SUMMARY
Brucella abortus, the etiological agent of bovine brucellosis, is an intracellular pathogen whose virulence is completely dependent on a type IV secretion system. This secretion system translocates effector proteins into the host cell to modulate the intracellular fate of the bacterium in order to establish a secure niche were it actively replicates. Although much has been done in understanding how this secretion system participates in the virulence process, few effector proteins have been identified to date. We describe here the identification of a type IV secretion substrate (SepA) that is only present in Brucella spp. and has no detectable homology to known proteins. This protein is secreted in a virB dependent manner in a two-step process involving a periplasmic intermediate and secretion is necessary for its function. The deletion mutant showed a defect in the early stages of intracellular replication in professional and non-professional phagocytes although it invades the cells more efficiently than the wild type parental strain. Our results indicate that, even though the mutant was more invasive, it had a defect in excluding the lysosomal marker Lamp-1 and was inactivated more efficiently during the early phases of the intracellular life cycle.
INTRODUCTION
Intracellular pathogens have evolved a variety of strategies to survive and multiply within the host cells. These mechanisms are very versatile and in many cases involve specialized secretion systems devoted to the secretion and translocation of effector proteins to the host cell that interfere or modulate the cellular response to favor the infectious process (Cossart et al., 2010, Ninio et al., 2007). Brucella spp are gram-negative facultative intracellular bacteria that cause brucellosis, a wide spread zoonosis that affects mainly domestic animals and humans, causing important economical and sanitary problems in many areas of the world (Corbel, 1997, Godfroid et al., 2011). One of the hallmarks of the infectious cycle of Brucella is its ability to survive and multiply in professional phagocytes like macrophages (Atluri et al., 2011), a process that requires the virB system, a type IV secretion system homologous to the Agrobacterium tumefaciens conjugation-like system that translocates the T-DNA into the plant cell (Alvarez-Martinez et al., 2009). This secretion system is necessary to modulate the intracellular trafficking of the bacteria in the host cell. In the early phases this system contributes to avoid the fusion of the Brucella containing vacuole (BCV) with the lysosomes and, in a second phase, redirects its fate to a calnexin-positive endoplasmic reticulum derived membrane niche were it can actively replicate (Celli et al., 2003, Comerci et al., 2001). The intracellular traffic of Brucella in non-professional and professional phagocytes has been extensively studied (Celli et al., 2003, Celli et al., 2005, Comerci et al., 2001, Pizarro-Cerda et al., 1999, Pizarro-Cerda et al., 1998). Shortly after its internalization, the BCV transiently acquires the lysosomal marker Lamp-1, which is subsequently excluded as the BCV is directed to its final replicative organelle. The BCV actively excludes the mature lysosomal marker Cathepsin-D, avoids maturation into an active lysosome and acquires endoplasmic reticulum derived membranes creating a suitable organelle for the replication of the bacteria.
Although much has been done on how the virB system affects the intracellular trafficking, the virulence process, and how this complex secretion system is transcriptionally regulated (Arocena et al., 2010, Arocena et al., 2012, Delrue et al., 2005, Sieira et al., 2010, Sieira et al., 2012, Sieira et al., 2004), very little is known on the identity of the secreted/translocated proteins as well as the molecular mechanisms they use to modulate the intracellular trafficking. To date eleven effectors have been identified and, of them, the activity of only one has been elucidated.
RicA was identified as a Brucella protein that interacts with the GTPase Rab2 and is translocated into the host cell in a virB dependent manner. This effector protein binds preferentially the GDP-bound Rab2, avoids the recruitment of this Rab to the BCV and affects the normal intracellular trafficking (de Barsy et al., 2011). An interesting observation made by the authors of this report is that, even though RicA is translocated into the host in a virB dependent manner, the protein is also secreted into the supernatant in bacteriological cultures in a virB independent manner, suggesting that their could be a different secretion system involved in this process. This raises the interesting point if the virB dependent translocation of effector proteins involves a two-step process with other secretion systems also participating.
Two effector proteins that were isolated identifying promoter regions similar to the virB promoter box are VceA and VceC. The authors of this report demonstrated, using a TEM1 β-lactamase reporter, that these proteins are translocated into the host cells in a virB dependent manner and, in one of these proteins (VceE), a C-terminal type four secretion signal was identified that is also functional in Legionella (de Jong et al., 2008).
Recently, Marchesini et. al. performed a bioinformatic and biochemical screening searching for novel virB effector proteins (Marchesini et al., 2011). They identified, using as a reporter the adenylate cyclase (Cya) from Bordetella pertussis, four new virB dependent translocated proteins. One of these proteins, Bab2_0123, was also detected as secreted in a virB dependent manner by immunoflourecence and biochemical fractionation. The authors demonstrated that the first 25 amino acids of the protein are necessary for translocation into the host cell and showed that the mutant had no detectable phenotype.
Very recently Myeni et. al. (Myeni et al., 2013) informed the identification of five new type IV effector proteins in B. abortus. The authors identified these proteins through a bioinformatic search and verified that they are translocated into the host cells using β-lactamase and CyaA adenylate cyclase reporter assays. The ectopic expression of several of them as well as single and combined deletion mutant strains indicate that Brucella modulates the secretory pathway using multiple effector proteins (Myeni et al., 2013).
We describe here the identification of a virB secretion substrate present in all Brucella species sequenced to date. We demonstrate by immunofluorescence that this protein is secreted in a virB dependent manner during the infection of host cells and that this secretion requires a periplasmic intermediate. We also show that the mutant strain has a significant defect in the early stages of the intracellular multiplication process and that is less effective in excluding the lysosomal marker Lamp-1. Surprisingly, even though the mutant strain is more efficiently inactivated during the first stages of the intracellular replication process, it invades the host cell more efficiently than the wild type suggesting that this gene might play a role in ”selecting” the phagocytosis route and, thus, modulating the final destination of the resulting phagosome. This hypothesis is discussed.
RESULTS
Identification of the gene product of Bab1_1492 as a secreted protein in Brucella abortus
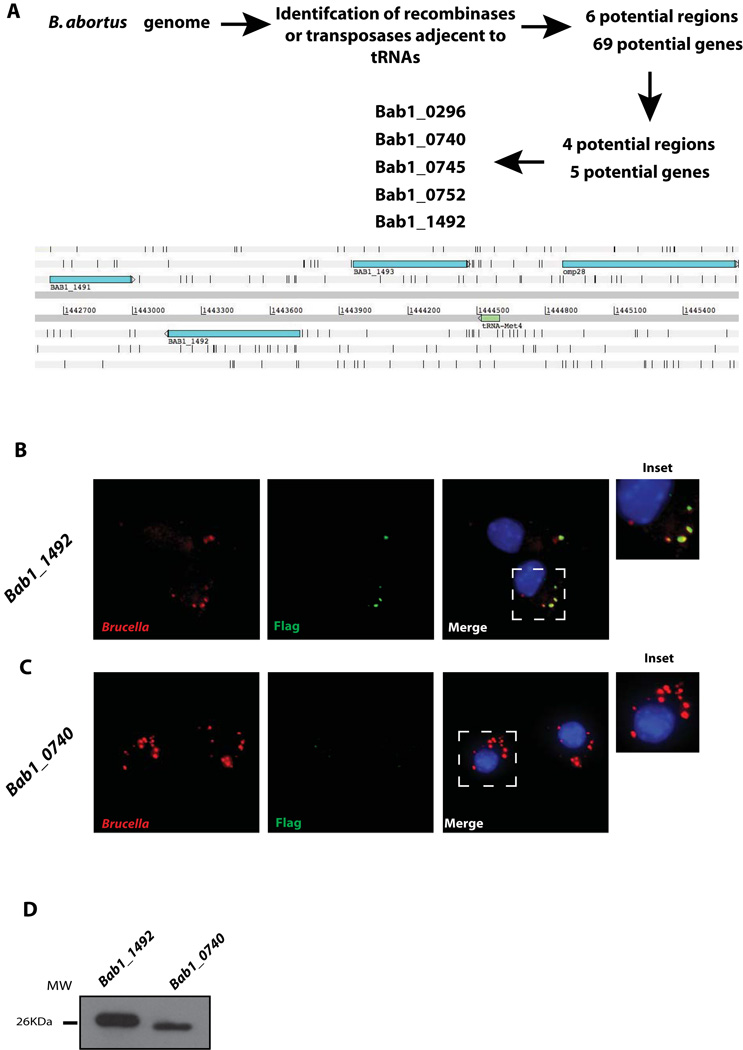
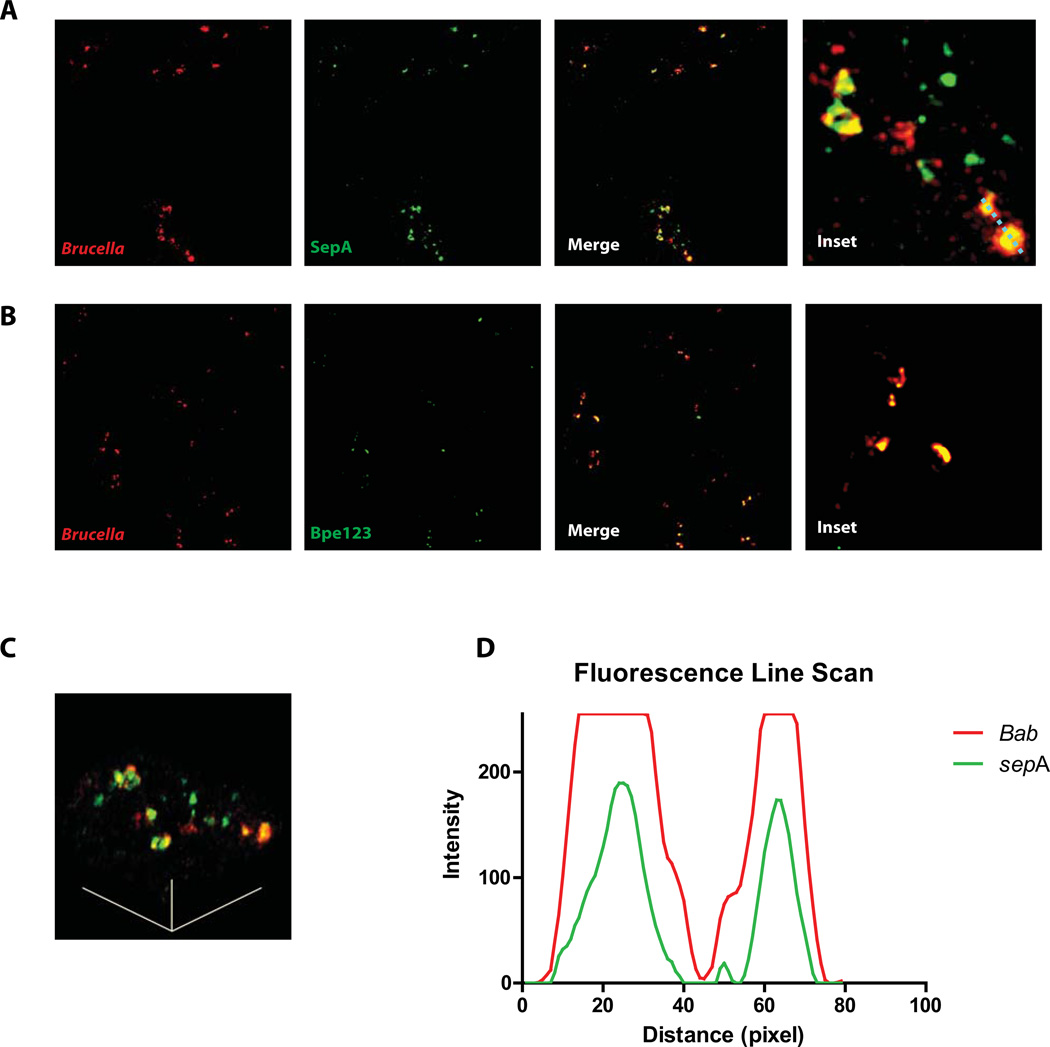
In order to identify novel virulence factors in B. abortus we performed a bioinformatic search seeking for potential horizontally transmitted regions identifying transposases or recombinases close to transfer tRNAs. This rational is based on the observation that, in many bacteria, horizontally transmitted pathogenic islands are located adjacent to tRNAs and encode enzymes involved in DNA processing. For this we used the program ARTEMIS (Rutherford et al., 2000) and searched both Brucella abortus 2308 chromosomes for putative transposases or recombinanses that were adjacent to genes coding for transfer tRNAs. Using this approach we identified several potentially horizontally transmitted regions and analyzed each one of them individually identifying the open reading frames and determining, for each, conservation among Brucella species, presence in other bacteria and domain structure. This methodology allowed us to select a group of potential proteins whose genes were cloned upstream of a 3xFLAG, subcloned in a Brucella replicative vector (pBBR1MCS-4) and introduced into a wild type B. abortus 23208 strain. The resulting clones were used to infect J774 A.1 cells and, at different times post-infection, the infected cells fixed and stained to detect the FLAG epitope and the bacteria. The objective of this approach was to identify gene products secreted by the bacteria during the infection of host cells. Figure 1A shows a schematic representation of the screening method utilized. Out of 5 candidates analyzed one was detected on the surface of the intracellular bacteria. Figure 1B shows that the protein product of the gene Bab1_1492 was detected on the bacterial surface during the infection of J774 A.1 cells but not the protein product of gene Bab1_0740 (Figure 1C) although both genes were expressed (Figure 1D). To further examine the secretion of the protein product of Bab1_1492, confocal images were obtained at 4 hrs post-infection and as a positive control a strain expressing a 3xFLAG fusion to the type IV effector Bpe123 was used (Marchesini et al., 2011). Figure 2A shows that at 4 hrs surface exposed Bab1_1492 was observed in most of the bacteria analyzed, while only 15 to 20 % of bacteria were positive for Bpe123 as has been informed (Del Giudice et al., 2013, Marchesini et al., 2011) (Figure 2B). A scanning of the fluorescence in both channels through a liner path indicated that both marks showed the same pattern (Figure 2D). These results demonstrate that the protein product of Bab1_1492 is secreted during infection, for this reason we named the gene sepA for Secreted Effector Protein A.
Figure 1. The protein product of Bab1_1492 is secreted during the infection of host cells.
A. Schematic representation of the screening method used to identify novel secreted virulence effectors in Brucella abortus. B. Immunofluorescence of J774 A.1 cells infected with a B. abortus strain coding for Bab1_1492-3xFLAG in a replicative vector at 4 hrs post-infection. Red, Brucella; green, Flag. C. Immunofluorescence of J774 A.1 cells infected with a B. abortus strain coding for Bab1_0740-3xFLAG in a replicative vector at 4 hrs post-infection. Red, Brucella; green, Flag. D. Western-blot with an anti-FLAG of the strains shown in panels B and C.
Figure 2. Confocal analysis of the secretion of the protein product of Bab1_1492.
Confocal images of J774 A.1 cells infected with the B. abortus strains coding for Bab1_1492-3xFLAG (A) or Bpe123-3xFLAG (B) in replicative vectors at 4 hrs post-infection. C. A 45° rotation view of the line traced in the inset of panel A. D. Quantification of the red and green fluorescence intensity over the line traced in the inset of panel A showing a correlation between both markers. Red, Brucella; green, FLAG.
Secretion of SepA is a two-step virB dependent process
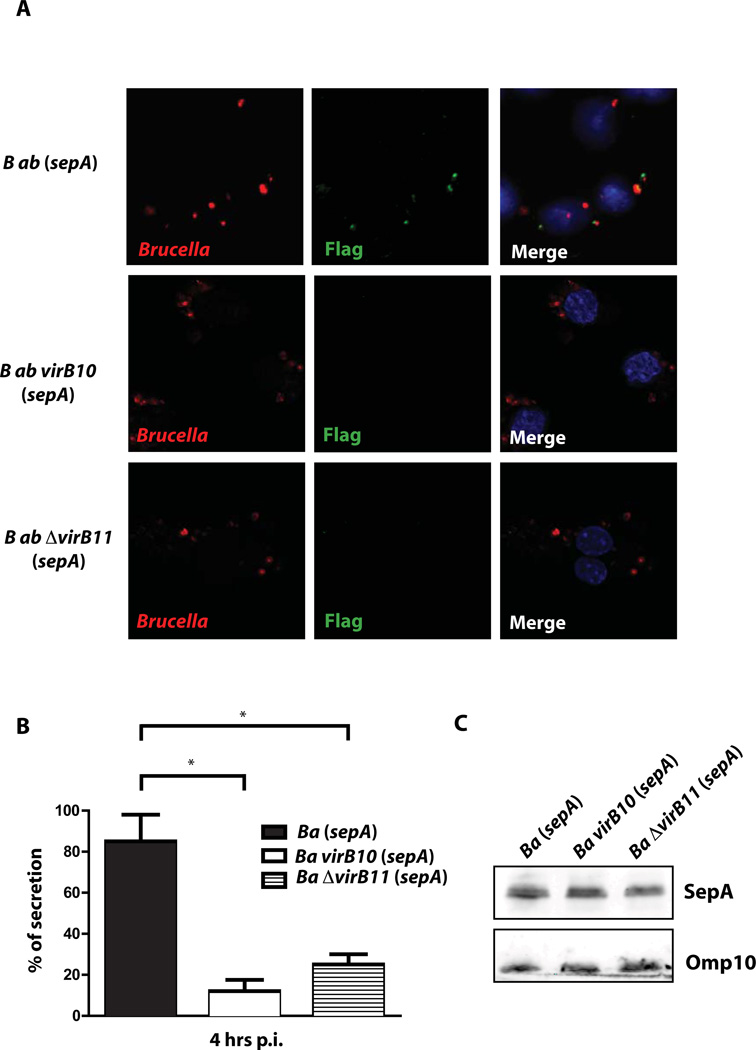
Because the type IV secretion system (virB) is one of the main virulence traits required for intracellular survival we asked if secretion of SepA was dependent on this system. For this we introduced the plasmid carrying the FLAG-tagged version of sepA in the virB10 and virB11 mutant strains and determined the number of intracellular bacteria displaying the protein on the surface. As can be observed in Figure 3A and 3B, secretion of SepA was significantly reduced in the virB mutant backgrounds measured as the number of bacteria showing surface exposed FLAG staining, although expression of the tagged protein was equivalent between strains (Figure 3C). To determine if SepA is secreted immediately after internalization we analyzed by immunofluorescence surface exposed SepA in the wild type and the virB10 mutant at 30 min post-infection. The results shown in Supplementary Figure 1 showed that SepA is secreted very early during infection and in a virB dependent manner. Altogether, these results demonstrate that secretion of SepA is dependent on a functional virB system.
Figure 3. Secretion of SepA is a virB dependent process.
A. Representative Immunofluorescence of J774 A.1 cells infected with B. abortus wild type, virB10 and ΔvirB11 mutant strains coding for SepA-3xFLAG in a replicative vector at 4 hrs post-infection. Red, Brucella; green, FLAG. B. Quantification of the percent of SepA secreting bacteria of the representative images shown in A. *P<0.05. C. Western-blot with an anti-FLAG of the strains showing equivalent levels of expression of SepA.
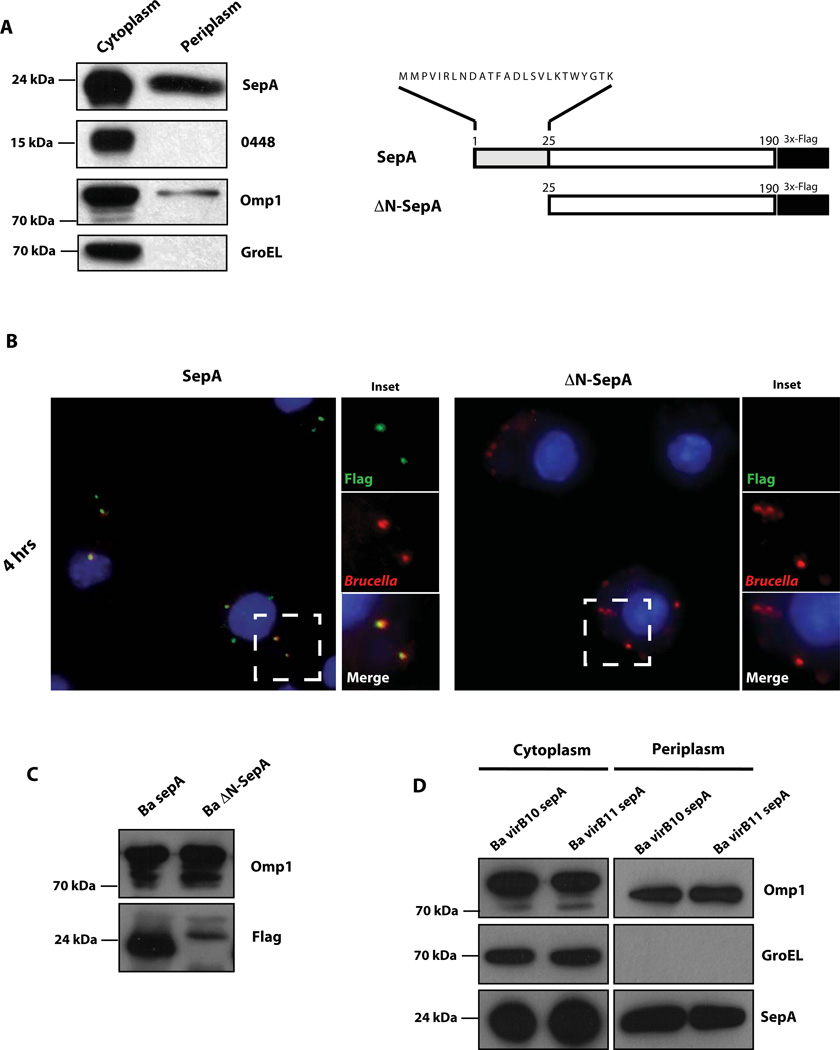
It has been informed that some type IV secretion substrates posses a canonical sec-dependent secretion signal that targets them to the periplasm or the inner membrane as a first secretion step (Burns, 2003, Nagai et al., 2001, Pantoja et al., 2002). Since the analysis of the protein product of sepA using different prediction programs for the detection of putative sec-dependent secretion signals were not conclusive, we decided to determine if this protein is targeted to the periplasm and, if so, if this intermediate state is necessary for the virB secretion. To test we generated a double-tagged strain carrying a chromosomal FLAG-tagged version of the gene Bab2_0448 coding for a cytoplasmic tellurite resistant protein that we are currently studying in the laboratory (unpublished results) in which we introduced the plasmid coding the sepA-3xFLAG. This strain was used to produce periplasmic and cytoplasmic fractions (see Experimental procedures) and the presence of SepA was determined in these fractions. As control proteins we used the presence of the protein product of Bab2_0448 and GroEL for cytoplasm and OMP1 for periplasm. As can be seen in Figure 4A, SepA was found partitioned in both the periplasm and the cytoplasm as was OMP1, while the cytoplasmic controls were only detected in the corresponding fraction (cytoplasmic). This result demonstrates that SepA is exported to the periplasm. In order to determine if this periplasmic intermediate is required for the virB secretion we analyzed the bacterial surface display of the construct lacking the first 25 amino acids of the protein (ΔN-SepA). As shown in Figure 4B, deletion of the amino terminal secretion signal abolished the surface display of SepA as did the absence of the virB system. Loading controls indicated that the deleted protein was expressed with lower levels than the wild type and that it migrated slightly less (Figure 4C). We do not have an explanation for this puzzling observation but speculate that maybe the truncated protein, because it cannot be transported to the periplasm, aggregates or associates with other proteins resistant in our gel conditions. In order to determine if in the virB mutant background SepA is still present in the periplasm we performed a periplamic extraction with the virB10 and virB11 strains expressing the SepA-3xFLAG fusion protein and determined the fractionation pattern of SepA in these strains. As can be observed in Figure 4D, SepA was found in the periplasm of these strains indicating that, even though it was not secreted to the bacterial surface, it retained a periplasmic localization. This result also demonstrates that the bacterial surface display of SepA is not the consequence of “leakage” of the periplasm of the extracellular space. Altogether these results indicate that SepA is targeted to the periplasm and that this intermediate state is required for the virB secretion.
Figure 4. Secretion of SepA involves a periplasmic intermediate.
A. Western-blot with an anti-FLAG, anti-GroEL and anti OMP1 monoclonal antibodies of periplasmic or cytoplasmic fractions of a periplasmic fractionation assay performed with B. abortus chromosomal Bab2_0448-3xFLAG tagged strain carrying the plasmid coding for SepA-3xFLAG. Schematic representation of the ΔN-SepA-3xFLAG (ΔN-SepA) construction carrying a N-terminal 25 amino acid deletion. B. Immunofluorescence of J774 A.1 cells infected with B. abortus strains carrying plasmids coding for SepA-3xFLAG (SepA) or ΔN-SepA-3xFLAG (ΔN-SepA) at 4 hrs post-infection. Red, Brucella; green, FLAG. C. Western-blot showing the expression levels of SepA and ΔN-SepA in Wild-type B. abortus. Loading control, OMP1. D. Western-blot with an anti-FLAG, anti-GroEL and anti OMP1 monoclonal antibodies of periplasmic or cytoplasmic fractions of a periplasmic fractionation assay performed with B. abortus virB10 and virB11 mutant strains carrying the plasmid coding for SepA-3xFLAG.
SepA participates in the early stages of intracellular replication
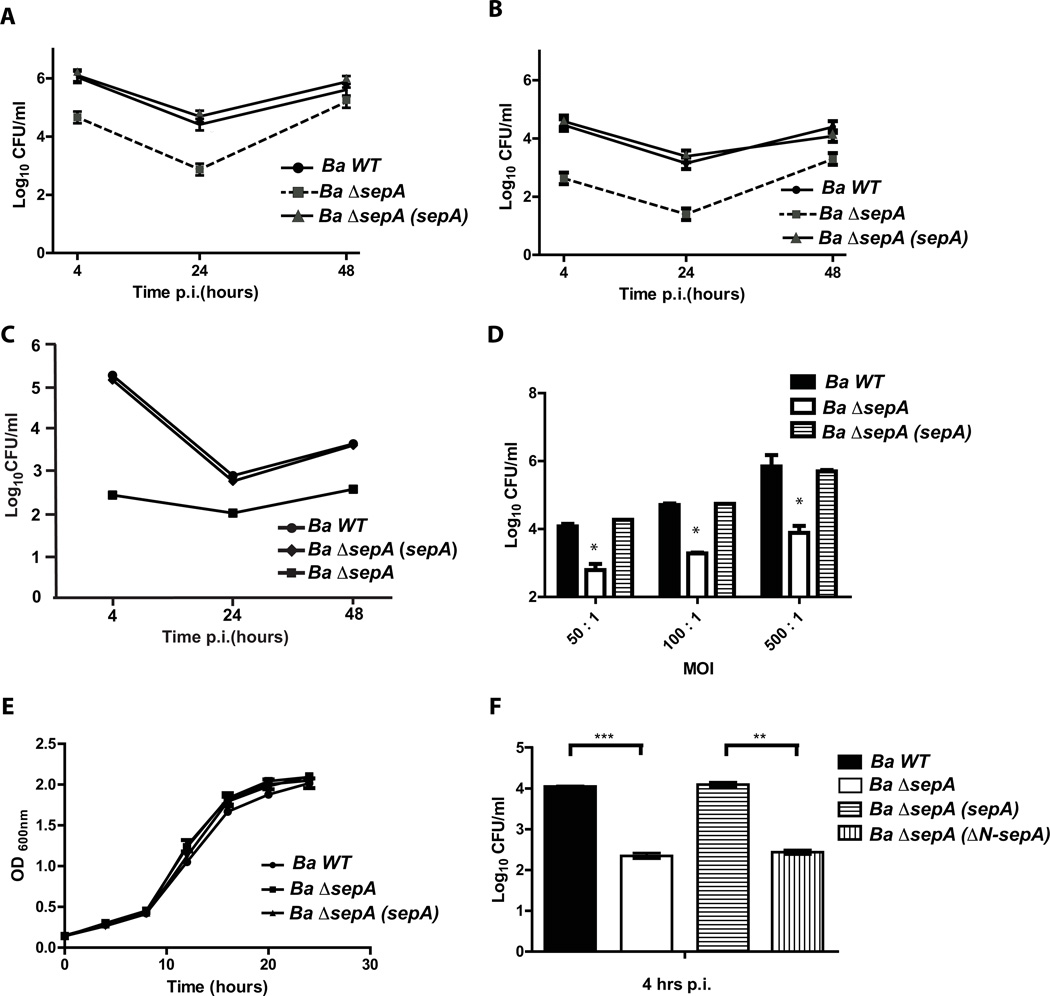
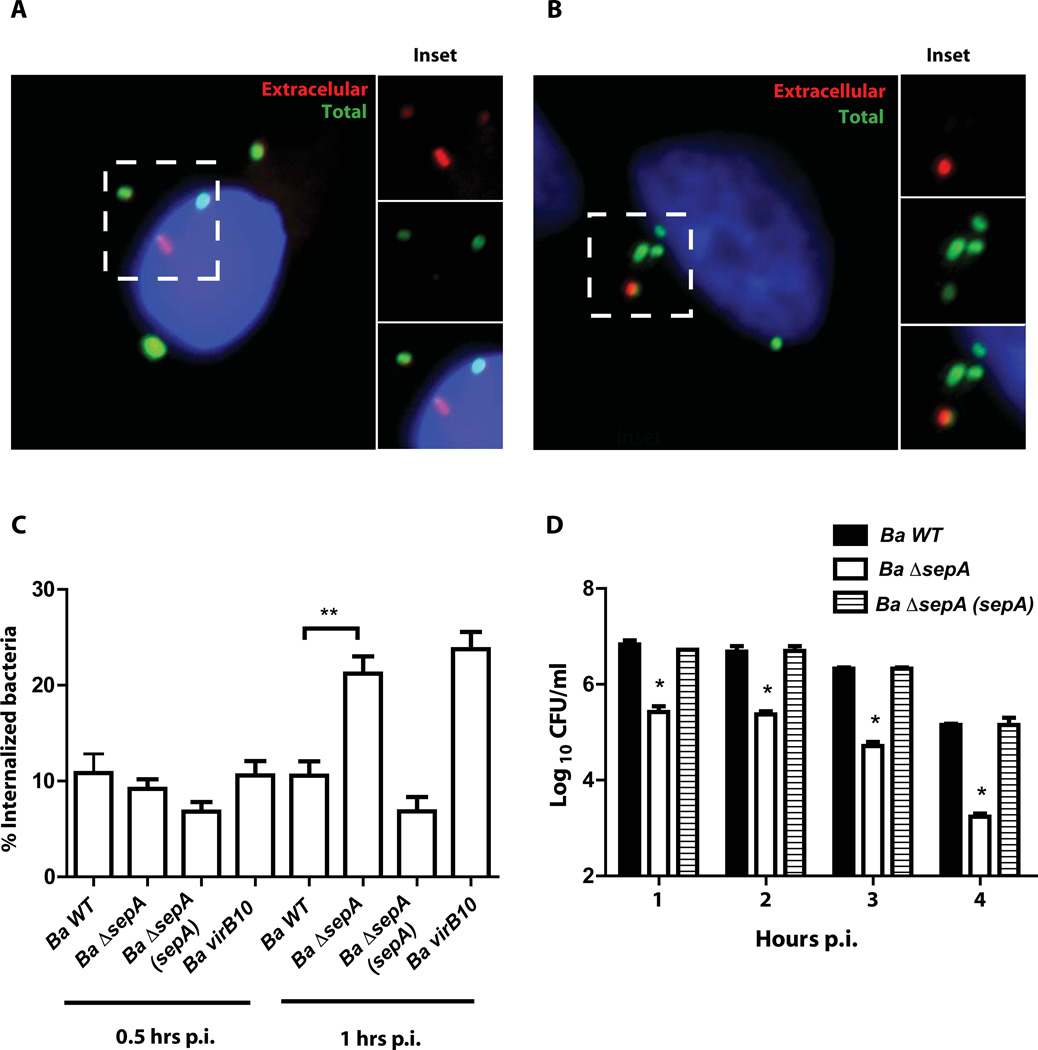
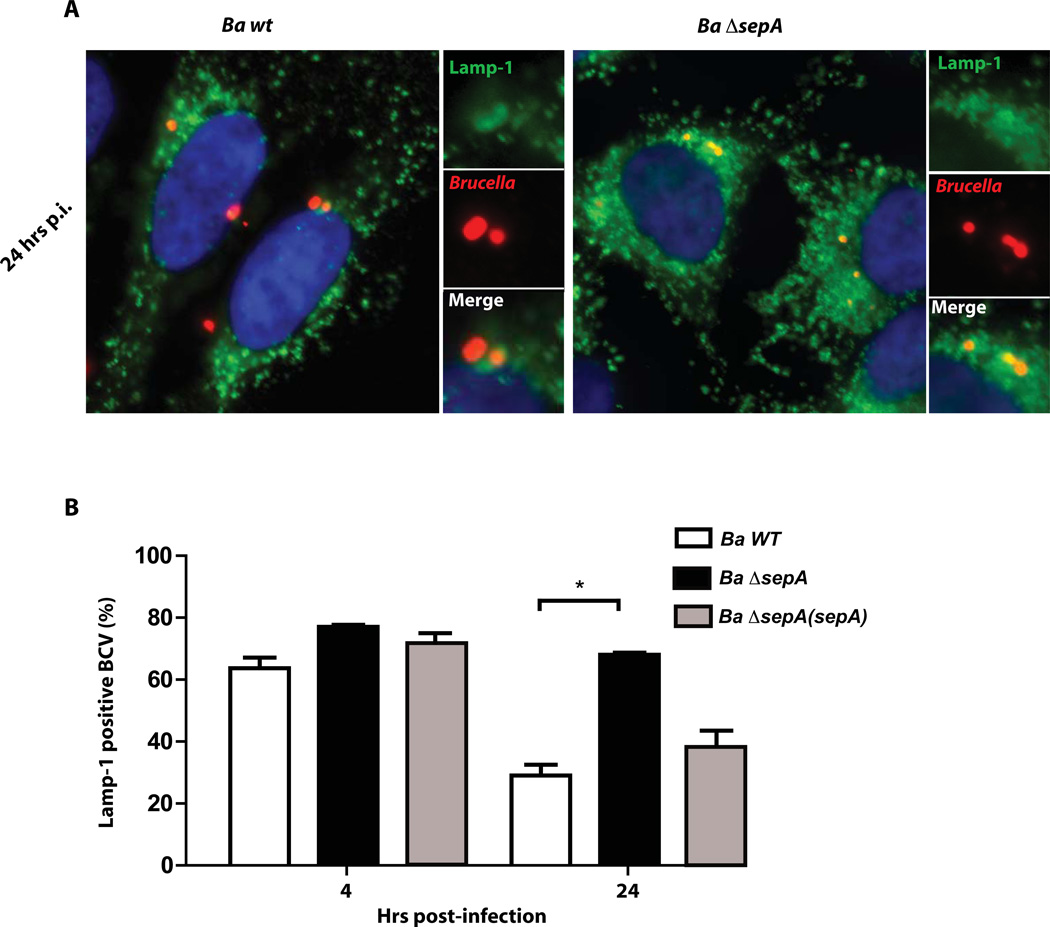
To determine if sepA plays a role during the intracellular life cycle we generated a clean deletion strain and analyzed the intracellular multiplication in HeLa, J774 A.1 cells and bone marrow derived macrophages (BMDM) with an antibiotic protection assay as described in Experimental procedures. As can be observed in Figure 5A, 5B and 5C the strain ΔsepA showed a significant reduction in the intracellular CFU at 4 and 24 hrs post-infection in both professional and non-professional phagocytes. At 48 hrs, even though the mutant had less viable counts than the wild type strain, the differences were less dramatic in HeLa and J774 A.1 cells indicating that the mutant had a defect in the early stages of the intracellular replication process but was able to replicate. In BMDM the mutant was unable to replicate at 48 hr post-infection indicating that the phenotype was more pronounced in these cells. The defects observed were not the consequence of a general replication phenotype as the mutant had an in vitro growth identical to the wild type parental strain (Figure 5D). To determine if the phenotype observed at 4 hrs post-infection was dependent on the multiplicity of infection (m.o.i.) we carried out an antibiotic protection assay on J774 A.1 cells with different multiplicity of infections and determined intracellular CFUs at that time point. The results indicated that the ΔsepA phenotype was not m.o.i. dependent (Figure 5E). As expected, we were unable to complement the ΔsepA mutant with the construction lacking the N-terminal periplasmic localization region indicating that the phenotype is dependent on the secretion of the protein (Figure 5F). The lower viable bacteria at 4 hrs post-infection could be result of a defect in the invasion or early intracellular survival. To discern between these possibilities, we initially determined the invasion capacity as described in Experimental procedures. The results, shown in Figure 6 indicated that invasion was not decreased in the mutant strongly suggesting that the defect was probably due a defect in the capacity to avoid the fusion of the BCV with the lysosomes. Moreover, an interesting observation was that, even though at 4 hrs post-infection the ΔsepA mutant showed a 10-fold reduction in the intracellular bacteria, it actually invaded the host cells more efficiently than the wild type (Figure 6C). To determine if at early time points the internalized bacteria were viable, we performed a gentamicin protection assay and recovered intracellular bacteria at 1, 2, 3 and 4 hrs post-infection and found that at all times tested the mutant had less viable counts (Figure 6D) that decreased over time in comparison to the wild type strain. These results strongly suggested that the mutant probably had a defect in the early stages of the intracellular trafficking. To test this we determined the co-localization of the lysosomal marker Lamp-1 with the BCV at 4 and 24 hrs post-infection in HeLa cells as described in Experimental procedures. As shown in Figure 7A and 7B, the BCV containing the ΔsepA mutant acquired the lysosomal marker Lamp-1 as the wild type at 4 hrs post-infection but was less efficient in excluding it at 24 hrs suggesting a defect in avoiding the maturation into an active lysosome. Even though the strain ΔsepA showed a defect in the initial phases of the intracellular life cycle the bacteria that were able to bypass this initial attack and replicated in an ER-like calnexin positive niche confirming that the defect was restricted to the initial phases of the intracellular cycle (data not shown).
Figure 5. SepA participates in the early stages of intracellular replication.
A. Intracellular replication of B. abortus wild type and ΔsepA deletion strains in HeLa cells determined by the gentamicin protection assay. B. Intracellular replication of B. abortus wild type and ΔsepA deletion strains in J774 A.1 cells determined by the gentamicin protection assay. C. Intracellular replication of B. abortus wild type and ΔsepA deletion strains in bone marrow derived macrophages determined by the gentamicin protection assay D. Viable bacteria of the wild type (Ba wt), Ba ΔsepA mutant and complemented strains at 4 hrs post-infection measured by an antibiotic protection assay in J774 A.1 cells with different multiplicity of infection ratios (50, 100 and 500). E. Growth curve of the wild type and the ΔsepA deletion strain in TSB. F. Complementation of the 4 hrs phenotype with the construction lacking the N-terminal periplasmic localization signal. **P<0.002 and ***P<0.001.
Figure 6. The ΔsepA mutant strain invades cells more efficiently.
A and B. In/out staining of the wild type strain (Ba wt) and the ΔsepA (Ba ΔsepA) mutant strain respectively at 1 hr post-infection in J774 A.1 cells. C. Quantification of the immunofluorescence shown in panels A and B. Ba virB10, assay performed with the B. abortus virB10 mutant. **P<0.002. D. Intracellular viable bacteria of the wild type mutant and complemented strains at 1, 2, 3 and 4 hrs post-infection determined by an antibiotic protection assay. *P<0.001.
Figure 7. The ΔsepA mutant strain is affected in its capacity to exclude the lysosomal marker Lamp-1.
A. Representative immunofluorescence images of Lamp-1 positive and negative BCVs of the wild type (Ba wt) and mutant (Ba ΔsepA) strains in infected HeLa cells at 24 hours post-infection. Red, Brucella; green, Lamp-1. B. Quantification of the percent of Lamp-1 positive BCVs at 4 and 24 hrs post-infection with the wild type and the ΔsepA mutant strains.
ΔsepA has no impact in the virulence in mice but over-expression of the gene causes a severe defect
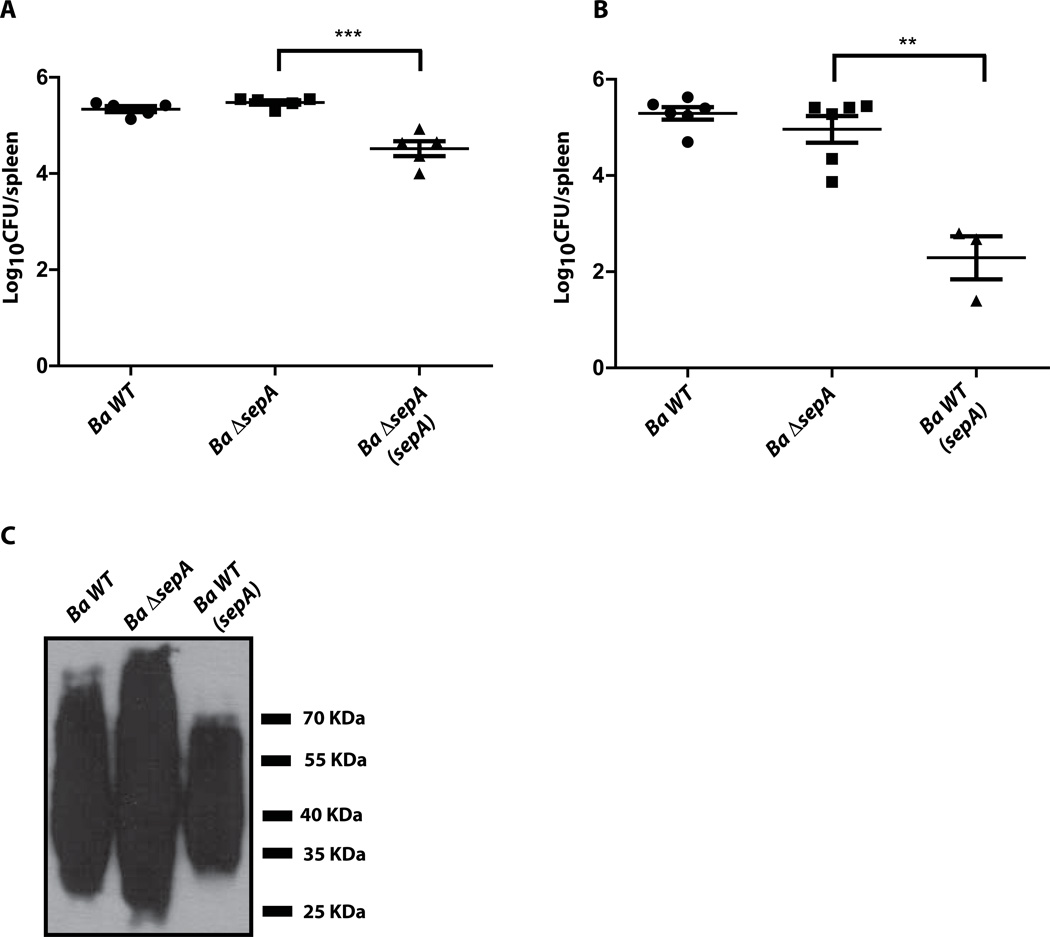
To determine if sepA plays a role in the virulence in the mouse model of infection, we infected intraperitoneally groups of five mice with the wild type 2308, the ΔsepA and the complemented strains and measured, at 15 days post-infection, the bacterial load in the spleens. As can be seen in Figure 8A, the mutant strain did not display any differences in the CFUs per spleen in comparison to the parental strain. An interesting observation was that the complemented strain colonized the spleens significantly less than the mutant and the parental strains suggesting that over-expression of the gene might cause a negative effect. To confirm this we introduced plasmid pBBR1-MCS4/F2-3xF over-expressing SepA in the wild type 2308 strain and evaluated its virulence in mice in comparison with the non-transformed wild type and the deletion strains. As can be observed in Figure 8B, over-expression of SepA dramatically reduced the bacterial load in the spleens of infected mice at 15 days post-infection and the mutant, as indicated before, was as virulent as the wild type parental strain. This effect was not the consequence of a defect in the viability of the strain as the strain grew as the parental strain (Supplementary Figure 2) and did not show any major membrane alterations evidenced by several indicators as for example LPS integrity (Figure 8C). These results strongly suggest that the expression of sepA is probably highly regulated and that an excess of the protein is detrimental for the virulence process although more work should be performed in order to understand the molecular basis of this defect.
Figure 8. Over-expression of SepA affects virulence in mice.
A and B. Spleen colonization at 15 days post-infection of mice intraperiteonally infected with the wild type (Ba wt), sepA mutant (Ba ΔsepA), complemented (Ba ΔsepA(sepA)) or SepA over-expressing (Ba wt(sepA)) strains. Bacterial load was determined as indicated in Experimental procedures. *P<0.002. C. Western-blot with a monoclonal anti-O-antigen antibody (M84) against whole bacteria showing that neither the mutant nor the over-expressing strains are rough.
DISCUSSION
Many intracellular pathogens have, as virulence strategies, the capacity to establish their multiplication niches inside professional phagocytes. To achieve this they have evolved complex secretion systems devoted to secrete and translocate into the host cells virulence factors that modulate the biological functions in their one benefit. Understanding at the molecular level this biochemical crosstalk has been one of the main focuses of the microbial pathogenesis field of the last decade. Brucella, the causative agent of Brucellosis, is an intracellular pathogen that replicates in macrophages and dendritic cells. The virB operon encodes one of the central virulence traits required for the intracellular lifestyle of the bacterium. This operon codes for a type IV secretion system responsible for the translocation of effector proteins that actively avoid the fusion of the Brucella containing vacuole with the lysosomes and recruit membranes from the endoplasmic reticulum to establish a competent replication niche (Celli et al., 2003, Comerci et al., 2001, O'Callaghan et al., 1999, Sieira et al., 2000). This macromolecular apparatus is essential for virulence and, to date, few secretion substrates have been identified (de Barsy et al., 2011, de Jong et al., 2008, Marchesini et al., 2011). We identify here a new virulence factor in Brucella abortus that is secreted in a virB dependent manner and we have named this gene sepA. We demonstrate by immunofluorescence that secretion of SepA occurs during the early phases of the intracellular life cycle of B. abortus, that this secretion is virB dependent and show that it involves a periplasmic intermediate. To date it is not clear the exact mechanism in type IV secretion systems. Even though it is highly accepted that secretion is a process in which the substrate is engaged in the cytoplasm, there have been reports of periplasmic and membrane proteins that are secreted in a type IV dependent process in some pathogens (Burns, 2003, Nagai et al., 2001, Pantoja et al., 2002). One of the canonical examples of these is the pertussis toxin, that assembles in the periplasm and is secreted in a type IV dependent manner (Burns, 2003). In the case of SepA we have clearly established that secretion to the surface of the bacteria is a process that depends on the virB system and on an N-terminal signal peptide that targets the protein to the periplasm indicating that its secretion is a two-step process. With the purpose of determining the role of sepA in the pathogenesis of B. abortus we constructed a deletion mutant and evaluated the intracellular replication capacity of the strain. Our results showed that the mutant had a defect in the early stages of the intracellular replication curve and that this alteration was the consequence of a reduced capacity to avoid the fusion of the BCV with the lysosomes. Interestingly, even though the mutant did not display any defects in its invasion it was significantly higher than that of the parental wild type strain. Consistently, the same observation was seen for the virB10 polar mutant. These results are, apparently, inconsistent with the reduced viable bacteria at 1, 2, 3 and 4 hrs post-infection. One hypothesis could be proposed here. Because it has been established that the route of entry in phagocytes can influence the final destination of the phagosome generated (Aderem et al., 1999), it could be possible that SepA is involved in the initial steps of the intracellular life cycle facilitating certain routes that will ultimately influence the initial composition and final destination of the BCV. In this model the type IV secretion system and some effectors would be involved in the very early stages of the intracellular life cycle selecting the internalization route of the bacterium. Even though the mutant is more invasive than the parental wild type strain, the BCV generated in the process are less efficient for evading the early phagosome-lysosome fusion. Mechanistically, this process could be either through a positive or a negative selection, this is SepA could facilitate phagocytosis through certain routes or inhibit others. We speculate that sepA participates in this early selection process and that its biological activity modifies the final destination of the BCV. At this stage the molecular mechanism used by SepA to promote infection is not known and the fact that the protein has no significant homology to any known proteins complicates this task significantly. Albeit the fact that a molecular mechanism is still lacking, to our knowledge this is the first Type IV secretion substrate in Brucella that shows a phenotype in cells altering the kinetics of Lamp-1 acquisition and exclusion.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
B. melitensis biovar abortus 2308 was used as a wild-type strain. B. abortus strains were cultured in Tryptic Soy Agar (TSA; Difco/Becton-Dickinson, Sparks, MD), or in Tryptic Soy Broth (TSB) at 37°C on a rotary shaker for 16–24 hrs. When needed, media were supplemented with 50µg/ml Kanamycin (Kmr), 5µg/ml Nalidixic Acid (Nalr), 100µg/ml Ampicillin (Ampr) or 3µg/ml Gentamicin (Gmr). Work with B. abortus was performed at the Biosafety Level 3 laboratory facility at the University of San Martín. E. coli strains were grown on Luria-Bertani (LB) agar and broth at 37°Covernight. Antibiotics, when required, were added at the following concentrations: 50 µg/ml Kanamycin or 100 µg/ml Ampicillin.
Recombinant DNA techniques
To generate plasmid pBBR1-MCS4-3xFlag, a fragment containing a 3xFLAG epitope was amplified from plasmid pBAD24-3xFLAG (Spano et al., 2008) using primers CC7-Flag (5’-CGCGGATCCACTCTAGAGATCGTCATCCTTGTAA-3’) and pbad/HindIII (5’-CCCAAGCTTGTTTCTCCATACCCGTTT-3’). The product was digested with BamHI-HindIII, and ligated into the same sites of plasmid pBBR1-MCS4 (Kovach et al., 1995).
For the construction of vectors expressing a C-terminal 3xFLAG-tagged version of genes Bab1_1492, Bab1_0296, Bab1_0752, Bab1_0740, Bab1_0745, the plasmid pBBR1-MCS4-3xFLAG was used. DNA fragments were amplified by PCR from B. abortus 2308 genomic DNA using primers F1492 (5’-CGGAATTCTTGAGCGGAGAGAGCC-3’) and R1492 (5’-CGCCATGGCGGCGGACGCCGGGCC-3’) for the Bab1_1492 gene; F0296 (5’-CGGAATTCTGAGACATGCGAATTAAG-3’) and R0296 (5’-CGCCATGGCAAGCTCCAAGCATCTAAT-3’) for the Bab1_0296 gene; F0752 (5’-TAGCTAGCTGTCGCGGAAGTCAGCC-3’) and R0752 (5’-GCCATGGCTAGGCGTCCAGACATTC-3’) for the Bab1_0752 gene; F0740 (5’-CGGAATTCTTTTGGTCGCTACGCGT-3’)- and R0740 (5’-CGCCATGGCGAATTTGTCTAGCAGGT-3’) for the Bab1_0740 gene and F0745 (5’-CGGAATTCAAATCAACGTTCAAATCTATC-3’) and R0745 (5’-CGCCATGGCATATTCCAAAATATTCCTTTG-3’) for the Bab1_0745 gene. The truncated version (amino acids 25 to190) of Bab1_1492 was generated, using primers FsNH2-1492 (5’-GGAATTCAGAGAGGGATGACCCCGAGCGAAACCATT-3’) and R1492 (5’-CGCCATGGCGGCGGACGCCGGGCC-3’). All the PCR products were digested with EcoRI-NcoI, except the fragment 0752 that was digested with EcoRI-NheI. The resulting fragments were cloned in pBBR1-MCS4-3xFlag in the same sites generating in frame fusions to the 3xFLAG epitope. The resulting plasmids were named pBBR1-MCS4/ F2-3xF (Bab1_1492), pBBR1-MCS4/ F2NH2-3xF (truncated version of Bab1_1492), pBBR1-MCS4/ 0296-3xF (Bab1_0296), pBBR1-MCS4/ 0752-3xF (Bab1_0752), PBBR1-MCS4/ 0740-3xF (Bab1_0740) and pBBR1-MCS4/ 0745-3xF (Bab1_0745). Plasmids expressing 3xFLAG tagged fusion proteins were introduced in B. abortus strains by biparental mating. The integrity of all constructs was confirmed by sequence analysis.
A B. abortus ΔsepA strain with a clean deletion of Bab1_1492 was constructed by standard recombinant DNA and allelic exchange procedures as previously described (Czibener et al., 2012). The flanking regions of Bab1_1492 were amplified by PCR using primers Del1492-1 (5’-CGGGATCCAGTTTCGCAGACGCGGTCCA-3’), Del1492-2 (5’-TATTCTCTCGTTGACCGCAA-3’), Del1492-3 (5’-GTCAACGAGAGAATACTTCCTGTTTCGAAGCCGTTT-3’) and Del1492-4 (5’-CCCAAGCTTTGAGCGGCCATTGCGTCAGG-3’). The PCR products were ligated using the recombinant PCR technique (Czibener et al., 2012), and the resulting DNA fragment digested with BamHI-HindIII and ligated to the same sites of suicide vector pK18mobsacB, giving rise to plasmid pK18mobsacB/ΔsepA. The plasmid was introduced in B. abortus strain by biparental mating, and the mutants Kms and SucR were confirmed by colony PCR with primers Del1492-1 and Del1492-4.
Intracellular replication curves
Gentamicin protection assays were performed as previously described (Ugalde et al., 2000). All tissue cultures were maintained at 37°C in a humidified atmosphere containing 5% CO2. HeLa and J774.A1 macrophage-like cell lines were maintained in Dulbecco modified Eagle medium (Gibco Life Technologies) and RPMI 1640 (Gibco Life technologies), respectively, supplemented with 5% fetal bovine serum (PAA labˊs GmBH, Pasching, Austria) and Streptomycin (50 µg/ml)/Penicillin (50 U/ml) (Gibco Life technologies). Cells were trypsinized, counted and seeded on 24-well plates in media, at a density of 5 × 104 cells per well, and were grown for 24 hrs. B. abortus infections were carried out at a multiplicity of infection (MOI) of 500:1 for HeLa cells and 100:1 for J774.A1 cells. Bacteria were centrifuged onto cells at 400 × g for 10 min. After 60 min wells were gently washed three times with phosphate-buffered saline (PBS) and incubated for 3 hrs with fresh medium containing 50 µg/ml Gentamicin to kill non-internalized bacteria. At 4 hrs post-infection, cells were washed three times with PBS, and fresh medium containing 10 µg/ml Gentamicin was added. Bone marrow derived macrophages (BMDM) were prepared as described in (Czibener et al., 2006). At the indicated times, infected cells were either washed three times with PBS and lysed with 500 µl 0.1% Triton X-100 in H2O (Sigma-Aldrich) or processed for immunoflourescence staining as described below. The intracellular CFU counts were determined by plating serial dilutions on TSA with the appropriate antibiotics.
Antibodies and reagents
All the antibodies were diluted in 10% horse serum (Gibco), 5% BSA (Sigma) and 0.1% saponin (Sigma), except for the anti-FLAG M2 monoclonal antibody (Sigma), that was diluted in 0.2 % Triton X-100 instead of 0.1% saponin for cell permeabilization. The primary antibodies used were rabbit anti-Brucella polyclonal antibody (1:1,500), mouse immunoglobulin G anti-O-antigen monoclonal antibody M-84 (1:1000) (Nielsen et al., 1995), mouse anti-human LAMP-1 H4A3 (1:400) (Developmental Studies Hybridoma Bank, Department of Biological Sciences, University of Iowa), and mouse anti-FLAG M2 monoclonal antibody (1:4000). The secondary antibodies used were Alexa Fluor 488 goat anti-rabbit or anti-mouse and Alexa Fluor 568 goat anti-rabbit or anti-mouse (Molecular Probes, Invitrogen Co.). For DNA staining, DAPI dye at 2 µg/ml (final concentration) was used.
Immunofluorescence microscopy
Eukaryotic cells were plated on glass coverslips (Marienfeld GmBH & co. KG) and infected as described above. For HeLa cells the MOI was 1000:1, and for J774.A1 cells the MOI was 500:1. At the indicated times, the coverslips were washed with PBS and the cells were fixed for 15 min in 4% paraformaldehyde (pH 7.4) at room temperature. After fixation, cells were washed three times with PBS and incubated for 10 min with PBS−50 mM NH4CL in order to quench free aldehyde groups. For the In/Out assay, the coverslips were incubated with rabbit anti-Brucella antibody for 45 min at room temperature, washed in PBS and incubated for 30 min with PBS−10% horse serum-5% BSA-0.1% saponin permeabilization solution. Then washed in PBS, and incubated for 45 min with mouse anti-O-antigen antibody M-84. After the second antibody the cells were washed with PBS and incubated for 45 min with Alexa Fluor 568 goat anti-rabbit, Alexa Fluor 488 goat anti-mouse secondary antibodies and the DAPI dye.
For the Lamp-1 co-localization assay, coverslips were incubated for 30 min with PBS−10% horse serum-5% BSA-0.1% saponin permeabilization solution, washed with PBS, and incubated for 45 min with mouse anti-human Lamp-1 H4A3 antibody. After 5 washes with PBS, they were incubated for 45 min with rabbit anti-Brucella antibody, washed again with PBS, and then incubated for 45 min with Alexa Fluor 568 goat anti-rabbit and Alexa Fluor 488 goat anti-mouse secondary antibodies.
For the Screening assays, the coverslips were incubated for 30 min with PBS−10% horse serum-5% BSA-0.1% saponin permeabilization solution, washed with PBS, and incubated for 45 min with a rabbit anti-Brucella antibody. After 5 washes with PBS, cells were incubated for 30 min with PBS-10% horse serum-5% BSA- 0.2% Triton X-100 permeabilization solution, washed again with PBS and incubated for 45 min with mouse anti-FLAG M2. Then washed with PBS, and finally incubated for 45 min with Alexa Fluor 568 goat anti-rabbit, Alexa Fluor 488 goat anti-mouse secondary antibodies and DAPI dye.
After immunofluorescence staining, all the coverslips were mounted onto slides with FluorSave (Calbiochem). Samples were examined on a Nikon microscope (Eclipse TE 2000) at a magnification of X600. The software MBF ImageJ v1.43m (Wayne Rasband, NIH, USA) was used to merge the microscopic images. Con-Focal images were acquired in an Olympus FV 1000 inverted microscope and images, as well as Z stacks, processed with the Fluoview FV100 software provided with the microscope.
Mouse infections
Mice infections were performed as previously described (Ugalde et al., 2003). Groups of 5 eight to nine week-old female BALB/c mice were intraperitoneally inoculated with 1 × 105 CFU of B. abortus 2308 wild type, ΔsepA, complemented or SepA overexpressing strains in PBS. At 2 weeks post-infection, spleens from infected mice were removed and homogenized in 2 ml of PBS. Serial dilutions from individualized spleens were plated on TSA with the appropriate antibiotics to quantify recovered CFU.
Periplasmic and cytoplasmic localization assay
For the periplasmic localization assays the B. abortus strains were cultivated in TSB for 16h – 24h at 37°C until an OD600nm 1 was reached 2.5 × 1010 bacterial cells were centrifuged 10 min at 3300×g. The pellets were washed with physiologic solution, centrifuged for 10 min at 3300×g and resuspended in 1 ml of 0.2M Tris-HCl pH 7.6.1 ml of 0.2M Tris-HCl pH 7.6, 1M Sucrose and 0.25% Zwitterion 3–16 solution was added to the cell suspension and incubated for 10 min at room temperature. The samples were centrifuged for 30 min at 8000×g, the pellets separated from the supernatants and stored at −20°C until used. The pellets and supernatants were processed for Western-Blot, using an anti-FLAG M2 monoclonal antibody (1:5000), anti GroEL (1:2000) and anti OMP-1 (1:2000) kindly provided by Dr. Axel Cloeckaert as primary antibodies, and goat anti-mouse HRP (1:2000) (Dako) as secondary antibody. All antibodies were diluted in Tris-buffered saline (TBS)- 5% non-fat milk- 0.05% Tween solution.
Supplementary Material
Acknowledgments
We thank Pablo Dosantos and Francisco Guaimas for confocal microscopy assistance. This work was supported by grants PICT-PRH08-160 and PICT-PRH08-230 to C.C. and J.E.U. respectively, NIAID grant AI078891 to J.E.U., and grants from the University of San Martín to C.C. and J.E.U. P.H.D and E.V. were supported by fellowships of the National Research Council of Argentina (CONICET). CC and J.E.U. are members of the CONICET.
Footnotes
The authors declare no conflict of interest.
REFERENCES
- Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arocena GM, Sieira R, Comerci DJ, Ugalde RA. Identification of the quorum-sensing target DNA sequence and N-Acyl homoserine lactone responsiveness of the Brucella abortus virB promoter. J Bacteriol. 2010;192:3434–3440. doi: 10.1128/JB.00232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arocena GM, Zorreguieta A, Sieira R. Expression of VjbR under nutrient limitation conditions is regulated at the post-transcriptional level by specific acidic pH values and urocanic acid. PLoS One. 2012;7:e35394. doi: 10.1371/journal.pone.0035394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atluri VL, Xavier MN, de Jong MF, den Hartigh AB, Tsolis RE. Interactions of the human pathogenic Brucella species with their hosts. Annu Rev Microbiol. 2011;65:523–541. doi: 10.1146/annurev-micro-090110-102905. [DOI] [PubMed] [Google Scholar]
- Burns DL. Type IV transporters of pathogenic bacteria. Curr Opin Microbiol. 2003;6:29–34. doi: 10.1016/s1369-5274(02)00006-1. [DOI] [PubMed] [Google Scholar]
- Celli J, de Chastellier C, Franchini DM, Pizarro-Cerda J, Moreno E, Gorvel JP. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J Exp Med. 2003;198:545–556. doi: 10.1084/jem.20030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J, Salcedo SP, Gorvel JP. Brucella coopts the small GTPase Sar1 for intracellular replication. Proc Natl Acad Sci U S A. 2005;102:1673–1678. doi: 10.1073/pnas.0406873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerci DJ, Martinez-Lorenzo MJ, Sieira R, Gorvel JP, Ugalde RA. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell Microbiol. 2001;3:159–168. doi: 10.1046/j.1462-5822.2001.00102.x. [DOI] [PubMed] [Google Scholar]
- Corbel MJ. Brucellosis: an overview. Emerg Infect Dis. 1997;3:213–221. doi: 10.3201/eid0302.970219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P, Roy CR. Manipulation of host membrane machinery by bacterial pathogens. Curr Opin Cell Biol. 2010;22:547–554. doi: 10.1016/j.ceb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czibener C, Sherer NM, Becker SM, Pypaert M, Hui E, Chapman ER, et al. Ca2+ and synaptotagmin VII-dependent delivery of lysosomal membrane to nascent phagosomes. J Cell Biol. 2006;174:997–1007. doi: 10.1083/jcb.200605004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czibener C, Ugalde JE. Identification of a unique gene cluster of Brucella spp. that mediates adhesion to host cells. Microbes Infect. 2012;14:79–85. doi: 10.1016/j.micinf.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Barsy M, Jamet A, Filopon D, Nicolas C, Laloux G, Rual JF, et al. Identification of a Brucella spp. secreted effector specifically interacting with human small GTPase Rab2. Cell Microbiol. 2011;13:1044–1058. doi: 10.1111/j.1462-5822.2011.01601.x. [DOI] [PubMed] [Google Scholar]
- de Jong MF, Sun YH, den Hartigh AB, van Dijl JM, Tsolis RM. Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol Microbiol. 2008;70:1378–1396. doi: 10.1111/j.1365-2958.2008.06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Giudice MG, Ugalde JE, Czibener C. A lysozyme-like protein in Brucella abortus is involved in the early stages of intracellular replication. Infect Immun. 2013;81:956–964. doi: 10.1128/IAI.01158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delrue RM, Deschamps C, Leonard S, Nijskens C, Danese I, Schaus JM, et al. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell Microbiol. 2005;7:1151–1161. doi: 10.1111/j.1462-5822.2005.00543.x. [DOI] [PubMed] [Google Scholar]
- Godfroid J, Scholz HC, Barbier T, Nicolas C, Wattiau P, Fretin D, et al. Brucellosis at the animal/ecosystem/human interface at the beginning of the 21st century. Prev Vet Med. 2011;102:118–131. doi: 10.1016/j.prevetmed.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, 2nd, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Marchesini MI, Herrmann CK, Salcedo SP, Gorvel JP, Comerci DJ. In search of Brucella abortus type IV secretion substrates: screening and identification of four proteins translocated into host cells through VirB system. Cell Microbiol. 2011;13:1261–1274. doi: 10.1111/j.1462-5822.2011.01618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myeni S, Child R, Ng TW, Kupko JJ, 3rd, Wehrly TD, Porcella SF, et al. Brucella Modulates Secretory Trafficking via Multiple Type IV Secretion Effector Proteins. PLoS Pathog. 2013;9:e1003556. doi: 10.1371/journal.ppat.1003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Roy CR. The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. EMBO J. 2001;20:5962–5970. doi: 10.1093/emboj/20.21.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KH, Kelly L, Gall D, Nicoletti P, Kelly W. Improved competitive enzyme immunoassay for the diagnosis of bovine brucellosis. Vet Immunol Immunopathol. 1995;46:285–291. doi: 10.1016/0165-2427(94)05361-u. [DOI] [PubMed] [Google Scholar]
- Ninio S, Roy CR. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 2007;15:372–380. doi: 10.1016/j.tim.2007.06.006. [DOI] [PubMed] [Google Scholar]
- O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli ML, Bourg G, Foulongne V, et al. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- Pantoja M, Chen L, Chen Y, Nester EW. Agrobacterium type IV secretion is a two-step process in which export substrates associate with the virulence protein VirJ in the periplasm. Mol Microbiol. 2002;45:1325–1335. doi: 10.1046/j.1365-2958.2002.03098.x. [DOI] [PubMed] [Google Scholar]
- Pizarro-Cerda J, Desjardins M, Moreno E, Akira S, Gorvel JP. Modulation of endocytosis in nuclear factor IL-6(−/−) macrophages is responsible for a high susceptibility to intracellular bacterial infection. J Immunol. 1999;162:3519–3526. [PubMed] [Google Scholar]
- Pizarro-Cerda J, Meresse S, Parton RG, van der Goot G, Sola-Landa A, Lopez-Goni I, et al. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect Immun. 1998;66:5711–5724. doi: 10.1128/iai.66.12.5711-5724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Sieira R, Arocena GM, Bukata L, Comerci DJ, Ugalde RA. Metabolic control of virulence genes in Brucella abortus: HutC coordinates virB expression and the histidine utilization pathway by direct binding to both promoters. J Bacteriol. 2010;192:217–224. doi: 10.1128/JB.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieira R, Arocena GM, Zorreguieta A, Comerci DJ, Ugalde RA. A MarR-Type Regulator Directly Activates Transcription from the Brucella abortus virB Promoter by Sharing a Redundant Role with HutC. J Bacteriol. 2012;194:6431–6440. doi: 10.1128/JB.01007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieira R, Comerci DJ, Pietrasanta LI, Ugalde RA. Integration host factor is involved in transcriptional regulation of the Brucella abortus virB operon. Mol Microbiol. 2004;54:808–822. doi: 10.1111/j.1365-2958.2004.04316.x. [DOI] [PubMed] [Google Scholar]
- Sieira R, Comerci DJ, Sanchez DO, Ugalde RA. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J Bacteriol. 2000;182:4849–4855. doi: 10.1128/jb.182.17.4849-4855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano S, Ugalde JE, Galan JE. Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host and Microbe. 2008 doi: 10.1016/j.chom.2007.11.001. In press. [DOI] [PubMed] [Google Scholar]
- Ugalde JE, Comerci DJ, Leguizamon MS, Ugalde RA. Evaluation of Brucella abortus phosphoglucomutase (pgm) mutant as a new live rough-phenotype vaccine. Infect Immun. 2003;71:6264–6269. doi: 10.1128/IAI.71.11.6264-6269.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugalde JE, Czibener C, Feldman MF, Ugalde RA. Identification and characterization of the Brucella abortus phosphoglucomutase gene: role of lipopolysaccharide in virulence and intracellular multiplication. Infect Immun. 2000;68:5716–5723. doi: 10.1128/iai.68.10.5716-5723.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.