Abstract
Tripartite motif (TRIM) proteins have been implicated in multiple cellular functions, including antiviral activity. Research efforts so far indicate that the antiviral activity of TRIMs relies, for the most part, on their function as E3-ubiquitin ligases. A substantial number of the TRIM-family members have been demonstrated to mediate innate immune cell signal transduction and subsequent cytokine induction. In addition, a subset of TRIMs has been shown to restrict viral replication by directly targeting viral proteins. Although the body of work on the cellular roles of TRIM E3 ubiquitin ligases has rapidly grown over the last years, many aspects of their molecular workings and multi-functionality remain unclear. The antiviral function of many TRIMs seems to be conferred by specific isoforms, sub-cellular localization, and in cell-type specific contexts. Here we review recent findings on TRIM antiviral functions, current limitations and an outlook for future research.
Keywords: tripartite motif, E3-ubiquitin ligase, innate immunity, restriction factors, interferon, ubiquitin, antiviral response
Introduction
In mammals the immune response is comprised of both innate and adaptive mechanisms. The innate response is the first line of defense against incoming pathogens and is crucial for controlling infection in the time it takes to mount an effective adaptive response 1. Critical innate immune responses against viruses include constitutively expressed proteins with intrinsic anti-microbial properties as well as the inducible type I interferon (IFN-I) system [2], [3], and [4]. The inducible antiviral response is initiated when pathogen associated molecular patterns (PAMPs) are recognized by pattern recognition receptors (PRRs), including Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) NOD-like receptors (NLRs) and C-type lectin receptors (CLRs) 5. Engagement of PRRs triggers downstream signaling pathways through different adaptor proteins that transmit downstream signals converging at the level of the IκB (IKK) and IKK-related kinases [6] and [7].
The classical IKKs (IKKα/β) are responsible for the activation of nuclear factor kappa B (NF-κB) and subsequent pro-inflammatory cytokine synthesis [8] and [9], whereas the IKK-related kinases (TBK1/IKKε) phosphorylate the transcription factors IFN-regulatory factor (IRF) 3 and IRF7 required for IFN-I production [10] and [11]. Upon binding of IFN-I to its receptor, activation of JAK1 and TYK2 kinases results in phosphorylation of the transcription factors STAT1 and STAT2 to form a complex with IRF-9 known as IFN-stimulated gene factor 3 (ISGF3) [4] and [12]. This complex translocates to the nucleus and binds IFN-stimulated response elements (ISREs), ultimately resulting in the expression of a large set of IFN inducible genes (ISGs) which can directly interfere with the viral replication cycle [13] and [14].
TRIM proteins as immune regulators
The tripartite motif (TRIM) proteins constitute a family in humans of over 70 distinct protein members. They derive their name from the fact that they share three conserved N-terminal domains: a Really Interesting New Gene (RING) domain, one or two B-Boxes (B1/B2) and a coiled-coil (CC) domain (Figure 1) [15], [16], [17], and [18]. This tripartite motif is often referred to as the RBBC. Members of the TRIM family have been long predicted to be part of innate immune pathways. One of the main underlying notions for this prediction is the fact that the number of TRIMs has rapidly expanded very recently in evolution [19], [20], [21], [22], [23], and [24]. The large number of TRIM genes in higher eukaryotes and the sequence homology shared by its members suggests a rapid evolution of this family by gene duplications [19], [21], and [25].
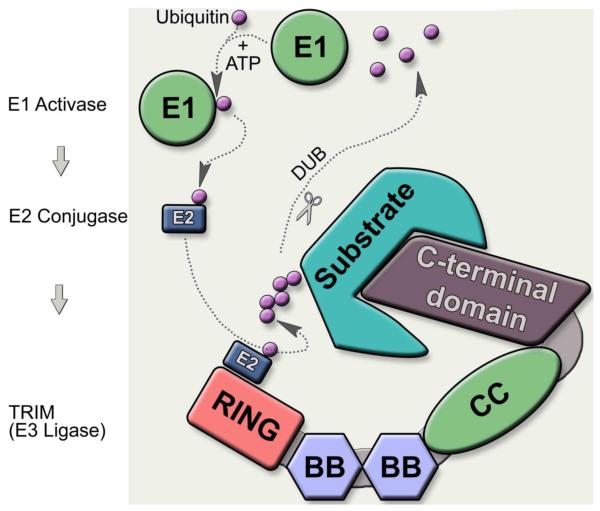
Figure 1. Model of TRIM E3-ubiquitin ligase function.
Ubiquitin conjugation requires an E1 activating enzyme in the presence of ATP and mono-ubiquitin. The E2-conjugating enzyme then forms an intermediate thioester with ubiquitin. TRIMs act as E3-ligases and confer specificity to the reaction. TRIMs recognizing the E2 through the RING domain and interact with the substrate, in general, through the C-terminal region. Deubiquitinases (DUB) hydrolyze poly-ubiquitin chains which are then recycled.
The evolutionary time frame of this expansion coincided with the emergence of traits specific for the adaptive immune system, suggesting that TRIM proteins may have evolved as an integral part of the machinery to regulate the increasingly complex immune system and fine tune cross-talk between innate and adaptive immune branches. For comparison, while humans have 73 TRIM genes, fruit flies have only seven 19. Interestingly, jawed fish who have very well-developed innate immune systems, also have many TRIM genes (in most species >100-120 genes) [22], [23] and [24]. In contrast to higher mammalian species, fish are free-living organisms from early embryonic stages and for that reason very heavily rely on their robust innate immune system for survival 26. In line with the notion that TRIM proteins may be important components of the immune system, recent studies have shown that an increasing number of TRIMs can mediate antiviral activity. TRIM proteins with these demonstrated immune functions did exert their function either by directly interfering with key steps in viral life cycles or indirectly as regulators of antiviral cell signaling [19], [25], [27], [28] and [29].
However, TRIM proteins do not merely have immune-related functions. In fact, many TRIM proteins were shown to be involved in a wide range of molecular functions, ranging from transcriptional regulation to post-translational modification in the context of various cellular processes such as apoptosis, cell differentiation, development, oncogenesis, etc. 30. Interestingly, several TRIM proteins have already been implicated in more than one cellular process, indicating that like other proteins, some of them may be multi-functional and/or fulfilling cell-type specific functions. In line with this notion, the majority of TRIMs seem to be non-ubiquitously expressed in different cell types at the mRNA level [31] and [32]. Moreover, for most TRIMs multiple alternatively spliced mRNAs have been reported 29, suggesting that different protein isoforms may add to additional diversity in regulation, cell specificity and protein function.
What unites all TRIMs is the fact that their domain organization and structural homology are predicted to confer ligating activity for ubiquitin and ubiquitin-like post-translational modifiers. Most of the reported cellular functions of TRIM proteins suggest that the ability to catalyze ubiquitin is an important functional requirement, including for immune regulation.
TRIM proteins as E3-ubiquitin ligases
The conserved RBBC domains in TRIM proteins suggest that this minimal structure was selectively maintained to carry out a function as ligating enzymes of the post-translational modifier ubiquitin. Ubiquitin (Ub) is a conserved 76 amino acid protein important in a wide variety of cellular functions. The free C-terminal glycine residue of ubiquitin can be conjugated to lysine residues of specific substrate proteins 33. In turn, Ub itself contains seven lysines (K6, K11, K27, K29, K33, K48, K63) on which poly-ubiquitin chains can be formed when the C-terminal glycine residue of one ubiquitin molecule is conjugated to a lysine residue of another ubiquitin molecule.
Ubiquitin chains linked through different lysines have specific cellular functions 34. Proteins covalently modified with lysine 48 (K48)-linked poly-ubiquitin are usually targeted for degradation by the proteasome. In contrast, protein modification with K63-linked poly-ubiquitin is involved in activation of antiviral signaling pathways 34. In addition, unanchored K63-linked poly-ubiquitin chains have also been proposed to activate kinases involved in signaling pathways in a proteasomal degradation-independent manner [35] and [36]. Like all post-translational modifications, the process of ubiquitin-conjugation can be reversed. Mono-ubiquitin and poly-ubiquitin chains can then be removed from the target protein by deubiquitinases (DUBs) which are critical for the dynamic regulation of the protein ubiquitination process (Figure 1).
Ubiquitin conjugation requires an E1 activating enzyme and ATP as the energy source, an E2-conjugating enzyme and an E3-ligase, which confers specificity by transferring ubiquitin to the target protein (Figure 1). It is believed that the RING domain of TRIM proteins confers E3 ligase activity by facilitating interaction with E2 enzymes, while determining target specificity through their unique C-terminal domains (Figure 1). A recent screen of interactions between 26 E2-conjugases and 42 TRIM proteins by a yeast two hybrid system 37 suggested that TRIMs have a preference for the UBE2D and UBE2E classes of E2 conjugases. However, this study did not identify the E2 enzymes UBC13/UEV1A and UBC5C previously reported to play critical roles in immune signaling by TRIM5 and TRIM25, respectively [36] and [38]. It is well known that E2/E3 interactions are often transient and notoriously challenging to detect. Hence, although the UBE2D/E enzymes may be often used by various TRIM proteins, other relevant combinations exist and may require special techniques to identify.
So far, the majority of TRIMs that have been characterized in detail have been found to promote either K48- or K63-linked covalent poly-ubiquitination of proteins. This could be in part due to the current lack of reagents available to identify other types of poly-ubiquitin linkages. Recent work has also shown that TRIMs can catalyze the synthesis of unanchored (non-covalently-bound) K63-linked poly-ubiquitin chains (TRIM5 38, TRIM25 36). Since the type of poly-ubiquitin linkage maybe defined by the E2 conjugase 33, it will be interesting to see if future studies will identify TRIMs involved in the synthesis of other types of poly-ubiquitin linkages and therefore involved in yet unidentified functions.
In addition to a role as ubiquitin E3 ligases, several TRIM proteins have been reported to also function as E3 ligases for other ubiquitin-like molecules such as SUMO and the IFN-inducible protein ISG15 (e.g. TRIM28 and TRIM25 [39] and [40]). Little is known about how conjugation of different ubiquitin-like molecules by the same TRIM proteins is regulated and what functional implications this has for protein regulation. The interesting possibility exists that ubiquitin and other ubiquitin-like modifiers compete for conjugation on the same target proteins, which would allow for an additional layer to manipulate the magnitude and direction of e.g. cell signaling. The existence and relevance of this possibility remains however to be determined.
Structure of TRIM proteins
The RING domain, which is composed of 40-60 amino acids, is a cysteine-histidine-rich domain that binds two zinc atoms in a unique cross-braced metal ligation scheme [17] and [41]. It is generally accepted that the RING domain confers E3-ubiquitin ligase activity by specifically interacting with and promoting E2-dependent ubiquitin conjugation [30] and [42]. However, some studies have also shown that this domain is involved in additional protein-protein interactions [42] and [43]. A large number of TRIMs with antiviral functions require the RING domain for their antiviral activity, either for direct ubiquitination of viral products or for innate immune signaling [28], [29], [38], [44], [45], [46] and [47].
The B-Box domain is also a cysteine-histidine zinc-binding motif and are found exclusively in TRIM proteins [30] and [48]. The molecular structures of the B1 and B2 domains of TRIM18/MID1 were recently solved and showed striking conserved structural features with the RING domain [49] and [50]. Although much remains unknown about B-Box function and structure in other TRIM proteins, this may suggest that the B-Box could also confer E3 ubiquitin ligase activity 50. In support of this hypothesis, TRIM16 (also called or estrogen-responsive B box protein; EBBP), which does not contain a RING domain, was found to have auto-ubiquitination activity 51.
The coiled-coil (CC) domain, a hyper-helical region predicted by bioinformatics, has been shown to be necessary and sufficient for homo-dimerization/oligomerization in a large number of TRIM proteins [52], [53] and [54]. In addition, the formation of heterodimers may add diversification to their biological functions 55. Mutation and deletion experiments have revealed that the CC domain may also be important for the formation of sub-cellular structures including cytoplasmic or nuclear bodies 54.
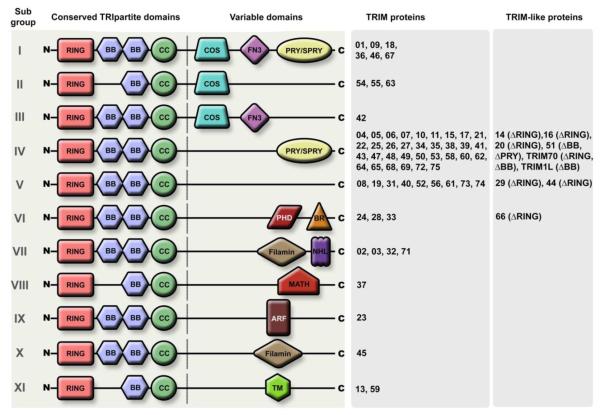
The N-terminal RBCC feature is followed by one or more specific C-terminal domains which cluster the TRIMs in eleven sub-groups and can determine function by recruiting unique functional partners (Figure 2) 25. The most common C-terminal TRIM domain consists of so-called PRY-SPRY motifs and is often referred to as the B30.2 domain. This domain has been proposed to be involved in protein-protein interactions and/or RNA binding 25.
Figure 2. TRIM subgroup classification.
TRIM members are classified based on their C-terminal domain composition as defined by Ozato et al 19. Adapted from Versteeg et al. 29
Interestingly, the B30.2 domain containing TRIMs expanded most dramatically in recent evolution, suggesting that this domain has conferred a strong selective advantage 20. In this context, the B30.2 domain has been implicated in the ability of TRIMs to restrict the replication of certain viruses [25] and [56]. Experimental evidence is consistent with a broad role for the B30.2 domain in innate immune recognition of retroviruses. In particularly TRIM5α has been implicated to be an important species-specific restriction factor for retroviruses 3. Both B30.2 mutagenesis studies, as well as sequence analysis of TRIM5α from related primates suggest that the differences in antiviral activities are defined by patches in the B30.2 domain 57.
Direct antiviral function of TRIM proteins
TRIM5α as a retroviral restriction factor
The short arm of human chromosome 11 contains a genetic hot-spot on which the following TRIM genes are localized: TRIM3/5/6/21/22/34/66/68 (Figure 3). The genes at this chromosomal position have been under heavy positive selective pressure and several have been identified as critical viral restriction factors, the best studied of which is TRIM5α in the context of retroviral infection. Over the years, the TRIM family members have been increasingly recognized as potential viral restriction factors. This was initiated by the discovery that African green monkeys and macaques specifically restrict HIV-1 infection through their TRIM5α protein 58. Since then other members of the TRIM family have also been found to have some antiviral function. This has led to the suggestion that the entire family of TRIMs may be a component of an innate or intrinsic immune response to viruses [3], [25], [59], [60] and [61]. Most of the viruses that have been studied and found to be affected by TRIM proteins are retroviruses. It remains however unclear whether this is because these viruses have provided a constant and strong evolutionary pressure or whether these viruses are simply the most widely studied in this context [25], [62] and [63].
Figure 3. TRIMs located on human chromosome 11.
Schematic representation of a cluster of TRIMs closely related to TRIM5 that map to human chromosome 11. The TRIMs with known antiviral activity are indicated.
The restriction activity of TRIM proteins can take place at different stages of viral replication including viral entry, transcription of viral genes or viral release from the cells (Figure 4) [25] and [63]. The direct antiviral role requires a direct interaction between the TRIM protein and a target viral protein resulting in interference of viral function. As a result, viral restriction factors show evidence of evolutionary positive selection at the binding sites 64.
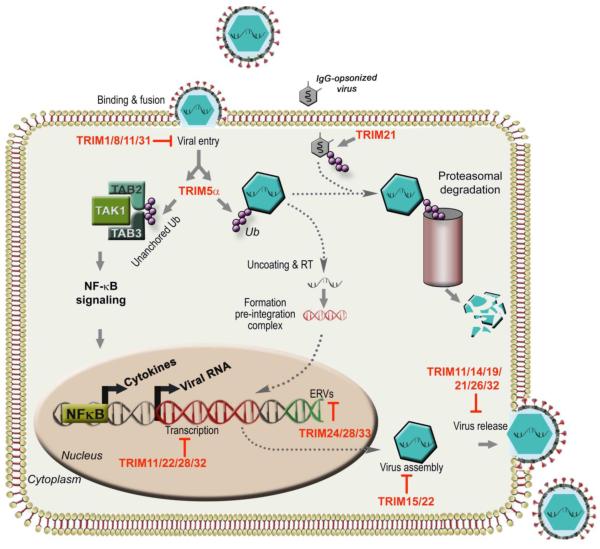
Figure 4. Direct antiviral activity of TRIMs.
Stages of retroviral replication cycle targeted by different TRIMs. TRIM5α blocks HIV-1 replication by targeting the capsid and preventing uncoating of the virus. At the same time, TRIM5α has been proposed to stimulate innate immune signaling upon capsid recognition. By catalyzing the synthesis of unanchored K63-linked poly-ubiquitin chains that bind and activate TAK1-dependent NF-κB. TRIM1/8/11/31 inhibit entry, between uncoating and viral gene integration. TRIM24/28/33 inhibit endogenous retrovirus (ERV) gene transcription. TRIM11/22/28/32 have been reported to inhibit retrovirus gene transcription. TRIM11/14//19/21/26/32 inhibit release and TRIM15/22 inhibit assembly. TRIM21 recognizes and targets antibody-opsonized viruses for proteasomal degradation.
One of the best characterized TRIMs with direct antiviral activity is TRIM5α, which is the largest isoform encoded by the TRIM5 gene and the only one containing the B30.2 domain required for its anti-HIV-1 function [56] and [65]. The human TRIM5 gene was first shown to encode a restriction activity to the N-tropic form of Murine leukemia virus (N-MLV) and was initially named Fv1-like as a result of its similarity to the restriction activity previously described for the murine Friend-virus-susceptibility factor 1 (Fv1) 66. Later studies showed that TRIM5α restriction is species-specific since African green monkey TRIM5α could restrict HIV-1, HIV-2, EIAV (equine infectious anemia virus), N-MLV and SIVmac while TRIM5α from macaques was only able to restrict HIV-1 [25], [58], [67], [68], [69], [70] and [71].
The detailed molecular mechanism of TRIM5α viral restriction is still not well understood. Nevertheless, it is clear that the inhibition occurs at early stages, immediately after entry in the cells and before reverse transcription [58] and [72]. Electron microscope imaging studies suggested that TRIM5α binds to the HIV-1 capsid in a hexagonal lattice and mimics the surface of the viral capsid [73], [74] and [75].
TRIM5α has E3 ubiquitin ligase activity and can be auto-ubiquitinated [76] and [77] leading to a rapid proteasome-dependent degradation 78. Therefore, TRIM5α turnover by the proteasome may target virions for degradation since TRIM5α complexed with HIV-1 virions has been found associated with proteasomal subunits 79. However, other studies reported that proteasome inhibitors did not rescue HIV-1 infectivity and TRIM5-mediated ubiquitination of HIV-1 capsid has not been detected 75. Alternatively, TRIM5α may promote rapid uncoating of incoming HIV-capsids 62, since expression of restrictive TRIM5α correlates with a decrease in the amount of particulate capsid in the cytosol 80. This rapid uncoating of the viral capsid results in inhibition of viral reverse transcription 81. Others have proposed that TRIM5α-mediated restriction depends on events that occur before capsid disassembly. Their data indicated that different retroviral core components could be either disassembled or degraded by the proteasome, and this could be blocked by proteasome inhibition without rescuing infectivity 82.
Although the precise mechanism of TRIM5α-mediated restriction may still not be fully understood, it is now clear that TRIM5α plays multiple roles in direct inhibition of viral replication. Interestingly, an additional role of TRIM5α as pattern-recognition receptor for the HIV-1 capsid lattice with the associated ability to trigger NF-κB signaling was recently proposed 38. Evidence of TRIM5-mediated restriction also comes from genetic studies showing constant genetic changes and evolutionary positive selective pressure.
In this regard, the TRIM5 gene and its closely related homologue TRIM22 are not found in rodents, and reciprocally its phylogenetically related homologues Trim12 and Trim30 are only found in mice but not in humans 83. Although TRIM5 does not exist in rodents, a cluster of closely related genes which includes Trim6/12/30/34, has evolved under positive selection in both primates and rodents suggesting important antiviral roles of the TRIMs in this cluster in both primates and rodents 84. In line with this hypothesis, these highly homologous TRIMs (Trim6/21/30/34) were found to be highly up-regulated upon virus infection in an IFN-I-dependent manner in murine macrophages and dendritic cells 85, suggesting that they play a role in pathogen restriction. Although these murine TRIMs did not show antiviral activity against a panel of retroviruses which included HIV-1, SIVmac, N-MLV, B-MLV, FIV, EIAV, MPMV 84, it remains to be seen if endogenous retroviruses or other species-restricted viruses maybe targeted by these murine TRIMs.
Additional genetic analysis shows that the primate TRIM5/6/22/34 cluster might have evolved by tandem gene duplications. TRIM5 and TRIM22 have been under strong positive selection in primates and gone through more dynamic genetic variations including complete loss in other species, whereas TRIM6 and TRIM34 have remain more static during evolution [57] and [86]. The positive selection on TRIM5 is more evident in the B30.2 domain since a single amino acid change in the human TRIM5α confers ability to restrict HIV-1, indicating that small changes during evolution have had a great impact on cross-species infection 56.
Therefore, the ability of TRIM5α to target the capsids of specific retroviruses is dependent on sequences in the TRIM5α B30.2 domain [56] and [65]. This B30.2 domain is present in almost all primates and has been under high positive selective pressure to restrict retroviruses in a species-specific manner. Interestingly, owl monkeys and certain macaque species have gained retrotranspositional insertions of a cyclophilin A (CypA) pseudogene in their TRIM5 gene 87. Since cellular CypA interacts with the retroviral capsid, the TRIM-CypA protein interacts efficiently with incoming retroviral capsids, resulting in potent viral restriction. Interestingly, an ancient common ancestor simian primate TRIMCyp gene (named TRIMCypA3), which was probably active against retroviruses, lost its antiviral activity via eight amino acid changes over the course of evolution due to lack of selective pressure 163. These evolutionary variations in how TRIM5 proteins from different species recognize and bind incoming viral capsids underpins the importance of this process for viral restriction.
TRIM21 facilitates cytoplasmic detection of antibody-opsonized, non-enveloped viruses
TRIM21 has been implicated in immune regulation for many years. The first indication for an immune-related function came from the observation that autoantibodies against TRIM21 (also called Ro52) were found in patients with auto-immune diseases including Sjögren’s syndrome and Systemic lupus erythematosus (SLE) 88. Follow-up studies, identified that TRIM21 contains IgG Fc-binding capabilities in its B30.2 domain, and that thereby TRIM21 can act as an Fc antibody receptor in the cytoplasm of cells [89], [90] and [91].
Accumulating evidence suggests that TRIM21 indeed plays a relevant role in the detection and response to antibody-opsonized viruses. Enveloped viruses leave any bound antibodies outside the cell upon fusion with the host membrane. Since TRIM21 is located in the cytoplasm, these are not the anticipated virus types to bring antibodies into the cytoplasm for recognition. In line with this, TRIM21 was found to mediate an intracellular antiviral immune response to antibody-opsonized adenovirus, which is a non-enveloped virus and thus carries any opsonizing antibodies into the cytoplasm 92. The authors demonstrated that TRIM21 targeted the virus for ubiquitin-dependent proteasomal degradation before translation of viral genes. These data suggest that humoral immunity can provide protection through an intracellular TRIM21-mediated mechanism that targets viruses for degradation 92.
In a follow-up study by the same group, it was found that in addition to the direct inhibition of viral replication by TRIM21-antibody recognition, TRIM21 recognition of intracellular antibodies bound to non-enveloped viruses and bacteria also promoted innate immune signaling leading to the production of antiviral cytokines 44. Mechanistically, antibody sensing by TRIM21 stimulated the synthesis of K63-linked poly-ubiquitin chains that activate TAK1-mediated NF-κB signaling 44. In line with this model, in vivo studies have shown that Trim21−/− mice are more susceptible to mouse adenovirus-1 infection as compared to WT mice. Moreover, antisera obtained from adenovirus infected mice protected WT mice in passive transfer experiments whereas they did not protect Trim21−/− mice 93.
Although these studies propose an interesting and novel model of viral inhibition by TRIM21, the connection in vivo between intracellular inhibition of viral replication by proteasomal degradation on the one hand and innate immune signaling on the other, is still unclear. This is exemplified by the fact that no differences were found in cytokine production between WT and Trim21−/− virus-infected mice in the study by Vaysburd et al., despite the fact that several other tissue culture based studies have indicated a role for TRIM21 in immune cell signaling 93. In this context it should be noted that studies on TRIM21 have been controversial since TRIM21 knockout mice generated by two different groups demonstrated strikingly different effects of TRIM21 removal on cytokine expression [94] and [95].
One study found no difference in survival or levels of cytokines upon LPS treatment, although Trim21−/− murine embryonic fibroblasts produced higher levels of NF-κB-dependent cytokines 94. In contrast, a different study with an independently generated Trim21−/− mouse strain found that their Trim21−/− mice developed systemic auto-immunity associated with increased levels of cytokines involved in the development of Th17 cells: IL-6, IL-12/IL-23p40, and IL-17 95. Although the reasons for these discrepancies are still unclear, it could be due to different strategies used to generate these knockout mice as previously proposed [96] and [97]. However, it is also plausible that the differences are due to TRIM21 isoforms that are targeted in these different knockout mice and the role that they might play in specific cell types, conditions, and/or stimuli. This current state of events exemplifies the challenging aspects of studying TRIM proteins in vitro and in vivo.
Direct antiviral activity of TRIM22
TRIM22 (also called STAF50) is phylogenetically related to TRIM5 and has been reported to exert antiviral activity against RNA viruses, such as hepatitis B virus (HBV), encephalomyocarditis virus (ECMV), and HIV-1 98. TRIM22 has been suggested to control levels of HIV-1 virus by down-regulating the HIV-1 Long Terminal Repeat (LTR)-directed transcription [99] and [100]. In addition, TRIM22 interacts with the HIV-1 Gag protein and interferes with its trafficking to the plasma membrane 101. TRIM22 inhibits EMCV by interacting with the EMCV 3C protease -which is important for processing the viral poly-protein and inhibition of host defense- and potentiating its ubiquitination 102. TRIM22 also interacts with the nucleoprotein (NP) protein of influenza virus and promotes its ubiquitin-proteasome dependent degradation, resulting in inhibition of viral replication 103.
The role of TRIM19 in nuclear body formation and antiviral activity
TRIM19 (also called PML; promyelocytic leukemia protein) is an essential component of the nuclear bodies (NB). NBs are highly organized nuclear structures composed of many proteins and have been shown to be sites of transcriptional regulation [104], [105] and [106]. Moreover, several viral components have been found to localize to these NBs (Figure 5) [107] and [108].
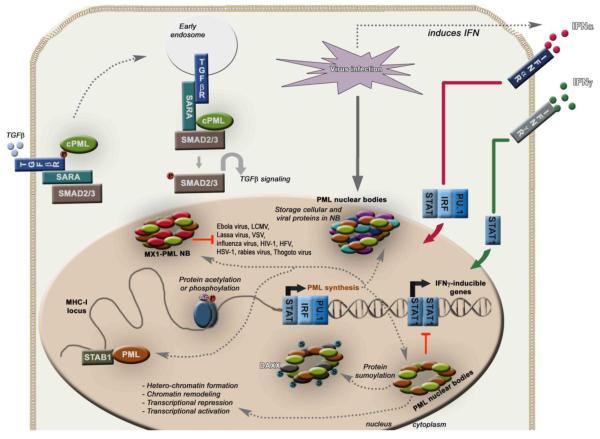
Figure 5. The role of TRIM19/PML in antiviral defense, signal transduction and induction by IFNs.
Mechanisms of TRIM19 up-regulation upon viral infection via IFN production. TRIM19 is an important effector of the IFN-mediated antiviral response. TRIM19 exerts its functions by interacting with other proteins including SP100 and DAXX to form nuclear bodies (NB). These protein complexes in NB are important in regulation of transcription, storage of proteins, sumoylation and antiviral functions. TRIM19 can regulate chromatin remodeling of the MHC-1 locus, and inhibit replication of viruses like influenza virus, Thogoto virus, herpes simplex virus-1 (HSV-1), Ebola virus, lymphocytic choriomeningitis virus (LCMV), Lassa virus, vesicular stomatitis virus (VSV), rabies virus, HIV-1, human foamy virus (HFV). TRIM19 also negatively regulates IFNγ signaling by inhibiting STAT1 transcriptional activity. The cytoplasmic isoform of TRIM19 (cPML) binds to the TGFβ receptor and serves as an adaptor molecule to recruit SARA and SMAD2/3 for TGFβ signaling.
The study of TRIM19 is complicated by the fact that some antiviral molecules also associate with the NBs (e.g. Mx1, SP100). Nevertheless, there is ample evidence that human TRIM19 itself can inhibit a large number of viruses including herpes simplex virus-1 (HSV-1), Ebola virus, lymphocytic chorio-meningitis virus (LCMV), Lassa virus, influenza virus, vesicular stomatitis virus (VSV), rabies virus, HIV-1, and human foamy virus (HFV) (Figure 5) 109. For example, LCMV, EMCV and rabies have been reported to replicate to higher levels in cells lacking TRIM19, and the antiviral effect of IFN against HSV-1 and HFV is reduced in TRIM19−/− cells 109. In addition, TRIM19 expression is known to be induced by type-I IFNs which also leads to an increase in size and numbers of NBs [110], [111] and [112]. Moreover, TRIM19 is highly up-regulated in macrophages and DCs during viral infections in an IFN-I dependent manner 85, supporting its role in anti-microbial defense.
The physiological role of TRIM19 during viral infections in vivo is still poorly defined. However, one study has shown that TRIM19 knockout mice are more susceptible to lethal immunopathology by LCMV and exhibit higher levels of VSV replication 113, although the mechanism of inhibition has not been addressed. Data from a different study indicated that TRIM19 regulates γ-herpesvirus latency in vivo. Lytic viral reactivation was reported to be higher in peritoneal cells from TRIM19 knockout mice as compared to WT counterparts, whereas no difference was observed during acute infection 114.
The fact that some viruses have developed strategies to disrupt the integrity of the PML-NBs supports a role of TRIM19 and NBs in anti-viral function. For example, LCMV encodes an 11 kDa RING finger protein called Z protein which associates with PML-NBs and induces relocation of TRIM19 to the cytoplasm, where this complex bind to eIF4E to inhibit translation [25] and [108]. Despite strong evidence supporting the role of TRIM19 as an antiviral effector, there is no compelling evidence that TRIM19 has been under positive selection or evolutionary pressure 115. This suggests that TRIM19 antiviral functions may be through indirect mechanisms, which may include the action of other antiviral proteins localized in the NBs, or by TRIM19-mediated regulation of antiviral cytokines. Therefore, more studies are needed to clarify the potential physiological role of TRIM19 as an antiviral effector, specially using relevant physiological contexts such as primary cells and in vivo systems. One of the complicating factors in directly addressing the role of TRIM19 is that seven different isoforms are expressed and may direct different cellular processes.
Additional TRIM proteins with direct antiviral activity
A recent study identified TRIM56 as a potential restriction factor for pestiviral infection. TRIM56 was found associated with the N-terminal protease (Npro) of bovine viral diarrhea virus (BVDV), a pestiviral interferon antagonist which targets IRF-3 for proteasome-dependent degradation. Inhibition of BVDV replication required the TRIM56 RING domain and its E3-ubiquitin ligase activity, but did not inhibit the replication of VSV or hepatitis C virus 116. In addition, TRIM56 has also been shown to play a role in innate immune signaling [45] and [117].
TRIM79α, a rodent specific IFN-inducible gene, was recently shown to inhibit tick-borne encephalitis virus (TBEV), a flavivirus that causes encephalitis in humans 118. TRIM79α targets the RNA-dependent RNA polymerase NS5 for lysosomal degradation. The authors proposed that TRIM79α acts alone to establish an efficient IFN-mediated antiviral response as TRIM79α knockdown resulted in reduced antiviral effects of IFNβ. However, the role of TRIM79α in the IFN signaling pathway remained unaddressed 118.
In addition to these TRIMs, some studies have been performed to screen larger numbers of TRIMs for antiviral activity against different viruses in vitro (Summarized in Figure 4). One study used overexpressed TRIMs in transduction assays for viral restriction activity against a selected group of GFP-expressing retroviruses including HIV-1, HIV2, SIVmac, EIAV (equine infectious anemia virus), MLV and prototypic foamy virus (PFV). TRIM1, TRIM5 and TRIM34 showed weak but specific inhibition of HIV-2/SIVmac, and TRIM34 also inhibited EIAV 63. In contrast, human TRIM1 restricted MLV but not HIV-1 70.
A different study using a larger panel of TRIMs could discriminate between viral restriction at the early stage (before viral gene transcription) or late stage of viral replication 119. In this study, mouse TRIM8/10/11/56 and human TRIM11/26/31 inhibited HIV entry, whereas human TRIM25/26/62 and mouse TRIM8/25/31/56 affected N-MLV entry. In terms of the inhibitory effect of viral release, this analysis identified the human TRIM proteins 15/26/32 and the mouse TRIM proteins 11/25/27/56 as factors specifically affecting HIV release from cells, but not viral gene expression 119.
In another recent study, 38 human TRIMs were tested for antiviral effects on Hepatitis B virus (HBV) replication in human hepatoma cells (HepG2) by overexpression 120. RT-PCR showed that overexpression of TRIM5/6/11/14/25/26/31/41 reduced the amount of HBV mRNA in HepG2 cells. These TRIMs inhibited the transcriptional activity of the HBV promoter enhancer. Further experiments focused on TRIM41 and mutations in its RING domain suggested that the antiviral effect relied on its E3 ubiquitin ligase activity. Deletion of the C-terminal SPRY domain also abrogated its antiviral function 120. Although these studies are interesting, they must be carefully interpreted, since they are prone to overexpression artifacts and their physiological relevance is still unaddressed. Nevertheless, these studies provide a framework for future work on TRIM-mediated antiviral function. In addition, these reports underscore the importance of studying other viruses in addition to retroviruses, as it is clear that TRIMs may have broader antiviral functions.
The role of bromo-domain containing TRIMs in transcriptional regulation and viral restriction
TRIM24, 28 and 33 belong to a subfamily of TRIM proteins that contain a bromo-domain (BRD) and a plant homeodomain (PHD) in the C-terminal region of the protein. The BRD can recognize acetyllysines on histones and can serve as a mechanism for regulating chromatin remodeling and transcriptional activation [121] and [122].
Accordingly, these TRIMs associate with chromatin regions in the nucleus 54, and have been shown to play positive and negative roles in transcriptional processes. TRIM24 (also called TIF-1α) forms complexes with TRIM28 (also called KAP1 or transcriptional intermediary factor; TIF-1β) and certain Kruppel-associated box (KRAB) motif-containing zinc finger repressors to inhibit transcription by a mechanism involving histone deacetylation and chromatin remodeling 123. TRIM33 is involved in erythroid differentiation, but has also been shown to have some silencing activity of gene promoters. The mechanism of TRIM33-mediated promoter repression seems to be independent of KRAB-motif containing repressors or the chromatin remodeling protein HP1 124. TRIM66 (also called TIF1δ) is a TRIM-like protein with a C-terminal BRD. Although it lacks a RING domain, it has also been shown to have a deacetylase-dependent transcriptional repression activity 125. This TRIM can form homodimers and can bind the HP1 indicating that it may function in a similar way to TRIM24 and TRIM28.
In addition to the repressive mechanism described above, TRIM28 can also inhibit transcription by binding histone methyltransferases 126. This negative role of TRIM28 on gene transcription has important implications in silencing of retroviral transcription. For example, replication of murine leukaemia virus (MLV) is restricted in embryonic carcinoma and embryonic stem cells in which TRIM28 is highly expressed. During MLV infection, the proviral DNA is integrated in the host genome but is subsequently silenced by formation of a complex with histone methyltransferases, histone deacetylase and HP1 family members. This facilitates methylation of histone H1, promotes chromatin condensation; especially the primer binding site (PBS) of MLV is a major target of this repression [127] and [128]. Similarly, TRIM28 together with the H3K9 methyltransferase ESET (also called SETDB1 or KMT1E) and HP1, mediates silencing of endogenous retroviruses in embryonic stem cells, highlighting the importance of TRIM28-mediated proviral silencing [129] and [130]. Finally, TRIM28 has also been reported to restrict HIV-1 replication by binding the acetylated HIV-1 integrase, which is required for viral cDNA integration into the host genome 131.
TRIM24 and TRIM33, which can form large multi-protein complexes with TRIM28, have also been reported to inhibit endogenous retroviruses 132. In TRIM24 knockout hepatocytes there was an accumulation of virus-like 30S cDNA elements in the cytoplasm which activated IFN responses similar to the ones observed upon exogenous viral infection 132. In addition to these direct antiviral roles, TRIM28 has been shown to negatively regulate IFN-I production by promoting IRF7 sumoylation 40. Moreover, both TRIM24 and TRIM28 were found to inhibit STAT1 signaling [133] and [134], suggesting that these TRIMs may also play a regulatory role during viral infection by inhibiting type-I IFN responses. In contrast, TRIM66 overexpression was demonstrated to promote RIG-I-dependent IFN induction 29. Together, these findings indicate that although many of the BRD-containing TRIMs inhibit viral replication by controlling transcriptional events, they may also mediate other processes.
Relevance of TRIM transcript variants and isoforms
So far the study of TRIM antiviral function has been complicated by the fact that many of the family members produce alternatively spliced transcripts that express different isoforms. The isoforms of a single TRIM protein may share the same RBCC motif but differ in their C terminus, potentially allowing them to recruit different sets of proteins. Alternatively, TRIM isoforms may also lack any of the domains in the RBCC which would potentially lack E3 ligase activity or oligomerization capabilities and may thus function as negative regulators during the immune response.
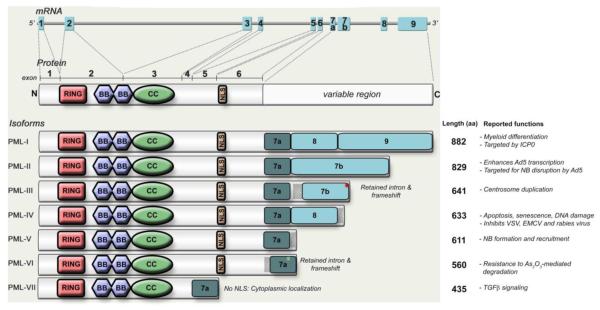
Bioinformatics analysis suggests that almost all TRIMs (90%) have more than one splice variant and about half of TRIM splice forms lack important domains 29. A good example is the human TRIM19 protein which has numerous transcript variants leading to seven different isoforms detected in cells (Figure 6)106. The function of TRIM19 and its isoforms has been extensively studied and reviewed in detail elsewhere [109] and [135]. One of the important points to be made is that indeed specific TRIM19 isoforms have been shown to have different functions upon viral infection. For example only PML IV, which lacks a region is the C-terminus encoded by exon 7b, inhibits varicella-zoster virus (VZV), EMCV and rabies virus, whereas none of the other isoforms do [136], [137] and [138]. In contrast, a cytoplasmic isoform of TRIM19 has also been reported to be expressed and to have a function in TGFβ cytokine signaling 139, as well as to play an important role in the antiviral response against herpes simplex virus-1 (HSV-1) 140.
Figure 6. TRIM19/PML isoforms.
TRIM19 isoforms, depicted here as PML I – PML VII are shown according to the nomenclature described by Jensen et al 162. All TRIM19/PML isoforms retain the first 3 exons which encode for the RBCC motif: RING (R), B-box (B1 and B2), and the coiled-coil domain (CC). The isoforms I to VII PMLI to PMLVII differ by alternative splicing of exons 7 to 9. The cytoplasmic form of TRIM19 (cPML) is the result of alternative splicing of exons 4–6. Adapted from Nisole et al. 135.
Another example of the functional difference of TRIM isoforms is the case of the antiviral activity of TRIM5α. This protein is the product of the TRIM5 gene which has a total of 5 isoforms. TRIM5α is the only isoform that contains the C-terminal B30.2 domain required for viral restriction [56] and [65]. Notably, TRIM5γ, TRIM5δ, TRIM5κ and TRIM5Ι (TRIM5-iota) isoforms, which lack the B30.2 domain, can act as dominant negative forms to TRIM5α by formation of non-functional dimers in overexpression assays [58], [141] and [164]. It appears that endogenous levels of TRIM5Ι are sufficient to exert a dominant negative effect since specific knockdown of TRIM5Ι in human cell lines increases TRIM5α antiviral activity 164. Interestingly, although the TRIM5δ isoform does not restrict viral replication, it retains its capacity to activate IFN and NF-κB signaling to similar levels as TRIM5α [29] and [38], suggesting that the mechanism of TRIM5 immune activation differs to its direct viral restriction activity.
Although the precise mechanism by which TRIM5δ activates immune signaling is unknown, it is most probably independent of virus the ability to act as a virus sensor since this isoform lacks the B30.2 domain required for binding to the viral capsid. This is consistent with the fact that TRIM5α can enhance innate immune signaling upon LPS stimulation downstream of TLR4 38, indicating that in some conditions the signaling function of TRIM5 isoforms is independent of virus sensing. TRIM5δ retains the RING domain shown to confer E3-ubiquitin ligase activity, hence making it conceivable that it activates immune signaling through a ubiquitin-dependent mechanism as has been shown for TRIM5α 38.
To understand the potential role of the TRIM proteins and address some of the discrepancies found between observations from different experimental settings, it will be necessary to identify the TRIM isoforms present in cells and how their expression and function is regulated during the stimuli being studied. Similarly, the effects on different isoforms during knock-out and knock-down studies should be carefully addressed the pinpoint residual effects of non-targeted isoforms.
Cellular localization and compartmentalization
Some TRIMs have been previously characterized with respect to their sub-cellular localization and their capacity to form or associate with specific compartments, such as nuclear bodies or microtubules. In addition, many TRIMs have been shown to localize in cytoplasmic bodies which do not co-localize with any commonly used cellular markers for sub-cellular compartments 54. Interestingly, several TRIMs relocalized from their cytoplasmic body localization upon virus infection 29. In many cases these cytoplasmic structures have not been well characterized and it may be that the formation of these dots solely dependents on the capacity of these TRIMs to oligomerize [54] and [55].
In an extensive study of TRIM cellular localization, Reymond et al. investigated the sub-cellular localization of a large number of TRIMs 54. The TRIMs found in the cytoplasm were either associated with filaments or concentrated in the form of cytoplasmic bodies, occasionally located around the nucleus. Nuclear TRIM proteins (TRIM8/19/30/32) localized mostly to NBs of which TRIM19 is the main component. The members of the bromodomain-containing subfamily (TRIM24/28/33) were found associated with specific chromatin regions 54, consistent with the proposed role of this domain in transcriptional regulation 142. Although this study provided an excellent insight in possible localization patterns of TRIMs, there is always concern that the formation of these cellular structures are the result of over-expression artifacts or protein aggregates. Unfortunately studies of the endogenous TRIM proteins have been complicated by the lack of specific antibodies.
A different study identified a sub-group of TRIMs which shares an identical domain arrangement (RBCC-COS-FN3-B30.2 domains; TRIM1/9/18/36/42/46/67) and co-localize to microtubules. Binding to the microtubules is mediated by the COS domain, suggesting functional similarities between the members of this subgroup 143. Interestingly, with exception of TRIM46, all COS domain containing TRIMs (which also include TRIM54/55/63 but do not contain the SPRY domain) enhanced RIG-I mediated signaling pathways 29. In addition, a few of these TRIMs were found up-regulated in differentiated CD4+ T cells 85. Therefore it is tempting to speculate that localization of COS-containing TRIMs to the microtubules, might be important for innate immune signaling, particularly in CD4+ T cells.
TRIMs in antiviral innate immune signaling
Increasing evidence indicates that TRIMs can act as antiviral factors indirectly by stimulating cytokine signaling pathways culminating in the induction of many antiviral ISGs (Figure 7) [19], [27], [28], [29], [96] and [144]. Many TRIMs have been found to play negative regulatory roles in immune signaling. In most cases this is consistent with mechanisms involving ubiquitin-dependent degradation of targeted proteins, although not all of them act by proteasome-dependent mechanisms (TRIM11/21/27/30/38/59; Summarized in Figure 7) [40], [145], [146], [147], [148], [149], [150], [151], [152], [153], [154], [155] and [156].
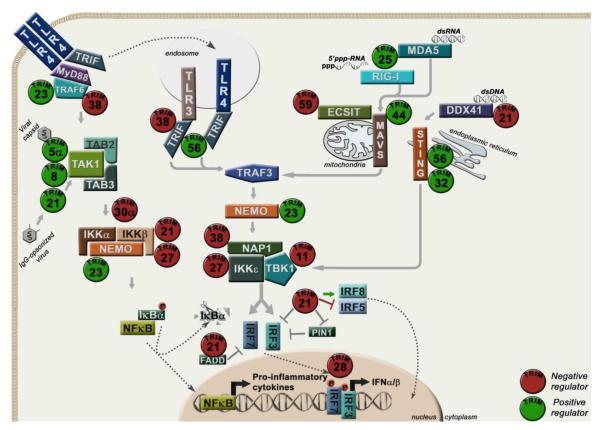
Figure 7. TRIMs in innate immune signaling.
Upon microbial infection, pattern recognition receptors (TLRs and RLRs) recognize pathogen products to trigger downstream signaling pathways to induce IFN-I and pro-inflammatory cytokines. TLR3 recognizes double stranded RNA (dsRNA), TLR4 recognizes bacterial lipopolysaccharide (LPS), RIG-I recognizes single stranded RNA (ssRNA), MDA5 recognizes dsRNA and DDX41 recognizes double stranded DNA (dsDNA). In the RIG-I/MDA5 pathway, TRIM25 ubiquitinates RIGI for binding to MAVS. TRIM44 stabilizes MAVS. TRIM21 can act as both a positive and negative regulator of IFNβ. TRIM27 inhibits NF-κB and IFN activation mediated by IKKα/β/ε. TRIM30 degrades TAB2-TAB3 to inhibit production of IL-6 and TNFα. TRIM23 enhances NEMO function by promoting K63-linked ubiquitination. TRIM21 is involved in the inhibition of IKKβ and IRF7 and positive or negative regulation of IRF3 and IRF8. TRIM32 and TRIM56 ubiquitinate STING to promote dsDNA signaling. TRIM56 also positively regulates TRIF-mediated signaling whereas TRIM38 inhibits it. TRIM21 negatively regulates IFN production upon dsDNA signaling by ubiquitinating and targeting DDX41 for degradation. TRIM21 and TRIM28 inhibit IRF7 activation. TRIM5α catalyzes the synthesis of unanchored K63-linked poly-ubiquitin chains that bind and activate TAK1 kinase. TRIM21 acts as an antibody receptor bound to viruses to activate TAK1. TRIMs in green and red represent positive and negative regulatory functions, respectively.
However, an increasing number of TRIMs has been found to promote synthesis of K63-linked poly-ubiquitin chains that have activating functions. One of the best characterized TRIMs involved in promoting immune signaling is TRIM25 (also called EFP). During viral replication, viral RNA bearing 5′-triphosphates is recognized by the RNA sensor RIG-I. The N-terminal caspase recruitment domains (CARDs) of RIG-I undergo K63-linked ubiquitination induced by TRIM25, which allows RIG-I binding to MAVS (also called IPS-1, Cardif, or VISA) and downstream signaling to produce IFN-I46. In addition, TRIM25 also catalyzes the synthesis of unanchored K63-ubiquitin chains, which are not covalently attached to any protein, and can bind and activate RIG-I 36. The importance of TRIM25 in antiviral signaling is supported by the fact that the NS1 protein of influenza A viruses directly antagonizes TRIM25 activity 157. This antagonism is species-specific, since NS1 protein encoded by H5N1 avian virus preferentially targeted the chicken TRIM25 orthologue for inhibition of IFN production in chicken cells 158, further highlighting the potential for viral adaptation by targeting TRIMs.
Other TRIMs that positively regulate innate signaling pathways include TRIM5α, which catalyzes the synthesis of unanchored K63-linked ubiquitin chains that activate the TAK1 kinase complex for downstream AP-1 and NF-κB signaling 38. Within the same pathway TRIM8 has been reported to mediate K63-linked poly-ubiquitination of TAK1 for downstream NF-κB activation 159. Moreover, it has been reported that TRIM23 ubiquitinates the ubiquitin-binding protein NEMO for IRF3 and NF-kB activation 47. Furthermore, TRIM32 and TRIM56 have been implicated in signaling down-stream of cytoplasmic dsDNA detection in the ubiquitination of STING thereby mediating its downstream IFN induction [45] and [160]. TRIM44 interacts with MAVS and was proposed to stabilize MAVS by preventing its ubiquitination and subsequent degradation 161, and TRIM56 also promotes TLR3 dependent signaling by interacting with TRIF [45] and [117] (Figure 7).
Interestingly, several TRIMs have been reported to fulfill both positive and negative regulatory functions in the same cell signaling pathways. As indicated above (Figure 6) different isoforms of the same protein may have different functions. However, isoforms may not account for all seemingly contradicting roles some TRIMs may have. It has been known for some time that many TRIMs are expressed in a cell-type specific manner [31], [32] and [85], which could be one of the factors influencing functions of individual proteins. Moreover, various TRIMs are regulated at the transcriptional, stability and localization level during e.g. virus infection 29, suggesting that the exact cellular context in which the relevance of individual TRIMs are addressed may be crucial.
As the levels of certain TRIMs change in response to cytokines, their signaling may also change. This would be particularly relevant for proteins with opposing roles in related pathways, to skew the pathways in one direction while inhibiting the other. One of the TRIM proteins for which this may be relevant is e.g. TRIM38, which was recently reported to play an inhibitory role in the TLR3/4 pathway [153], [154] and [155], while enhancing RIG-I-mediated signaling 29.
It will be interesting to see if the role of these TRIMs in immune signaling is connected to direct antiviral roles or direct interaction with viruses. The increasing number of TRIMs shown to be involved in innate immune signaling as well as direct antiviral function further strengthens the hypothesis that TRIMs evolved as a component of the immune response.
Concluding remarks and future perspectives
It has become clear that TRIM proteins play important roles in immune regulation and microbial restriction. However, the specific molecular mechanisms of action are just starting to emerge. Probably one of the most complex and challenging aspects to study TRIM function has been to elucidate how TRIMs are activated upon virus infection and how their expression and localization is regulated in different cell types. Most of the studies on TRIMs have been performed in cell lines and by overexpression/knockdown experiments. There is still a lack of understanding of the dominant roles of TRIMs in vivo and their physiological relevance.
Although studies in vivo may also be challenging as exemplified by the controversial role of TRIM21 using knockout mice, other in vivo strategies may prove useful to demonstrate antiviral physiological roles of TRIMs in animals. For example, the rhesus macaque TRIM5 gene has multiple alleles that encode for polymorphisms in the B30.2 domain with either permissive or restrictive antiviral phenotypes 165. By using different TRIM5 genotypes in a model of cross-species transmission it was shown that TRIM5α restrictive alleles were able to attenuate transmission of sooty mangabey SIV in rhesus macaques and exerted selective pressure resulting in viral capsid adaptive mutants 166. This model of cross-species transmission using different TRIM5 genotypes has also proved useful to study evolution of SIV in vivo 167. Therefore, it will be important to identify polymorphisms in other TRIM members that may facilitate studies on viral restriction in vivo.
As it is becoming clear that shorter forms of TRIMs lacking the important RING domain may act as dominant negative forms, it will be essential to generate technologies to knockout specific transcript variants in vivo. Another important aspect of TRIM study is the type of ubiquitin chains that TRIMs synthesize. An increasing number of TRIMs has been discovered to catalyze the synthesis of unanchored poly-ubiquitin chains with functions in antiviral immunity. It remains to be seen if TRIMs may comprise a family of E3 ligases that specializes in these types of ubiquitin chains, in which lysine-branched topologies these poly-ubiquitin chains exist, and ultimately what their physiological roles are.
Highlights.
The TRIM family of proteins inhibits replication of various viruses
TRIMs act as E3-ubiquitin ligases for activation of innate immune signaling
TRIMs evolved as a component of the immune response
Acknowledgments
Research in A-GS lab is supported by NIAID grants R01AI046954, U19AI083025, U54AI057158, U19AI106754, P01AI090935, R01DA033773, U01AI095611, U19AI089987, NIAID contract HHSN272201000054C and by the NIAID funded CEIRS network under a contract for a Center of Research in Influenza Pathogenesis (CRIP, HHSN266200700010C).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–26. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 2.Blanco-Melo D, Venkatesh S, Bieniasz PD. Intrinsic cellular defenses against human immunodeficiency viruses. Immunity. 2012;37:399–411. doi: 10.1016/j.immuni.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol. 2004;5:1109–15. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 4.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 5.Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3:920–40. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–8. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–3. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 8.Pham AM, Tenoever BR. The IKK Kinases: Operators of Antiviral Signaling. Viruses. 2010;2:55–72. doi: 10.3390/v2010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 1997;91:243–52. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 10.Hemmi H, Takeuchi O, Sato S, Yamamoto M, Kaisho T, Sanjo H, Kawai T, Hoshino K, Takeda K, Akira S. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J Exp Med. 2004;199:1641–50. doi: 10.1084/jem.20040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–51. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 12.Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 13.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–25. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–5. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy BA, Etkin LD. A unique bipartite cysteine-histidine motif defines a subfamily of potential zinc-finger proteins. Nucleic Acids Res. 1991;19:6330. doi: 10.1093/nar/19.22.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy BA, Etkin LD, Freemont PS. A novel zinc finger coiled-coil domain in a family of nuclear proteins. Trends Biochem Sci. 1992;17:344–5. doi: 10.1016/0968-0004(92)90308-v. [DOI] [PubMed] [Google Scholar]
- 17.Saurin AJ, Borden KL, Boddy MN, Freemont PS. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–14. [PubMed] [Google Scholar]
- 18.Freemont PS. The RING finger. A novel protein sequence motif related to the zinc finger. Ann N Y Acad Sci. 1993;684:174–92. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- 19.Ozato K, Shin DM, Chang TH, Morse HC., 3rd TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–60. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhodes DA, de Bono B, Trowsdale J. Relationship between SPRY and B30.2 protein domains. Evolution of a component of immune defence? Immunology. 2005;116:411–7. doi: 10.1111/j.1365-2567.2005.02248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han K, Lou DI, Sawyer SL. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet. 2011;7:e1002388. doi: 10.1371/journal.pgen.1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boudinot P, van der Aa LM, Jouneau L, Du Pasquier L, Pontarotti P, Briolat V, Benmansour A, Levraud JP. Origin and evolution of TRIM proteins: new insights from the complete TRIM repertoire of zebrafish and pufferfish. PLoS One. 2011;6:e22022. doi: 10.1371/journal.pone.0022022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sardiello M, Cairo S, Fontanella B, Ballabio A, Meroni G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol Biol. 2008;8:225. doi: 10.1186/1471-2148-8-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Aa LM, Levraud JP, Yahmi M, Lauret E, Briolat V, Herbomel P, Benmansour A, Boudinot P. A large new subset of TRIM genes highly diversified by duplication and positive selection in teleost fish. BMC Biol. 2009;7:7. doi: 10.1186/1741-7007-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 26.Rombout JH, Huttenhuis HB, Picchietti S, Scapigliati G. Phylogeny and ontogeny of fish leucocytes. Fish Shellfish Immunol. 2005;19:441–55. doi: 10.1016/j.fsi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Kawai T, Akira S. Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins. EMBO Mol Med. 2011;3:513–27. doi: 10.1002/emmm.201100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uchil PD, Hinz A, Siegel S, Coenen-Stass A, Pertel T, Luban J, Mothes W. TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J Virol. 2013;87:257–72. doi: 10.1128/JVI.01804-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, Inn KS, Fernandez-Sesma A, Jung J, Garcia-Sastre A. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38:384–98. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;27:1147–57. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 31.Wu C, Macleod I, Su AI. BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 2013;41:D561–5. doi: 10.1093/nar/gks1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trempe JF. Reading the ubiquitin postal code. Curr Opin Struct Biol. 2011;21:792–801. doi: 10.1016/j.sbi.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 34.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 35.Chen J, Chen ZJ. Regulation of NF-kappaB by ubiquitination. Curr Opin Immunol. 2013;25:4–12. doi: 10.1016/j.coi.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–30. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Napolitano LM, Jaffray EG, Hay RT, Meroni G. Functional interactions between ubiquitin E2 enzymes and TRIM proteins. Biochem J. 2011;434:309–19. doi: 10.1042/BJ20101487. [DOI] [PubMed] [Google Scholar]
- 38.Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, Bisiaux A, Albert ML, Strambio-De-Castillia C, Mothes W, Pizzato M, Grutter MG, Luban J. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–5. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou W, Zhang DE. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J Biol Chem. 2006;281:3989–94. doi: 10.1074/jbc.M510787200. [DOI] [PubMed] [Google Scholar]
- 40.Liang Q, Deng H, Li X, Wu X, Tang Q, Chang TH, Peng H, Rauscher FJ, 3rd, Ozato K, Zhu F. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. J Immunol. 2011;187:4754–63. doi: 10.4049/jimmunol.1101704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freemont PS. RING for destruction? Curr Biol. 2000;10:R84–7. doi: 10.1016/s0960-9822(00)00287-6. [DOI] [PubMed] [Google Scholar]
- 42.Kentsis A, Borden KL. Construction of macromolecular assemblages in eukaryotic processes and their role in human disease: linking RINGs together. Curr Protein Pept Sci. 2000;1:49–73. doi: 10.2174/1389203003381478. [DOI] [PubMed] [Google Scholar]
- 43.Borden KL. RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem Cell Biol. 1998;76:351–8. doi: 10.1139/bcb-76-2-3-351. [DOI] [PubMed] [Google Scholar]
- 44.McEwan WA, Tam JC, Watkinson RE, Bidgood SR, Mallery DL, James LC. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat Immunol. 2013;14:327–36. doi: 10.1038/ni.2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2012;33:765–76. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 46.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 47.Arimoto K, Funami K, Saeki Y, Tanaka K, Okawa K, Takeuchi O, Akira S, Murakami Y, Shimotohno K. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc Natl Acad Sci U S A. 2010;107:15856–61. doi: 10.1073/pnas.1004621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borden KL, Martin SR, O’Reilly NJ, Lally JM, Reddy BA, Etkin LD, Freemont PS. Characterisation of a novel cysteine/histidine-rich metal binding domain from Xenopus nuclear factor XNF7. FEBS Lett. 1993;335:255–60. doi: 10.1016/0014-5793(93)80741-c. [DOI] [PubMed] [Google Scholar]
- 49.Massiah MA, Matts JA, Short KM, Simmons BN, Singireddy S, Yi Z, Cox TC. Solution structure of the MID1 B-box2 CHC(D/C)C(2)H(2) zinc-binding domain: insights into an evolutionarily conserved RING fold. J Mol Biol. 2007;369:1–10. doi: 10.1016/j.jmb.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 50.Massiah MA, Simmons BN, Short KM, Cox TC. Solution structure of the RBCC/TRIM B-box1 domain of human MID1: B-box with a RING. J Mol Biol. 2006;358:532–45. doi: 10.1016/j.jmb.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 51.Bell JL, Malyukova A, Holien JK, Koach J, Parker MW, Kavallaris M, Marshall GM, Cheung BB. TRIM16 acts as an E3 ubiquitin ligase and can heterodimerize with other TRIM family members. PLoS One. 2012;7:e37470. doi: 10.1371/journal.pone.0037470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao T, Borden KL, Freemont PS, Etkin LD. Involvement of the rfp tripartite motif in protein-protein interactions and subcellular distribution. J Cell Sci. 1997;110(Pt 14):1563–71. doi: 10.1242/jcs.110.14.1563. [DOI] [PubMed] [Google Scholar]
- 53.Cainarca S, Messali S, Ballabio A, Meroni G. Functional characterization of the Opitz syndrome gene product (midin): evidence for homodimerization and association with microtubules throughout the cell cycle. Hum Mol Genet. 1999;8:1387–96. doi: 10.1093/hmg/8.8.1387. [DOI] [PubMed] [Google Scholar]
- 54.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. The tripartite motif family identifies cell compartments. Embo J. 2001;20:2140–51. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Napolitano LM, Meroni G. TRIM family: Pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life. 2012;64:64–71. doi: 10.1002/iub.580. [DOI] [PubMed] [Google Scholar]
- 56.Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15:73–8. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- 57.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A. 2005;102:2832–7. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–53. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 59.Goff SP. Host factors exploited by retroviruses. Nat Rev Microbiol. 2007;5:253–63. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- 60.Sokolskaja E, Luban J. Cyclophilin, TRIM5, and innate immunity to HIV-1. Curr Opin Microbiol. 2006;9:404–8. doi: 10.1016/j.mib.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 61.Towers GJ, Goff SP. Post-entry restriction of retroviral infections. AIDS Rev. 2003;5:156–64. [PubMed] [Google Scholar]
- 62.Towers GJ. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology. 2007;4:40. doi: 10.1186/1742-4690-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang F, Hatziioannou T, Perez-Caballero D, Derse D, Bieniasz PD. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology. 2006;353:396–409. doi: 10.1016/j.virol.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 64.Johnson WE. Rapid adversarial co-evolution of viruses and cellular restriction factors. Curr Top Microbiol Immunol. 2013;371:123–51. doi: 10.1007/978-3-642-37765-5_5. [DOI] [PubMed] [Google Scholar]
- 65.Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79:3139–45. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Towers G, Bock M, Martin S, Takeuchi Y, Stoye JP, Danos O. A conserved mechanism of retrovirus restriction in mammals. Proc Natl Acad Sci U S A. 2000;97:12295–9. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hatziioannou T, Perez-Caballero D, Yang A, Cowan S, Bieniasz PD. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc Natl Acad Sci U S A. 2004;101:10774–9. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keckesova Z, Ylinen LM, Towers GJ. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc Natl Acad Sci U S A. 2004;101:10780–5. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perron MJ, Stremlau M, Song B, Ulm W, Mulligan RC, Sodroski J. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc Natl Acad Sci U S A. 2004;101:11827–32. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yap MW, Nisole S, Lynch C, Stoye JP. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci U S A. 2004;101:10786–91. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–73. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 72.Munk C, Brandt SM, Lucero G, Landau NR. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc Natl Acad Sci U S A. 2002;99:13843–8. doi: 10.1073/pnas.212400099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Diaz-Griffero F. Caging the beast: TRIM5alpha binding to the HIV-1 core. Viruses. 2011;3:423–8. doi: 10.3390/v3050423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ganser-Pornillos BK, Chandrasekaran V, Pornillos O, Sodroski JG, Sundquist WI, Yeager M. Hexagonal assembly of a restricting TRIM5alpha protein. Proc Natl Acad Sci U S A. 2011;108:534–9. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grutter MG, Luban J. TRIM5 structure, HIV-1 capsid recognition, and innate immune signaling. Curr Opin Virol. 2013;2:142–50. doi: 10.1016/j.coviro.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Diaz-Griffero F, Li X, Javanbakht H, Song B, Welikala S, Stremlau M, Sodroski J. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology. 2006;349:300–15. doi: 10.1016/j.virol.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 77.Xu L, Yang L, Moitra PK, Hashimoto K, Rallabhandi P, Kaul S, Meroni G, Jensen JP, Weissman AM, D’Arpa P. BTBD1 and BTBD2 colocalize to cytoplasmic bodies with the RBCC/tripartite motif protein, TRIM5delta. Exp Cell Res. 2003;288:84–93. doi: 10.1016/s0014-4827(03)00187-3. [DOI] [PubMed] [Google Scholar]
- 78.Rold CJ, Aiken C. Proteasomal degradation of TRIM5alpha during retrovirus restriction. PLoS Pathog. 2008;4:e1000074. doi: 10.1371/journal.ppat.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lukic Z, Hausmann S, Sebastian S, Rucci J, Sastri J, Robia SL, Luban J, Campbell EM. TRIM5alpha associates with proteasomal subunits in cells while in complex with HIV-1 virions. Retrovirology. 2011;8:93. doi: 10.1186/1742-4690-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A. 2006;103:5514–9. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roa A, Hayashi F, Yang Y, Lienlaf M, Zhou J, Shi J, Watanabe S, Kigawa T, Yokoyama S, Aiken C, Diaz-Griffero F. RING domain mutations uncouple TRIM5alpha restriction of HIV-1 from inhibition of reverse transcription and acceleration of uncoating. J Virol. 2012;86:1717–27. doi: 10.1128/JVI.05811-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kutluay SB, Perez-Caballero D, Bieniasz PD. Fates of retroviral core components during unrestricted and TRIM5-restricted infection. PLoS Pathog. 2013;9:e1003214. doi: 10.1371/journal.ppat.1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Si Z, Vandegraaff N, O’Huigin C, Song B, Yuan W, Xu C, Perron M, Li X, Marasco WA, Engelman A, Dean M, Sodroski J. Evolution of a cytoplasmic tripartite motif (TRIM) protein in cows that restricts retroviral infection. Proc Natl Acad Sci U S A. 2006;103:7454–9. doi: 10.1073/pnas.0600771103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tareen SU, Sawyer SL, Malik HS, Emerman M. An expanded clade of rodent Trim5 genes. Virology. 2009;385:473–83. doi: 10.1016/j.virol.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rajsbaum R, Stoye JP, O’Garra A. Type I interferon-dependent and - independent expression of tripartite motif proteins in immune cells. Eur J Immunol. 2008;38:619–30. doi: 10.1002/eji.200737916. [DOI] [PubMed] [Google Scholar]
- 86.Sawyer SL, Emerman M, Malik HS. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog. 2007;3:e197. doi: 10.1371/journal.ppat.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Newman RM, Hall L, Kirmaier A, Pozzi LA, Pery E, Farzan M, O’Neil SP, Johnson W. Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 2008;4:e1000003. doi: 10.1371/journal.ppat.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wahren M, Tengner P, Gunnarsson I, Lundberg I, Hedfors E, Ringertz NR, Pettersson I. Ro/SS-A and La/SS-B antibody level variation in patients with Sjogren’s syndrome and systemic lupus erythematosus. J Autoimmun. 1998;11:29–38. doi: 10.1006/jaut.1997.0173. [DOI] [PubMed] [Google Scholar]
- 89.Rhodes DA, Trowsdale J. TRIM21 is a trimeric protein that binds IgG Fc via the B30.2 domain. Mol Immunol. 2007;44:2406–14. doi: 10.1016/j.molimm.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 90.James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc Natl Acad Sci U S A. 2007;104:6200–5. doi: 10.1073/pnas.0609174104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Keeble AH, Khan Z, Forster A, James LC. TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proc Natl Acad Sci U S A. 2008;105:6045–50. doi: 10.1073/pnas.0800159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mallery DL, McEwan WA, Bidgood SR, Towers GJ, Johnson CM, James LC. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21) Proc Natl Acad Sci U S A. 2010;107:19985–90. doi: 10.1073/pnas.1014074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vaysburd M, Watkinson RE, Cooper H, Reed M, O’Connell K, Smith J, Cruickshanks J, James LC. Intracellular antibody receptor TRIM21 prevents fatal viral infection. Proc Natl Acad Sci U S A. 2013;110:12397–401. doi: 10.1073/pnas.1301918110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yoshimi R, Chang TH, Wang H, Atsumi T, Morse HC, 3rd, Ozato K. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappaB-dependent cytokine expression in fibroblasts. J Immunol. 2009;182:7527–38. doi: 10.4049/jimmunol.0804121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Espinosa A, Dardalhon V, Brauner S, Ambrosi A, Higgs R, Quintana FJ, Sjostrand M, Eloranta ML, Ni Gabhann J, Winqvist O, Sundelin B, Jefferies CA, Rozell B, Kuchroo VK, Wahren-Herlenius M. Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL-23-Th17 pathway. J Exp Med. 2009;206:1661–71. doi: 10.1084/jem.20090585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McNab FW, Rajsbaum R, Stoye JP, O’Garra A. Tripartite-motif proteins and innate immune regulation. Curr Opin Immunol. 2011;23:46–56. doi: 10.1016/j.coi.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 97.Ozato K, Yoshimi R, Chang TH, Wang H, Atsumi T, Morse HC., 3rd Comment on “Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappa B-dependent cytokine expression in fibroblasts”. J Immunol. 2009;183:7619. doi: 10.4049/jimmunol.0990103. author reply 720-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hattlmann CJ, Kelly JN, Barr SD. TRIM22: A Diverse and Dynamic Antiviral Protein. Mol Biol Int. 2012;2012:153415. doi: 10.1155/2012/153415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tissot C, Mechti N. Molecular cloning of a new interferon-induced factor that represses human immunodeficiency virus type 1 long terminal repeat expression. J Biol Chem. 1995;270:14891–8. doi: 10.1074/jbc.270.25.14891. [DOI] [PubMed] [Google Scholar]
- 100.Kajaste-Rudnitski A, Marelli SS, Pultrone C, Pertel T, Uchil PD, Mechti N, Mothes W, Poli G, Luban J, Vicenzi E. TRIM22 inhibits HIV-1 transcription independently of its E3 ubiquitin ligase activity, Tat, and NF-kappaB-responsive long terminal repeat elements. J Virol. 2011;85:5183–96. doi: 10.1128/JVI.02302-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barr SD, Smiley JR, Bushman FD. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 2008;4:e1000007. doi: 10.1371/journal.ppat.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Eldin P, Papon L, Oteiza A, Brocchi E, Lawson TG, Mechti N. TRIM22 E3 ubiquitin ligase activity is required to mediate antiviral activity against encephalomyocarditis virus. J Gen Virol. 2009;90:536–45. doi: 10.1099/vir.0.006288-0. [DOI] [PubMed] [Google Scholar]
- 103.Di Pietro A, Kajaste-Rudnitski A, Oteiza A, Nicora L, Towers GJ, Mechti N, Vicenzi E. TRIM22 inhibits influenza A virus infection by targeting the viral nucleoprotein for degradation. J Virol. 2013;87:4523–33. doi: 10.1128/JVI.02548-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stuurman N, de Graaf A, Floore A, Josso A, Humbel B, de Jong L, van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J Cell Sci. 1992;101(Pt 4):773–84. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- 105.Stuurman N, Floore A, Colen A, de Jong L, van Driel R. Stabilization of the nuclear matrix by disulfide bridges: identification of matrix polypeptides that form disulfides. Exp Cell Res. 1992;200:285–94. doi: 10.1016/0014-4827(92)90174-7. [DOI] [PubMed] [Google Scholar]
- 106.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–16. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 107.Everett RD. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene. 2001;20:7266–73. doi: 10.1038/sj.onc.1204759. [DOI] [PubMed] [Google Scholar]
- 108.Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007;89:819–30. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 109.Geoffroy MC, Chelbi-Alix MK. Role of promyelocytic leukemia protein in host antiviral defense. J Interferon Cytokine Res. 2011;31:145–58. doi: 10.1089/jir.2010.0111. [DOI] [PubMed] [Google Scholar]
- 110.Chelbi-Alix MK, Pelicano L, Quignon F, Koken MH, Venturini L, Stadler M, Pavlovic J, Degos L, de The H. Induction of the PML protein by interferons in normal and APL cells. Leukemia. 1995;9:2027–33. [PubMed] [Google Scholar]
- 111.Lavau C, Marchio A, Fagioli M, Jansen J, Falini B, Lebon P, Grosveld F, Pandolfi PP, Pelicci PG, Dejean A. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene. 1995;11:871–6. [PubMed] [Google Scholar]
- 112.Stadler M, Chelbi-Alix MK, Koken MH, Venturini L, Lee C, Saib A, Quignon F, Pelicano L, Guillemin MC, Schindler C, et al. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene. 1995;11:2565–73. [PubMed] [Google Scholar]
- 113.Bonilla WV, Pinschewer DD, Klenerman P, Rousson V, Gaboli M, Pandolfi PP, Zinkernagel RM, Salvato MS, Hengartner H. Effects of promyelocytic leukemia protein on virus-host balance. J Virol. 2002;76:3810–8. doi: 10.1128/JVI.76.8.3810-3818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sewatanon J, Liu H, Ling PD. PML protein modulates establishment and maintenance of latent gamma-herpesvirus infection in peritoneal cells. J Virol. 2013 doi: 10.1128/JVI.01696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ortiz M, Bleiber G, Martinez R, Kaessmann H, Telenti A. Patterns of evolution of host proteins involved in retroviral pathogenesis. Retrovirology. 2006;3:11. doi: 10.1186/1742-4690-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang J, Liu B, Wang N, Lee YM, Liu C, Li K. TRIM56 is a virus- and interferon-inducible E3 ubiquitin ligase that restricts pestivirus infection. J Virol. 2011;85:3733–45. doi: 10.1128/JVI.02546-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shen Y, Li NL, Wang J, Liu B, Lester S, Li K. TRIM56 is an essential component of the TLR3 antiviral signaling pathway. J Biol Chem. 2012;287:36404–13. doi: 10.1074/jbc.M112.397075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Taylor RT, Lubick KJ, Robertson SJ, Broughton JP, Bloom ME, Bresnahan WA, Best SM. TRIM79alpha, an interferon-stimulated gene product, restricts tick-borne encephalitis virus replication by degrading the viral RNA polymerase. Cell Host Microbe. 2011;10:185–96. doi: 10.1016/j.chom.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Uchil PD, Quinlan BD, Chan WT, Luna JM, Mothes W. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 2008;4:e16. doi: 10.1371/journal.ppat.0040016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang S, Guo JT, Wu JZ, Yang G. Identification and Characterization of Multiple TRIM Proteins That Inhibit Hepatitis B Virus Transcription. PLoS One. 2013;8:e70001. doi: 10.1371/journal.pone.0070001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–8. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 122.Dyson MH, Rose S, Mahadevan LC. Acetyllysine-binding and function of bromodomain-containing proteins in chromatin. Front Biosci. 2001;6:D853–65. doi: 10.2741/dyson. [DOI] [PubMed] [Google Scholar]
- 123.Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. Embo J. 1999;18:6385–95. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Venturini L, You J, Stadler M, Galien R, Lallemand V, Koken MH, Mattei MG, Ganser A, Chambon P, Losson R, de The H. TIF1gamma, a novel member of the transcriptional intermediary factor 1 family. Oncogene. 1999;18:1209–17. doi: 10.1038/sj.onc.1202655. [DOI] [PubMed] [Google Scholar]
- 125.Khetchoumian K, Teletin M, Mark M, Lerouge T, Cervino M, Oulad-Abdelghani M, Chambon P, Losson R. TIF1delta, a novel HP1-interacting member of the transcriptional intermediary factor 1 (TIF1) family expressed by elongating spermatids. J Biol Chem. 2004;279:48329–41. doi: 10.1074/jbc.M404779200. [DOI] [PubMed] [Google Scholar]
- 126.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–32. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ellis J, Hotta A, Rastegar M. Retrovirus silencing by an epigenetic TRIM. Cell. 2007;131:13–4. doi: 10.1016/j.cell.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 128.Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 129.Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–31. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 130.Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, Spitz F, Constam DB, Trono D. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–40. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 131.Allouch A, Di Primio C, Alpi E, Lusic M, Arosio D, Giacca M, Cereseto A. The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe. 2011;9:484–95. doi: 10.1016/j.chom.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 132.Herquel B, Ouararhni K, Martianov I, Le Gras S, Ye T, Keime C, Lerouge T, Jost B, Cammas F, Losson R, Davidson I. Trim24-repressed VL30 retrotransposons regulate gene expression by producing noncoding RNA. Nat Struct Mol Biol. 2013;20:339–46. doi: 10.1038/nsmb.2496. [DOI] [PubMed] [Google Scholar]
- 133.Kamitani S, Ohbayashi N, Ikeda O, Togi S, Muromoto R, Sekine Y, Ohta K, Ishiyama H, Matsuda T. KAP1 regulates type I interferon/STAT1-mediated IRF-1 gene expression. Biochem Biophys Res Commun. 2008;370:366–70. doi: 10.1016/j.bbrc.2008.03.104. [DOI] [PubMed] [Google Scholar]
- 134.Tisserand J, Khetchoumian K, Thibault C, Dembele D, Chambon P, Losson R. Tripartite motif 24 (Trim24/Tif1alpha) tumor suppressor protein is a novel negative regulator of interferon (IFN)/signal transducers and activators of transcription (STAT) signaling pathway acting through retinoic acid receptor alpha (Raralpha) inhibition. J Biol Chem. 2011;286:33369–79. doi: 10.1074/jbc.M111.225680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nisole S, Maroui MA, Mascle XH, Aubry M, Chelbi-Alix MK. Differential Roles of PML Isoforms. Front Oncol. 2013;3:125. doi: 10.3389/fonc.2013.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Maroui MA, Pampin M, Chelbi-Alix MK. Promyelocytic leukemia isoform IV confers resistance to encephalomyocarditis virus via the sequestration of 3D polymerase in nuclear bodies. J Virol. 2011;85:13164–73. doi: 10.1128/JVI.05808-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Reichelt M, Wang L, Sommer M, Perrino J, Nour AM, Sen N, Baiker A, Zerboni L, Arvin AM. Entrapment of viral capsids in nuclear PML cages is an intrinsic antiviral host defense against varicella-zoster virus. PLoS Pathog. 2011;7:e1001266. doi: 10.1371/journal.ppat.1001266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Blondel D, Kheddache S, Lahaye X, Dianoux L, Chelbi-Alix MK. Resistance to rabies virus infection conferred by the PMLIV isoform. J Virol. 2010;84:10719–26. doi: 10.1128/JVI.01286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lin HK, Bergmann S, Pandolfi PP. Cytoplasmic PML function in TGF-beta signalling. Nature. 2004;431:205–11. doi: 10.1038/nature02783. [DOI] [PubMed] [Google Scholar]
- 140.McNally BA, Trgovcich J, Maul GG, Liu Y, Zheng P. A role for cytoplasmic PML in cellular resistance to viral infection. PLoS One. 2008;3:e2277. doi: 10.1371/journal.pone.0002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Passerini LD, Keckesova Z, Towers GJ. Retroviral restriction factors Fv1 and TRIM5alpha act independently and can compete for incoming virus before reverse transcription. J Virol. 2006;80:2100–5. doi: 10.1128/JVI.80.5.2100-2105.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–5. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 143.Short KM, Cox TC. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J Biol Chem. 2006;281:8970–80. doi: 10.1074/jbc.M512755200. [DOI] [PubMed] [Google Scholar]
- 144.Rajsbaum R, Garcia-Sastre A. Viral evasion mechanisms of early antiviral responses involving regulation of ubiquitin pathways. Trends Microbiol. 2013;21:421–9. doi: 10.1016/j.tim.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lee Y, Song B, Park C, Kwon KS. TRIM11 negatively regulates IFNbeta production and antiviral activity by targeting TBK1. PLoS One. 2013;8:e63255. doi: 10.1371/journal.pone.0063255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Higgs R, Ni Gabhann J, Ben Larbi N, Breen EP, Fitzgerald KA, Jefferies CA. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J Immunol. 2008;181:1780–6. doi: 10.4049/jimmunol.181.3.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Young JA, Sermwittayawong D, Kim HJ, Nandu S, An N, Erdjument-Bromage H, Tempst P, Coscoy L, Winoto A. Fas-associated death domain (FADD) and the E3 ubiquitin-protein ligase TRIM21 interact to negatively regulate virus-induced interferon production. J Biol Chem. 2011;286:6521–31. doi: 10.1074/jbc.M110.172288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang Z, Bao M, Lu N, Weng L, Yuan B, Liu YJ. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat Immunol. 2013;14:172–8. doi: 10.1038/ni.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wada K, Niida M, Tanaka M, Kamitani T. Ro52-mediated monoubiquitination of IKK{beta} down-regulates NF-{kappa}B signalling. J Biochem. 2009;146:821–32. doi: 10.1093/jb/mvp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kong HJ, Anderson DE, Lee CH, Jang MK, Tamura T, Tailor P, Cho HK, Cheong J, Xiong H, Morse HC, 3rd, Ozato K. Cutting edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J Immunol. 2007;179:26–30. doi: 10.4049/jimmunol.179.1.26. [DOI] [PubMed] [Google Scholar]
- 151.Shi M, Deng W, Bi E, Mao K, Ji Y, Lin G, Wu X, Tao Z, Li Z, Cai X, Sun S, Xiang C, Sun B. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat Immunol. 2008;9:369–77. doi: 10.1038/ni1577. [DOI] [PubMed] [Google Scholar]
- 152.Kondo T, Watanabe M, Hatakeyama S. TRIM59 interacts with ECSIT and negatively regulates NF-kappaB and IRF-3/7-mediated signal pathways. Biochem Biophys Res Commun. 2012;422:501–7. doi: 10.1016/j.bbrc.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 153.Xue Q, Zhou Z, Lei X, Liu X, He B, Wang J, Hung T. TRIM38 negatively regulates TLR3-mediated IFN-beta signaling by targeting TRIF for degradation. PLoS One. 2012;7:e46825. doi: 10.1371/journal.pone.0046825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhao W, Wang L, Zhang M, Wang P, Yuan C, Qi J, Meng H, Gao C. Tripartite motif-containing protein 38 negatively regulates TLR3/4- and RIG-I-mediated IFN-beta production and antiviral response by targeting NAP1. J Immunol. 2012;188:5311–8. doi: 10.4049/jimmunol.1103506. [DOI] [PubMed] [Google Scholar]
- 155.Zhao W, Wang L, Zhang M, Yuan C, Gao C. E3 ubiquitin ligase tripartite motif 38 negatively regulates TLR-mediated immune responses by proteasomal degradation of TNF receptor-associated factor 6 in macrophages. J Immunol. 2012;188:2567–74. doi: 10.4049/jimmunol.1103255. [DOI] [PubMed] [Google Scholar]
- 156.Zha J, Han KJ, Xu LG, He W, Zhou Q, Chen D, Zhai Z, Shu HB. The Ret finger protein inhibits signaling mediated by the noncanonical and canonical IkappaB kinase family members. J Immunol. 2006;176:1072–80. doi: 10.4049/jimmunol.176.2.1072. [DOI] [PubMed] [Google Scholar]
- 157.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–49. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villan E, Garcia-Sastre A, Gack MU. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 2012;8:e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li Q, Yan J, Mao AP, Li C, Ran Y, Shu HB, Wang YY. Tripartite motif 8 (TRIM8) modulates TNFalpha- and IL-1beta-triggered NF-kappaB activation by targeting TAK1 for K63-linked polyubiquitination. Proc Natl Acad Sci U S A. 2011;108:19341–6. doi: 10.1073/pnas.1110946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang J, Hu MM, Wang YY, Shu HB. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J Biol Chem. 2012;287:28646–55. doi: 10.1074/jbc.M112.362608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yang B, Wang J, Wang Y, Zhou H, Wu X, Tian Z, Sun B. Novel function of Trim44 promotes an antiviral response by stabilizing VISA. J Immunol. 2013;190:3613–9. doi: 10.4049/jimmunol.1202507. [DOI] [PubMed] [Google Scholar]
- 162.Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001;20:7223–33. doi: 10.1038/sj.onc.1204765. [DOI] [PubMed] [Google Scholar]
- 163.Malfavon-Borja R, Wu LI, Emerman M, Malik HS. Birth, decay, and reconstruction of an ancient TRIMCyp gene fusion in primate genomes. Proc Natl Acad Sci U S A. 2013;110:E583–92. doi: 10.1073/pnas.1216542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Battivelli E, Migraine J, Lecossier D, Matsuoka S, Perez-Bercoff D, Saragosti S, Clavel F, Hance AJ. Modulation of TRIM5alpha activity in human cells by alternatively spliced TRIM5 isoforms. J Virol. 2011;85:7828–35. doi: 10.1128/JVI.00648-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wilson SJ, Webb BL, Maplanka C, Newman RM, Verschoor EJ, Heeney JL, Towers GJ. Rhesus macaque TRIM5 alleles have divergent antiretroviral specificities. J Virol. 2008;82:7243–7. doi: 10.1128/JVI.00307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kirmaier A, Wu F, Newman RM, Hall LR, Morgan JS, O’Connor S, Marx PA, Meythaler M, Goldstein S, Buckler-White A, Kaur A, Hirsch VM, Johnson WE. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wu F, Kirmaier A, Goeken R, Ourmanov I, Hall L, Morgan JS, Matsuda K, Buckler-White A, Tomioka K, Plishka R, Whitted S, Johnson W, Hirsch VM. TRIM5 alpha drives SIVsmm evolution in rhesus macaques. PLoS Pathog. 2013;9:e1003577. doi: 10.1371/journal.ppat.1003577. [DOI] [PMC free article] [PubMed] [Google Scholar]