Abstract
Dengue is the most prevalent arthropod-borne virus, with at least 40% of the world's population at risk of infection each year. In Australia, dengue is not endemic, but viremic travelers trigger outbreaks involving hundreds of cases. We compared the susceptibility of Aedes aegypti mosquitoes from two geographically isolated populations to two strains of dengue virus serotype 2. We found, interestingly, that mosquitoes from a city with no history of dengue were more susceptible to virus than mosquitoes from an outbreak-prone region, particularly with respect to one dengue strain. These findings suggest recent evolution of population-based differences in vector competence or different historical origins. Future genomic comparisons of these populations could reveal the genetic basis of vector competence and the relative role of selection and stochastic processes in shaping their differences. Lastly, we show the novel finding of a correlation between midgut dengue titer and titer in tissues colonized after dissemination.
Introduction
Human infection with dengue viruses (DENVs) causes a spectrum of disease that ranges from classical dengue fever to the life-threatening dengue hemorrhagic fever. DENVs, designated serotypes 1–4, are RNA viruses belonging to the genus Flavivirus and vectored by mosquitoes, most commonly Aedes aegypti.1–4 In the past 20 years, dengue has emerged to become the most prevalent arthropod-borne virus affecting humans today. This exponential increase in disease incidence has brought with it significant health, social, and economic problems.5,6 It has also led to an increase in genetic diversity in DENV populations, enhancing the probability that new strains will emerge with enhanced pathogenic properties.7
The evolutionary history of dengue is very recent, with the four serotypes originating approximately 1,000 years ago.8 Selection within the vertebrate host and the vector9 as well as stochastic processes have shaped the key differences between dengue serotypes and strains.10 Recent genomic approaches have shown how selection within the human can affect virus transmission and subsequently, the amount of circulating interhost variation in virus strains.11 In contrast, selection in the vector is thought to explain the displacement of the American DENV type 2 (DENV-2) genotype (AM) by the Southeast Asian DENV-2 genotype (SEA) in the Western Hemisphere, because SEA strains produce higher midgut infection rates and disseminate to the salivary glands earlier, leading to an increase in vectorial capacity.12–14 Also, recent studies have revealed how mosquitoes from across geographic regions vary in their ability to transmit genetically distinct dengue strains.15 These vector genotype × virus genotype (G × G) interactions could allow coevolution, where viruses with a long history of circulation in an area become well-adapted to infecting local mosquitoes. Abundant genetic variation and plasticity in vector competence of mosquitoes is a prerequisite for local adaptation.16–19
Although dengue is not endemic to Australia, epidemics occur when travelers infected with the virus enter Australia and are bitten by local mosquitoes.20,21 The magnitude of outbreaks in far north Queensland (where A. aegypti are found) ranges from tens to hundreds of cases, including the largest outbreak in 2008–2009 of up to 1,000 cases across Cairns and Townsville.22 Although A. aegypti has a widespread distribution in north Queensland, the majority of epidemic activity has occurred in the cities of Cairns and Townsville. Despite the presence of the vector, local transmission has not been reported in many locations since the first one-half of the 20th century.20,22 Although anthropogenic factors, such as the number of viremic travelers visiting a given location, could influence the risk of local transmission, differences in local disease incidence may, in part, be explained by degree of compatibility between particular virus and vector genotypes.
In Australia, most of the variation in mosquito:dengue compatibility is likely to result from variation in the virus, because A. aegypti has only recently been introduced into Australia in the last 200 years23; therefore, it harbors limited genetic variation.24 Also, because dengue is not endemic, selection pressure on the vector from the virus is most likely rare and episodic rather than constant. A previous study examining vector competence in Australian A. aegypti revealed variation in the trait, although the vast majority of difference was between Torres Strait Island and mainland Australian populations.25 Here, we explore the vector competence of A. aegypti collected from a wild population in Cairns and a geographically distinct population in Rockhampton, a city without dengue activity for at least the last 50 years. We used two different low-passage DENV-2 strains: one strain isolated from a patient in Australia during a large Townsville outbreak in 1992 (92-T) and another strain from an Australian soldier infected with DENV during deployment to Timor-Leste in the year 2000 (ET-300). Although both populations were highly susceptible to ET-300, the Rockhampton population was especially susceptible to 92-T. This finding highlights the recent evolution of vector competence genotypes within Australia, but also, it suggests that other factors (human, ecological, etc.) may be more important in determining disease incidence given the low rates of dengue in Rockhampton. We also highlight a novel finding that, beyond a threshold of infection in the midgut, there is a correlation between midgut titer and titer in subsequently infected tissues after dissemination. This predictive relationship may be a general feature of mosquito:dengue associations.
Materials and methods
Mosquitoes.
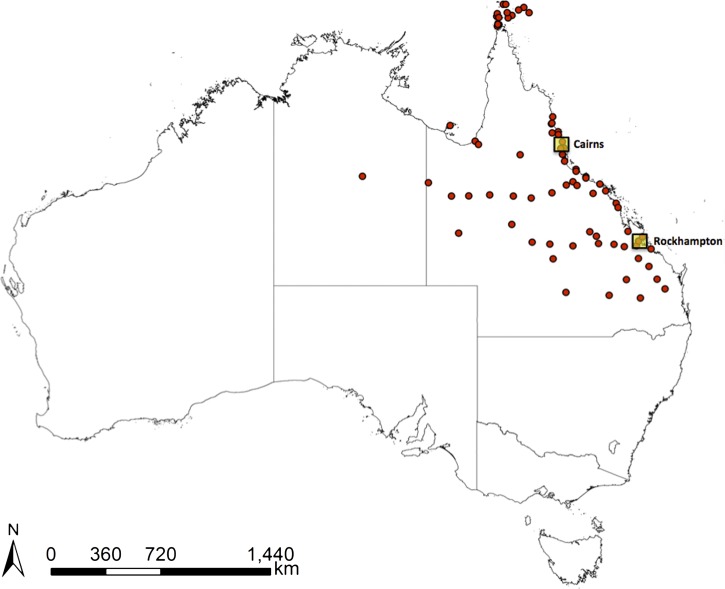
Laboratory populations were derived from A. aegypti eggs collected in ovitraps in Cairns and Rockhampton in late 2010 (Figure 1).26 All subsequent experimental work was carried out on the two laboratory lines between generations 3–5 from field isolation to preserve genetic diversity. After hatching, larvae were reared at a standard density of 150 individuals per 3 L distilled water in 30 × 40 × 8-cm plastic trays and fed fish food (Tetramin Tropical Tablets; Tetra, Melle, Germany) until pupation. Pupae were transferred to 30 × 30 × 30-cm cages to allow adult emergence. All mosquitoes were maintained in a controlled environment insectary at 25°C with ~70% relative humidity and 12:12-hour light:dark cycles. Adults were maintained on a 10% sucrose diet.27
Figure 1.
Map of Australia showing the locations of Cairns and Rockhampton as well as sites where A. aegypti has been collected since 1980.26
Viruses.
Two DENV-2 strains were used in the experiments. The 92-T strain of DENV was isolated from an infected patient during the 1992 outbreak in Townsville and had been passaged seven times in A. albopictus C6/36 cells since isolation to generate sufficiently high titers for infection. The ET-300 strain of dengue virus was isolated from a patient in 2000 in Timor-Leste and passaged five times in C6/36 cells before experimental work (GenBank accession number EF440433.1). To propagate the two virus strains, C6/36 cells were grown at 26°C in RPMI 1640 media (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 1× Glutamax (Invitrogen), and 25 mM HEPES buffer (Sigma Aldrich, St. Louis, MO). Cells were allowed to form monolayers in large T175 flasks, and then, they inoculated with virus and maintained in media containing only 2% FBS. At day 7 post-infection, virus was collected by harvesting the cell culture media and centrifuged at 3,200 × g for 15 minutes at 4°C. Virus stocks were stored at −80°C in single-use aliquots. Dengue virus stocks were titrated using plaque assays and adjusted to a final titer of 106 plaque-forming units (pfus) per milliliter for feeding experiments.28
DENV infection.
For infection, 5- to 7-day-old female A. aegypti were deprived of sucrose for 24 hours and then allowed to feed on a blood meal consisting of defibrinated sheep blood mixed with an equal volume of DENV through a section of desalted porcine intestine stretched over a water-jacketed membrane feeding apparatus for 1 hour. Blood-engorged mosquitoes were sorted the following day and placed in 250-mL cups in groups of 8–10 females per cup. At 7, 10, 11, 12, 13, 14, and 16 days post-infection (dpi), midgut and head tissues were dissected and used as proxies of infection and dissemination, respectively. Only mosquitoes with DENV-positive midguts were progressed for dissemination in the head, and hence, the infection percentages in these two tissues are not directly comparable. Sample size collected per time point per treatment per virus strain ranged from 5 to 20 individuals, with a mean of 13 individuals. The entire experiment was repeated to generate a second independent replicate.
DENV quantification.
Tissue samples were homogenized using a mini Bead Beater (BioSpec Products Inc., Bartlesville, OK), and RNA was extracted from mosquito tissues using Trizol (Invitrogen) according to the manufacturer's instruction. RNA was resuspended in a final volume of 25 μL Mili Q H2O. A two-step approach was used for cDNA synthesis of DENV-2 RNA and subsequent quantification using quantitative polymerase chain reaction (qPCR).
The first-strand synthesis reaction contained 1.25 μL 10 μM DENV-2 reverse primer (5′-CGTTCTGTGCCTGGAATGATG-3′),29 0.4 μL 10 μM deoxynucleoside triphosphates mix, 8 μL template RNA, and 5.85 μL diethyl phosphorocyanidate-treated H2O to a final volume of 15.5 μL. Reactions were heated to denature double-stranded RNA (dsRNA) complexes (86°C for 15 minutes) and placed on ice for 5 minutes. A total of 4 μL First-Strand Buffer of 250 mM Tris·HCL, 375 mM KCL, and 15 mM MgCl2 and 0.5 μL 200 U/μL Superscript III RT Enzyme (Invitrogen) were added to obtain a final reaction volume of 20 μL. cDNA was synthesized at 25°C for 10 minutes, 42°C for 50 minutes, and 95°C for 10 minutes for reverse transcriptase inactivation and stored at −20°C. qPCR was performed on a Roche Lightcycler 480. Each reaction mixture contained 5 μL 2 × Lightcycler 480 Probe Master (Roche, Basel, Switzerland), 0.5 μL 10 μM forward primer (-5′ AAGGACTAGAGGTTAGAGGAGACCC-3′) and reverse primer (5′-CGTTCTGTGCCTGGAATGATG-3′-), 0.5 μL 10 μM hydrolysis probe (5′-FAM-AACAGCATATTGACGCTGGGAGAGACCAGA-BHQ1-3′) targeting the 3′ untranslated region (UTR) of dengue virus,29 1.5 μL H2O, and 2 μL cDNA in a final volume of 10 μL. The samples were analyzed under the following cycling conditions: 95°C for 5 minutes followed by 40 cycles at 95°C for 10 seconds, 60°C for 15 seconds, and 72°C for 1 second, and subsequently, 40°C for 10 seconds. The amplification efficiency of these primers did not differ for the two DENV strains.
Viral titer was expressed as dengue virus copy number per tissue using absolute quantification. A 107-bp fragment from the 3′ UTR region of the DENV (that is, in the same region that the primers amplify) was amplified and cloned into the pGEM-T vector system (Promega, Madison, WI). The plasmid was transformed into Escherichia coli, extracted using phenol-chloroform, and linearized by restriction enzyme digest. The copy number of the linearized plasmid was measured using the NanoDrop spectrophotometer. A standard curve of 107, 105, 104, 103, 102, and 50 DENV copies was constructed from a serial dilution. The limit of detection was set at 1,000 copies for this study, because it is the last dilution of the standard curve that amplified at least 95% of the time. The concentrations of DENV in the samples were extrapolated from the standard curve and expressed as concentration per tissue by back calculating to the initial concentration of RNA.30
Data analysis.
The proportion of mosquitoes with DENV-2 infection was analyzed with a binomial regression model with experimental replicate, mosquito population, and DENV-2 strain as categorical predictors and dpi as a continuous predictor. Viral titer data were analyzed using a general linear model with replicate, mosquito population, and DENV strain as categorical predictors and dpi as a continuous predictor. Models were run separately for midgut versus head. When the factor of experimental replicate was significant, simple main effects models were run blocked by experiment to assist with interpretation of results. Statistical analysis was performed with the software Statistica 8.0 (Statsoft, Inc., Tulsa, OK). Spearman's non-parametric r test was used to test for a statistically significant correlation between viral titer in the midgut and head in GraphPad Prism, version 5.00 (San Diego, CA).
Results
Geographically isolated mosquito populations from Cairns and Rockhampton were orally infected with either ET-300 (Timor-Leste isolate) or 92-T (Townsville outbreak strain) of DENV-2. Midgut infection was used as a measure of susceptibility to DENV-2 infection, and head infection was used as a measure of dissemination. DENV titers were obtained from both tissues. There is some variability between replicate experiments that is not surprising given the variability in virus activity between preparations and the interaction between the environment and mosquito physiology. We have sought to focus our conclusions only on those findings common to both replicate experiments.
Susceptibility to infection and dissemination.
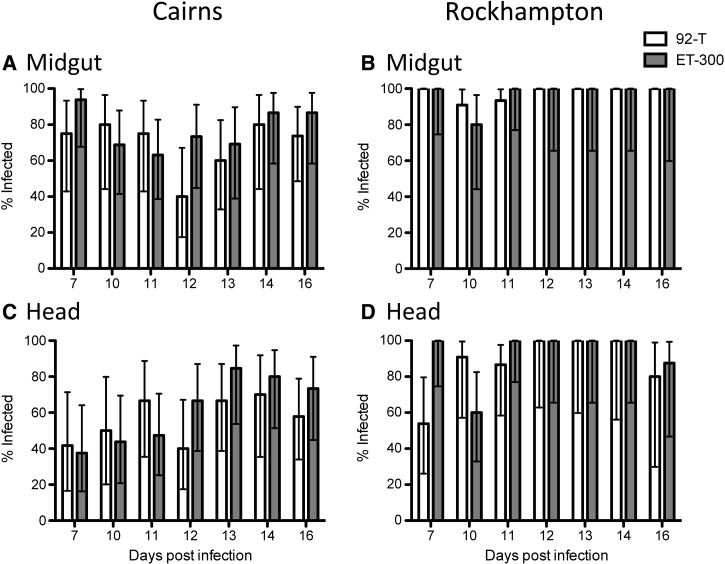
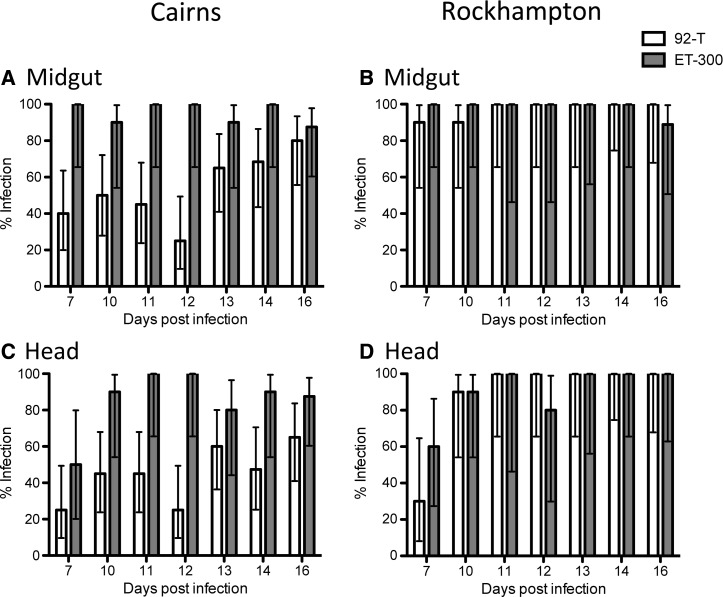
In the midgut, the three-way interaction of replicate experiment × mosquito population × DENV strain was significant (Table 1), requiring examination of a main effects model blocked by experimental replicate to interpret the data (Table 2). In both replicate experiments, there was a significant effect of mosquito population, with Rockhampton being more susceptible than Cairns (Figures 2 and 3). Midgut infection frequencies at 14 dpi were 100% for Rockhampton, whereas they were only 80–88% for Cairns (Figures 2 and 3). There is a marginal effect of dengue strain in replicate 2 only, with 92-T being less infective than ET-300 (Figure 3 and Table 2). In the head tissue, the three-way interaction was also significant (Table 1) along with all of the factors in the main effects blocked design (Table 2). Similar to the midgut, Rockhampton mosquitoes exhibited higher dissemination in the heads than Cairns mosquitoes (Figures 2 and 3). Infection rates in the head for Rockhampton were 100% at 14 dpi (both experiments) and 70–80% or 40–80% for Cairns, depending on the virus strain and replicate experiment. In Cairns, both viruses exhibited lower infection and dissemination than in Rockhampton, but this finding was especially the case for 92-T. In summary, Rockhampton mosquitoes exhibited greater infection and dissemination rates than Cairns mosquitoes, but the magnitude of the effect varied with respect to virus strain and replicate experiment. Cairns mosquitoes were particularly less susceptible to infection with ET-300.
Table 1.
Summary of statistical analysis of viral infection as a function of mosquito population, DENV strain, and replicate experiment for midgut and head separately
| df | Midgut | Head | |||
|---|---|---|---|---|---|
| F | P value | F | P value | ||
| dpi | 1 | 3.2 | 0.073 | 44.9 | < 0.001 |
| Experiment | 1 | 0.1 | 0.79 | 0.1 | 0.72 |
| Mosquito population | 1 | 80.3 | < 0.001 | 93.7 | < 0.001 |
| DENV strain | 1 | 22.6 | < 0.001 | 21.6 | < 0.001 |
| Experiment × mosquito population | 1 | 0.1 | 0.77 | 3.2 | 0.073 |
| Experiment × DENV strain | 1 | 8.6 | < 0.01 | 3.3 | 0.070 |
| Mosquito population × DENV strain | 1 | 20.4 | < 0.001 | 7.4 | < 0.01 |
| Experiment × mosquito population × DENV strain | 1 | 7.7 | < 0.01 | 12.2 | < 0.001 |
dpi is specified as a continuous predictor; df = degrees of freedom; F = F-ratio.
Table 2.
Summary of statistical analysis of main effects model for viral infection as a function of mosquito population and DENV strain blocked by replicate experiment for midgut and head separately
| df | Replicate 1 | Replicate 2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Midgut | Head | Midgut | Head | ||||||
| χ2 | P value | χ2 | P value | χ2 | P value | χ2 | P value | ||
| Mosquito population | 1 | 24.00 | < 0.001 | 31.75 | < 0.001 | 11.58 | 0.001 | 15.79 | < 0.001 |
| DENV strain | 1 | 0.35 | 0.54 | 6.17 | 0.013 | 5.46 | 0.019 | 8.43 | 0.004 |
| Mosquito population × DENV strain | 1 | 0.81 | 0.77 | 4.13 | 0.042 | 3.06 | 0.080 | 8.77 | 0.003 |
dpi is specified as a continuous predictor; df = degrees of freedom; F = F-ratio.
Figure 2.
Replicate experiment 1. (A and B) Infection rate for midgut and (C and D) dissemination rate for head tissues of mosquito populations from (A and C) Cairns or (B and D) Rockhampton infected with either 92-T (white) or ET-300 (grey). Each data point represents 7–19 females.
Figure 3.
Replicate experiment 2. (A and B) Infection rate for midgut and (C and D) dissemination rate for head tissues of mosquito populations from (A and C) Cairns or (B and D) Rockhampton infected with either 92-T (white) or ET-300 (grey). Each data point represents 5–20 females.
DENV titer.
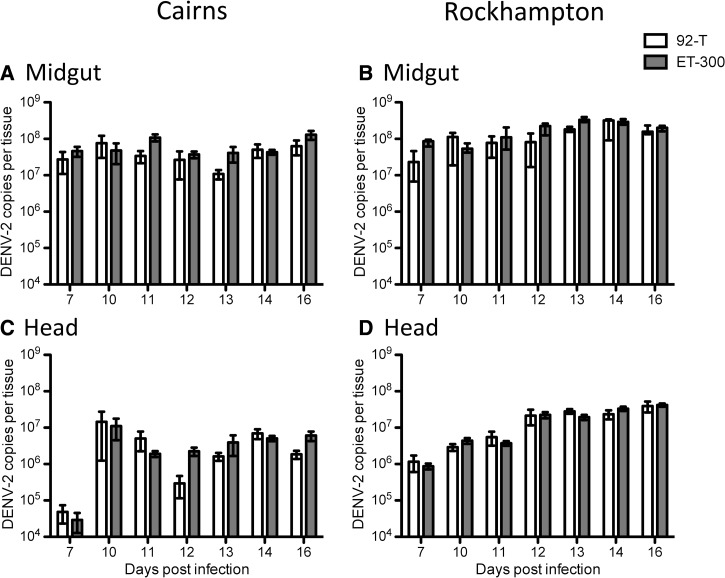
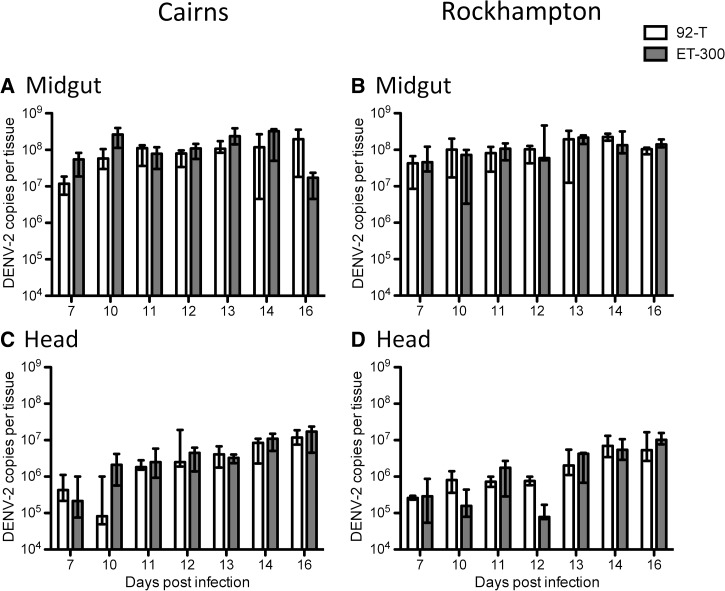
As with infection rates, there were differences between replicate experiments and several significant interactions (Table 3) requiring subsequent examination by models blocked by replicate. In the midgut, there was an overarching effect of DENV strain (Table 3), with ET-300 growing to higher titers regardless of replicate experiment or mosquito population (Figures 4 and 5). In replicate 1, Rockhampton mosquitoes exhibited slightly higher titers than Cairns mosquitoes (Figure 4 and Table 3). Head titer was less variable than midgut and exhibited some differences between replicate experiments. In replicate 1, mosquito population was the only factor in dictating titer, whereas in replicate 2, it was DENV strain (Figures 4 and 5 and Table 4). In summary, titer seemed to be less predictably affected by mosquito population or DENV strain than susceptibility and dissemination.
Table 3.
Summary of statistical analysis of viral titer as a function of mosquito population, DENV strain, and replicate experiment for midgut and head separately
| df | Midgut | Head | |||
|---|---|---|---|---|---|
| F | P value | F | P value | ||
| dpi | 1 | 76.0 | < 0.001 | 59.2 | < 0.001 |
| Experiment | 1 | 13.8 | < 0.001 | 6.4 | 0.012 |
| Mosquito population | 1 | 20.9 | < 0.001 | 26.8 | < 0.001 |
| DENV strain | 1 | 22.0 | < 0.001 | 0.00 | 0.99 |
| Experiment × mosquito population | 1 | 41.2 | < 0.001 | 17.6 | < 0.001 |
| Experiment × DENV strain | 1 | 0.00 | 0.952 | 0.22 | 0.63 |
| Mosquito population × DENV strain | 1 | 0.42 | 0.837 | 7.4 | < 0.01 |
| Experiment × mosquito population × DENV strain | 1 | 3.09 | 0.079 | 0.63 | 0.42 |
dpi is specified as a continuous predictor.
Figure 4.
Replicate experiment 1. Viral titer of (A and B) midgut and (C and D) head tissues of mosquito populations from (A and C) Cairns or (B and D) Rockhampton infected with either 92-T (white) or ET-300 (grey). Medians and interquartile ranges are plotted. Each data point represents 7–19 females.
Figure 5.
Replicate experiment 2. Viral titer of (A and B) midgut and (C and D) head tissues of mosquito populations from (A and C) Cairns or (B and D) Rockhampton infected with either 92-T (white) or ET-300 (grey). Medians and interquartile ranges are plotted. Each data point represents 5–20 females.
Table 4.
Summary of statistical analysis of main effects model for viral titer as a function of mosquito population and DENV strain blocked by replicate experiment for midgut and head separately
| df | Replicate 1 | Replicate 2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Midgut | Head | Midgut | Head | ||||||
| χ2 | P value | χ2 | P value | χ2 | P value | χ2 | P value | ||
| Mosquito population | 1 | 94.7 | < 0.001 | 66.39 | < 0.001 | 1.1 | 0.24 | 0.37 | 0.54 |
| DENV strain | 1 | 18.2 | < 0.001 | 0.19 | 0.65 | 7.9 | < 0.01 | 0.11 | 0.74 |
| Mosquito population × DENV strain | 1 | 2.1 | 0.14 | 0.001 | 0.98 | 1.4 | 1.13 | 1.38 | 0.24 |
dpi is specified as a continuous predictor.
Viral dissemination is associated with midgut viral titer.
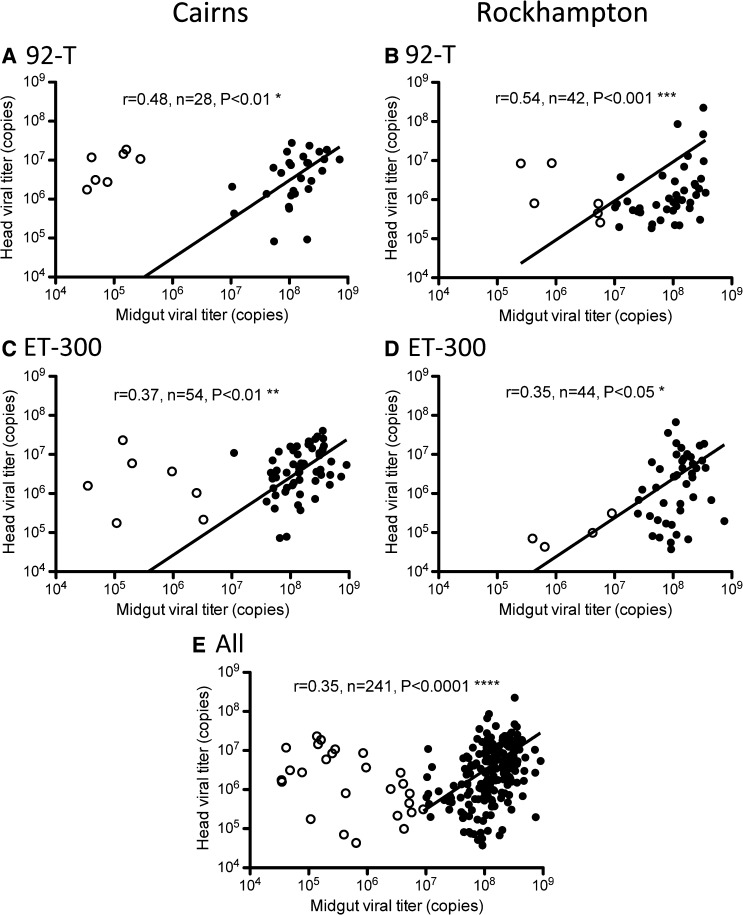
Overall, virus did not disseminate in 23% (63/275) of the mosquitoes that developed a midgut infection. In those mosquitoes with dissemination, the midgut viral titer was then plotted against head viral titer according to mosquito population and dengue strain (Figure 6A–D) or with all data combined (Figure 6E) for all individuals with disseminated infections. At lower midgut viral titers (Figure 6, hollow circles) (from 104 to 107), the amount of virus in the midgut was not positively correlated with the viral titer in the head. Mosquitoes that had midgut infections of high titer (Figure 6, solid circles) (> 107 DENV copies), however, also had higher viral titers in the head. Our data suggest that there is a threshold titer in the infected midgut above which the virus disseminates and influences virus titer in the head tissue.
Figure 6.
Viral titer of midgut versus head pooled across dpi for Cairns mosquitoes infected with (A) 92-T and (C) ET-300 and Rockhampton mosquitoes infected with (B) 92-T and (D) ET-300. All the individuals from both mosquito populations and DENV strains from the experiment are shown in E. Linear least-square fit lines are plotted. Spearman's non-parametric r test is used to test for a correlation between midgut and head titer. Individuals with midgut viral titer < 107 (hollow circles) were excluded from the analysis.
Discussion
We compared the susceptibility of two geographically distinct populations (> 1,000 km apart) of A. aegypti from north Queensland to a local outbreak strain of DENV-2 isolated from Australia in 1992 and a DENV strain from neighboring Timor-Leste by examining midgut infection and dissemination rates as well as quantifying viral titer. Surprisingly, mosquitoes from Rockhampton, where no confirmed cases of locally acquired dengue had been reported since recording began,20 were more easily infected by DENV than mosquitoes from Cairns, particularly in response to the 92-T dengue strain.
These findings are significant, because they indicate that, in a short period of time, differences in vector competence have evolved in Australian A. aegypti populations that may have been aided by multiple incursions of the vector.23 They also support the findings of a mosaic structure of mosquito × dengue interaction over geographic space as shown in Thailand.31 It is possible that the differences between populations are the result of random genetic drift, pleiotropic responses caused by selection for other traits, or greater selection for resistance in the vector in an outbreak-prone region. Recent studies of transcriptional profiles of mosquitoes infected with DENV show substantial changes in gene expression for innate immunity-, stress response-, metabolism-, and oxidoreductive process-associated genes, suggesting that the insect reacts to the presence of the infection and is mounting an active and possibly costly response against it.32,33 A meta-analysis of a range of studies examining mosquito survival, in contrast, showed that the impact of dengue infection was minor.31 Evolution of resistance in the Australian context is even more difficult to imagine given the rarity of outbreaks22 and the low frequency of dengue-infected mosquitoes in wild populations (0.1%) during epidemics.34 Nonetheless, a genomic comparison of mosquitoes from these two populations might help to identify variants that underpin differences in vector competence for dengue and then, determine if they have been driven by selection or more random population processes, such as different historical origins or processes like drift.
The absence of dengue cases in Rockhampton is counter to expectation given the level of susceptibility measured in this study, suggesting that other factors are more important in determining dengue incidence in these communities. Viremic humans are the most likely sources of importation of DENV throughout the world.4 There are several sources of evidence. It is often observed that dengue infections occur in clusters; if a member of the household becomes infected, other members of the household are likely to be infected as well.35,36 This finding suggests that population density and physical proximity between households are key determinants of outbreaks. A study on the genetic structure of mosquito populations from different islands of French Polynesia showed that the circulation of dengue is unlikely to be caused by the movement of infected mosquitoes but rather, is likely caused by the transportation of people.37 Cairns city is a regional hub, with large numbers of travelers arriving from overseas for tourism and using it as a transit point to other destinations within Australia. There are, hence, more opportunities for local A. aegypti to bite humans carrying dengue than in other sites in Australia. Furthermore, the genetic structure of A. aegypti populations in Australia is likely to be influenced by a combination of human population density, insect control, and ecological factors.38,39 The reasons why Cairns and Townsville (but not Rockhampton) remain receptive to dengue outbreaks may also be partially attributed to larger resident populations of A. aegypti.22,26
For both DENV strains, we found that DENV midgut titers above a baseline of 107 copies/mL can be predictive of titers in secondary tissues and virus dissemination. This finding is novel, because previous reports have found no clear relationship between dengue midgut titer and titer in the commonly used proxy for dissemination (heads).40 However, in a parallel experimental system with Venezuelan equine encephalitis virus, viral diversity did not experience a bottleneck at the midgut level when infected in high titers but did at low titers.41 This finding and our data suggest greater stochasticity in the lower titer ranges and highlight the need to understand the behavior of different titers from low to high when interpreting experimental findings.
ACKNOWLEDGMENTS
The authors thank Nichola Kenny for assistance with mosquito rearing and Alison Carrasco for assistance with PCR.
Footnotes
Financial support: This work was supported by grants from the National Health and Medical Research Council Australia.
Authors' addresses: Yixin H. Ye, Tat Siong Ng, Thomas Walker, and Elizabeth A. McGraw, School of Biological Sciences, Monash University, Clayton, VIC, Australia, E-mails: henry.ye@monash.edu, tatsiong.ng@uqconnect.edu.au, thomas_walker79@hotmail.com, and beth.mcgraw@monash.edu. Francesca D. Frentiu, Institute for Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, Australia, E-mail: francesca.frentiu@qut.edu.au. Andrew F. van den Hurk, Virology, Queensland Health Forensic and Scientific Services, Coopers Plains, QLD, Australia, E-mail: andrew_hurk@health.qld.gov.au. Scott L. O'Neill, Faculty of Science, School of Biological Sciences, Monash University, Clayton, VIC, Australia, and Institute of Molecular Biosciences, University of Queensland, St. Lucia, QLD, Australia, E-mail: scott.oneill@monash.edu. Nigel W. Beebe, School of Biological Sciences, University of Queensland, St. Lucia, QLD, Australia, E-mail: n.beebe@uq.edu.au.
References
- 1.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martinez E, Nathan MB, Pelegrino JL, Simmons C, Yoksan S, Peeling RW. Dengue: a continuing global threat. Nat Rev Microbiol. 2010;8:S7–S16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kyle JL, Harris E. Global spread and persistence of dengue. Annu Rev Microbiol. 2008;62:71–92. doi: 10.1146/annurev.micro.62.081307.163005. [DOI] [PubMed] [Google Scholar]
- 3.Ross TM. Dengue virus. Clin Lab Med. 2010;30:149–160. doi: 10.1016/j.cll.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halstead SB. Dengue virus-mosquito interactions. Annu Rev Entomol. 2008;53:273–291. doi: 10.1146/annurev.ento.53.103106.093326. [DOI] [PubMed] [Google Scholar]
- 5.Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gubler DJ. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002;10:100–103. doi: 10.1016/s0966-842x(01)02288-0. [DOI] [PubMed] [Google Scholar]
- 7.Holmes EC, Burch SS. The causes and consequences of genetic variation in dengue virus. Trends Microbiol. 2000;8:74–77. doi: 10.1016/s0966-842x(99)01669-8. [DOI] [PubMed] [Google Scholar]
- 8.Vasilakis N, Weaver SC. The history and evolution of human dengue emergence. Adv Virus Res. 2008;72:1–76. doi: 10.1016/S0065-3527(08)00401-6. [DOI] [PubMed] [Google Scholar]
- 9.Chevillon C, Failloux AB. Questions on viral population biology to complete dengue puzzle. Trends Microbiol. 2003;11:415–421. doi: 10.1016/s0966-842x(03)00206-3. [DOI] [PubMed] [Google Scholar]
- 10.Holmes EC. Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J Virol. 2003;77:11296–11298. doi: 10.1128/JVI.77.20.11296-11298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parameswaran P, Charlebois P, Tellez Y, Nunez A, Ryan EM, Malboeuf CM, Levin JZ, Lennon NJ, Balmaseda A, Harris E, Henn MR. Genome-wide patterns of intrahuman dengue virus diversity reveal associations with viral phylogenetic clade and interhost diversity. J Virol. 2012;86:8546–8558. doi: 10.1128/JVI.00736-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson JR, Rico-Hesse R. Aedes aegypti vectorial capacity is determined by the infecting genotype of dengue virus. Am J Trop Med Hyg. 2006;75:886–892. [PMC free article] [PubMed] [Google Scholar]
- 13.Armstrong PM, Rico-Hesse R. Differential susceptibility of Aedes aegypti to infection by the American and Southeast Asian genotypes of dengue type 2 virus. Vector-Borne Zoonotic Dis. 2001;1:159–168. doi: 10.1089/153036601316977769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armstrong PM, Rico-Hesse R. Efficiency of dengue serotype 2 virus strains to infect and disseminate in Aedes aegypti. Am J Trop Med Hyg. 2003;68:539–544. doi: 10.4269/ajtmh.2003.68.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambrechts L, Chevillon C, Albright RG, Thaisomboonsuk B, Richardson JH, Jarman RG, Scott TW. Genetic specificity and potential for local adaptation between dengue viruses and mosquito vectors. BMC Evol Biol. 2009;9:160. doi: 10.1186/1471-2148-9-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett KE, Olson KE, Munoz MD, Fernandez-Salas I, Farfan-Ale JA, Higgs S, Black WC, Beaty BJ. Variation in vector competence for dengue 2 virus among 24 collections of Aedes aegypti from Mexico and the United States. Am J Trop Med Hyg. 2002;67:85–92. doi: 10.4269/ajtmh.2002.67.85. [DOI] [PubMed] [Google Scholar]
- 17.Gubler DJ, Nalim S, Tan R, Saipan H, Suliantisaroso J. Variation in susceptibility to oral infection with dengue viruses among geographic strains of Aedes aegypti. Am J Trop Med Hyg. 1979;28:1045–1052. doi: 10.4269/ajtmh.1979.28.1045. [DOI] [PubMed] [Google Scholar]
- 18.Vazeille-Falcoz M, Mousson L, Rodhain F, Chungue E, Failloux AB. Variation in oral susceptibility to dengue type 2 virus of populations of Aedes aegypti from the islands of Tahiti and Moorea, French Polynesia. Am J Trop Med Hyg. 1999;60:292–299. doi: 10.4269/ajtmh.1999.60.292. [DOI] [PubMed] [Google Scholar]
- 19.Rosen L, Roseboom LE, Gubler DJ, Lien JC, Chaniotis BN. Comparative susceptibility of mosquito species and strains to oral and parenteral infection with dengue and Japanese encephalitis viruses. Am J Trop Med Hyg. 1985;34:603–615. doi: 10.4269/ajtmh.1985.34.603. [DOI] [PubMed] [Google Scholar]
- 20.Hanna JN, Ritchie SA. Outbreaks of dengue in North Queensland, 1990–2008. Commun Dis Intell. 2009;33:32–33. doi: 10.33321/cdi.2009.33.5. [DOI] [PubMed] [Google Scholar]
- 21.Van Den Hurk AF, Craig SB, Tulsiani SM, Jansen CC. Emerging tropical diseases in Australia. Part 4. Mosquitoborne diseases. Ann Trop Med Parasitol. 2010;104:623–640. doi: 10.1179/136485910X12851868779984. [DOI] [PubMed] [Google Scholar]
- 22.Queensland-Health . Dengue in North Queensland. 2012. http://www.health.qld.gov.au/dengue/outbreaks/previous.asp Available at. Accessed July 20, 2013. [Google Scholar]
- 23.Beebe NW, Whelan PI, van den Hurk A, Ritchie S, Cooper RD. Genetic diversity of the dengue vector Aedes aegypti in Australia and implications for future surveillance and mainland incursion monitoring. Commun Dis Intell. 2005;29:299–304. doi: 10.33321/cdi.2005.29.32. [DOI] [PubMed] [Google Scholar]
- 24.Endersby NM, Hoffmann AA, White VL, Lowenstein S, Ritchie S, Johnson PH, Rapley LP, Ryan PA, Nam VS, Yen NT, Kittayapong P, Weeks AR. Genetic structure of Aedes aegypti in Australia and Vietnam revealed by microsatellite and exon primed intron crossing markers suggests feasibility of local control options. J Med Entomol. 2009;46:1074–1083. doi: 10.1603/033.046.0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knox TB, Kay BH, Hall RA, Ryan PA. Enhanced vector competence of Aedes aegypti (Diptera: Culicidae) from the Torres Strait compared with mainland Australia for dengue 2 and 4 viruses. J Med Entomol. 2003;40:950–956. doi: 10.1603/0022-2585-40.6.950. [DOI] [PubMed] [Google Scholar]
- 26.Beebe NW, Cooper RD, Mottram P, Sweeney AW. Australia's dengue risk driven by human adaptation to climate change. PLoS Negl Trop Dis. 2009;3:e429. doi: 10.1371/journal.pntd.0000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreira LA, Saig E, Turley AP, Ribeiro JM, O'Neill SL, McGraw EA. Human probing behavior of Aedes aegypti when infected with a life-shortening strain of Wolbachia. PLoS Negl Trop Dis. 2009;3:e568. doi: 10.1371/journal.pntd.0000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frentiu FD, Robinson J, Young PR, McGraw EA, O'Neill SL. Wolbachia-mediated resistance to dengue virus infection and death at the cellular level. PLoS One. 2010;5:e13398. doi: 10.1371/journal.pone.0013398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warrilow D, Northill JA, Pyke A, Smith GA. Single rapid TaqMan fluorogenic probe based PCR assay that detects all four dengue serotypes. J Med Virol. 2002;66:524–528. doi: 10.1002/jmv.2176. [DOI] [PubMed] [Google Scholar]
- 30.Kien DTH, Tuan TV, Hanh TNT, Chau TNB, Huy HLA, Wills BA, Simmons CP. Validation of an internally controlled one-step real-time multiplex RT-PCR assay for the detection and quantitation of dengue virus RNA in plasma. J Med Virol. 2011;177:168–173. doi: 10.1016/j.jviromet.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lambrechts L, Scott TW. Mode of transmission and the evolution of arbovirus virulence in mosquito vectors. Proc Biol Sci. 2009;276:1369–1378. doi: 10.1098/rspb.2008.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye YH, Chenoweth SF, McGraw EA. Effective but costly, evolved mechanisms of defense against a virulent opportunistic pathogen in Drosophila melanogaster. PLoS Pathog. 2009;5:e1000385. doi: 10.1371/journal.ppat.1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoon IK, Getis A, Aldstadt J, Rothman AL, Tannitisupawong D, Koenraadt CJ, Fansiri T, Jones JW, Morrison AC, Jarman RG, Nisalak A, Mammen MP, Jr, Thammapalo S, Srikiatkhachorn A, Green S, Libraty DH, Gibbons RV, Endy T, Pimgate C, Scott TW. Fine scale spatiotemporal clustering of dengue virus transmission in children and Aedes aegypti in rural Thai villages. PLoS Neglect Trop Dis. 2012;6:e1730. doi: 10.1371/journal.pntd.0001730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neff JM, Morris L, Gonzalez R, Coleman PH, Lyss SB, Negron H. Dengue fever in a Puerto Rican community. Am J Epidemiol. 1967;86:162–184. doi: 10.1093/oxfordjournals.aje.a120722. [DOI] [PubMed] [Google Scholar]
- 36.Halstead SB, Nimmannitta S, Margiotta MR. Dengue and Chikungunya virus infection in man in Thailand, 1962–1964: II. Observations on disease in outpatients. Am J Trop Med Hyg. 1969;18:972–983. doi: 10.4269/ajtmh.1969.18.972. [DOI] [PubMed] [Google Scholar]
- 37.Failloux AB, Darius H, Pasteur N. Genetic differentiation of Aedes aegypti, the vector of dengue virus in French Polynesia. J Am Mosq Control Assoc. 1995;11:457–462. [PubMed] [Google Scholar]
- 38.Huber K, Loan LL, Chantha N, Failloux AB. Human transportation influences Aedes aegypti gene flow in Southeast Asia. Acta Trop. 2004;90:23–29. doi: 10.1016/j.actatropica.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 39.Paupy C, Vazeille-Falcoz M, Mousson L, Rodhain F, Failloux AB. Aedes aegypti in Tahiti and Moorea (French Polynesia): isoenzyme differentiation in the mosquito population according to human population density. Am J Trop Med Hyg. 2000;62:217–224. doi: 10.4269/ajtmh.2000.62.217. [DOI] [PubMed] [Google Scholar]
- 40.Bosio CF, Beaty BJ, Black WC., 4th Quantitative genetics of vector competence for dengue-2 virus in Aedes aegypti. Am J Trop Med Hyg. 1998;59:965–970. doi: 10.4269/ajtmh.1998.59.965. [DOI] [PubMed] [Google Scholar]
- 41.Forrester NL, Guerbois M, Seymour RL, Spratt H, Weaver SC. Vector-borne transmission imposes a severe bottleneck on an RNA virus population. PLoS Pathog. 2012;8:e1002897. doi: 10.1371/journal.ppat.1002897. [DOI] [PMC free article] [PubMed] [Google Scholar]