at 150 V for 1 h and 200 V for 45 min. After destaining the gel, the lane was then divided into ∼30 slices, and an in-gel tryptic digest was performed as per the protocol described above.
Sm-TSP-2 Immunoprecipitation
BS3 was used to cross-link the teguments of ∼180 worm pairs as described above. The teguments were then isolated by the standard freeze/thaw/vortex protocol. The resulting tegument pellet was resuspended in lysis buffer (0.5 m urea, 0.2% (w/v) SDS, 1.0% (v/v) Triton X-100, 1 mm dithiothreitol (DTT), PBS-0.1% Tween 20 (PBST)). A polyclonal anti-Sm-TSP-2 antibody raised against recombinant Sm-TSP-2-EC2 was used to precipitate protein complexes containing Sm-TSP-2, and antibodies to REX, a ring stage-specific protein from Plasmodium falciparum (30), were used as a negative control. Protein G Sepharose beads (BioVision) were washed 3 times with PBST before antibody binding in PBST at 4 °C for 1 h under rotation. The beads were washed twice more with PBST before the addition of cross-linked tegument proteins and incubation overnight at 4 °C under rotation. The unbound fraction was collected, and the beads washed three times with PBST. Bound proteins were eluted by adding 4 × Laemmli sample buffer and incubating at 95 °C for 10 min. Eluted proteins were fractionated by SDS-PAGE using a 4% stacking and 12% resolving gel. The gels were stained using Coomassie Brilliant Blue and destained in 25:10:65 methanol/acetic acid/water (v/v/v), and in-gel tryptic digest was performed.
In-gel Tryptic Digest and Mass Spectroscopy
SDS-PAGE gel lanes were divided into ∼30 slices, and each slice was cut into small pieces. Each gel slice was processed independently and was first destained twice by incubation in 50% acetonitrile, 200 mm NH4HCO3 for 45 min at 37 °C for 30 min at 37 °C and then dried using a vacuum centrifuge. The gel pieces were resuspended in 20 mm DTT and reduced for 1 h at 65 °C. DTT was removed, and the samples was alkylated by the addition of 50 mm iodoacetamide and incubation in darkness at 37 °C for 40 min. Gel pieces were washed twice in 25 mm NH4HCO3 for 15 min and completely dried in a vacuum centrifuge. Gel pieces were rehydrated with 20 μl of trypsin reaction buffer (40 mm NH4HCO3, 10% acetonitrile) containing 20 μg/ml trypsin (Sigma) for 20 min at room temperature. An additional 50 μl of trypsin reaction buffer was added to the samples and incubated overnight at 37 °C. The digest supernatant was removed from the gel slices, and residual peptides were washed from the gel slices by incubating 3 times with 0.1% formic acid for 45 min at 37 °C. The original supernatant and extracts were combined and dried in a vacuum centrifuge. The tryptic peptides were resuspended in 12 μl of 5% formic acid before mass spectral analysis.
Protein Identification Using Tandem Mass Spectroscopy
Tryptic fragments from in-gel digests were chromatographically separated on a Dionex Ultimate 3000 HPLC using an Agilent Zorbax 300SB-C18 (3.5 μm, 150 mm × 75 μm) column and a linear gradient of 0–80% solvent B over 60 min. A flow rate of 0.3 μl/min was used for all experiments. The mobile phase consisted of solvent A (0.1% formic acid (aqueous)) and solvent B (80/20 acetonitrile, 0.1% formic acid (aqueous)). Eluates from the reverse phase HPLC column were directly introduced into the NanoSpray II ionization source of a 5600 MS/MS System (AB Sciex) operated in positive ion electrospray mode. All analyses were performed using Information Dependant Acquisition. Analyst 2.0 (Applied Biosystems) was used for data analysis and peak list generation. Briefly, the acquisition protocol consisted of the use of an Enhanced Mass Spectrum scan as the survey scan. The three most abundant ions detected over the background threshold were subjected to examination using an Enhanced Resolution scan to confirm the charge state of the multiply charged ions. The ions with a charge state of +2, +3, or with unknown charge were then subjected to collision-induced dissociation using a rolling collision energy dependent upon the m/z and the charge state of the ion. Enhanced Product Ion scans were acquired resulting in full product ion spectra for each of the selected precursors, which were then used in subsequent database searches.
X! Tandem Searches
Searches were performed using Version 12.10.01.1 of X! Tandem (31) with a 0.1-Da tolerance on the precursor and 0.1-Da tolerance on the product ions, allowing for methionine oxidation and carbamidomethylation as fixed and variable modifications, respectively, one missed cleavage, charge states +2 and +3, and trypsin as the enzyme. All experiments were searched against the Uniprot S. mansoni proteome data set, downloaded on the Feb. 10, 2013 (11,712 sequences). Searches were also made against the SwissProt database (as of the May 23, 2011) to control for contamination. Any peptide matching a non-schistosome protein was removed from the peptides attributed to identifications from the S. mansoni database. The criteria for accepting protein identifications were as follows: (i) the identification needed to contain at least two peptides with an e value less than 0.05, and (ii) at least one significant peptide was unique to the identified protein. In the case of protein identifications relying on the same peptide identifications, the protein with the highest score was used.
RESULTS
Recombinantly Expressed Sm-TSP2-EC2 Showed Well Defined Secondary Structure
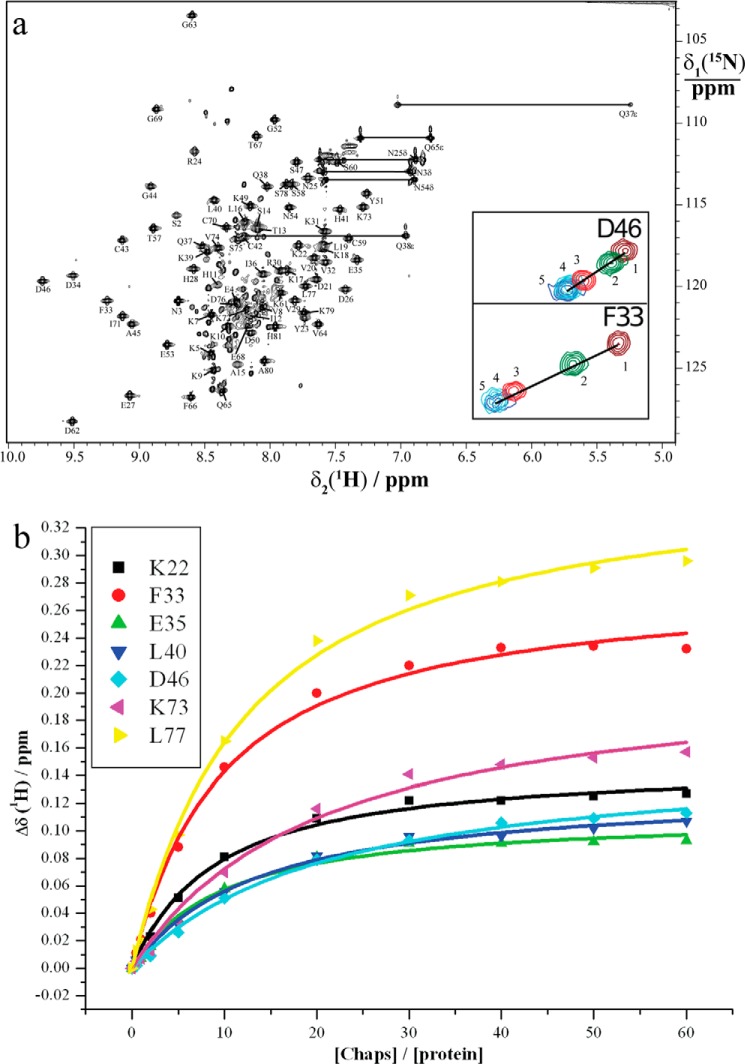
Periplasmic expression of Sm-TSP-2-EC2 resulted in the production of 7 mg of 13C,15N-labeled protein from 500 ml of minimal M9 medium. Mass spectroscopy of the expressed protein confirmed the anticipated mass based on the amino acid sequence of the Sm-TSP-2-EC2 expression construct (9.2 KDa for a 15N-labeled protein). NMR data recorded using 1.4 mm Sm-TSP-2-EC2 protein in water at pH 5.2 showed well dispersed resonances in the 15N HSQC spectrum, suggesting that the protein was correctly folded. However, mild-line broadening was observed for a small number of resonances, indicative of either oligomeric assemblies or conformational exchange. As the human tetraspanin hCD81-LEL exists as a homodimer in solution (32), 5 mm CHAPS, a zwitterionic detergent commonly used for disaggregation of protein complexes (33), was added to Sm-TSP-2-EC2. This reduced line broadening in affected residues, suggesting disaggregation of protein complexes though still consistent with conformational stabilization. Given the well dispersed resonances for Sm-TSP-2-EC2 in water and the high degree of similarity in chemical shifts for the majority of resonances recorded in 5 mm CHAPS, spectra recorded under the latter conditions were used to assign all backbone resonances using standard triple-resonance methods (34) (Fig. 1a). Chemical shift analysis of α and β carbons indicated the presence of three major helical regions spanning residues Lys-10–Lys-22 (helix A), Glu-27–Leu-40 (helix B), and Cys-70–Lys-79 (helix C), and these were confirmed using predicted ϕ and ψ angles in TALOS+ (20). These major elements of secondary structure correspond to the three-helix conserved region described in the hCD81-LEL crystal structure (helices A, B, and E in that structure).
FIGURE 1.
15N HSQC spectrum of Sm-TSP-2-EC2. a, 15N HSQC spectrum of a 1.4 mm solution of Sm-TSP-2-EC2 in H2O at 298 K in the presence of 5 mm CHAPS. The cross-peaks of the backbone amides are assigned with the one-letter amino acid code and the sequence number of the residue. Horizontal lines connect pairs of cross-peaks from the side-chain amides of Asn and Gln. Folded peaks from arginine side chains are labeled with a star. Gly-1, Ser-2, and Asn-3 were introduced by the protein expression construct. Insets are amide cross-peaks for residues Phe-33 and Asp-46 corresponding to protein:CHAPS ratios of 1:0 (1), 1:5 (2), 1:20 (3), 1:40 (4), and 1:60 (5). b, binding affinity of Sm-TSP-2-EC2 to CHAPS measured by NMR spectroscopy at 298 K. Kd values obtained from the chemical-shift changes of Lys-22, Phe-33, Glu-35, Leu-40, Asp-46, Lys-73, and Leu-77 were 0.86 ± 0.08, 0.92 ± 0.09, 0.89 ± 0.11, 1.23 ± 0.11, 2.02 ± 0.19, 1.94 ± 0.30, and 1.16 ± 0.10 mm, respectively, indicating an upper limit of about 2.00 mm.
Sm-TSP-2-EC2 Possesses a Conserved and Variable Region When Compared with hCD81-LEL
The solution structure of Sm-TSP-2-EC2, in the presence of 5 mm CHAPS, was calculated using 1705 non-redundant NOE upper-distance limits, 11 hydrogen bonds restraints, and 129 TALOS+-derived dihedral angle restraints (Table 1). The 20 conformers with the lowest final target function values converged into a single well defined structure with a backbone root mean square deviation of 0.35 ± 0.07 Å (Fig. 2a) for residues Lys-9–Lys-79. Analysis of the lowest energy structures with MolProbity (23) showed that the structure was of a high stereochemical quality as reflected in a low clash score, high Ramachandran plot quality, and favorable side-chain rotamers (Table 1). The average MolProbity score of 1.90 places the 20 models in the ensemble in the 81st percentile relative to all other structures ranked using this algorithm. The consensus structure of Sm-TSP-2-EC2 has the same mushroom-like fold possessed by hCD81-LEL (10), with a “stem” composed of helices A, B, and C and a variable region, between residues His-41 and Gly-69 that has a well defined structure but few secondary structure motifs apart from a small β-hairpin between residues Val-64 and Lys-61 (Fig. 2b). The variable region is folded tightly back upon itself and is reinforced by two disulfide bonds formed between the four conserved cysteine residues characteristic of the TSPs. Despite the conserved disulfide motif, the variable region of Sm-TSP-2-EC2 shows the greatest structural deviation from the hCD81-LEL structure with the variable regions of the two molecules exhibiting very little conservation of secondary or tertiary structure (Fig. 3). Initial structure calculations, in which the disulfide topology of Sm-TSP-2-EC2 was left undefined, showed that only one disulfide topology was possible based on the observed NOEs, and this topology corresponded to that observed in characterized TSPs, including hCD81-LEL.
TABLE 1.
NMR and refinement statistics for protein structures
| Protein | |
|---|---|
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOEa | 1705 |
| Intra-residue | 491 |
| Inter-residue | |
| Sequential (|i − j| = 1) | 448 |
| Medium range (|i − j| ≤ 4) | 351 |
| Long range (|i − j| > 5) | 415 |
| Intermolecular | N/A |
| Hydrogen bondsb | 11 |
| Total dihedral angle restraints | |
| φ | 64 |
| ψ | 65 |
| Structure statistics | |
| Cyana target function (Å2) | 0.46 ± 0.05 |
| Violations (mean and S.D.) | |
| Distance constraints (>0.1Å) | 1.25 ± 0.97 |
| Dihedral angle constraints (>5°) | 0 ± 0 |
| Maximum dihedral angle violation (°) | 3.28 |
| Maximum distance constraint violation (Å) | 0.32 |
| Stereochemical qualityc | |
| Residues in most favored | |
| Ramachandran region, % | 92.95 ± 1.93 |
| Ramachandran outlier, % | 0 ± 0 |
| Unfavorable side chain rotamers, % | 13.63 ± 2.49 |
| Clashscore, all atoms | 0.36 ± 0.40 |
| Overall MolProbity score | 1.90 ± 0.15 |
| Average pairwise r.m.s. deviationd (Å) | |
| Heavy | 0.85 ± 0.09 |
| Backbone | 0.35 ± 0.07 |
a Non-redundant NOE upper-distance limits.
b Two restraints used per hydrogen bond.
c Stereochemical quality was assessed using MolProbity. Clashscore is the number of steric overlaps (>0.4 Å) per 1000 atoms.
d Root mean square deviation values were calculated over the well defined regions of the structure (residues 9–79) among the ensemble of 20 structures.
FIGURE 2.
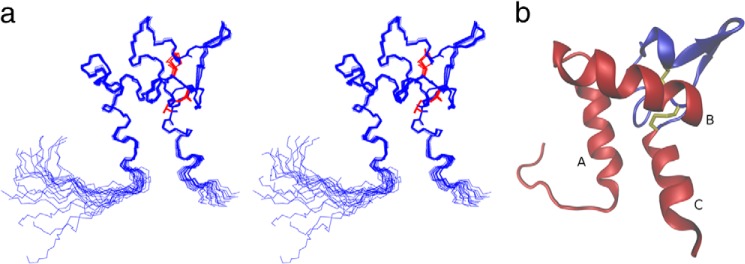
Structure of Sm-TSP-2-EC2. a, stereo view of the 20 lowest energy structures calculated for Sm-TSP-2-EC2. The protein backbone is shown in blue, and the two disulfide bonds are shown in red. b, structure of Sm-TSP-2-EC2 depicted as a ribbon diagram. The three major helices composing the constant domain of the protein are labeled A, B, and C and disulfide bonds are highlighted in yellow. Residues in the constant, or stem, region are colored red and in the variable, or head, region are colored blue.
FIGURE 3.
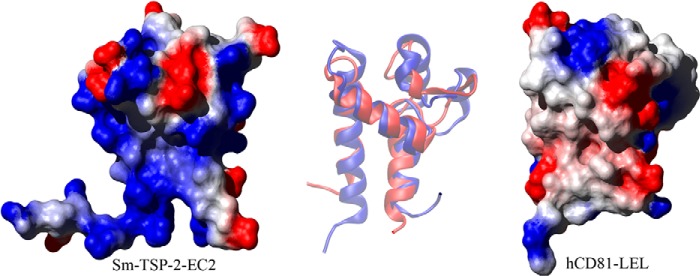
Comparison of Sm-TSP-2-EC2 and hCD81-LEL. A comparison of the structures of Sm-TSP-2-EC2 and hCD81-LEL showing the surface electrostatic potential of the two molecules (produced using MOLMOL; Ref. 51). Both proteins are orientated so that the stem region and helices A and C of the Sm-TSP-2-EC2 structure are in the background with helix B bridging the two stem helices. The variable region is protruding to the top right of both molecules. In hCD81-LEL the constant domain exhibits a low polarity surface, and this area has been proposed as the interaction interface between homodimers of the molecule. This region in Sm-TSP-2-EC2 contrasts strongly with hCD81-LEL, showing a strongly positive electrostatic potential caused by the presence of multiple Lys residues in helix A of the EC2 domain. At center is an overlay of a schematic depiction of each molecule orientated in the same way as the surface diagrams. The overlay shows Sm-TSP-2-EC2 in red and hCD81-LEL in blue and highlights the contrast between the structural conservation in the stem region of the two TSPs and the diversity shown in the head region.
Intramolecular Interactions Stabilize the Structure of Sm-TSP-2-EC2
Slowly exchanging amide protons were used to determine potential hydrogen bonds in Sm-TSP-2-EC2. After 6 h in D2O, amide protons from 20 residues were still identifiable by the presence of a NH resonance in a 15N HSQC spectrum (data not shown). Analysis of the structures confirmed that the majority of these amides are involved in hydrogen bonds stabilizing the major elements of secondary structure of Sm-TSP-2-EC2, including three helical hydrogen bonds in helix C and five helical hydrogen bonds in helix B. Interestingly no slow exchanging amides were observed in helix A, suggesting it to be flexible in solution, compared with helices B and C. In the head region, hydrogen bonds between the HN of Tyr-51 and O of Pro-48 as well as HN of Ser-47 and a side chain OD of Asp-50 stabilize a small 310 helical region, and reciprocal HN-O hydrogen bonds between Val-64 and Lys-61 stabilize a small β-hairpin. In addition, a hydrogen bond between HN of Ile-71 and the O of Ala-45 further stabilizes the head region by anchoring it to the beginning of helix C.
The Electrostatic Potential of Sm-TSP-2-EC2 Shows Marked Differences to hCD81-LEL
Analysis of the surface electrostatic potential of Sm-TSP-2-EC2 shows a distinct low polarity patch in the head region of the molecule. Three Pro residues (Pro-48, Pro-55, and Pro-56), Cys-59, Val-64, and Phe-66 form the hydrophobic patch, which is circled by charged residues, including Lys-49, Glu-53, and Asp-62. The conformation of the loop is stabilized primarily by the disulfide bond between Cys-43 and Cys-59, which draws the center of the loop toward the core of the molecule (Fig. 4). Gly-44 and Gly-52 in the head region possess backbone conformations only accessible by Gly residues (as indicated by their Ramachandran ϕ, ψ pairs: 54.8 ± 3.8°, −178.1 ± 10.0°, and 113.3 ± 4.0°, −102.4 ± 5.3° respectively) due to their increased conformational freedom. Interestingly, this Gly pair and their conformation was also observed in hCD81-LEL (11), and Gly-44 together with its two preceding Cys residues form an absolutely conserved motif of all TSPs, suggesting that the Gly residue and its conformational flexibility is required for the tight folding of the head region. The surface electrostatic potential of Sm-TSP-2-EC2 also shows a strongly positive region formed primarily by a Lys-rich region in helix A (Fig. 3). This region of the molecule corresponds to the proposed dimerization interface of hCD81-LEL, which in hCD81-LEL is a large non-polar patch (10). This difference in the electrostatic potential of the two molecules makes it an unlikely interface for dimerization of Sm-TSP-2.
FIGURE 4.
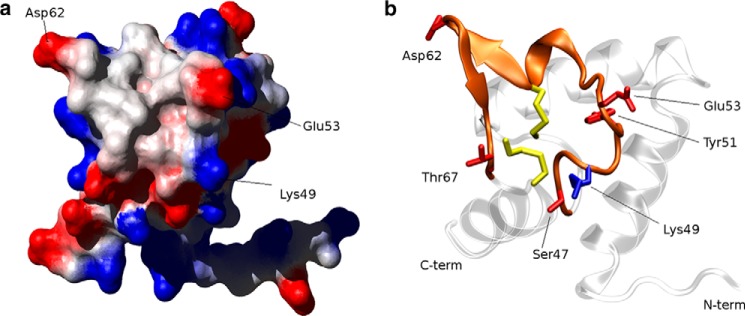
Exposed hydrophobic patch in the head region of Sm-TSP-2-EC2. a, surface electrostatic potential of the variable region of Sm-TSP-2-EC2 showing a non-polar region ringed by the charged or polar residues Lys-49, Glu-53, Asp-62, and Thr-67. b, ribbon diagram of Sm-TSP-2-EC2, in the same orientation as a, highlighting the extended loop forming the non-polar patch and the disulfide bonds that hold the loop structure in position.
Sm-TSP-2-EC2 Is Monomeric in Solution
Given the strongly positive electrostatic potential of the area of Sm-TSP-2-EC2 corresponding to the proposed dimerization interface of hCD81-LEL, we addressed the question of where, or if, Sm-TSP-2-EC2 formed dimers? Despite the absence of a low polarity patch, mild line-broadening in initial spectra, recorded in H2O, was still consistent with oligomeric protein assemblies, although conformational exchange could cause similar effects. Likewise, sharpening of resonances upon the introduction of 5 mm CHAPS could indicate disruption of protein complexes but is also consistent with the stabilization of parts of the protein by interactions with CHAPS molecules. Therefore, to determine the source of the line broadening and whether Sm-TSP-2-EC2 oligomerized in solution, the diffusion co-efficient of Sm-TSP-2-EC2 in water was calculated and compared with that of the internal standard dioxane. From this comparison a hydrodynamics radius of (16.8 ± 0.5) × 10−10 Å was measured for Sm-TSP-2-EC2 (25). This value is consistent with a protein of 71 to 88 amino acids, suggesting that Sm-TSP-2-EC2 is monomeric in solution and that the EC2 domain alone does not mediate dimerization of Sm-TSP-2. Apart from resonances originating from unfolded protein, 15N HSQC spectra recorded before and after purification by HPLC were similar, suggesting that purification under denaturing conditions did not lead to a monomeric state (data not shown).
The α-Helices of the Constant Region Mediate Contacts with CHAPS
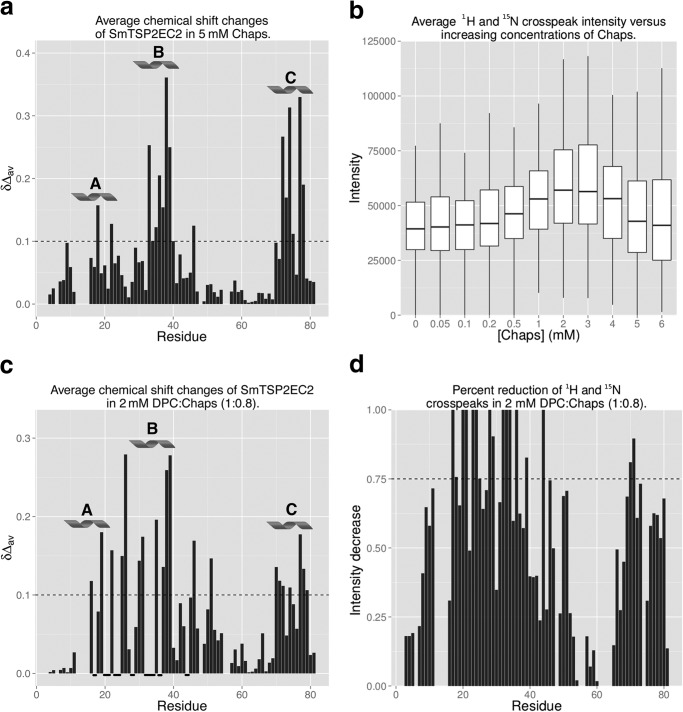
The concentration of CHAPS used was well below its CMC (∼9 mm in pure water; Ref. 35), and accordingly, it is likely that CHAPS molecules were interacting with specific parts of Sm-TSP-2-EC2 and stabilizing conformational fluctuations. To map these interaction sites, Sm-TSP-2-EC2 in water was titrated with increasing concentrations of CHAPS, and chemical shift and peak intensity changes were monitored in 15N HSQC experiments (see Fig. 1a for examples). Analysis of chemical shifts showed two well defined areas of increased perturbation that corresponded to helices B and C (Fig. 5a). In particular, residues 33–40 and 71–77 (excluding residue 75) all showed average 1H and 15N chemical shift changes >0.10 ppm. Binding affinities were calculated for some amino acids with a perturbation greater than 0.10 ppm, and these yielded an affinity of Sm-TSP-2-EC2 for CHAPS of 1–2 mm at 25 °C (Fig. 1b). In the same titrations, 1H and 15N cross-peak intensities increased with increasing concentrations of CHAPS until a concentration of 3 mm. After this, concentration intensities decreased as the concentration of CHAPS was increased (Fig. 5b). As Sm-TSP-2-EC2 is monomeric in solution, the reduction in line broadening at low concentrations suggests that CHAPS may stabilize conformational fluctuations, most likely in the stem region of the molecule where chemical shift changes are the greatest. Given that under physiological conditions the EC2 domain would be anchored to the membrane by the rest of the molecule, this conformational flexibility is probably an artifact of examining the EC2 domain in isolation from its transmembrane domains.
FIGURE 5.
Sm-TSP-2-EC2 interactions with micelle forming lipids. a, average chemical shift changes for Sm-TSP-2-EC2 after the addition of 5 mm CHAPS. Residues comprising helices B and C displayed the greatest amount of chemical shift variation, suggesting that CHAPS interacts primarily with residues in these helices. Averaged 1H and 15N chemical shift changes Δδav was calculated as (0.1ΔδN2 +ΔδH2)1/2 where ΔδN and ΔδH denote the chemical shift changes in ppm observed in the 15N and 1H dimensions of the 15N HSQC spectrum. b, average amide cross-peak intensity in increasing concentrations of CHAPS. Average intensity increased initially but began to decrease as the concentration approached the CMC of CHAPS. c, average chemical shift changes for Sm-TSP-2-EC2 in 2 mm DPC:CHAPS (1:0.8). As was the case in CHAPS titrations, residues comprising helices B and C displayed the greatest amount of chemical shift variation, suggesting primary sites of interaction were within these areas of the molecule. In some cases residues in these areas were broadened beyond detection, and these have been marked with a bold underline along the x axis. d, reductions in 1H and 15N cross-peak intensity for Sm-TSP-2-EC2 in 2 mm DPC:CHAPS (1:0.8). The decreasing intensities are calculated as (|Iprotein − Icomplex|/Iprotein), where Iprotein and Icomplex denote the peak intensity of Sm-TSP-2-EC2 without and with DPC:CHAPS (1:0.8) mixture. Significant line broadening was observed in the majority of residues with areas corresponding to helical regions A, B, and C displaying the greatest amount of broadening. In a and c, regions comprising helices A, B, and C are labeled and marked with a helix symbol, and a dashed line marks a chemical shift δ of 0.1. In d, a dashed line marks a 75% reduction in signal intensity.
The α-Helices of the Constant Region Interact with DPC:CHAPS Micelles
Despite an extracellular domain, the TSP EC2 domain is still thought to interact with the surface of the plasma membrane (32). The schistosome tegument has a cholesterol to phospholipid molar ratio of 0.7–1.1 (27); thus to examine the interactions of Sm-TSP-2-EC2 with micelles approximating the lipid composition of the schistosome, tegument titrations were conducted using a 1:0.8 mixture of DPC and CHAPS to mimic cholesterol (28). At concentrations below the CMC of DPC (∼1.2 mm; Ref. 36) small chemical shift variations (<0.10 ppm) were observed, mostly in residues from the helices comprising the constant region of the molecule. However, significant line broadening was observed for the same residues, indicating intermediate exchange interactions with the detergent. At concentrations of 2 mm and above, significant chemical shift changes were observed in spin systems from the helical constant domain of Sm-TSP-2-EC2 (Fig. 5c), and line broadening was observed in >80% of the spin systems (Fig. 5d), with 13% of the amide cross-peaks broadened beyond detection. This broadening is consistent with a decrease in the T2 relaxation time of a micelle/protein complex and, when combined with the chemical shift changes, suggests the interaction of Sm-TSP-2-EC2 with DPC:CHAPS micelles via the helical constant domain. Similar chemical shift perturbations and line broadening was observed upon the addition of DPC only, with 0.4 mm DPC sufficient to broaden some cross-peaks beyond detection (data not shown).
Sm-TSP-2 Forms Homodimers in TEMs
Although cross-linking studies of the human TSPs, CD9, CD81, and CD151 suggest that the primary units of TEMs are TSP homodimers, heterodimeric assemblies have also been observed (37). Accordingly, to determine whether Sm-TSP-2 interacts with other schistosome TSPs, potential protein constituents of Sm-TSP-2-mediated TEMs were identified using tandem mass spectroscopy (MS/MS) after chemical cross-linking of surface membrane proteins on whole worms with the lipid insoluble BS3. Initial experiments comparing the migration of proteins from cross-linked and control tegument preparations using SDS-PAGE showed that Sm-TSP-2 exhibited a significantly slower migration after BS3 treatment (Fig. 6), suggesting the successful cross-linking of Sm-TSP-2 with other TEM proteins. Incubation of cross-linked proteins with anti-Sm-TSP-2 antibodies and subsequent SDS-PAGE and MS/MS of the bound fraction led to the identification of six proteins occurring in each of two replicates. Analysis of SDS-PAGE gels of proteins incubated with a control antibody provided no significant identifications from a S. mansoni sequence database. Interacting proteins were identified in the same band as Sm-TSP-2 and included calpain (G4VAG2), annexin (G4VL68), heat shock protein 70 (G4V8L4), alkaline phosphatase (G4VJ94), and actin (G4VSW6) (Table 2). No schistosome TSP other than Sm-TSP-2 was identified, suggesting that Sm-TSP-2 forms homodimers in the schistosome tegument.
FIGURE 6.
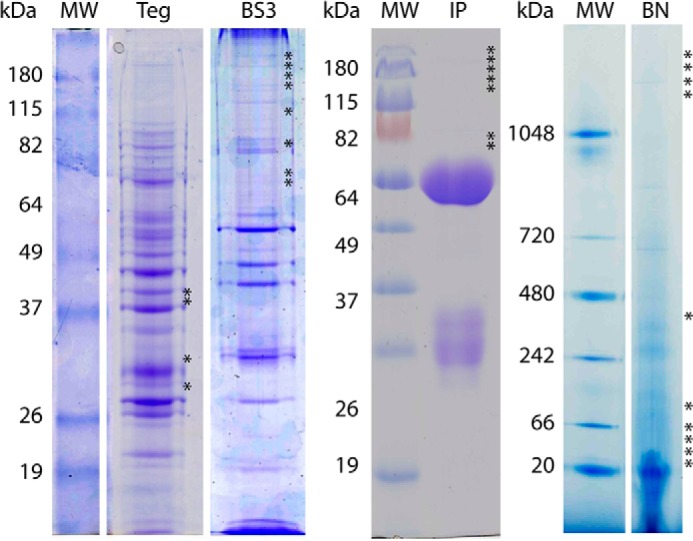
Representative gels from BS3 cross-linking and immunoprecipitation experiments. S. mansoni tegument protein preparations not reacted with BS3 were analyzed using SDS-PAGE (Teg) and non-denaturing BN-PAGE (BN). Crude BS3 cross-linked tegument preparations and were analyzed using SDS-PAGE (BS3). Elutes from immunoprecipitation experiments using BS3 cross-linked tegument preparations and anti-Sm-TSP-2 polyclonal antibodies were analyzed using SDS-PAGE (IP). Lanes were divided into 33 bands, and the protein constituents of the bands were determined using tandem mass spectroscopy. Gel bands in which Sm-TSP-2 was identified are marked with an asterisk. In BS3, BN-PAGE, and IP experiments Sm-TSP-2 exhibited slower migration through the gel, indicating interactions with proteins co-identified in the relevant bands. Marker lanes are denoted with MW, and the apparent molecule masses are shown (kDa).
TABLE 2.
Potential members of Sm-TSP-2 mediated TEMs
Mass spectral identification of potential binding partners from non-denaturing BN-PAGE gels and IP experiments. Column headings are: Accession, Uniprot accession; Score, the X! Tandem protein score; SC, number of unique peptides attributed to the protein identification; CO, percent cover of the protein provided by the identified peptides; Description, protein description; IP1, IP2, and BN-PAGE, asterisk (*) denotes identification of protein in the first or second immunoprecipitation experiment or in BN-PAGE experiments. Mass spectrometry identification data are from IP experiments; Loc., asterisk (*) denotes identification of the protein on the surface of S. mansoni using membrane-impermeable biotin (reported in Braschi and Wilson (38)).
| Accession | Score | SC | CO | Description | IP1 | IP2 | BN-PAGE | Loc. |
|---|---|---|---|---|---|---|---|---|
| G4VL68 | −92.7 | 11 | 0.32 | Annexin | * | * | * | * |
| G4VJ94 | −14.6 | 3 | 0.08 | Alkaline phosphatase | * | * | * | |
| G4VSW6 | −18.1 | 3 | 0.09 | Actin | * | * | * | |
| G4VAG2 | −21.1 | 3 | 0.02 | Calpain (CO2 family) | * | * | ||
| G4VJU0 | −19.3 | 3 | 0.10 | Fructose-bisphosphate aldolase | * | * | ||
| G4LV54 | −12.4 | 2 | 0.01 | Fer-1-related (Dysferlin) | * | * | * | * |
| G4V8L4 | −8.6 | 2 | 0.04 | Heat shock protein 70 | * | * | * | |
| O96368 | −14.2 | 2 | 0.11 | sm29 | * | * | * |
BN-PAGE Experiments Validated Immunoprecipitation Results
Non-denaturing blue native PAGE (BN-PAGE) was also used to fractionate partially solubilized protein complexes from the S. mansoni tegument. A number of detergents of differing solubilizing strength, including n-dodecyl-β-d-maltoside and digitonin, were used, and the resulting protein mixtures were fractionated using BN-PAGE. In BN-PAGE gels of n-dodecyl-β-d-maltoside-treated teguments Sm-TSP-2 was identified in bands 1–4 as well as in bands 14, 19, and 21–24. The apparent molecular mass of bands 19 and 21–24 approximated the molecular mass of native Sm-TSP-2 (∼26 kDa); however bands 1–4 and 14 indicated a substantial shift in the apparent molecular mass of Sm-TSP-2. In bands 1–4, 8 proteins were co-identified with Sm-TSP-2, including all of the proteins identified in both IP experiments with the exception of alkaline phosphatase and calpain. Another two proteins, Sm29 (O96368) and fructose-bisphosphate aldolase (G4VJU0), were identified in the BN-PAGE as well as one of the IP experiments, and these were added to the final list of eight potential interaction partners presented in Table 2. Consistent with the IP experiments, no other schistosome TSP was identified in bands containing Sm-TSP-2.
DISCUSSION
As scaffold proteins, TSPs play an important role in a range of biological processes due to their close association with proteins that mediate these processes. The TSP EC2 domain appears to play the primary role in the formation of these protein complexes termed TEMs (2), and accordingly an understanding of the mechanisms by which this domain mediates contacts between other proteins, other TSPs, and the plasma membrane is of importance for understanding how TEMs assemble and function. In S. mansoni, the crucial role of the TSPs is emphasized by the effectiveness of Sm-TSP-2 as a vaccine target. The primary goal of this study was to understand the structural basis by which this domain contributes to the formation of TEMs in the schistosome tegument and to identify potential protein complexes that form around Sm-TSP-2. The three-dimensional structure of Sm-TSP-2-EC2 reported here along with a number of potential protein constituents of the Sm-TSP-2 TEM provides insight into the role of the EC2 domain in the formation of TEMs in the schistosome tegument. Furthermore, these findings support a functional role of Sm-TSP-2 in maintaining tegumental integrity and mediating dynamic processes occurring at the surface of the membrane. Moreover, the structure reported here is the first solution structure of the EC2 domain and suggests a model in which the conserved region mediates contacts with the plasma membrane while the head region provides structural variability for the mediation of contacts with other proteins in the TEMs. Along with the crystal structure of hCD81-LEL (10, 32), the structure presented here is the only structural information available for these ubiquitous proteins.
As with hCD81-LEL, Sm-TSP-2-EC2 adopts a mushroom-like fold comprising a stem and “head” region. The stem regions of the two molecules display structural similarity in both the length of the helices and in their orientation to the rest of the molecule (Fig. 3). Conversely, the head region exhibits significant structural variation. This confirms predictions based on extensive homology modeling (11) that the stem and head region represent, respectively, conserved and variable regions. Titrations of Sm-TSP-2-EC2 with a mixture of DPC:CHAPS suggests that this organization might reflect a functional dichotomy in which the constant domain mediates interactions with the plasma membrane, whereas the variable region provides the structural diversity required for interacting with a variety of protein partners. Chemical shifts and intensity changes in DPC:CHAPS titrations support a model in which all three helices of the stem region of Sm-TSP-2-EC2 mediate contacts with the plasma membrane. In DPC:CHAPS, at concentrations greater than the CMC of DPC (∼1.2 mm for DPC; Ref. 36), line broadening of a majority of residues indicates the formation of protein:micelle complexes and chemical shift analysis indicates interactions predominantly with residues of helices A, B, and C (Fig. 5). In similar studies of hCD81-LEL, peak intensities also showed the interaction of the protein with DPC:CHAPS micelles, but specific interactions were mapped only to helix E (helix C in Sm-TSP-2-EC2) and with a small number of residues from the variable head region (32). Unlike Sm-TSP-2-EC2, hCD81-LEL was shown to be a dimer in this study, and corresponding parts of the molecule were thus not surface-exposed and available for membrane interactions.
However, although cross-linking studies of the human TSPs, CD9, CD81, and CD151 clearly show that the primary units of TEMs are TSP homodimers (37), the role of the EC2 domain in dimerization has not been fully elucidated. In hCD81-LEL, a low polarity region in the constant domain of hCD81-LEL has been proposed as a potential dimerization interface in CD81 (10). This interface mediates the dimerization of hCD81-LEL in the crystals used for x-ray diffraction, and a subsequent study also found that hCD81-LEL was dimerized in solution (32). In the current study, Sm-TSP-2-EC2 was found to be monomeric in solution, and the region of the molecule corresponding to the proposed dimerization interface in hCD81-LEL had a strongly positive surface potential, making it an unlikely homodimer interface. Furthermore, no evidence for the presence of other schistosome TSPs was identified in Sm-TSP-2 TEMs using chemical cross-linking or BN-PAGE experiments, suggesting that although homodimers are the primary units of the TEMs forming around Sm-TSP-2, the EC2 domain of Sm-TSP-2 does not mediate the dimerization. Although different dimerization mechanisms may be at work, there is evidence that the dimerized crystal may not be physiologically relevant. First, in the crystal structure, the molecules essentially point in opposite directions, and it is hard to reconcile this arrangement with a model also incorporating the plasma membrane (32). Moreover, mutation of residues in the proposed interface does not prevent oligomerization of the CD81 in mammalian cells (12). In other TSPs, chemical cross-linking of TSP homodimers at cysteine residues of the second, third, and fourth transmembrane domains (37) suggests that these domains form the dimerization interface. When combined with the evidence presented here, it appears that the EC2 domain does not play a major role in oligomerization of TSPs.
In total, eight proteins were identified as potential members of Sm-TSP-2 mediated TEMs. As BS3 will cross-link any proteins with free amines in close proximity, the identification of a protein in association with Sm-TSP-2 in an IP experiments does not establish that the protein directly interacts with Sm-TSP-2. A more meaningful interpretation is to consider these proteins as potential members of a protein complex formed around Sm-TSP-2, with second-order interactions also captured by cross-linking. An examination of the proteins identified in the cross-linking experiments (Table 2) shows that half have been previously identified at the surface of S. mansoni using membrane-impermeable biotin labeling (38) and, on the evidence presented here, are potential members of the exposed TEM in schistosomes. Of the remaining proteins, whereas calpain and HSP70 have been putatively identified as membrane-associated in S. mansoni (38) and S. japonicum (39), the presence of both actin and fructose-bisphosphate aldolase suggests vertical interactions were also captured. Although BS3 is membrane-impermeable, limited penetration of the membranocalyx and/or plasma membrane could have occurred across damaged membranes or by active transport. Internalized BS3 could, therefore, link either Sm-TSP-2 or dysferlin, both of which span the plasma membrane, to cytoplasmic proteins. For instance, in humans calpain is known to interact with dysferlin during membrane repair (40), and the interaction is thought to occur at the cytoplasmic face of the plasma membrane. Accordingly, given a similar interaction in schistosomes, even limited penetration of BS3 could have resulted in the cross-linking of the two proteins in this study. The phenotype of adult worms subjected to RNA interference against Sm-TSP-2 has been described (9), and knockdown of the putative TEM members identified here will provide insight into the significance of the interaction for the function of the Sm-TSP-2 TEM. The identification of TEM proteins that are crucial for TEM function is of particular importance for the development of multivalent vaccine antigens targeting Sm-TSP-2 TEMs.
In humans TSP binding partners include integrins, immunoglobulin superfamily members, growth factor receptors, transmembrane proteinases, G-protein-coupled receptors and cytoplasmic signaling molecules (41). None of these proteins were identified as potential TEM members in this study, and of those that were identified none has been previously associated with TEMs. In S. mansoni, knockdown of Sm-TSP-2 in the immature stage of the worm results in large scale degradation of the tegument (9). Given the essential role of Sm-TSP-2 in schistosome tegument integrity, it is interesting that several of the potential TEM proteins that were identified have roles in membrane repair and fusion. For instance strong evidence links members of the ferlin family to a role in the regulation and triggering of Ca2+-dependent membrane fusion events (42). Similarly, annexin, dysferlin, and calpain have all independently been shown to participate in membrane repair (43–45), and in the case of calpain this has been shown specifically in schistosomes (46). Both calpain and annexin interact with dysferlin (40, 47), potentially explaining why all three proteins were identified as potential TEM proteins in this work.
Recently the TSPs have been suggested to play a key role in the formation and budding of small vesicles called exosomes. A large quantity of TSPs are released from the cell in exosomes (3, 48–50), and it has been suggested that TSPs orchestrate changes in the plasma membrane leading to the formation and budding of exosomes from the cell (4). Although schistosome exosomes have not yet been described, they have been found in related species Echinostoma caproni and Fasciola hepatica (52) as well as in the nematode Caenorhabditis elegans (53). Using Exocarta (54), all proteins identified as potential members of a Sm-TSP-2 TEM had homologues that are known constituents of exosomes, the sole exception being Sm29, a protein unique to schistosomes. Given the role of the TSPs in exosome formation and budding and the proteins identified here as potential members of the Sm-TSP-2 TEM, it is quite possible that Sm-TSP-2 will be found to play a role in exosomal processes.
Apart from Sm-TSP-2, several other proteins identified as potential members of Sm-TSP-2 TEMs are themselves potential vaccine antigens for schistosomiasis. Three have been shown to be promising vaccine antigens, including Sm29 (55), calpain (56), and fructose-bisphosphate aldolase (57). Annexin is also currently under active investigation as a vaccine antigen (58). The susceptibility of schistosomes to vaccines targeting the proteins identified as members of TEMs might indicate that the disruption of the TEMs is an underlying mechanism of the protection afforded by these vaccines. If this is the case, the TEMs represent an attractive target for multivalent vaccines targeting multiple TEM antigens. The identification of members of TEMs formed by other schistosome TSPs might also provide further vaccine or drug targets. This is particularly so in the case of Sm-TSP-3, the most abundantly expressed TSP in schistosomula (59), the larval stage that develops once the schistosome parasite invades a new human host. The newly transformed schistosomulum is widely viewed as the most susceptible stage to antibody-mediated damage (60), and identification of proteins forming complexes with Sm-TSP-3 will be of great interest for the development of new vaccine antigens specifically targeting the schistosomulum.
CONCLUSIONS
In this work we have characterized the tertiary structure of the EC2 domain of Sm-TSP-2-EC2 and shown that it shares the mushroom-like topology of hCD81-LEL. Furthermore, we suggest that the stem region of the molecule does not provide a dimerization interface but rather mediates interactions with the plasma membrane while leaving the variable domain free for interactions with other protein partners. Structural conservation of the constant domain means this model is likely applicable to TSPs in general. The schistosome tegument is a dynamic and complex syncytial cytoplasm critical for the survival of the parasite in its mammalian host. Sm-TSP-2 is vital for tegument formation, repair, and maintenance and thus is likely essential for the dynamic processes occurring across the surface of the plasma membrane. The identification of proteins with known roles in membrane repair as potential members of Sm-TSP-2 TEMs reinforces the idea that Sm-TSP-2 maintains the integrity of the schistosome tegument and for mediating process occurring at the surface of the schistosome. Although further work is required to elucidate the exact mechanisms by which these processes occur, the identification of a subset of proteins interacting with Sm-TSP-2 TEMs widens the pool of vaccine and drug targets known to be essential for maintain the in vivo integrity of the parasite. The importance of Sm-TSP-2 for schistosome survival is reflected in its status as one of the few schistosomiasis vaccine antigens under active development, and the current study provides further insight into the function of this protein and why it appears to be the Achilles heel of one of humanity's most pernicious foes.
Acknowledgments
This work used the infrastructure provided by the Australian Government through the Linkage Infrastructure, Equipment, and Facilities scheme from the Australian Research Council. The technical support of Mary Duke (QIMR) in maintaining the schistosome lifecycle is acknowledged.
This work was supported by National Health and Medical Research Council, Australia (NHMRC) Project Grant 613682.
The atomic coordinates and structure factors (code 2m7z) have been deposited in the Protein Data Bank (http://wwpdb.org/).
S. Shouldice and J. Martin, unpublished data.
- TSP
- tetraspanin
- TEM
- tetraspanin-enriched microdomain
- CMC
- critical micelle concentration
- DPC
- dodecylphosphocholine
- BS3
- bis(sulfosuccinimidyl)suberate
- BN-PAGE
- non-denaturing blue native PAGE
- MBP
- maltose-binding protein
- TEV
- tobacco etch virus
- HBSS
- Hanks' balanced salt solution
- HSQC
- heteronuclear single quantum correlation
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- IP
- immunoprecipitation.
REFERENCES
- 1. Hemler M. E. (2008) Targeting of tetraspanin proteins. Potential benefits and strategies. Nat. Rev. Drug Discov. 7, 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hemler M. E. (2005) Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6, 801–811 [DOI] [PubMed] [Google Scholar]
- 3. Schorey J. S., Bhatnagar S. (2008) Exosome function. From tumor immunology to pathogen biology. Traffic 9, 871–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rana S., Zöller M. (2011) Exosome target cell selection and the importance of exosomal tetraspanins. A hypothesis. Biochem. Soc. Trans. 39, 559–562 [DOI] [PubMed] [Google Scholar]
- 5. Hotez P. J., Bethony J. M., Diemert D. J., Pearson M., Loukas A. (2010) Developing vaccines to combat hookworm infection and intestinal schistosomiasis. Nat. Rev. Microbiol. 8, 814–826 [DOI] [PubMed] [Google Scholar]
- 6. van der Werf M. J., de Vlas S. J., Brooker S., Looman C. W., Nagelkerke N. J., Habbema J. D., Engels D. (2003) Quantification of clinical morbidity associated with schistosome infection in sub-Saharan Africa. Acta Trop. 86, 125–139 [DOI] [PubMed] [Google Scholar]
- 7. Tran M. H., Pearson M. S., Bethony J. M., Smyth D. J., Jones M. K., Duke M., Don T. A., McManus D. P., Correa-Oliveira R., Loukas A. (2006) Tetraspanins on the surface of Schistosoma mansoni are protective antigens against schistosomiasis. Nat. Med. 12, 835–840 [DOI] [PubMed] [Google Scholar]
- 8. Loukas A., Gaze S., Mulvenna J. P., Gasser R. B., Brindley P. J., Doolan D. L., Bethony J. M., Jones M. K., Gobert G. N., Driguez P., McManus D. P., Hotez P. J. (2011) Vaccinomics for the major blood feeding helminths of humans. OMICS 15, 567–577 [DOI] [PubMed] [Google Scholar]
- 9. Tran M. H., Freitas T. C., Cooper L., Gaze S., Gatton M. L., Jones M. K., Lovas E., Pearce E. J., Loukas A. (2010) Suppression of mRNAs encoding tegument tetraspanins from Schistosoma mansoni results in impaired tegument turnover. PLoS Pathog. 6, e1000840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kitadokoro K., Bordo D., Galli G., Petracca R., Falugi F., Abrignani S., Grandi G., Bolognesi M. (2001) CD81 extracellular domain 3D structure. Insight into the tetraspanin superfamily structural motifs. EMBO J. 20, 12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Seigneuret M., Delaguillaumie A., Lagaudrière-Gesbert C., Conjeaud H. (2001) Structure of the tetraspanin main extracellular domain. A partially conserved fold with a structurally variable domain insertion. J. Biol. Chem. 276, 40055–40064 [DOI] [PubMed] [Google Scholar]
- 12. Drummer H. E., Wilson K. A., Poumbourios P. (2005) Determinants of CD81 dimerization and interaction with hepatitis C virus glycoprotein E2. Biochem. Biophys. Res. Commun. 328, 251–257 [DOI] [PubMed] [Google Scholar]
- 13. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP, and related tools. Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 14. Stols L., Gu M., Dieckman L., Raffen R., Collart F. R., Donnelly M. I. (2002) A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 25, 8–15 [DOI] [PubMed] [Google Scholar]
- 15. Liu H., Naismith J. H. (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marley J., Lu M., Bracken C. (2001) A method for efficient isotopic labeling of recombinant proteins. J. Biomol. NMR 20, 71–75 [DOI] [PubMed] [Google Scholar]
- 17. Mobli M., Maciejewski M. W., Gryk M. R., Hoch J. C. (2007) An automated tool for maximum entropy reconstruction of biomolecular NMR spectra. Nat. Methods 4, 467–468 [DOI] [PubMed] [Google Scholar]
- 18. Mobli M., Stern A. S., Bermel W., King G. F., Hoch J. C. (2010) A non-uniformly sampled 4D HCC(CO)NH-TOCSY experiment processed using maximum entropy for rapid protein sidechain assignment. J. Magn. Reson. 204, 160–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Löhr F., Hänsel R., Rogov V. V., Dötsch V. (2007) Improved pulse sequences for sequence specific assignment of aromatic proton resonances in proteins. J. Biomol. NMR 37, 205–224 [DOI] [PubMed] [Google Scholar]
- 20. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+. A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) The CCPN data model for NMR spectroscopy. Development of a software pipeline. Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 22. Güntert P. (2004) Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 23. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilkins D. K., Grimshaw S. B., Receveur V., Dobson C. M., Jones J. A., Smith L. J. (1999) Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry 38, 16424–16431 [DOI] [PubMed] [Google Scholar]
- 25. Altieri A. S., Hinton D. P., Byrd R. A. (1995) Association of biomolecular systems via pulsed-field gradient NMR self-diffusion measurements. J. Am. Chem. Soc. 117, 7566–7567 [Google Scholar]
- 26. Jones J. A., Wilkins D. K., Smith L. J., Dobson C. M. (1997) Characterisation of protein unfolding by NMR diffusion measurements. J. Biomol. NMR 10, 199–203 [Google Scholar]
- 27. Allan D., Payares G., Evans W. H. (1987) The phospholipid and fatty acid composition of Schistosoma mansoni and of its purified tegumental membranes. Mol. Biochem. Parasitol. 23, 123–128 [DOI] [PubMed] [Google Scholar]
- 28. Bader R., Lerch M., Zerbe O. (2008) NMR of Membrane-Associated Peptides and Proteins. in Protein Science Encyclopedia, pp. 95–120, Wiley-VCH Verlag GmbH & Co., Weinheim, Germany [Google Scholar]
- 29. van Balkom B. W., van Gestel R. A., Brouwers J. F., Krijgsveld J., Tielens A. G., Heck A. J., van Hellemond J. J. (2005) Mass spectrometric analysis of the Schistosoma mansoni tegumental sub-proteome. J. Proteome Res. 4, 958–966 [DOI] [PubMed] [Google Scholar]
- 30. Hawthorne P. L., Trenholme K. R., Skinner-Adams T. S., Spielmann T., Fischer K., Dixon M. W., Ortega M. R., Anderson K. L., Kemp D. J., Gardiner D. L. (2004) A novel Plasmodium falciparum ring stage protein, REX, is located in Maurer's clefts. Mol. Biochem. Parasitol. 136, 181–189 [DOI] [PubMed] [Google Scholar]
- 31. Craig R., Beavis R. C. (2004) TANDEM. Matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 32. Rajesh S., Sridhar P., Tews B. A., Fénéant L., Cocquerel L., Ward D. G., Berditchevski F., Overduin M. (2012) Structural basis of ligand interactions of the large extracellular domain of tetraspanin CD81. J. Virol. 86, 9606–9616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tao H., Liu W., Simmons B. N., Harris H. K., Cox T. C., Massiah M. A. (2010) Purifying natively folded proteins from inclusion bodies using sarkosyl, Triton X-100, and CHAPS. Biotechniques 48, 61–64 [DOI] [PubMed] [Google Scholar]
- 34. Bax A. (2011) Triple resonance three-dimensional protein NMR. Before it became a black box. J. Magn. Reson. 213, 442–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Partearroyo M. A., Goñi F. M., Katime I., Alonso A. (1988) Micellar properties of the zwitterionic bile derivative CHAPS. Biochem. Int. 16, 259–265 [PubMed] [Google Scholar]
- 36. Lauterwein J., Bösch C., Brown L. R., Wüthrich K. (1979) Physicochemical studies of the protein-lipid interactions in melittin-containing micelles. Biochim. Biophys. Acta 556, 244–264 [DOI] [PubMed] [Google Scholar]
- 37. Kovalenko O. V., Yang X., Kolesnikova T. V., Hemler M. E. (2004) Evidence for specific tetraspanin homodimers. Inhibition of palmitoylation makes cysteine residues available for cross-linking. Biochem. J. 377, 407–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Braschi S., Wilson R. A. (2006) Proteins exposed at the adult schistosome surface revealed by biotinylation. Mol. Cell. Proteomics 5, 347–356 [DOI] [PubMed] [Google Scholar]
- 39. Mulvenna J., Moertel L., Jones M. K., Nawaratna S., Lovas E. M., Gobert G. N., Colgrave M., Jones A., Loukas A., McManus D. P. (2010) Exposed proteins of the Schistosoma japonicum tegument. Int. J. Parasitol. 40, 543–554 [DOI] [PubMed] [Google Scholar]
- 40. Huang Y., Verheesen P., Roussis A., Frankhuizen W., Ginjaar I., Haldane F., Laval S., Anderson L. V., Verrips T., Frants R. R., de Haard H., Bushby K., den Dunnen J., van der Maarel S. M. (2005) Protein studies in dysferlinopathy patients using llama-derived antibody fragments selected by phage display. Eur. J. Hum. Genet. 13, 721–730 [DOI] [PubMed] [Google Scholar]
- 41. Charrin S., le Naour F., Silvie O., Milhiet P.-E., Boucheix C., Rubinstein E. (2009) Lateral organization of membrane proteins. Tetraspanins spin their web. Biochem. J. 420, 133–154 [DOI] [PubMed] [Google Scholar]
- 42. Martens S., McMahon H. T. (2008) Mechanisms of membrane fusion. Disparate players and common principles. Nat. Rev. Mol. Cell Biol. 9, 543–556 [DOI] [PubMed] [Google Scholar]
- 43. Bansal D., Miyake K., Vogel S. S., Groh S., Chen C. C., Williamson R., McNeil P. L., Campbell K. P. (2003) Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature 423, 168–172 [DOI] [PubMed] [Google Scholar]
- 44. McNeil A. K., Rescher U., Gerke V., McNeil P. L. (2006) Requirement for annexin A1 in plasma membrane repair. J. Biol. Chem. 281, 35202–35207 [DOI] [PubMed] [Google Scholar]
- 45. Mellgren R. L., Zhang W., Miyake K., McNeil P. L. (2007) Calpain is required for the rapid, calcium-dependent repair of wounded plasma membrane. J. Biol. Chem. 282, 2567–2575 [DOI] [PubMed] [Google Scholar]
- 46. Siddiqui A. A., Zhou Y., Podesta R. B., Karcz S. R., Tognon C. E., Strejan G. H., Dekaban G. A., Clarke M. W. (1993) Characterization of Ca2+-dependent neutral protease (calpain) from human blood flukes, Schistosoma mansoni. Biochim. Biophys. Acta 1181, 37–44 [DOI] [PubMed] [Google Scholar]
- 47. Lennon N. J., Kho A., Bacskai B. J., Perlmutter S. L., Hyman B. T., Brown R. H., Jr. (2003) Dysferlin interacts with annexins A1 and A2 and mediates sarcolemmal wound-healing. J. Biol. Chem. 278, 50466–50473 [DOI] [PubMed] [Google Scholar]
- 48. Lakkaraju A., Rodriguez-Boulan E. (2008) Itinerant exosomes. Emerging roles in cell and tissue polarity. Trends Cell Biol. 18, 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hemler M. E. (2003) Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19, 397–422 [DOI] [PubMed] [Google Scholar]
- 50. Zöller M. (2006) Gastrointestinal tumors. Metastasis and tetraspanins. Z. Gastroenterol. 44, 573–586 [DOI] [PubMed] [Google Scholar]
- 51. Koradi R., Billeter M., Wüthrich K. (1996) MOLMOL. A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 52. Marcilla A., Trelis M., Cortés A., Sotillo J., Cantalapiedra F., Minguez M. T., Valero M. L., Sánchez del Pino M. M., Muñoz-Antoli C., Toledo R., Bernal D. (2012) Extracellular vesicles from parasitic helminths contain specific excretory/secretory proteins and are internalized in intestinal host cells. PLoS ONE 7, e45974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liégeois S., Benedetto A., Garnier J. M., Schwab Y., Labouesse M. (2006) The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J. Cell Biol. 173, 949–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mathivanan S., Simpson R. J. (2009) ExoCarta. A compendium of exosomal proteins and RNA. Proteomics 9, 4997–5000 [DOI] [PubMed] [Google Scholar]
- 55. Cardoso F. C., Macedo G. C., Gava E., Kitten G. T., Mati V. L., de Melo A. L., Caliari M. V., Almeida G. T., Venancio T. M., Verjovski-Almeida S., Oliveira S. C. (2008) Schistosoma mansoni tegument protein Sm29 is able to induce a Th1-type of immune response and protection against parasite infection. PLoS Negl. Trop. Dis. 2, e308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang R., Yoshida A., Kumagai T., Kawaguchi H., Maruyama H., Suzuki T., Itoh M., El-Malky M., Ohta N. (2001) Vaccination with calpain induces a Th1-biased protective immune response against Schistosoma japonicum. Infect. Immun. 69, 386–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Marques H. H., Zouain C. S., Torres C. B., Oliveira J. S., Alves J. B., Goes A. M. (2008) Protective effect and granuloma down-modulation promoted by RP44 antigen a fructose 1,6 bisphosphate aldolase of Schistosoma mansoni. Immunobiology 213, 437–446 [DOI] [PubMed] [Google Scholar]
- 58. Hofmann A., Osman A., Leow C. Y., Driguez P., McManus D. P., Jones M. K. (2010) Parasite annexins. New molecules with potential for drug and vaccine development. Bioessays 32, 967–976 [DOI] [PubMed] [Google Scholar]
- 59. Gobert G. N., Tran M. H., Moertel L., Mulvenna J., Jones M. K., McManus D. P., Loukas A. (2010) Transcriptional changes in Schistosoma mansoni during early schistosomula development and in the presence of erythrocytes. PLoS Negl. Trop. Dis. 4, e600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Loukas A., Tran M., Pearson M. S. (2007) Schistosome membrane proteins as vaccines. Int. J. Parasitol. 37, 257–263 [DOI] [PubMed] [Google Scholar]