Abstract
Appropriate timing of flowering is crucial for crop yield and the reproductive success of plants. Flowering can be induced by a number of molecular pathways that respond to internal and external signals such as photoperiod, vernalization or light quality, ambient temperature and biotic as well as abiotic stresses. The key florigenic signal FLOWERING LOCUS T (FT) is regulated by several flowering activators, such as CONSTANS (CO), and repressors, such as FLOWERING LOCUS C (FLC). Chromatin modifications are essential for regulated gene expression, which often involves the well conserved MULTICOPY SUPRESSOR OF IRA 1 (MSI1)-like protein family. MSI1-like proteins are ubiquitous partners of various complexes, such as POLYCOMB REPRESSIVE COMPLEX2 or CHROMATIN ASSEMBLY FACTOR 1. In Arabidopsis, one of the functions of MSI1 is to control the switch to flowering. Arabidopsis MSI1 is needed for the correct expression of the floral integrator gene SUPPRESSOR OF CO 1 (SOC1). Here, we show that the histone-binding protein MSI1 acts in the photoperiod pathway to regulate normal expression of CO in long day (LD) photoperiods. Reduced expression of CO in msi1-mutants leads to failure of FT and SOC1 activation and to delayed flowering. MSI1 is needed for normal sensitivity of Arabidopsis to photoperiod, because msi1-mutants responded less than wild type to an intermittent LD treatment of plants grown in short days. Finally, genetic analysis demonstrated that MSI1 acts upstream of the CO-FT pathway to enable an efficient photoperiodic response and to induce flowering.
Keywords: Arabidopsis, flowering time, chromatin, MSI1, photoperiod, FLOWERING LOCUS T (FT), CONSTANS (CO)
Introduction
The reproductive success of plants depends on the appropriate time to flower, which is of great agronomic relevance in crops. Flowering can be induced by a number of molecular pathways that respond to internal and external signals. Major genetic pathways controlling flowering time have been characterized based on the phenotype of Arabidopsis thaliana flowering time mutants in different growth conditions. These pathways include the photoperiod pathway, which responds to seasonal changes in day length, and the vernalization pathway, which responds to prolonged exposure to cold. The autonomous and gibberellin-pathways mediate the response to endogenous signals. Additionally, light quality, ambient temperature, and biotic as well as abiotic stresses can contribute to floral induction in plants (for review see: Jarillo and Piñeiro, 2011; Srikanth and Schmid, 2011).
The different pathways converge on pathway integrators, a set of genes that strongly promote flowering such as FLOWERING LOCUS T (FT), SUPPRESSOR OF CONSTANS 1 (SOC1/AGL20) or the FT homolog TWIN SISTER OF FT (TSF). Mutants in these genes have late flowering phenotypes (Kardailsky et al., 1999; Kobayashi et al., 1999; Borner et al., 2000; Lee et al., 2000; Samach et al., 2000; Yamaguchi et al., 2005). The pathway integrators FT and TSF are antagonistically regulated by the floral repressor FLOWERING LOCUS C (FLC) (Michaels and Amasino, 1999) and the floral activator CONSTANS (CO) (Yanovsky and Kay, 2002).
The nuclear zinc finger transcription factor CO is the key activator in the photoperiod pathway to promote expression of FT and TSF (Suarez-Lopez et al., 2001; Valverde et al., 2004). CO protein is stable in the light and rapidly degraded in the dark. CO is regulated by the circadian clock and accumulates diurnally late in the day in long day (LD) conditions. In contrast, CO peaks during the night in SD where protein degradation prevents CO accumulation. FT is directly regulated by CO and follows the clock-regulated expression pattern of CO in LD (Suarez-Lopez et al., 2001; Valverde et al., 2004). Regulation of FT expression can occur also independently of CO and the photoperiodic pathway such as due to decreased red to far red light ratios in the shade avoidance response (SAR). SAR is mediated through the key regulator of the light-quality pathway phytochrom B by post-transcriptional repression of the FT-activator PHYTOCHROME AND FLOWERING TIME 1 (PFT1) (Cerdan and Chory, 2003; Halliday and Whitelam, 2003; Bäckström et al., 2007). In several species, such as Arabidopsis, tomato, tobacco and rice, the FT protein or its homologs are known to move from leaves into the shoot apical meristem (SAM) where it induces the switch to flowering (Corbesier et al., 2007) by inducing the expression of the downstream targets SOC1 and APETALA 1 (AP1) (reviewed in Zeevaart, 2008).
The floral integrators FT, SOC1, and TSF are commonly repressed by the potent flowering repressor FLC. Vernalization or the autonomous pathway of floral promotion establish low FLC levels and thus favor flowering. In contrast to other flowering time pathways, the autonomous pathway does not represent a linear genetic pathway and involves RNA-binding proteins (FCA, FPA, FLK), RNA processing proteins (FY) and chromatin regulators (FVE/MSI4, FLD) (for review see Simpson, 2004).
The autonomous pathway gene FVE is needed to establish repressive chromatin at FLC and encodes a MULTICOPY SUPRESSOR OF IRA 1 (MSI1)-like chromatin-adaptor protein (Ausin et al., 2004; Kim et al., 2004; Jeon and Kim, 2011). MSI1-like proteins belong to a subfamily of WD-40 repeat proteins that are subunits in several chromatin remodeling complexes (for review see Hennig et al., 2005). The single Drosophila MSI1-like protein p55 is a core subunit of Polycomb Group Repressive Complex 2 (PRC2), of Chromatin Assembly Factor 1, of histone deacetylase complexes and of other chromatin-associated protein complexes (for review see: Hennig et al., 2005). In contrast to flies, Arabidopsis has five MSI1-like proteins (MSI1-5). While Arabidopsis MSI4 was suggested to act in histone deacetylation (Ausin et al., 2004; Kim et al., 2004; Jeon and Kim, 2011), Arabidopsis MSI1 was shown to be part of Chromatin Assembly Factor 1 and PRC2-like complexes (Kaya et al., 2001; Köhler et al., 2003; Exner et al., 2006; Schönrock et al., 2006a; De Lucia et al., 2008; Derkacheva et al., 2013). MSI1 functions in the FERTILISATION INDEPENDENT SEED (FIS)-PRC2 complex, which silences target genes during gametophyte and early seed development (Köhler et al., 2003; Guitton et al., 2004; Guitton and Berger, 2005); the VERNALIZATION (VRN)-PRC2 complex, which is required for epigenetic repression of FLC and acceleration of flowering by extended cold (De Lucia et al., 2008; Derkacheva et al., 2013) and the EMF-complex, which suppresses precocious flowering by repressing FT and AGL19 but which contributes also to repression of FLC (Yoshida et al., 2001; Schönrock et al., 2006a; Jiang et al., 2008). Because of the propeller-like structure of the WD40-domain, MSI1 and other MSI1-like proteins can possibly participate in additional chromatin-modifying complexes. Indeed, MSI1 was found to interact with LHP1 connecting plant PRC2 with LHP1 to establish repressive H3K27 methylation marks (Derkacheva et al., 2013). Further, MSI1 interacts with the CUL4-DDB1 complex and the Retinoblastoma-related protein to control imprinting in Arabidopsis (Jullien et al., 2008; Dumbliauskas et al., 2011). Chromatin-based mechanisms have recently emerged as a major means of control for many cellular processes including flowering time. In particular, the importance of chromatin-based regulation for control of FLC is well documented (Zografos and Sung, 2012).
Previously, we found that MSI1 represses drought stress responses (Alexandre et al., 2009) and is needed for timely flowering and for normal expression of SOC1 (Bouveret et al., 2006). Here we demonstrate that MSI1 functions during floral transition by establishing normal expression of the flowering activator CO and subsequently of the florigen FT and TSF. We also show that delayed up-regulation of gene expression of the floral integrator genes correlates with the delay in flowering in a msi1-mutant. Our data suggest that MSI1 is needed for the efficient activation of CO, thus allowing full activity of the photoperiodic pathway for floral induction.
Materials and methods
Plant material
Arabidopsis thaliana ecotype Columbia (Col) was used throughout this study. T-DNA insertion lines phyB (SALK_022035, Mayfield et al., 2007; Ruckle et al., 2007), pft1-2 (SALK_129555, Kidd et al., 2009), flc-6 (SALK_41126, Schönrock et al., 2006a) and msi1-5 (WiscDs Lox302B08) were obtained from NASC and confirmed by PCR. Seeds of FRI-Col, esd1-10, ft-10 and soc1-2 have been described (Lee and Amasino, 1995; Lee et al., 2000; Yoo et al., 2005; Martin-Trillo et al., 2006) and were kindly provided by J. Jarillo (FRI-Col, esd1-10), D. Weigel (ft-10), I. Lee (soc1-2), B. Ayres (co-1). The mutant co-1 (accession La-0, Redei, 1962) was backcrossed into Col. The line msi1-tap1 (accession Col) has been described before (Bouveret et al., 2006). Double mutants were identified among progeny of appropriate crosses by PCR with gene-specific primers (Supplementary Table 1).
To construct plants that ectopically overexpress FT (35S::FT), the full-length coding sequence was inserted into the binary destination vector pK7WG2 (Karimi et al., 2002) downstream of the cauliflower mosaic virus (CaMV) 35S promoter and transformed into msi1-tap1 plants. Transformants were selected on kanamycin plates and genotyped by PCR. Hemizygous T2-generation plants of three independent T1 lines were analyzed for flowering time.
Growth conditions and flowering time
Seeds were sterilized and plants were grown on Murashige and Skoog (MS) basal salt medium (Duchefa, Brussels, Belgium) after stratification at 4°C for 2–3 days. Plants were analyzed on plates or transferred to soil (“Einheitserde,” H. Gilgen optima-Werke, Arlesheim, Switzerland) 10 days after germination. Plants were kept in Conviron growth chambers with mixed cold fluorescent and incandescent light (130 μmol m−2 s−1, 21 ± 2°C) under (LD, 16 h light) or short-day (SD, 8 h light) photoperiods or were raised in green houses [LD: 14 h light, 19°C/10 h dark, 14°C; SD: 8 h light, 20°C/16 h dark, 20°C; supplemented with mercury vapor lamps (Sylvania Lighting S.A., Meyrin, Switzerland) to a maximum of 150 μmol m−2 s−1]. Flowering time was scored as described (Möller-Steinbach et al., 2010).
RNA isolation and quantitative RT-PCR (qRT-PCR)
Total RNA was extracted as previously described (Hennig et al., 2003; Leroy et al., 2007; Alexandre et al., 2009). 1 μ g RNA treated with DNase I (Promega, Dübendorf, Switzerland) was transcribed into cDNA using a RevertAid First Strand cDNA Synthesis Kit (Fermentas, Nunningen, Switzerland) according to manufacturer's instructions. qRT-PCR with gene-specific primers (Supplementary Table 2) was performed on three technical replicates with the Fast Start Universal Probe Master (Rox) reagent and the Universal Probe Library set (UPL) (Roche Diagnostics, Rotkreuz, Switzerland) according to the manufacturer's instructions and results were normalized to PP2A as described (Exner et al., 2009). Shown is one of at least two independent biological experiments with similar results.
Results
MSI1 functions independently of light quality
Previously, we had reported that MSI1 antisense lines and msi1 mutants partially complemented with untagged pMSI1::MSI1 or tagged pMSI1::MSI1:TAP constructs were late flowering (Bouveret et al., 2006). This suggests that undisturbed MSI1 levels are needed for normal flowering promotion. Homozygous msi1 null mutants are lethal (Köhler et al., 2003; Guitton et al., 2004). Here, we analyzed heterozygous plants of the original msi1-1 and a novel msi1-5 allele and found that both flowered later than wild type under LD but not SD (Supplementary Figure 1). Similarly, msi1-1−/− plants partially complemented with a pMSI1::MSI1:HA construct were late flowering (Supplementary Figure 1). Therefore the dose of MSI1 is important for flowering time. The observation of a late flowering phenotype for msi1-mutants and transgenic lines motivated us to investigate the genetic pathway(s) in which MSI1 acts to affect flowering. Because the flowering delay was considerably more severe for the msi1-1−/− pMSI1::MSI1:TAP (msi1-tap) line than for heterozygous msi1 mutants, we used the msi1-tap1 line in subsequent experiments. Unlike heterozygous msi1 mutants, msi1-tap1 flowered much later than wild type in SD (Bouveret et al., 2006). This is consistent with the generally milder late flowering phenotype of heterozygous msi1 mutants. The normal flowering in SD may suggest that a single wild-type MSI1 allele can largely suffice for normal MSI1 functionin SD. It remains to be tested whether MSI1 requirements are lower in SD or whether a potential compensatory mechanism can more efficiently up-regulate the remaining MSI1 allele in SD than in LD.
We then tested a potential function of MSI1 in the light quality pathway, which functions through phytochromes and independently of the circadian system. PHYB, PHYD, and PHYE repress FT expression and therefore flowering, with PHYB having the major role in this process (Kim et al., 2008). The PHYB target PFT1 was proposed to directly activate both, CO and FT expression, while it simultaneously acts as negative regulator of phytochrome signaling by inactivation of PHYB protein (Cerdan and Chory, 2003; Wollenberg et al., 2008). Null alleles of phyB and pft, which are early and late flowering, respectively, were introduced into the msi1-tap1 background and flowering time was analyzed in LD. The loss of PHYB in the phyB msi1-tap1 double mutant led to flowering with 15 rosette leaves (RL) in LD, which was intermediate between the phyB single mutant (4 RL) and msi1-tap1 (19 RL, Table 1). This result suggests an additive interaction between MSI1 and PHYB. The loss of PFT1 in the pft1 msi1-tap1 double mutant resulted in a synergistic delay in flowering (41 RL) compared to the pft and msi1-tap1 single mutants (14 and 17 RL respectively, Table 1), suggesting likewise independent effects of MSI1 and PFT1 in flowering promotion but likely on the same common targets. Together, these results propose a function of MSI1 independent of the light quality pathway in floral induction.
Table 1.
Flowering time of double mutants of msi1-tap1 and different flowering time mutants in LD.
| Rosette leaves | Days to bolting | ||
|---|---|---|---|
| 1 | Col | 12.1 ± 0.4 | 28.8 ± 0.6 |
| msi1-tap1 | 19.2 ± 0.7 | 36.5 ± 0.6 | |
| phyB | 4.3 ± 0.2 | 18.9 ± 0.3 | |
| phyB msi1-tap1 | 15.4 ± 0.6 | 30.9 ± 0.7 | |
| 2 | Col | 9.1 ± 0.2 | 26.5 ± 0.2 |
| msi1-tap1 | 17.4 ± 0.6 | 33.7 ± 0.5 | |
| pft1 | 14.4 ± 0.3 | 32.7 ± 0.5 | |
| pft1 msi1-tap1 | 41.3 ± 3.7 | 59.8 ± 3.4 | |
| 3 | Col | 7.3 ± 0.4 | 20.8 ± 0.9 |
| msi1-tap1 | 19.6 ± 1.4 | 40.1 ± 1.3 | |
| Col FRI | 72.7 ± 3.1 | 79.4 ± 2.7 | |
| ColFRI msi1-tap1 | 84.6 ± 3.1a | 143.8 ± 12.2a | |
| flc | 6.2 ± 0.1 | 21.0 ± 1.1 | |
| flc msi1-tap1 | 16.9 ± 0.8 | 34.3 ± 0.5 | |
| 4 | Col | 10.4 ± 0.5 | 27.8 ± 0.5 |
| msi1-tap1 | 19.9 ± 0.5 | 36.9 ± 0.5 | |
| esd1 | 5.0 ± 0.1 | 24.0 ± 0.0 | |
| esd1 msi1-tap1 | 11.9 ± 0.4 | 33.3 ± 0.3 | |
| 5 | Col | 7.1 ± 0.4 | 23.9 ± 0.6 |
| msi1-tap1 | 16.5 ± 0.8 | 39.0 ± 0.6 | |
| fca | 68.6 ± 2.5 | 75.9 ± 4.0 | |
| fca msi1-tap1 | 57.0 ± 1.1a | 289.6 ± 41a |
Shown are mean value ± SE (n ≥ 14).
Analysis of flowering time was stopped when ~1/3 of the plants had died before bolting. Number of rosette leaves and days until death or termination of the experiment are shown.
MSI1 functions independently of the floral repressor FLC on flowering
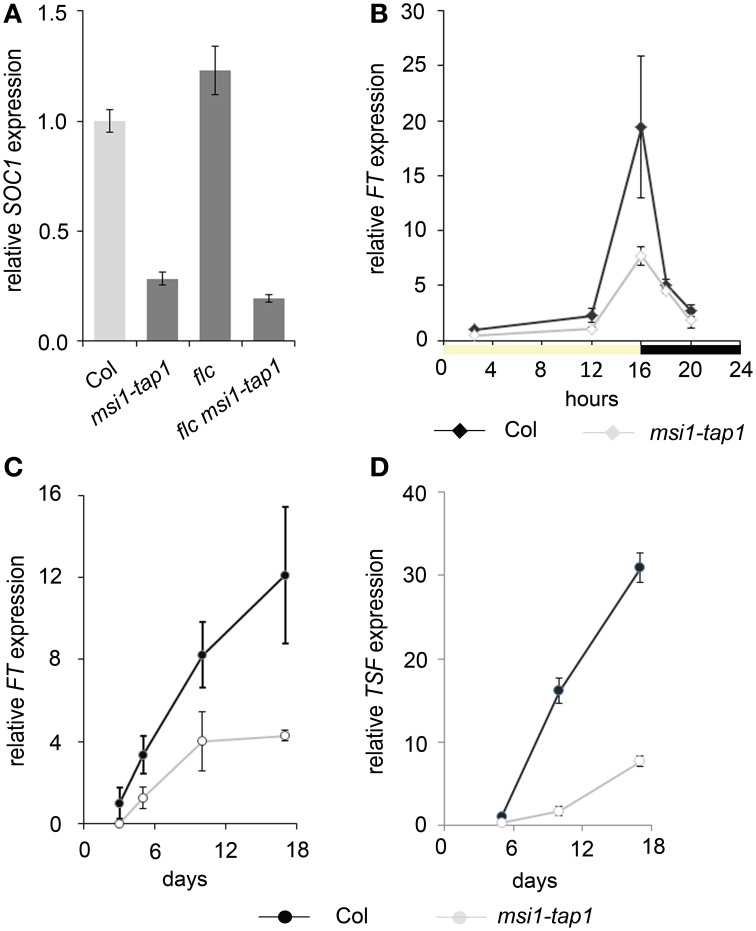
Next, we investigated the potential role of the flowering repressor FLC in MSI1 effects on flowering time. FLC is one of the main regulators of flowering time in Arabidopsis, and altered flowering time is often caused by altered FLC expression. Consistent with earlier observations (Bouveret et al., 2006), analysis of flc msi1-tap1 double mutants suggested that late flowering of msi1-tap1 was independent of FLC, as evident from the largely unaffected late flowering of msi1-tap1 flc plants (Table 1). Previously, we observed reduced SOC1 expression in msi1-tap1 (Bouveret et al., 2006). Here, we tested whether the reduced expression of SOC1 was independent of FLC. SOC1 expression in the double mutant was as low as in msi1-tap1, suggesting that the flc mutation could not lift SOC1 repression in msi1-tap1 plants (Figure 1A). Because MSI1 was recently shown to be involved in FLC control as part of plant PRC2 complexes (De Lucia et al., 2008; Derkacheva et al., 2013), we performed additional genetic tests of a potential role of FLC in msi1-tap1 late flowering. The active FRI-allele of the late flowering Arabidopsis accession San Feliu (Sf2) crossed into Columbia (Col FRI), was introgressed into msi1-tap1. As previously reported (Lee et al., 1993; Clarke and Dean, 1994), Col FRI flowered very late, possibly due to high FLC expression (Table 1). Col FRI msi1-tap1 plants flowered much later (85 RL) than either parent (73 and 20 RL for Col FRI and msi1-tap1, respectively). Some of the Col FRI msi1-tap1 plants were not able to flower at all and died without completing their life cycle. This additive delay in flowering suggests an independent role of MSI1 and FRI in flowering. Previously, we found a strongly synergistic interaction between MSI1 and FVE (Bouveret et al., 2006). FVE is part of the autonomous pathway, which represses FLC, and genes in this pathway were grouped in two epistasis groups. While FVE represents one of the two groups, FCA is a gene from the second group. Here, we tested the genetic interaction between MSI1 and FCA. The fca msi1-tap1 double mutants were extremely delayed in flowering. They ceased to produce leaves without starting to bolt or flower leading to a smaller rosette leave number than for fca. After an extended period of developmental inactivity they eventually died (Table 1). The strongly synergistic interaction suggests that MSI1 and FCA do not function in the same genetic pathway to control flowering time.
Figure 1.
Expression of SOC1, FT, and TSF is reduced in msi1-tap1 in LD. (A) SOC1-expression in 9-day-old seedlings of Col (light gray), msi1-tap1, flc, and flc msi1-tap1 (dark gray) in LD at EOD (end of day). (B) Diurnal expression pattern of FT in 10-day-old seedlings. X-axis represents hours after start of the light period (yellow box—light period, black box—dark period). (C,D) Gene expression kinetics of FT (C) and TSF (D) in Col and msi1-tap1 at the end of the day (EOD) at 5, 10, and 17 days after germination (DAG). Values are relative expression ± SE (n = 3). Black lines, Col; gray lines, msi1-tap1.
Another activator of FLC is EARLY IN SHORT DAYS1 (ESD1, also known as SUPPRESSOR OF FRIGIDA 3 and ACTIN RELATED PROTEIN 6). Mutations in ESD1 hasten flowering through reduced FLC expression in LD and SD (Martin-Trillo et al., 2006; Choi et al., 2007; Lazaro et al., 2008). An early flowering esd1 mutant allele was crossed into msi1-tap1. In LD, the esd1 msi1-tap1 double mutant flowered intermediate (12 RL) to both parents esd1 and msi1-tap1 (5 and 20 RL, respectively) disclosing an additive effect between ESD1 and MSI1 on flowering (Table 1). These data suggest that ESD1 and MSI1 function in separate genetic pathways. Together, these results firmly established that MSI1 can function independently of FLC to affect flowering time in LD.
Changes in MSI1 lead to reduced levels of FT and TSF
FT and its homolog TSF are activators of SOC1 (Yamaguchi et al., 2005; Yoo et al., 2005). Increased FT expression was found in msi1-tap1 suppressor mutants, which rescued the msi1-tap1 late flowering phenotype (Exner et al., 2009, 2010). In LD-grown wild-type Arabidopsis, CO activates FT at the end of the day (Suarez-Lopez et al., 2001; Yanovsky and Kay, 2002). To test whether the diurnal rhythm of FT was affected in msi1-tap1, FT expression was profiled throughout the light-dark cycle in seedlings (Figure 1B). In wild type, FT had its expression peak toward the end of the light and beginning of the dark period as previously reported. The FT expression in msi1-tap1 followed the same pattern as in wild type, but expression values were lower, especially at the end of the day (EOD), when expression of FT was reduced by up to 50%. Additionally, we tested whether MSI1 affected the temporal activation of FT or its homolog TSF (Figures 1C,D). Under LD conditions, FT and TSF levels increased steadily in wild type between 3 and 17 days. In msi1-tap1, FT transcripts started to accumulate similarly to wild type but the increase was much slower leading to considerably reduced FT levels. The accumulation of TSF transcripts was even stronger reduced in msi1-tap1 leading to 70% lower levels than in wild type at 17 days after germination. These results demonstrate that normal MSI1-function is needed for typical activation of FT and its homolog TSF in LD.
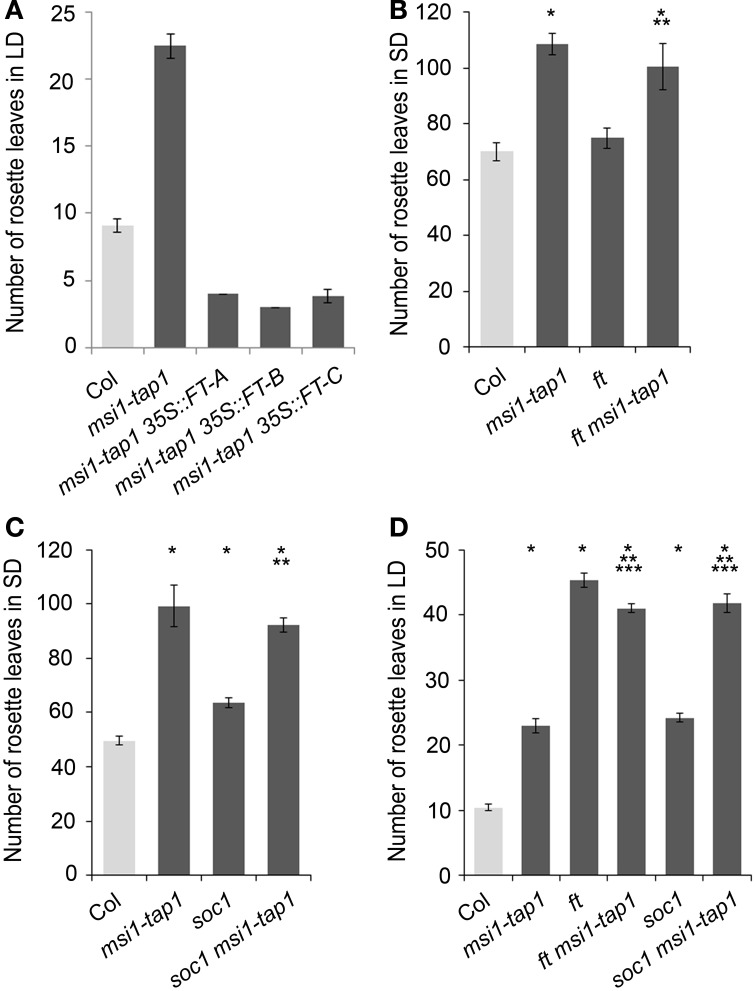
To test whether higher FT expression can be sufficient to suppress the late flowering phenotype of msi1-tap1, a 35S::FT transgene was introduced into msi1-tap1. The FT over-expression caused extremely early flowering (Figure 2A), which is consistent with the notion that reduced FT expression contributed to the late flowering of msi1-tap1.
Figure 2.
Flowering time analysis of msi1-tap1 and double mutants. (A) Flowering time of Col, msi1-tap1 and msi1-tap1 35S::FT lines in LD. The 35S::FT suppresses the late flowering of msi1-tap1. (B) Flowering time of Col, msi1-tap1, ft, and ft msi1-tap1 in SD. (C) Flowering time of Col, msi1-tap1, soc1, and soc1 msi1-tap1 in SD. (D) Flowering time of Col, msi1-tap1, ft, ft msi1-tap1, soc1 and soc1 msi1-tap1 in LD. Light gray bars represent wild type Col. Shown are means ± SE of rosette leaves (n ≥ 14). Significance of difference was tested using t-tests. Asterisks denote differences that were significant at p < 0.05 to WT (*), single mutant (**), or msi1-tap1 (***).
To substantiate that delayed activation of FT and therefore of SOC1 was responsible for the late flowering of msi1-tap1, a ft mutant allele was crossed into msi1-tap1 for flower time measurements. The double mutant soc1 msi1-tap1, which was already described in LD before (Bouveret et al., 2006), was included into the analysis (Figures 2B–D; Supplementary Figure 2). Under SD conditions, ft flowered similar to wild type, and the ft msi1-tap1 line flowered similar to msi1-tap1, confirming that FT does not play a major role under these conditions (Figure 2B) (Yanovsky and Kay, 2002; Corbesier et al., 2007). In contrast to FT, SOC1 functions in induction of flowering in SD (Borner et al., 2000) and the soc1 single mutant flowered later than wild type (Figure 2C). While the soc1 msi1-tap1 line needed longer until flowering than either parent, it produced a similar number of RL as the msi1-tap1 parent suggesting that delayed activation of SOC1 contributes at least partially to the late flowering of msi1-tap1 in SD. Thus, during flowering induction in SD, MSI1 and SOC1 appear to function partially in the same genetic pathway.
Under LD conditions, both ft and soc1 flowered later than wild type consistent with their roles in photoperiodic flowering (Borner et al., 2000). The double mutant soc1 msi1-tap1 exhibited an additive late flowering phenotype (42 RL) compared to the msi1-tap1 and soc1 parents (23 and 24 RL, respectively, Figure 2D) confirming earlier results (Bouveret et al., 2006). The ft msi1-tap1 line flowered with 42 RL similar to the ft parent (45 RL) supporting the notion that reduced FT expression is the main reason for late flowering of msi1-tap1 in LD (Figure 2D). In summary, MSI1 affects full activation of FT, TSF and SOC1 expression to promote timely flowering.
MSI1 function is connected to the photoperiod pathway
Because CO is a main activator of FT, SOC1, and TSF (Suarez-Lopez et al., 2001; Hepworth et al., 2002; Yamaguchi et al., 2005), we asked whether reduced expression of FT, SOC1, and TSF in msi1-tap1 was caused by defects in CO regulation. CO is under strong circadian and diurnal control (for review see Searle and Coupland, 2004), and CO expression in msi1-tap1 was tested throughout an entire light-dark cycle. This experiment revealed that CO expression was considerably lower in msi1-tap1 than in wild type (Figure 3A). The CO expression in wild type showed the previously reported peak toward the end of the day and beginning of the dark. Similarly, this expression pattern was observed for msi1-tap1 suggesting that diurnal regulation was not grossly altered. This conclusion was supported by normal diurnal cycling of CCA1 and TOC1, two components of the central circadian oscillator. However, under the tested conditions, CCA1 and TOC1 showed lower amplitudes of peak expression values in msi1-tap (Figure 3B). Further, we analyzed the CO transcript levels at different developmental time points until 17 days after germination (Figure 3C). Under our conditions, CO increased steadily in wild type during 10 days after germination. In msi1-tap1, CO transcripts started to accumulate similarly to wild type but the increase was slower leading to considerably reduced CO levels. Together, the expression data suggest the hypothesis that MSI1 affects expression of FT, TSF, and SOC1 and flowering time in LD via CO.
Figure 3.
Gene expression of CO and GI is changed in msi1-tap1 in LD. Diurnal gene expression of CO (A) and CCA1 and TOC1 (B) in 10-day-old seedlings in LD. X-axis represents hours after start of the light period (yellow box—light period, black box—dark period). Black lines, Col; gray lines, msi1-tap1. (C,D) Gene expression kinetics of CO (C) and GI (D) in msi1-tap1 at EOD at 5, 10, and 17 DAGs. Values are relative expression ± SE (n = 3).
To test genetically whether reduced CO expression was responsible for delayed flowering of msi1-tap1, a co mutant allele was introduced into the msi1-tap1 line. Consistent with earlier findings (Koornneef et al., 1991; Robson et al., 2001), the co mutant was late flowering in LD. While msi1-tap1 delayed flowering substantially in the CO wild-type background, it only slightly delayed flowering of a co mutant (Figure 4A) suggesting that late flowering in msi1-tap1 is caused mainly by effects on CO. The similar flowering time of ft msi1-tap1 and the ft co msi1-tap1 triple mutant (Figure 4A) further supported the notion of an epistatic genetic interaction between MSI1 and CO.
Figure 4.
MSI1 is needed for normal sensitivity of the photoperiod. (A) Flowering time of Col, msi1-tap1, co, co msi1-tap1, co ft msi1-tap1, and ft msi1-tap1 in LD expressed in mean number of RL ± SE (n ≥ 14). Significance of difference was tested using t-tests. Asterisks denote differences that were significant at p < 0.05 to WT (*) or co (**). (B) Flowering time of Col, msi1-tap1, and heterozygous msi1-1in SD and after single LD treatments. Plants were grown continuously in SD (light gray bars) or exposed to 1 or 3 LD (16 h light/8 h dark; dark gray and black columns, respectively) at 45 DAG. LD treatments were performed by extension of the light period and shortening of the following dark period. Flowering time is expressed in DTB ± SE (n ≥ 14). Asterisks denote significant differences to the corresponding SD-only control (t-test, p < 0.05).
Because GIGANTEA (GI) is a major activator of CO expression (Imaizumi et al., 2005; Sawa et al., 2007), we tested whether GI expression was altered in msi1-tap1. At 5 d after germination, when CO levels did not differ between WT and msi1-tap1, GI was also not affected (Figure 3D). In contrast, at 10 d and 17 d, not only CO but also GI expression was substantially reduced in msi1-tap1.
Together, these data suggest that MSI1 acts on flowering in response to the photoperiod through GI and CO on FT.
MSI1 is needed for normal sensitivity of the photoperiod pathway
To further test the importance of MSI1 in the photoperiodic pathway, we performed a SD-LD-SD shift experiment. In SD, FT is only very weakly activated due to immediate destabilization of CO protein after synthesis, and flowering is very much delayed. A brief LD experience can be sufficient to activate FT and induce flowering if the photoperiodic pathway functions normally (Corbesier et al., 1996; King et al., 2008). We cultivated plants under SD conditions interrupted by 1 or 3 days of LD at 45 days after germination and measured flowering time (Figure 4B). Under these conditions, Col was highly sensitive to the additional LD exposures, and flowering was accelerated by about 3 weeks. In contrast, the effect on flowering time of msi1-tap1 was minor and not statistically significant. Heterozygous msi1-1 mutants reacted like wild type to three additional LDs but showed a reduced response to a single additional LD (Figure 4B). Together, these results demonstrate that MSI1 is needed for normal sensitivity of the photoperiodic pathway.
Discussion
In plants, flowering at the right time is determined by several endogenous and external signals. One of the genes affecting flowering is MSI1. Late flowering was observed in lines expressing either tagged (TAP-, GFP-, HA) or untagged MSI1 under a 2 kb MSI1-promoter fragment in a msi1 mutant background (Bouveret et al., 2006; Alexandre et al., 2009; this work). In addition, MSI1-antisense lines and heterozygous msi1 mutants were late flowering, together establishing that normal MSI1 function is needed for normal timing of flowering. The MSI1-TAP construct did not affect flowering in WT plants nor did these plants in any other way differ from WT. Similarly, when a 35S::MSI1 construct was introduced into msi1-tap1, the late flowering was suppressed (Bouveret et al., 2006). Therefore, we consider it unlikely that the late flowering of msi1-tap1 plants was caused by a dominant negative effect of the fusion protein. Here, we used this line as a tool to dissect the function of the essential MSI1 gene in flowering time control.
One signal affecting flowering is light quality, which gives information about competition by neighboring plants and is sensed mainly by phytochromes, in particular PHYB (for review see Thomas, 2006). In light-quality sensing, PHYB functions via PFT1 both to activate CO and to activate FT in a CO-independent way (Cerdan and Chory, 2003; Iñigo et al., 2012a,b). In addition, PHYB has also functions in photoperiod sensing (for review see Thomas, 2006). Genetic interaction analysis between PHYB and MSI1 showed an additive flowering time phenotype suggesting a function of MSI1 independent from the light quality pathway to promote flowering. Similarly, PFT1 and MSI1 did not show an epistatic interaction, suggesting that both genes function in distinct genetic pathway. The finding that PFT1 and MSI1 showed a tendency for a synergistic genetic interaction with a greater than additive flowering delay, is consistent with the notion that both genes commonly affect CO and FT in flowering time control. Interestingly, MSI1 and PFT1 both affect not only flowering time but also drought stress responses (Alexandre et al., 2009; Elfving et al., 2011). Because PFT1 is a subunit of the Mediator complex (Bäckström et al., 2007), future studies should aim to test whether MSI1 and Mediator share direct targets.
In Arabidopsis, FLC is a major repressor of flowering and mutants deficient in FLC repression are often late flowering. FLC is repressed both by vernalization and also by the autonomous pathway to allow flowering even without vernalization (Baurle et al., 2007). Previously, it was shown that MSI1 functions both in the major FLC-dependent vernalization pathway and in a FLC-independent vernalization pathway that regulates AGL19 (Schönrock et al., 2006a; De Lucia et al., 2008; Derkacheva et al., 2013). Here we find that MSI1 can affect flowering independent of vernalization and of FLC. This conclusion is based on genetic interaction studies between (i) MSI1 and FLC, (ii) MSI1 and FRI, an FLC-activator (Michaels and Amasino, 2001), (iii) MSI1 and FCA or FVE, two FLC repressors from the autonomous pathway, and (iv) MSI1 and ESD1/SUF3/ARP6, a FLC-activator and putative subunit of the SWR1 complex (Martin-Trillo et al., 2006; Choi et al., 2007; Lazaro et al., 2008).
In summary, MSI1 affects flowering time independent of the light quality pathway and of FLC.
The late flowering of msi1-tap1 could be explained as a consequence of reduced expression of FT, SOC1, and TSF. Genetic interaction analysis showed epistatic effects of FT with MSI1, demonstrating that MSI1 functions through the main flowering time integrators to promote flowering. FT, which is a major activator of SOC1, is in turn activated by CO in the photoperiod pathway to promote flowering in LD (Putterill et al., 1995). Strict diurnal regulation of CO protein levels is controlled by several complex pathways coupled to the core circadian oscillator and light conditions (for review see Andres and Coupland, 2012). CO is repressed in the morning by PHYB and activated in the evening by GI. The diurnal expression pattern of CO appeared not significantly altered in msi1-tap1, where CO still shows an expression peak late in the day. The level of detectable CO mRNA, however, was substantially reduced in msi1-tap1. The lower abundance of CO mRNA is associated with reduced GI expression. Together, reduced GI expression in msi1-tap1 could cause the reduced CO expression that in turn delays activation of FT and eventually SOC1and could explain the delay in flowering.
Although GI is thought to function mainly by directly activating CO, GI can also directly activate FT and accelerate flowering in the absence of CO (Sawa and Kay, 2011). Notably, msi1-tap1 did not cause any further delay of ft mutants but could slightly delay co mutants. These observations are consistent with a model in which reduced GI expression in msi1-tap1 does not only affect flowering via reduced CO levels but also directly via compromising FT activation.
Here, we studied the role of MSI1 in flowering under LD conditions and identified its function upstream of the photoperiodic CO-FT module. However, MSI1 has also a function for flowering under SD conditions, and this is independent of FT. Flowering in SD depends on SOC1 (Borner et al., 2000). We found not only that SOC1 expression is reduced in msi1-tap1 plants but also that MSI1 and SOC1 show a genetic interaction in SD suggesting that under these conditions MSI1 affects flowering by contributing to normal SOC1 expression. It remains to be tested which other flowering time genes are affected by MSI1 and contribute to the late flowering phenotype of msi1-tap1 in SD.
This work and earlier studies have established that MSI1 affects flowering in multiple pathways (Figure 5). First, MSI1 represses flowering via its functions in the EMF-PRC2-complex to represses AGL19 prior to vernalization and FT prior to photoperiodic activation (Schönrock et al., 2006a; Jiang et al., 2008). Second, MSI1 favors flowering via its function in the VRN-PRC2 complex to repress FLC after vernalization (De Lucia et al., 2008; Derkacheva et al., 2013). Here, we have shown that MSI1 affects flowering in a third way—by contributing to CO expression MSI1 allows to rapidly respond to photoperiod. The relative importance of these diverse functions will depend on conditions, such as LD vs. SD or with or without vernalization treatment. Given that the histone adaptor MSI1 may be part of additional complexes, it is possible that MSI1 affects flowering in even other ways.
Figure 5.
Model of MSI1 function in flowering time control. Scheme of the different pathways MSI1 is acting in to affect flowering time. First, MSI1 represses flowering via its functions in the EMF-PRC2-complex to represses AGL19 prior to vernalization and FT prior to photoperiodic activation (Schönrock et al., 2006a; Jiang et al., 2008). Second, MSI1 favors flowering via its function in the VRN-PRC2 complex to repress FLC after vernalization (De Lucia et al., 2008; Derkacheva et al., 2013). Third, MSI1 affects flowering through the photoperiodic GI-CO-FT pathway.
Author contributions
Yvonne Steinbach conceived and carried out the experiments and analyzed the data. Yvonne Steinbach and Lars Hennig planned the study and wrote the manuscript. All authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mena Nater and André Imboden for help with flowering time experiments. For providing seeds we thank: J. Jarillo (FRI-Col, esd1-10), D. Weigel (ft-10), I. Lee (soc1-2), B. Ayres (co-1). We thank Drs. E. Truernit and K. Kölling for critical reading of the manuscript. We thank Prof. Sam Zeeman for continuous support and providing access to infrastructure. Yvonne Steinbach was supported by an SNF Marie-Heim-Vögtlin grant. This work was also supported by grants from the Carl-Tryggers-Foundation and the Knut-and-Alice-Wallenberg foundation to Lars Hennig.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00077/abstract
Characterization of a new msi1 allele and the transgenic msi1-HA lines. (A) Insertion site of the WiscDsLox302B08 T-DNA in the second exon of MSI1-gene locus (AT5G58230). (B) Scheme of the pMSI1::MSI1:HA transgene containing the MSI1 cDNA fused to a triple HA-tag at the C-terminus under control of 2 kb of the MSI1-promoter. (C) Flowering time of heterozygous msi1-1 and msi1-5 mutants in LD (gray bars) and SD (dark bars) in RL ± SE (n ≥ 14). (D) Flowering time of msi1-HA lines expressed in RL ± SE (n ≥ 14). (E) Seed abortion rate in heterozygous msi1-5 mutants and homozygous msi1-HA lines. Siliques of msi1-5 contained about 50% normal and 50% aborted seeds, similar to the embryo lethal phenotype of the msi1-1 mutant (Köhler et al., 2003). The pMSI1::MSI1-HA construct can rescue the seed abortion phenotype similar to pMSI1::msi1-tap1 (Bouveret et al., 2006).
Flowering time of msi1-tap1 and double mutants. (A-C) Flowering time in LD of (A) Col, msi1-tap1, soc1, soc1 msi1-tap1. (B) Col, msi1-tap1, ft, ft msi1-tap1. (C) Col, msi1-tap1, co, co msi1-tap1, ft co msi1-tap1 and ft msi1-tap1. (D,E) Flowering time in SD of (D) Col, msi1-tap1, soc1, soc1 msi1-tap1, (E) Col, msi1-tap1, ft, ft msi1-tap1. Values are shown in mean DTB ± SE (n ≥ 14).. Significance of difference was tested using t-tests. Asterisks denote differences that were significant at p < 0.05 to WT (*), of the double mutants to the appropriate single mutant (**) or msi1-tap1 (***).
Primers used for genotyping and cloning.
qRT-PCR primers used in this study. Shown are the forward and reverse primers with the appropriate Universal probe library (UPL) (Roche) probe number.
References
- Alexandre C., Möller-Steinbach Y., Schönrock N., Gruissem W., Hennig L. (2009). Arabidopsis MSI1 is required for negative regulation of the response to drought stress. Mol. Plant 2, 675–687 10.1093/mp/ssp012 [DOI] [PubMed] [Google Scholar]
- Andres F., Coupland G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639 10.1038/nrg3291 [DOI] [PubMed] [Google Scholar]
- Ausin I., Alonso-Blanco C., Jarillo J. A., Ruiz-Garcia L., Martinez-Zapater J. M. (2004). Regulation of flowering time by FVE, a Retinoblastoma-associated protein. Nat. Genet. 36, 162–166 10.1038/ng1295 [DOI] [PubMed] [Google Scholar]
- Bäckström S., Elfving N., Nilsson R., Wingsle G., Björklund S. (2007). Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26, 717–729 10.1016/j.molcel.2007.05.007 [DOI] [PubMed] [Google Scholar]
- Baurle I., Smith L., Baulcombe D. C., Dean C. (2007). Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science 318, 109–112 10.1126/science.1146565 [DOI] [PubMed] [Google Scholar]
- Borner R., Kampmann G., Chandler J., Gleissner R., Wisman E., Apel K., et al. (2000). A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 24, 591–599 10.1046/j.1365-313x.2000.00906.x [DOI] [PubMed] [Google Scholar]
- Bouveret R., Schönrock N., Gruissem W., Hennig L. (2006). Regulation of flowering time by Arabidopsis MSI1. Development 133, 1693–1702 10.1242/dev.02340 [DOI] [PubMed] [Google Scholar]
- Cerdan P. D., Chory J. (2003). Regulation of flowering time by light quality. Nature 423, 881–885 10.1038/nature01636 [DOI] [PubMed] [Google Scholar]
- Choi K., Park C., Lee J., Oh M., Noh B., Lee I. (2007). Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134, 1931–1941 10.1242/dev.001891 [DOI] [PubMed] [Google Scholar]
- Clarke J. H., Dean C. (1994). Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Mol. Gen. Genet. 242, 81–89 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Gadisseur I., Silvestre G., Jacqmard A., Bernier G. (1996). Design in Arabidopsis thaliana of a synchronous system of floral induction by one long day. Plant J. 9, 947–952 10.1046/j.1365-313X.1996.9060947.x [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033 10.1126/science.1141752 [DOI] [PubMed] [Google Scholar]
- De Lucia F., Crevillen P., Jones A. M., Greb T., Dean C. (2008). A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. U.S.A. 105, 16831–16836 10.1073/pnas.0808687105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkacheva M., Steinbach Y., Wildhaber T., Mozgova I., Mahrez W., Nanni P., et al. (2013). Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 32, 2073–2085 10.1038/emboj.2013.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbliauskas E., Lechner E., Jaciubek M., Berr A., Pazhouhandeh M., Alioua M., et al. (2011). The Arabidopsis CUL4–DDB1 complex interacts with MSI1 and is required to maintain MEDEA parental imprinting. EMBO J. 30, 731–743 10.1038/emboj.2010.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfving N., Davoine C., Benlloch R., Blomberg J., Brännström K., Müller D., et al. (2011). The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc. Natl. Acad. Sci. U.S.A. 108, 8245–8250 10.1073/pnas.1002981108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V., Aichinger E., Shu H., Wildhaber T., Alfarano P., Caflisch A., et al. (2009). The Chromodomain of LIKE HETERO-CHROMATIN PROTEIN 1 is essential for H3K27me3 binding and function during Arabidopsis development. PLoS ONE 4:e5335 10.1371/journal.pone.0005335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V., Alexandre C., Rosenfeldt G., Alfarano P., Nater M., Caflisch A., et al. (2010). A gain-of-function mutation of Arabidopsis cryptochrome1 promotes flowering. Plant Physiol. 154, 1633–1645 10.1104/pp.110.160895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V., Taranto P., Schonrock N., Gruissem W., Hennig L. (2006). Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development 133, 4163–4172 10.1242/dev.02599 [DOI] [PubMed] [Google Scholar]
- Guitton A. E., Berger F. (2005). Loss of function of MULTICOPY SUPPRESSOR OF IRA 1 produces nonviable parthenogenetic embryos in Arabidopsis. Curr. Biol. 15, 750–754 10.1016/j.cub.2005.02.066 [DOI] [PubMed] [Google Scholar]
- Guitton A. E., Page D. R., Chambrier P., Lionnet C., Faure J. E., Grossniklaus U., et al. (2004). Identification of new members of fertilisation independent seed polycomb group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131, 2971–2981 10.1242/dev.01168 [DOI] [PubMed] [Google Scholar]
- Halliday K. J., Whitelam G. C. (2003). Changes in photoperiod or temperature alter the functional relationships between phytochromes and reveal roles for phyD and phyE. Plant Physiol. 131, 1913–1920 10.1104/pp.102.018135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L., Bouveret R., Gruissem W. (2005). MSI1-like proteins: an escort service for chromatin assembly and remodeling complexes. Trends Cell Biol. 15, 295–302 10.1016/j.tcb.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Hennig L., Taranto P., Walser M., Schönrock N., Gruissem W. (2003). Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130, 2555–2565 10.1242/dev.00470 [DOI] [PubMed] [Google Scholar]
- Hepworth S. R., Valverde F., Ravenscroft D., Mouradov A., Coupland G. (2002). Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 21, 4327–4337 10.1093/emboj/cdf432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T., Schultz T. F., Harmon F. G., Ho L. A., Kay S. A. (2005). FKF1 F-Box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297 10.1126/science.1110586 [DOI] [PubMed] [Google Scholar]
- Iñigo S., Alvarez M. J., Strasser B., Califano A., Cerdán P. D. (2012a). PFT1, the MED25 subunit of the plant Mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. Plant J. 69, 601–612 10.1111/j.1365-313X.2011.04815.x [DOI] [PubMed] [Google Scholar]
- Iñigo S., Giraldez A. N., Chory J., Cerdán P. D. (2012b). Proteasome-mediated turnover of Arabidopsis MED25 is coupled to the activation of FLOWERING LOCUS T transcription. Plant Physiol. 160, 1662–1673 10.1104/pp.112.205500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo J. A., Piñeiro M. (2011). Timing is everything in plant development. The central role of floral repressors. Plant Sci. 181, 364–378 10.1016/j.plantsci.2011.06.011 [DOI] [PubMed] [Google Scholar]
- Jeon J., Kim J. (2011). FVE, an Arabidopsis homologue of the retinoblastoma-associated protein that regulates flowering time and cold response, binds to chromatin as a large multiprotein complex. Mol. Cells. 32, 227–234 10.1007/s10059-011-1022-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Wang Y., Wang Y., He Y. (2008). Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis polycomb repressive complex 2 components. PLoS ONE 3:e3404 10.1371/journal.pone.0003404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien P. E., Mosquna A., Ingouff M., Sakata T., Ohad N., Berger F. (2008). Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol. 6:e194 10.1371/journal.pbio.0060194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V. K., Ahn J. H., Dagenais N., Christensen S. K., Nguyen J. T., et al. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965 10.1126/science.286.5446.1962 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 10.1016/S1360-1385(02)02251-3 [DOI] [PubMed] [Google Scholar]
- Kaya H., Shibahara K. I., Taoka K. I., Iwabuchi M., Stillman B., Araki T. (2001). FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104, 131–142 10.1016/S0092-8674(01)00197-0 [DOI] [PubMed] [Google Scholar]
- Kidd B. N., Edgar C. I., Kumar K. K., Aitken E. A., Schenk P. M., Manners J. M., et al. (2009). The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21, 2237–2252 10.1105/tpc.109.066910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Hyun Y., Park J. Y., Park M. J., Park M. K., Kim M. D., et al. (2004). A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat. Genet. 36, 167–171 10.1038/ng1298 [DOI] [PubMed] [Google Scholar]
- Kim S. Y., Yu X., Michaels S. D. (2008). Regulation of CONSTANS and FLOWERING LOCUS T expression in response to changing light quality. Plant Physiol. 148, 269–279 10.1104/pp.108.122606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R. W., Hisamatsu T., Goldschmidt E. E., Blundell C. (2008). The nature of floral signals in Arabidopsis. I. Photosynthesis and a far-red photoresponse independently regulate flowering by increasing expression of FLOWERING LOCUS T (FT). J. Exp. Bot. 59, 3811–3820 10.1093/jxb/ern231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962 10.1126/science.286.5446.1960 [DOI] [PubMed] [Google Scholar]
- Köhler C., Hennig L., Bouveret R., Gheyselinck J., Grossniklaus U., Gruissem W. (2003). Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22, 4804–4814 10.1093/emboj/cdg444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Hanhart C. J., Van Der Veen J. H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66 10.1007/BF00264213 [DOI] [PubMed] [Google Scholar]
- Lazaro A., Gomez-Zambrano A., Lopez-Gonzalez L., Pineiro M., Jarillo J. A. (2008). Mutations in the Arabidopsis SWC6 gene, encoding a component of the SWR1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J. Exp. Bot. 59, 653–666 10.1093/jxb/erm332 [DOI] [PubMed] [Google Scholar]
- Lee H., Suh S. S., Park E., Cho E., Ahn J. H., Kim S. G., et al. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376 10.1101/gad.813600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Amasino R. M. (1995). Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol. 108, 157–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Bleecker A., Amasino R. (1993). Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol. Gen. Genet. 237, 171–176 [DOI] [PubMed] [Google Scholar]
- Leroy O., Hennig L., Breuninger H., Laux T., Köhler C. (2007). Polycomb group proteins function in the female gametophyte to determine seed development in plants. Development 134, 3639–3648 10.1242/dev.009027 [DOI] [PubMed] [Google Scholar]
- Martin-Trillo M., Larazo A., Poethig R. S., Gomez-Mena C., Piñeiro M. A., Martinez-Zapater J. M., et al. (2006). EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development 133, 1241–1252 10.1242/dev.02301 [DOI] [PubMed] [Google Scholar]
- Mayfield J. D., Folta K. M., Paul A. L., Ferl R. J. (2007). The 14-3-3 Proteins μ and υ influence transition to flowering and early phytochrome response. Plant Physiol. 145, 1692–1702 10.1104/pp.107.108654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D., Amasino R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D., Amasino R. M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant. Cell 13, 935–941 10.1105/tpc.13.4.935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller-Steinbach Y., Alexandre C., Hennig L. (2010). Flowering time control. Methods Mol. Biol. 655, 229–237 10.1007/978-1-60761-765-5_15 [DOI] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857 10.1016/0092-8674(95)90288-0 [DOI] [PubMed] [Google Scholar]
- Redei G. P. (1962). Supervital mutants of Arabidopsis. Genetics 47, 443–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F., Costa M. M., Hepworth S. R., Vizir I., Pineiro M., Reeves P. H., et al. (2001). Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28, 619–631 10.1046/j.1365-313x.2001.01163.x [DOI] [PubMed] [Google Scholar]
- Ruckle M. E., Demarco S. M., Larkin R. M. (2007). Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19, 3944–3960 10.1105/tpc.107.054312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S. E., Ditta G. S., Schwarz-Sommer Z., Yanofsky M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive develop-ment of Arabidopsis. Science 288, 1613–1616 10.1126/science.288.5471.1613 [DOI] [PubMed] [Google Scholar]
- Sawa M., Kay S. A. (2011). GIGANTEA directly activates FLOWERING LOCUS T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 108, 11698–11703 10.1073/pnas.1106771108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M., Nusinow D. A., Kay S. A., Imaizumi T. (2007). FKF1 and GIGANTEA complex formation is required for Day-Length measurement in Arabidopsis. Science 318, 261–265 10.1126/science.1146994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönrock N., Bouveret R., Leroy O., Borghi L., Köhler C., Gruissem W., et al. (2006a). Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev. 20, 1667–1678 10.1101/gad.377206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönrock N., Exner V., Probst A., Gruissem W., Hennig L. (2006b). Functional genomic analysis of CAF-1 mutants in Arabidopsis thaliana. J. Biol. Chem. 281, 9560–9568 10.1074/jbc.M513426200 [DOI] [PubMed] [Google Scholar]
- Searle I., Coupland G. (2004). Induction of flowering by seasonal changes in photoperiod. EMBO J. 23, 1217–1222 10.1038/sj.emboj.7600117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G. G. (2004). The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant Biol. 7, 570–574 10.1016/j.pbi.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Srikanth A., Schmid M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68, 2013–2037 10.1007/s00018-011-0673-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Lopez P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120 10.1038/35074138 [DOI] [PubMed] [Google Scholar]
- Thomas B. (2006). Light signals and flowering. J. Exp. Bot. 57, 3387–3393 10.1093/jxb/erl071 [DOI] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006 10.1126/science.1091761 [DOI] [PubMed] [Google Scholar]
- Wollenberg A. C., Strasser B., Cerdan P. D., Amasino R. M. (2008). Acceleration of flowering during shade avoidance in Arabidopsis alters the balance between FLOWERING LOCUS C-mediated repression and photoperiodic induction of flowering. Plant Physiol. 148, 1681–1694 10.1104/pp.108.125468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A., Kobayashi Y., Goto K., Abe M., Araki T. (2005). TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 46, 1175–1189 10.1093/pcp/pci151 [DOI] [PubMed] [Google Scholar]
- Yanovsky M. J., Kay S. A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419, 308–312 10.1038/nature00996 [DOI] [PubMed] [Google Scholar]
- Yoo S. K., Chung K. S., Kim J., Lee J. H., Hong S. M., Yoo S. J., et al. (2005). CONSTANS Activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 139, 770–778 10.1104/pp.105.066928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N., Yanai Y., Chen L., Kato Y., Hiratsuka J., Miwa T., et al. (2001). EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13, 2471–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart J. A. (2008). Leaf-produced floral signals. Curr. Opin. Plant Biol. 11, 541–547 10.1016/j.pbi.2008.06.009 [DOI] [PubMed] [Google Scholar]
- Zografos B. R., Sung S. (2012). Vernalization-mediated chromatin changes. J. Exp. Bot. 63, 4343–4348 10.1093/jxb/ers157 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of a new msi1 allele and the transgenic msi1-HA lines. (A) Insertion site of the WiscDsLox302B08 T-DNA in the second exon of MSI1-gene locus (AT5G58230). (B) Scheme of the pMSI1::MSI1:HA transgene containing the MSI1 cDNA fused to a triple HA-tag at the C-terminus under control of 2 kb of the MSI1-promoter. (C) Flowering time of heterozygous msi1-1 and msi1-5 mutants in LD (gray bars) and SD (dark bars) in RL ± SE (n ≥ 14). (D) Flowering time of msi1-HA lines expressed in RL ± SE (n ≥ 14). (E) Seed abortion rate in heterozygous msi1-5 mutants and homozygous msi1-HA lines. Siliques of msi1-5 contained about 50% normal and 50% aborted seeds, similar to the embryo lethal phenotype of the msi1-1 mutant (Köhler et al., 2003). The pMSI1::MSI1-HA construct can rescue the seed abortion phenotype similar to pMSI1::msi1-tap1 (Bouveret et al., 2006).
Flowering time of msi1-tap1 and double mutants. (A-C) Flowering time in LD of (A) Col, msi1-tap1, soc1, soc1 msi1-tap1. (B) Col, msi1-tap1, ft, ft msi1-tap1. (C) Col, msi1-tap1, co, co msi1-tap1, ft co msi1-tap1 and ft msi1-tap1. (D,E) Flowering time in SD of (D) Col, msi1-tap1, soc1, soc1 msi1-tap1, (E) Col, msi1-tap1, ft, ft msi1-tap1. Values are shown in mean DTB ± SE (n ≥ 14).. Significance of difference was tested using t-tests. Asterisks denote differences that were significant at p < 0.05 to WT (*), of the double mutants to the appropriate single mutant (**) or msi1-tap1 (***).
Primers used for genotyping and cloning.
qRT-PCR primers used in this study. Shown are the forward and reverse primers with the appropriate Universal probe library (UPL) (Roche) probe number.