Abstract
Traditional dairy products are potential sources of a variety of microorganisms which participate in food poisoning. Staphylococcus aureus is a conspicuous example of toxigenic bacteria causative for food-borne diseases. Moreover, resistance to methicillin is a prominent index in food hygiene studies. In the present study, we have aimed at characterization and identification of enterotoxigenic methicillin resistant S. aureus (MRSA) isolated from traditional cheeses in Azerbaijan region in the northwest of Iran during 2012. A number of phenotypical and molecular assays were utilized for screening of S. aureus. Subsequently, the prevalence of the genes responsible for the five staphylococcal enterotoxins (SEA-SEE) and also methicillin resistance gene was assessed. The outcomes of phenotypical methods were in conformity with those of the molecular procedures. The results indicated that 16% of cheese samples were contaminated by S. aureus. 110 isolates were authenticated by both phenotypical and molecular methods. All of the mentioned isolates were positive for coa, nuc, and 16S rDNA primers. 21% of these isolates were mecA positive and 60.8% of these MRSA were positive for SEs. Regarding the frequent outbreaks of enterotoxigenic MRSA, new hygiene policies and management practices should be considered to increase food safety and avoid extra treatment costs.
1. Introduction
Food-borne diseases are a public health problem worldwide. To date, approximately 250 different food-borne diseases have been described while bacteria were responsible for the two-thirds of them. Among these bacteria, Staphylococcus aureus is one of the most common agents in food poisoning outbreaks [1, 2]. S. aureus is a Gram and coagulase positive cocci, which belongs to the Staphylococcaceae family. This bacterium is a major causative pathogen of clinical or subclinical mastitis of dairy milk and milk products [3, 4]. There are frequent reports of prevalence of S. aureus in extensively used traditional cheeses [5, 6]. Staphylococcal Enterotoxins (SEs), the main causatives for food poisoning, are a group of single chain, low molecular mass proteins and are produced during all phases of growth [7]. Based on serological classification, to date several SEs have been recognized such as SEA, SEB, SEC, SED, and SEE [8]. SEs are highly thermostable and resistant to most proteolytic enzymes and different environmental conditions. The detection of SEs is known as a reliable means for the confirmation of staphylococcal outbreaks and determination of the enterotoxigenicity of strains [9]. On the other hand, due to the intensive use of antibiotics in public health and animal breeding, antibiotic resistance in pathogens including the genus Staphylococcus has become an increasing medical problem during the last decades [10, 11]. Moreover, the methicillin resistant S. aureus (MRSA) should be also considered resistant to all penicillins, cephalosporins, cephems, and other β-lactams [12]. Therefore, preventing the prevalence of resistant strains seems to be of a central importance. Accordingly, a variety of sufficient techniques should be employed for surveying the presence of MRSA and related toxins in foodstuff. Nowadays, molecular biology techniques are considered as important tools in the microbiological studies [9]. Traditional methods such as immunological procedures require detectable amounts of toxins. In addition, the detection of enterotoxins by the mentioned methods is generally more complicated and time consuming than the molecular methods and sometimes they are not practical for detection and identification of a large group of SEs [13]. In the present study both phenotypical and molecular procedures were employed for assessment of enterotoxigenicity and methicillin resistance of S. aureus in traditional cheeses in the East Azerbaijan, Iran.
2. Materials and Methods
2.1. Samples Collection and Culture
Traditional cheeses in Azerbaijan region are generally produced from goat or sheep's fatty milk with a temperature of 30°C by adding commercial rennet [14]. In the present study, one hundred samples of 20 grams were collected from local producers in the rural areas of East Azerbaijan from January through December 2012. For the primary culture, 50 μL of homogenized samples was surface-plated on nutrient agar and incubated at 37°C for 24–48 h under aerobic conditions. Colonies suspected as S. aureus were selected and transferred into individual tubes of nutrient broth. Colonies were sub-cultured on 5% sheep blood agar and mannitol salt agar (Himedia, India). Culture characteristics, Gram staining, and catalase and coagulase test utilizing fresh rabbit plasma were the criteria used for the identification of presumptive isolates. The carbohydrates differential fermentation was also used for confirmation of all coagulase positive (CP) isolates [15]. S. aureus ATCC 25923 and Escherichia coli ATCC 35218 served as reference strains for biochemical tests.
2.2. Methicillin Resistance Test
Resistance of staphylococcal isolates to methicillin was tested by disk diffusion assay according to the guidelines of the National Committee for Clinical Laboratory Standards (NCCLS, 2003) using Mueller Hinton agar [11].
2.3. DNA Extraction
A modified one-step protocol was utilized for genomic DNA extraction from overnight cultures of putative staphylococcal isolates [16]. Bacterial pellets in Eppendorf tubes were alternatively placed in liquid nitrogen and 60°C water bath for five to seven times. Then, 900 μL of CTAB lysis buffer (Applichem, A4150) was added and the microtubes were incubated in 65°C water bath for 45 min. 500 μL of phenol-chloroform-isoamyl alcohol in a ratio of 25 : 24 : 1 was added to the samples and the procedure was followed by 2-propanol precipitation. RNase-A (DNase-free) was added into tubes in final concentration of 20 μg/mL. The extracted DNA was evaluated by 0.8% agarose gel electrophoresis.
2.4. Primer Design for 16S Ribosomal RNA Gene
Several nucleotide sequences corresponding to staphylococcal 16S rDNA gene were acquired from NCBI database. ClustalX (ver. 1.81) and Mega (ver. 4) software were used for alignment of the mentioned sequences. Subsequently the Oligo5 software was utilized for designing of Staphylococcus 16S rDNA primers. The specifications of the mentioned primers, namely, ST16F and ST16R, are shown in Table 1. The mentioned primers can be used for identification of a wide range of staphylococci in genus level.
Table 1.
List of primers used in the present study.
| Gene | Primer | Primer sequence (5′ to 3′) | Size (bp) | Reference |
|---|---|---|---|---|
| Coagulase | coa-F | ATAGAGATGCTGGTACAGG | 674–917 | [15] |
| coa-R | GCTTCCGATTGTTCGATGC | |||
| Methicillin resistance | mecA-F | AAAATCGATGGTAAAGGTTGGC | 533 | [39] |
| mecA-R | AGTTCTGCAGTACCGGATTTGC | |||
| 16S rDNA | ST16-F | TTG CTT CTC TGA TGT TAG CG | 1412 | Designed |
| ST16-R | AAT CAT TTG TCC CAC CTT C | |||
| Nuclease | nuc-F | GCGATTGATGGTGATACGGTT | 276 | [32] |
| nuc-R | AGCCAAGCCTTGACGAACTAAAGC | |||
| Enterotoxin A | SEA-A2 | ATTAACCGAAGGTTCTGTAGA | 582 | [40] |
| SEA-U2 | TTGCGTAAAAAGTCTGAATT | |||
| Enterotoxin B | SEB-B2 | TTTTTCTTTGTCGTAAGATAA | 732 | |
| SEB-U1 | CCAACGTTTTAGCAGAGAAG | |||
| Enterotoxin C | SEC-C2 | TAAGTTCCCATTATCAAAGTG | 403 | |
| SEC-U1 | CCAACGTTTTAGCAGAGAAG | |||
| Enterotoxin D | SED-D2 | TAATGCTATATCTTATAGGG | 251 | |
| SED-U2 | TTGCGTAAAAAGTCTGAATT | |||
| Enterotoxin E | SEE-E2 | TAAACCAAATTTTCCGTG | 474 | |
| SEE-U2 | TTGCGTAAAAAGTCTGAATT |
2.5. PCR Amplification of 16S rDNA Gene
The designated primers were used for amplification of an approximately 1412 bp of 16S ribosomal RNA gene. PCR was carried out in a final volume of 50 μL in a Techne (TC-412) thermal cycler. Each reaction contained 40 ng of DNA, 0.5 μM of ST16 primers, and 25 μL of 2X Amplicon Master Mix. PCR program consisted of an initial denaturation step at 94°C for 4 min which was followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 45 s, extension at 72°C for 80 s, and eventually a final extension step of 72°C for 10 min.
2.6. Multiplex-PCR for Staphylococcal Enterotoxins (A–E)
Multiplex PCR was carried out in a final volume of 50 μL. Each reaction contained 50 ng of DNA, 0.5 μM of A2, B2, C2, D2, and E2 primers, 0.7 μM of U1 primer, and 0.9 μM of U2 primer (Table 1). 2X Amplicon Master Mix was used in the same quantity described for 16S ribosomal RNA gene. Initial denaturation was 3 min at 94°C, which was followed by 35 cycles of denaturation at 94°C for 50 s, annealing at 52°C for 55 s, and extension at 72°C for 80 s with a final extension step of 72°C for 10 min.
2.7. PCR Amplification of Coagulase, Nuclease, and Methicillin Resistance Genes
PCR for amplification of nuclease (nuc), coagulase (coa), and methicillin resistance (mecA) genes was performed in separate reactions. Primer sequences are indicated in Table 1. All PCR products were analyzed by 1% (w/v) agarose gel electrophoresis in TAE buffer stained with ethidium bromide.
3. Results
120 susceptible colonies were isolated from cheese samples and subjected to phenotypical and molecular authentication tests. Making use of biochemical assays, 110 colonies were found to be positive for coagulase. PCR for amplification of the entire 3′ repeat elements of coa gene resulted in one of 875, 660, 603, or 547 bp amplicons for all biochemically coa positive isolates (Figure 1). PCR with 16S-rDNA primers resulted in approximately 1412 bp amplicons for all 110 isolates. Evaluation of strains for nuc gene also revealed that all of the putative samples were positive for this trait. Eventually, 16 out of the 100 cheese samples (16%) examined were found to be contaminated with S. aureus.
Figure 1.
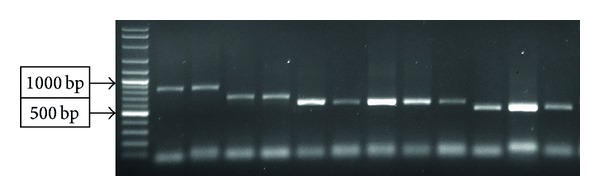
Examples of the four sizes of coa gene from representative S. aureus isolates.
Methicillin susceptibility test also indicated that 21% of S. aureus isolates (23 out of 110 isolates) were resistant to this antibiotic. Application of molecular methods in the present study showed that all of the MRSA phenotypes were positive for presence of mecA gene which is an important index for antibiotic resistance.
Furthermore, the multiplex PCR for SEs revealed that 14 isolates of these 23 methicillin resistant strains were positive for SEs. In this case eight (34.8%) of the isolates were positive for SEA, three (13%) were positive for the SEB gene, one (4.3%) was found to be SEC positive, none of samples were positive for SED, and two isolates (8.7%) contained the gene responsible for SEE.
4. Discussion
Methicillin resistant S. aureus strains, because of their high mortality, have become a major concern worldwide [17]. MRSA, including animal associated strains, have been frequently detected in dairy products such as raw milk or traditional cheeses [18–21]. Since the microbes of dairy products such as cheese are not being eliminated by heating or processing, they may serve as a vehicle to disperse MRSA [22].
Application of conventional criteria such as cultural and biochemical features for identification of S. aureus and MRSA isolates is slow and time consuming. In contrast, molecular methods have been proven as rapid and reliable means in the study of outbreaks and epidemiology of a wide range of microorganisms.
In the present study, 16S rDNA gene amplification was employed for characterization of the isolates. The deigned primers could be utilized for direct and rapid identification of a wide range of Staphylococcus isolates at genus level. Consequently, all susceptible strains were positive for 16S rDNA primers. Moreover, PCR on nuc gene for rapidly diagnosis of S. aureus [23] confirmed all presumptive strains. Molecular analysis of coa gene resulted in four sizes of products. The presence of different amplicon sizes is due to repetitions of 3′ elements of coa gene in various strains [24]. In a similar study performed in the north west of Iran, the four sizes were detectable [25]. The prevalence of S. aureus was about 16% in the present study. However, the occurrence of this microorganism in dairy products or other foodstuff is different among studies [24, 26–34]. Several factors such as source of sampling, geographical origin, sensitivity of the identification methods, and the quantity of samples can affect the outcomes. For instance, in one study S. aureus was found in 7.3% of commercial milk samples in Brazil [35]. In other study in Italy, 13.3% of fresh cheeses were contaminated with S. aureus [36]. The contamination degree also varies among different kinds of dairy products, while cream, cheese, and milk are arranged in a descending order from the viewpoint of contamination. It is deduced that further handling and manipulating of milk products could lead to more S. aureus contamination [37].
In analysis of methicillin resistance, molecular methods are also proved to be significantly useful. In one study, PCR based mecA gene amplification confirmed more than 99% of MRSA isolates [26]. In our study, mecA gene was detected in 21% of the S. aureus isolates (23 out of 110). In a similar study by Türkyılmaz, among 93 S. aureus strains isolated from bovine milk with mastitis in Aydin, Turkey, 16 were resistant to methicillin [12]. The high prevalence of resistance genes should be considered as a potential health risk for humans and livestock. Consequently, necessary precautions should be taken by governments and individuals to prevent the further spread of MRSA. Hygiene promotion and avoiding the unsupervised use of antibiotics seem to be elementary steps in this regard.
On the other hand, the investigation of the strains for enterotoxins is an important experiment in pathogenesis studies [38]. In our study, 60.8% of MRSA were positive for the presence of SEs.
In the study performed by Løvseth et al. in Brazil [35], strains isolated from 14.3% samples were enterotoxigenic. In another study, which included SEA and SEB genes, the PCR results proved that 15.6% of the S. aureus isolates possessed the SEA gene and 9.3% the SEB gene [37]. In brief, detecting the SE genes by molecular techniques could help understand the virulence mechanisms and pathogenic potential of this microorganism.
5. Conclusion
The data presented in this study represents the information about prevalence of methicillin resistant and enterotoxigenic Staphylococcus aureus isolates from traditional cheeses in the north west of Iran and highlights a strong need for epidemiological and pathogenic studies to monitor distribution, infectious species, and infection kinetics. This information should provide awareness and persuade the authorities to set new hygiene policies. Furthermore, it could be helpful in disease management practices and reducing the overall costs of medication. It also shows that molecular methods provide rapid, accurate, and economic means for S. aureus screening in food safety evaluations.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Le Loir Y, Baron F, Gautier M. Staphylococcus aureus and food poisoning. Genetics and Molecular Research. 2003;2(1):63–76. [PubMed] [Google Scholar]
- 2.Hennekinne J-A, de Buyser M-L, Dragacci S. Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiology Reviews. 2011;36(4):815–836. doi: 10.1111/j.1574-6976.2011.00311.x. [DOI] [PubMed] [Google Scholar]
- 3.Oliver SP, Jayarao BM, Almeida RA. Foodborne pathogens in milk and the dairy farm environment: food safety and public health implications. Foodborne Pathogens and Disease. 2005;2(2):115–129. doi: 10.1089/fpd.2005.2.115. [DOI] [PubMed] [Google Scholar]
- 4.Abdel-Rady A, Sayed M. Epidemiological studies on subclinical mastitis in dairy cows in Assiut Governorate. Veterinary World. 2009;2(10):373–380. [Google Scholar]
- 5.Pinto MS, de Carvalho AF, Pires ACDS, et al. The effects of nisin on Staphylococcus aureus count and the physicochemical properties of Traditional Minas Serro cheese. International Dairy Journal. 2011;21(2):90–96. [Google Scholar]
- 6.Borelli BM, Lacerda ICA, Brandão LR, et al. Identification of Staphylococcus spp. isolated during the ripening process of a traditional minas cheese. Arquivo Brasileiro de Medicina Veterinaria e Zootecnia. 2011;63(2):481–487. [Google Scholar]
- 7.Naffa RG, Bdour SM, Migdadi HM, Shehabi AA. Enterotoxicity and genetic variation among clinical Staphylococcus aureus isolates in Jordan. Journal of Medical Microbiology. 2006;55(2):183–187. doi: 10.1099/jmm.0.46183-0. [DOI] [PubMed] [Google Scholar]
- 8.Tokajian S, Haddad D, Andraos R, Hashwa F, Araj G. Toxins and antibiotic resistance in Staphylococcus aureus isolated from a major hospital in Lebanon. ISRN Microbiology. 2011;2011:9 pages. doi: 10.5402/2011/812049.812049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.da Cunha ML, Calsolari RA, Junior JP. Detection of enterotoxin and toxic shock syndrome toxin 1 genes in Staphylococcus, with emphasis on coagulase-negative staphylococci. Microbiology and Immunology. 2007;51(4):381–390. doi: 10.1111/j.1348-0421.2007.tb03925.x. [DOI] [PubMed] [Google Scholar]
- 10.Perreten V, Vorlet-Fawer L, Slickers P, Ehricht R, Kuhnert P, Frey J. Microarray-based detection of 90 antibiotic resistance genes of gram-positive bacteria. Journal of Clinical Microbiology. 2005;43(5):2291–2302. doi: 10.1128/JCM.43.5.2291-2302.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadigh-Eteghad S, Dehnad A, Shanebandi D, Khalili I, Razmarayii N, Namvaran A. Identification and characterization of a Streptomyces sp. isolate exhibiting activity against multidrug-resistant coagulase-negative Staphylococci. Veterinary Research Communications. 2011;35(8):477–486. doi: 10.1007/s11259-011-9491-9. [DOI] [PubMed] [Google Scholar]
- 12.Türkyilmaz S, Tekbiyik S, Oryasin E, Bozdogan B. Molecular epidemiology and antimicrobial resistance mechanisms of methicillin-resistant Staphylococcus aureus isolated from bovine milk. Zoonoses and Public Health. 2010;57(3):197–203. doi: 10.1111/j.1863-2378.2009.01257.x. [DOI] [PubMed] [Google Scholar]
- 13.Vasconcelos NG, da Cunha MLRS. Staphylococcal enterotoxins: molecular aspects and detection methods. Journal of Public Health and Epidemiology. 2010;2(3):29–42. [Google Scholar]
- 14.Shanehbandi D, Dafe A, Alizadeh A, et al. Motal cheese of Iranian nomadic tribes as an untouched source of potentially probiotic Lactobacilli. African Journal of Microbiology Research. 2013;7(22):2751–2756. [Google Scholar]
- 15.Quinn PJ, Markey BK, Leonard FC, Hartigan P, Fanning S, FitzPatrick ES. Veterinary Microbiology and Microbial Disease. 2nd edition. New York, NY, USA: John Wiley & Sons; 2011. [Google Scholar]
- 16.Wilson K. Current Protocols in Molecular Biology. New York, NY, USA: John Wiley & Sons; 2001. UNIT 2.4 preparation of genomic DNA from bacteria. [DOI] [PubMed] [Google Scholar]
- 17.Hookey JV, Richardson JF, Cookson BD. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. Journal of Clinical Microbiology. 1998;36(4):1083–1089. doi: 10.1128/jcm.36.4.1083-1089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saei HD, Ahmadi M, Mardani K, Batavani RA. Molecular typing of Staphylococcus aureus isolated from bovine mastitis based on polymorphism of the coagulase gene in the North West of Iran. Veterinary Microbiology. 2009;137(1-2):202–206. doi: 10.1016/j.vetmic.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Brakstad OG, Aasbakk K, Maeland JA. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. Journal of Clinical Microbiology. 1992;30(7):1654–1660. doi: 10.1128/jcm.30.7.1654-1660.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fagundes H, Barchesi L, Filho AN, Ferreira LM, Oliveira CAF. Occurrence of Staphylococcus aureus in raw milk produced in dairy farms in São Paulo state, Brazil. Brazilian Journal of Microbiology. 2010;41(2):376–380. doi: 10.1590/S1517-838220100002000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peton V, Le Loir Y. Staphylococcus aureus in veterinary medicine. Infection, Genetics and Evolution. 2013:S1567–S1348. doi: 10.1016/j.meegid.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 22.Wendlandt S, Schwarz S, Silley P. Methicillin-resistant Staphylococcus aureus: a food-borne pathogen? Annual Review of Food Science and Technology . 2013;4:117–139. doi: 10.1146/annurev-food-030212-182653. [DOI] [PubMed] [Google Scholar]
- 23.Giannatale ED, Prencipe V, Tonelli A, Marfoglia C, Migliorati G. Characterisation of Staphylococcus aureus strains isolated from food for human consumption. Veterinaria Italiana. 2011;47(2):165–173. [PubMed] [Google Scholar]
- 24.Mellmann A, Friedrich AW, Rosenkötter N, et al. Automated DNA sequence-based early warning system for the detection of methicillin-resistant Staphylococcus aureus outbreaks. PLoS Medicine. 2006;3(3, article e33) doi: 10.1371/journal.pmed.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon NH, Park KT, Jung WK, et al. Characteristics of methicillin resistant Staphylococcus aureus isolated from chicken meat and hospitalized dogs in Korea and their epidemiological relatedness. Veterinary Microbiology. 2006;117(2–4):304–312. doi: 10.1016/j.vetmic.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Hata E, Katsuda K, Kobayashi H, Uchida I, Tanaka K, Eguchi M. Genetic variation among Staphylococcus aureus strains from bovine milk and their relevance to methicillin-resistant isolates from humans. Journal of Clinical Microbiology. 2010;48(6):2130–2139. doi: 10.1128/JCM.01940-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hata E, Kobayashi H, Nakajima H, Shimizu Y, Eguchi M. Epidemiological analysis of Staphylococcus aureus isolated from cows and the environment of a dairy farm in Japan. The Journal of Veterinary Medical Science. 2010;72(5):647–652. doi: 10.1292/jvms.09-0452. [DOI] [PubMed] [Google Scholar]
- 28.Fosheim GE, Nicholson AC, Albrecht VS, Limbago BM. Multiplex Real-Time PCR assay for detection of methicillin-resistant Staphylococcus aureus and associated toxin genes. Journal of Clinical Microbiology. 2011;49(8):3071–3073. doi: 10.1128/JCM.00795-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imani Fooladi A, Tavakoli HR, Naderi A. Detection of enterotoxigenic Staphylococcus aureus isolates in domestic dairy products. Iranian Journal of Microbiology. 2010;2(3):137–142. [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson IG, Cooper JE, Gilmour A. Detection of enterotoxigenic Staphylococcus aureus in dried skimmed milk: use of the polymerase chain reaction for amplification and detection of staphylococcal enterotoxin genes entB and entC1 and the thermonuclease gene nuc . Applied and Environmental Microbiology. 1991;57(6):1793–1798. doi: 10.1128/aem.57.6.1793-1798.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker K, Roth R, Peters G. Rapid and specific detection of toxigenic Staphylococcus aureus: use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene. Journal of Clinical Microbiology. 1998;36(9):2548–2553. doi: 10.1128/jcm.36.9.2548-2553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma NK, Rees CED, Dodd CER. Development of a single-reaction multiplex PCR toxin typing assay for Staphylococcus aureus strains. Applied and Environmental Microbiology. 2000;66(4):1347–1353. doi: 10.1128/aem.66.4.1347-1353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehrotra M, Wang G, Johnson WM. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. Journal of Clinical Microbiology. 2000;38(3):1032–1035. doi: 10.1128/jcm.38.3.1032-1035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kikuchi K, Takahashi N, Piao C, Totsuka K, Nishida H, Uchiyama T. Molecular epidemiology of methicillin-resistant Staphylococcus aureus strains causing neonatal toxic shock syndrome-like exanthematous disease in neonatal and perinatal wards. Journal of Clinical Microbiology. 2003;41(7):3001–3006. doi: 10.1128/JCM.41.7.3001-3006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Løvseth A, Loncarevic S, Berdal KG. Modified multiplex PCR method for detection of pyrogenic exotoxin genes in staphylococcal isolates. Journal of Clinical Microbiology. 2004;42(8):3869–3872. doi: 10.1128/JCM.42.8.3869-3872.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katsuda K, Hata E, Kobayashi H, et al. Molecular typing of Staphylococcus aureus isolated from bovine mastitic milk on the basis of toxin genes and coagulase gene polymorphisms. Veterinary Microbiology. 2005;105(3-4):301–305. doi: 10.1016/j.vetmic.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Hwang SY, Park YK, Koo HC, Park YH. spa typing and enterotoxin gene profile of Staphylococcus aureus isolated from bovine raw milk in Korea. Journal of Veterinary Science. 2010;11(2):125–131. doi: 10.4142/jvs.2010.11.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cretenet M, Even S, Le Loir Y. Unveiling Staphylococcus aureus enterotoxin production in dairy products: a review of recent advances to face new challenges. Dairy Science & Technology. 2011;91(2):127–150. [Google Scholar]
- 39.Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. Journal of Clinical Microbiology. 1991;29(10):2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsieh J-M, Chen R-S, Tsai T-Y, Pan T-M, Chou C-C. Phylogenetic analysis of livestock oxacillin-resistant Staphylococcus aureus . Veterinary Microbiology. 2008;126(1–3):234–242. doi: 10.1016/j.vetmic.2007.07.013. [DOI] [PubMed] [Google Scholar]


