Abstract
Dengue virus (DENV) is the most important mosquito-transmitted flavivirus that is transmitted throughout the tropical and subtropical regions of the world. The primary mosquito vector of DENV in urban locations is Aedes aegypti. Key to understanding the transmission of DENV is the relationship between pathogen and vector. Accordingly, we report our preliminary characterization of the differentially expressed proteins from Ae. aegypti mosquitoes after DENV infection. We investigated the virus–vector interaction through changes in the proteome of the salivary glands of mosquitoes with disseminated DENV serotype 2 (DENV-2) infections using two-dimensional gel electrophoresis and identification by mass spectrometry. Our findings indicate that DENV-2 infection in the Ae. aegypti salivary gland alters the expression of structural, secreted, and metabolic proteins. These changes in the salivary gland proteome highlight the virally influenced environment caused by a DENV-2 infection and warrant additional investigation to determine if these differences extend to the expectorated saliva.
Introduction
Dengue virus (DENV) is the most important mosquito-transmitted flavivirus throughout the tropical and subtropical regions of the world. Among 2.5 billion people at risk for exposure, DENV is responsible for over 50 million cases each year in affected countries.1,2 The first epidemics of dengue fever were documented over 200 years ago and have been occurring with increasing frequency.3 Since the 1950s, dengue hemorrhagic fever and dengue shock syndrome have emerged as serious public health threats, becoming the leading causes of hospitalization among children in several Southeast Asian countries.4
The primary mosquito vector of DENV in urban locations is Aedes aegypti.5 Key to understanding the transmission of DENV is the relationship between pathogen and vector. Much research has been done to examine vector infection by and transmission of DENV strains, but comparatively little is known about the direct influence of that infection on transmission.6–10 Vector competence experiments provide empirical findings on the relationship between infection and transmission potential; however, these studies rarely examine the underlying mechanisms of any perceived associations. Recent efforts to understand the role of innate vector immunity in limiting infection have yielded such possibilities as RNA interference and Toll pathways as modifiers of infection progress or kinetics, but the impact of such responses on transmission remains to be described.11–14
To date, researchers have explored the composition of uninfected Ae. aegypti salivary glands at a transcriptional level and the expressed protein level and even discerned the function of various salivary components.15–23 Others have attempted to define the effect that DENV has on the salivary gland proteome in silico or by one-dimensional gel electrophoresis without identifying individual proteins by mass spectrometry.24,25 Similarly, another group has looked at the salivary glands of Ae. aegypti infected with an alphavirus.26 As for DENV, the invertebrate response to infection that might be relevant to the transmission event in vivo using two-dimensional gel electrophoresis and mass spectrometry-based protein identification has yet to be detailed.
Therefore, we report our preliminary characterization of the differentially expressed proteins from Ae. aegypti mosquitoes during a DENV infection. Specifically, we investigated the virus–vector interaction through changes in the proteome of the salivary glands of mosquitoes with disseminated DENV serotype 2 (DENV-2) infections. Thereby, only productive infections that might result in transmission are considered. In summary, after a disseminated infection with DENV-2, several proteins were found to be differentially expressed in these structures of Ae. aegypti that have putative roles in the transmission of virus during feeding.
Materials and Methods
Virus.
DENV-2, which was isolated from a human patient in Jakarta, Indonesia in 1978, was used for this experiment. It was inoculated on African green monkey kidney (vero) cells grown at 37°C and 5% CO2 in Medium-199 with Earle's salts, Penicillin/Streptomycin/Amphotericin B, and 10% fetal bovine serum. After 5 days, the supernatant was harvested and used at a concentration of 2.76×106 plaque-forming units/mL.
Mosquitoes.
Laboratory strain Ae. aegypti Rockefeller were maintained under constant environmental conditions (28°C with a 16:8-hour light:dark photoperiod). The mosquitoes were allowed to feed on citrated bovine blood mixed with either (1) DENV-2–infected vero cell culture media or (2) non-infected control vero cell culture media at a ratio of 1:1 through a Hemotek feeding device (Discovery Workshops, Lancashire, England). After feeding, blood-fed females were sorted and kept at the same environmental conditions as above. After a 10-day extrinsic incubation period, mosquitoes were dissected, and their salivary glands were removed and stored individually at −80°C. Legs were removed and placed in 900 μL BA-1 diluent for viral RNA extraction and detection by quantitative reverse transcription polymerase chain reaction (qRT-PCR).27 Only the salivary glands from the mosquitoes with disseminated infections confirmed by qRT-PCR were combined to create a pool of infected salivary gland extract in 200 μL phosphate-buffered saline.
Two-dimensional gel electrophoresis.
The samples were subsequently taken through a ReadyPrep 2-D Cleanup Kit (Bio-Rad, Hercules, CA) and resuspended in two-dimensional rehydration buffer (Bio-Rad) consisting of 8 M urea, 2% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate hydrate (CHAPS), 50 mM dithiothreitol, 0.2% Bio-Lyte 3/10 ampholytes, and 0.001% bromophenol blue. Protein concentrations were determined with a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE); 30 μg protein diluted in the previously mentioned two-dimensional rehydration buffer to a final volume of 200 μL were loaded per sample per 11-cm (pH 3–10) non-linear immobilized pH gradient (IPG) strip (Bio-Rad) overlaid with approximately 2 mL molecular biology-grade mineral oil (Bio-Rad) to prevent evaporation. First-dimension electrophoresis conditions were 12-hour passive rehydration of the sample and the IPG strip followed by 15 minutes at 250 volts (v) and a rapid ramp up to 8,000 v over the course of 2.5 hours; subsequently, the strips were focused at 8,000 v until a total of 35,000 v/hours had been achieved. Then, a 500-v hold was implemented for approximately 30 minutes until the equilibration buffers were prepared. After the first-dimension isoelectric focusing, strips were equilibrated in three washes for 15 minutes each to remove excess mineral oil, reduce, and alkylate the proteins, respectively. The first buffer consisted of 6 M urea, 4% sodium dodecyl sulfate (SDS), 0.375 M Tris, and 20% glycerol, whereas the second and third buffers were the same as above but with the addition of 130 mM dithiothreitol for the second buffer and 130 mM iodoacetimide for the third buffer. The strips were then rinsed by briefly dipping into a graduated cylinder containing 25 mM Tris, 192 mM glycine, and 0.1% SDS at pH 8.3 (Tris-Glycine-SDS [TGS]) to remove all traces of the equilibration buffers; they were carefully loaded into the top well of a 12.5% Tris·HCl pre-cast second-dimension Criterion gel (Bio-Rad) and overlaid with molten overlay agarose consisting of 0.5% agarose in TGS buffer with 0.003% bromophenol blue (Bio-Rad). The gels were then loaded into a Dodeca (Bio-Rad) second-dimension rig loaded with TGS buffer and run for 2 hours at 100 v. The gels were then fixed in a 10% methanol and 7% acetic acid solution for 30 minutes, stained overnight with SYPRO Ruby (Molecular Probes, Eugene, OR), and destained for 30 minutes with 10% methanol and 7% acetic acid. The gels were then imaged on a Gel Doc XR Imager (Bio-Rad) using Quantity One 4.6.1 software (Bio-Rad) under the ultraviolet light setting and an exposure time of 20 seconds, and the resulting images were analyzed with PDQuest 8.0.1 Image Analysis Software (Bio-Rad).
Image analysis.
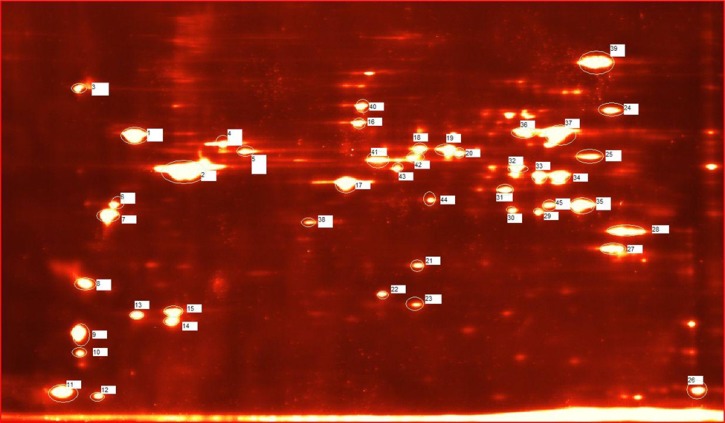
For each treatment group (infected and non-infected), duplicate two-dimensional gels were matched by each protein spot present to characterize the relative abundance of individual protein spots between each gel in a given treatment group. Gels were normalized according to PDQuest's normalization setting Total Quantity in Valid Spots. As per PDQuest's user manual, in this normalization method, the raw quantity of each spot in a member gel is divided by the total quantity of all the spots in that gel that have been included in the synthetic master image according to the normalization formula: normalized spot quantity = (raw spot quantity×scaling factor)/normalization factor. The default scaling factor 106 was chosen to provide parts per million (ppm) values to the spot intensity measurements. This normalization method assumes that few protein spots change within the experiment and that the changes average out across the whole gel. This method allowed determination of the changes in protein spot abundance caused by our experimental treatment. The relative expression levels of the individual spots were evaluated by calculating a fold change value per spot as the infected spot intensity divided by the uninfected control spot intensity to determine the change relative to infection with DENV-2. A representative gel image has been provided with the spots excised for mass spectrometry analysis marked and numbered for reference (Figure 1).
Figure 1.
Analyzed protein spots. Representative Ae. aegypti salivary gland extract two-dimensional gel image (12.5% Tris·HCl in TGS buffer using a pH 3–10 non-linear IPG strip) with the spots cut and numbered according to the MS data included in Supplementary Information.
Mass spectrometry.
After image analysis, representative gels containing all of the spots of interest from each group (infected and uninfected) were sent to the Harvard School of Public Health's Proteomics Research Center in the Department of Genetics and Complex Diseases for liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis using an LTQ Orbitrap Mass Spectrometer (Thermo Scientific). Differentially regulated spots of interest were manually cut from the gel that contained the spot with the highest intensity to maximize subsequent MS success (i.e., up-regulated spots were cut from the infected gel, and down-regulated spots were cut from the uninfected gel). Protein identification from spectra was performed using the SEQUEST-Sorcerer algorithm (Version 4.0.4) and the Sorcerer IDA2 search engine (Version 3.5 RC2; Sage-N Research, Thermo Scientific) on an Ae. aegypti-specific database downloaded from VectorBase.28 Additionally, results were searched against a non-redundant DENV-specific database (taxon ID 11052), and no DENV proteins were identified within the spots analyzed. Detailed peptide sequences, accession numbers, scores, and other MS-related data are included in Supplementary Information.
Results
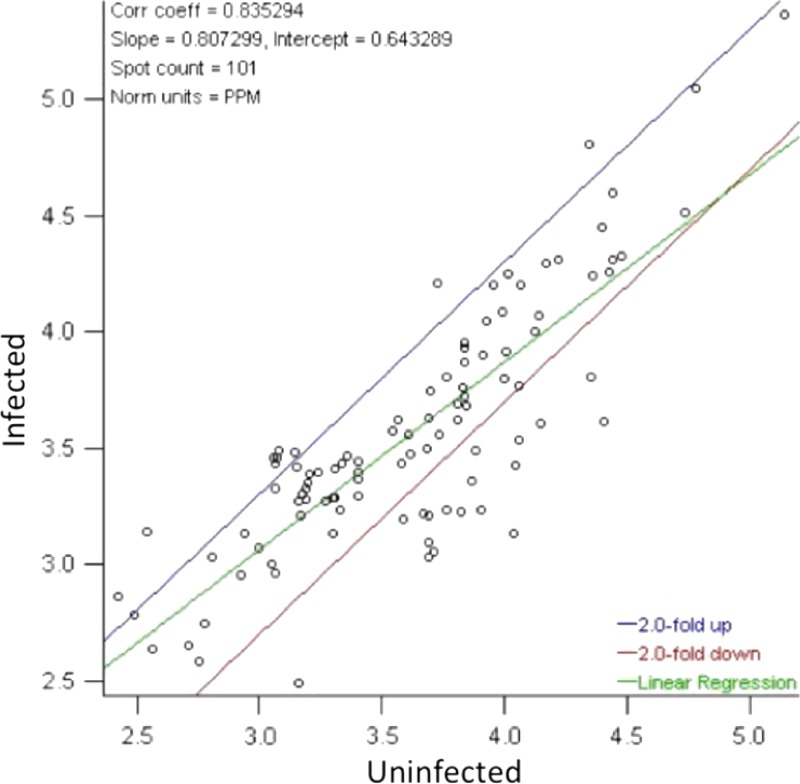
After normalization by the total quantities present in valid spots for each gel, a plot of the relative spot intensity was constructed (Figure 2). This plot includes the average linear relationship of spot intensity between the DENV-2 infected and non-infected control groups as well as bounds at the twofold regulation difference in either direction. Overall, the infected group had 20% less spot intensity than the uninfected group (regression slope = 0.807), showing a global trend to down-regulation of protein spots; 15 spots were found to be up-regulated in DENV-2–infected salivary glands (twofold or greater increase compared with the non-infected control group), and 65 spots were found to be down-regulated (twofold or greater decrease compared with the non-infected control group).
Figure 2.
Regression of normalized protein spot intensities comparing infected with uninfected Ae. aegypti salivary gland protein extracts. Protein spots located outside of the twofold differential regulation bounds (blue and red lines) correspond to a greater than twofold change in protein spot intensity between the treatments. The average linear relationship of the infected:uninfected protein spot intensities is given by the linear regression line (green) and indicates that infected salivary gland extract (SGE) has 20% less spot intensity than the uninfected group (regression slope = 0.807).
Among the up-regulated protein spots, 13 spots were below the limit of detection for MS analysis and therefore, were not analyzed. The remaining two spots with sufficient intensity for detection by LC-MS/MS were found to consist of the following proteins: phosphoglycerate kinase, troponin T, enolase, pyruvate dehydrogenase, isocitrate dehydrogenase, fructose bisphosphate aldolase, and ubiquinol cytochrome C reductase (Table 1).
Table 1.
Altered proteins with accession numbers and fold change information
| GenBank identification | Protein name | Spot | Spot fold change |
|---|---|---|---|
| gi|108879368| | Phosphoglycerate kinase | 20 | +3.01 |
| gi|108882176| | Troponin T | 20 | +3.01 |
| gi|108882996| | Enolase | 20 | +3.01 |
| gi|108869893| | Pyruvate dehydrogenase | 33 | +2.89 |
| gi|108873693| | Isocitrate dehydrogenase | 33 | +2.89 |
| gi|108878478| | Fructose-bisphosphate aldolase | 33 | +2.89 |
| gi|108879066| | Ubiquinol cytochrome-C reducatase | 33 | +2.89 |
| gi|108873586| | AaeL_AAEL010228 (30 kD allergen; aegyptin) | 8 | –3.50 |
| gi|108881783| | Tropomyosin | 3, 6, 7, 8 | –2.96, −3.20, −3.56, −3.50 |
| gi|108881993| | Myosin | 8 | –3.50 |
| gi|108869797| | Histone H4 | 3 | –2.96 |
| gi|108874452| | Actin | 3, 13, 15 | –2.96, −4.64, −7.98 |
| gi|108881105| | ATP synthase | 3, 13, 25, 27, 36 | –2.96, −4.64, −4.58, −4.53, −6.14 |
| gi|108871930| | Glutathione s-transferase | 13, 15 | –4.64, −7.98 |
| gi|108877202| | Dihydrolipamide dehydrogenase | 16 | –3.88 |
| gi|108880826| | Ubiquinol-cytochrome C reductase iron-sulfur subunit | 23 | –4.13 |
| gi|108873981| | Alanine aminotransferase | 36 | –6.14 |
| gi|108880696| | 4-Hydroxybutyrate CoA-transferase | 36 | –6.14 |
| gi|108877769| | D7 | 45 | –2.49 |
| gi|108880454| | Toll | 45 | –2.49 |
| gi|108875864| | Malate dehydrogenase | 27, 28 | –4.53, −3.36 |
| gi|108879274| | Electron transfer flavoprotein | 27 | –4.53 |
| gi|108882772| | Voltage-dependent anion channel | 27 | –4.53 |
| gi|108877302| | Succinyl-CoA synthetase | 28 | –3.36 |
| gi|108871873| | Citrate synthase | 25 | –4.58 |
| gi|108881521| | Mitochondrial NADH-dehydrogenase | 26 | –66.73 |
| gi|108879508| | NADH-ubuiquinone oxidoreductase 13 kD B-subunit | 26 | –66.73 |
| gi|108879930| | Cytochrome-C oxidase | 26 | –66.73 |
| gi|108883622| | AaeL_AAEL001082 (LSD-2) | 26 | –66.73 |
Fourteen of sixty-five down-regulated spots were within the limit of detection and thus, sequenceable. The following proteins were identified within the 14 spots and correspond to histone H4, actin, adenosine triphosphate (ATP) synthase, tropomyosin, hypothetical protein AaeL_AAEL010228 (a member of the 30 kD allergen family previously named aegyptin), myosin, gluthione s-transferase, ubiquinol cytochrome C reductase iron-sulfur subunit, dihydrolipoamide dehydrogenase, alanine aminotransferase, 4-hydroxybutrate CoA transferase, citrate synthase, D7, Toll, malate dehydrogenase, succinyl CoA-synthatase, electron transfer flavoprotein, voltage-dependent anion channel protein, mitochondrial reduced nicotinamide adenine dinucleotide (NADH) dehydrogenase, NADH-ubiquone oxoreducatase 13 kD subunit, cytochrome C oxidase, and conserved hypothetical protein AaeL_AAEL001082 (putatively identified as a lipid storage droplet-2 [LSD-2]) (Table 1).
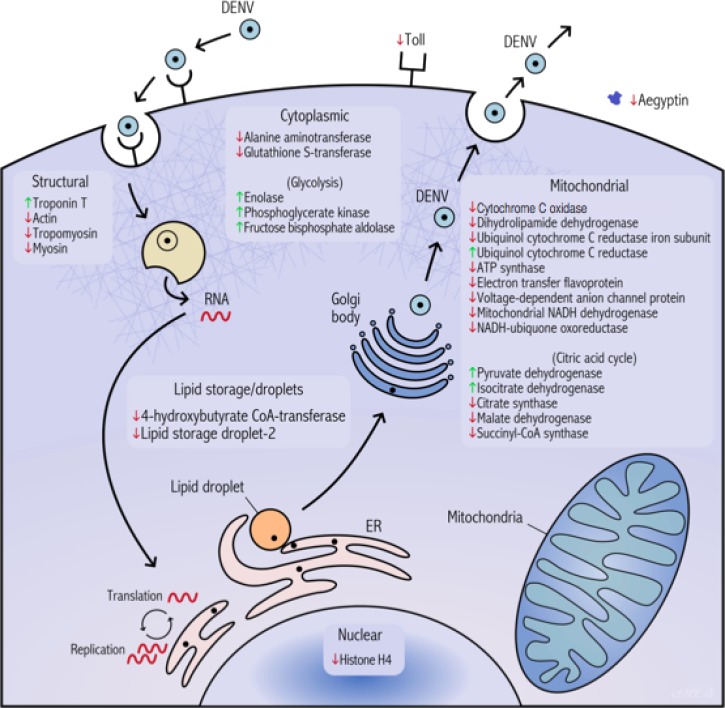
A graphical summary of these proteins and their respective changes is represented in Figure 3.
Figure 3.
Summary of proteomic changes in Ae. aegypti salivary glands in the context of a DENV infection at 10 DPE per os. The endoplasmic reticulum is abbreviated as ER.
In addition to the previously mentioned 16 differentially expressed spots that were detectable by MS, the remaining 33 most intense spots on the gel were also sequenced, and the identifications and data are in Supplemental Information. As has been previously reported by others, several spots corresponded to the same protein, which was interpreted as evidence of post-translational modifications.26 Additionally, the detection limitations of the MS analysis performed did not permit us to identify every single protein-level change detected during image analysis; therefore, there could be other protein-level changes occurring beyond the limit of our ability to identify.
Discussion
Differential gene expression during a viral infection is a complex process that is highly dependent on each study's experimental design with regard to time of analysis and detection methodology. In an attempt to bypass those constraints, we investigated the result of those interactions by quantifying protein changes at the expressed protein level by two-dimensional gel electrophoresis followed by image analysis. In our study, the number of proteins down-regulated compared with up-regulated in response to infection was more than four to one. In support of our findings, similar reductions have been seen by others in recent transcriptome analyses.29 Additionally, this finding contrasts with the findings of others who reported that transcript data obtained from Ae. aegypti were primarily up-regulated during infection.30 Differences between this study and our results can be attributed to several factors. Their study analyzed whole-mosquito carcasses, whereas we limited our analysis to excised salivary glands. Also, different strains of DENV-2 (1232 versus New Guinea C) were used. In addition, as others have noted in Drosophila melanogaster infected with Drosophila C virus, activation of host gene expression at the transcriptional level does not always provide evidence of equivalent translation of the corresponding proteins.12,20,31–33
Another functional explanation for our finding of a disparity between the number of down-regulated and up-regulated proteins may be the differences between salivary gland-specific analysis at the time point of 10 days post-exposure (DPE) and the generalized expression differences found throughout the carcass at that same time point. In this way, the different tissues in the carcass of the mosquito at 10 DPE were in varying states of viral infection (uninfected/infected/recovering) and therefore, had variable transcriptional responses to DENV infection, whereas the salivary glands themselves were the site of sustained viral infection. This finding is in accordance with the timetable of infection on various tissues examined by others, where at 10 days post-intrathoracic (i.t.) inoculation, the salivary glands were 100% positive for DENV-3 antigen; also, the compound eye and the levels of antigen were declining in the fat body and the brain, rising in the thoracic ganglion, and not yet present in the midgut (because of i.t.) and abdominal ganglion.34
This explanation is also supported by the work done by others, although different strains of mosquitoes (Rockefeller versus Chetumal/Rex-D/ D2S3) and DENV-2 (1232 versus Jam 1409) were used: at 10 DPE, the levels of viral antigen began to decline in the midgut along with a notable decrease at 7 DPE in the trachea, showing varying levels of viral infection in different tissues by 10 DPE.35 Also, the same study found that the salivary glands were infected as early as 4 DPE in the non-laboratory–adapted Chetumal mosquitoes and as late as 10 DPE in the laboratory-adapted Rex-D strain and that the percentage of infected salivary glands as well as the amount of viral antigen in the gland increased over time; these findings revealed a major DENV-2 tropism for this tissue, indicating that, at 10 DPE, the salivary glands were the site of active and sustained viral infection.35
Among the proteins that were found to be reduced in response to DENV-2 infection, of particular interest is spot 8 (consisting of tropomyosin and myosin) as well as the conserved hypothetical protein AaeL_AAEL010228 (VectorBase), a member of the glycine- and glutamate-(GE)-rich 30 kD allergen family that is also known as aegyptins.36,37 It may be that the reduction in aegyptin would also decrease the likelihood of an allergen-related immune response at the bite site if it is also reduced in the expectorated saliva. It has been shown that humans bitten by mosquitoes lacking the ability to expectorate saliva failed to produce wheals at the bite sites.38 Mosquito saliva induces numerous immunological reactions at the site of inoculation, including immunoglobulin E- (IgE-), IgG-, and T cell-mediated hypersensitivities, IgE-independent mast cell degranulation, and immune cell recruitment.39 A reduction in these inflammatory immune responses could benefit the establishment of DENV-2 within the vertebrate host.
In addition to the status of allergen, aegyptin has also been shown to bind to collagen and inhibit platelet aggregation, which facilitates blood feeding by aiding in the prevention of blood clots.37,40 A reduction in the expression of this protein could enhance the likelihood of a successful viral infection at the bite site by encouraging the mosquito to expectorate more saliva relative to an uninfected mosquito in an attempt to restore aegyptin levels to the point where it can disrupt the clotting process, consequently increasing the overall viral inoculum. Alternatively, increased clotting at the bite site from reduced aegyptin levels could lead to an unsuccessful feeding attempt despite successful viral deposition, because the non-replete female then would be forced to seek another host, where a subsequent transmission event could occur. Either of these scenarios linking virally induced protein changes to feeding disruption could be the physiological basis for the increase in probing and feeding as seen by others.34,41
Another down-regulated spot of interest (spot 45) was identified as having subunits of D7 and Toll. Although D7 was found in three additional spots (two spots containing only D7 and one spot containing only D7 and a hypothetical protein [AaeL_AAEL012032]), none of these spots was significantly altered, suggesting that the reduction seen in spot 45 is caused by the Toll subunit. Therefore, DENV-2 may be altering the innate mosquito response to the virus by reducing Toll. The Toll pathway has been implicated in resistance to infection by multiple DENV serotypes in multiple strains of Ae. aegypti.42 A reduction in the immune response against DENV-2 in the mosquito salivary glands could lead to increased viral titers in the expectorated saliva.
Spot 26 was also significantly reduced and shown to contain mitochondrial NADH dehydrogenase, NADH-ubiquone oxoreducatase 13 kD subunit, cytochrome C oxidase, and a conserved hypothetical protein (AaeL_AAEL001082). Although the composition of the spot is complex and it is impossible to determine which protein or proteins are responsible for the decrease in spot intensity, the hypothetical protein AaeL_AAEL001082 is an ortholog to the D. melanogaster protein LSD-2.36 The decreased expression of this protein would be notable, because DENV has been shown to have a tropism to the fat body of Ae. aegypti.43 If the reduction of this protein is seen throughout the mosquito, then this disruption of lipid-based energy homeostasis could result in an overall decrease in the energy stores of the mosquito, leading the mosquito to initiate additional host-seeking and feeding behavior and thereby, boosting DENV-2 viral fitness by additional transmission events. Alternatively, the reduction of this protein could also be because of the formation of vesicle packets from host cell lipid membranes, creating viral replication complexes as seen in another flavivirus infection (Kunjin virus).44
Throughout the salivary gland, there seems to be a trend to a decrease in membrane-bound, energy-related proteins, such as ATP synthase, electron transfer flavoprotein, voltage-dependent anion-channel protein, mitochondrial NADH dehydrogenase, NADH-ubiquone oxoreductase, and cytochrome C oxidase, perhaps because of viral disruption of cellular membranes.45 Other energy and/or lipid-related proteins found to be reduced were dihydrolipamide dehydrogenase, alanine aminotransferase, and 4-hydroxybutyrate CoA transferase.
Many of the remaining differentially expressed spots seem to contain proteins involved in metabolism, energy transport, and structural processes. For example, three up-regulated proteins—phosphoglycerate kinase, enolase, and fructose bisphosphate aldolase—are involved in glycolysis. In addition, isocitrate dehydrogenase and pyruvate dehydrogenase are components of the citric acid cycle, which were also increased. The enhanced expression of these particular metabolic proteins could be an attempt by the mosquito to compensate for the overall disruption of metabolism by the virus. However, three other proteins involved in the citric acid cycle (citrate synthase, malate dehydrogenase, and succinyl CoA-synthatase) were reduced, highlighting the importance for additional work on the differential regulation and function of these proteins. Some mosquito structural proteins were also found to have altered expression during DENV-2 infection. Troponin T, which binds with tropomyosin, was found to be up-regulated, whereas tropomyosin, myosin, and actin were down-regulated.46 The changes in these structural components could be caused by viral disruption of cellular scaffolding during infection.47 Histone H4 seems to be lowered, perhaps because of a reduction in normal host gene expression, causing fewer histone subunits to become uncoupled from their corresponding sections of nucleic acid.48 Glutathione S-transferase is reduced, perhaps because of its involvement in binding to and exporting exogenous toxins from infected cells.49–51 Finally, the conflicting finding of up-regulated ubiquinol cytochrome C reductase and down-regulated ubiquinol cytochrome C reductase's iron-sulfur subunit could be explained by virally induced disruption of iron metabolism leading to lack of iron substrate for synthesis of the iron-sulfur subunits.52
In summary, our findings indicate that DENV-2 infection in the Ae. aegypti salivary gland alters the expression of structural, secreted, and metabolic proteins. These changes may confer a fitness advantage on the virus by enhancing replication and dissemination in the mosquito or increasing the number of transmission events. Although this work is an important beginning, much information remains to be characterized. These salivary gland-specific changes (particularly, aegyptin) warrant additional investigation to determine if protein expression differences extend to the composition of expectorated saliva itself, thereby altering the viral inoculum and the resulting environment at the bite site.
Supplementary Material
Footnotes
Financial support: This work was supported by National Institutes of Health Grant P20GM103458.
Authors' addresses: Daniel M. Chisenhall, Berlin L. Londono, Rebecca C. Christofferson, Michael K. McCracken, and Christopher N. Mores, School of Veterinary Medicine, Department of Pathobiological Sciences, Louisiana State University, Baton Rouge, LA, E-mails: dchisenh@lsu.edu, berlin@lsu.edu, rcarri1@lsu.edu, mmccra4@lsu.edu, and cmores@lsu.edu.
References
- 1.Martina BE, Koraka P, Osterhaus AD. Dengue virus pathogenesis: an integrated view. Clin Microbiol Rev. 2009;22:564–581. doi: 10.1128/CMR.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weaver SC, Reisen WK. Present and future arboviral threats. Antiviral Res. 2010;85:328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gubler DJ. Dengue/dengue haemorrhagic fever: history and current status. Novartis Found Symp. 2006;277:3–16. doi: 10.1002/0470058005.ch2. [DOI] [PubMed] [Google Scholar]
- 4.Jarman RG, Holmes EC, Rodpradit P, Klungthong C, Gibbons RV, Nisalak A, Rothman AL, Libraty DH, Ennis FA, Mammen MP, Jr, Endy TP. Microevolution of dengue viruses circulating among primary school children in Kamphaeng Phet, Thailand. J Virol. 2008;82:5494–5500. doi: 10.1128/JVI.02728-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weaver SC, Vasilakis N. Molecular evolution of dengue viruses: contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect Genet Evol. 2009;9:523–540. doi: 10.1016/j.meegid.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alto BW, Reiskind MH, Lounibos LP. Size alters susceptibility of vectors to dengue virus infection and dissemination. Am J Trop Med Hyg. 2008;79:688–695. [PMC free article] [PubMed] [Google Scholar]
- 7.Alto BW, Lounibos LP, Mores CN, Reiskind MH. Larval competition alters susceptibility of adult Aedes mosquitoes to dengue infection. Proc Biol Sci. 2008;275:463–471. doi: 10.1098/rspb.2007.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knox TB, Kay BH, Hall RA, Ryan PA. Enhanced vector competence of Aedes aegypti (Diptera: Culicidae) from the Torres Strait compared with mainland Australia for dengue 2 and 4 viruses. J Med Entomol. 2003;40:950–956. doi: 10.1603/0022-2585-40.6.950. [DOI] [PubMed] [Google Scholar]
- 9.Bennett KE, Olson KE, Munoz Mde L, Fernandez-Salas I, Farfan-Ale JA, Higgs S, Black WC, 4th, Beaty BJ. Variation in vector competence for dengue 2 virus among 24 collections of Aedes aegypti from Mexico and the United States. Am J Trop Med Hyg. 2002;67:85–92. doi: 10.4269/ajtmh.2002.67.85. [DOI] [PubMed] [Google Scholar]
- 10.Black WC, 4th, Bennett KE, Gorrochotegui-Escalante N, Barillas-Mury CV, Fernandez-Salas I, de Lourdes Munoz M, Farfan-Ale JA, Olson KE, Beaty BJ. Flavivirus susceptibility in Aedes aegypti. Arch Med Res. 2002;33:379–388. doi: 10.1016/s0188-4409(02)00373-9. [DOI] [PubMed] [Google Scholar]
- 11.Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008;4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fragkoudis R, Attarzadeh-Yazdi G, Nash AA, Fazakerley JK, Kohl A. Advances in dissecting mosquito innate immune responses to arbovirus infection. J Gen Virol. 2009;90:2061–2072. doi: 10.1099/vir.0.013201-0. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez-Vargas I, Travanty EA, Keene KM, Franz AW, Beaty BJ, Blair CD, Olson KE. RNA interference, arthropod-borne viruses, and mosquitoes. Virus Res. 2004;102:65–74. doi: 10.1016/j.virusres.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez-Vargas I, Scott JC, Poole-Smith BK, Franz AW, Barbosa-Solomieu V, Wilusz J, Olson KE, Blair CD. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito's RNA interference pathway. PLoS Pathog. 2009;5:e1000299. doi: 10.1371/journal.ppat.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Champagne DE, Smartt CT, Ribeiro JM, James AA. The salivary gland-specific apyrase of the mosquito Aedes aegypti is a member of the 5′-nucleotidase family. Proc Natl Acad Sci USA. 1995;92:694–698. doi: 10.1073/pnas.92.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valenzuela JG, Pham VM, Garfield MK, Francischetti IM, Ribeiro JM. Toward a description of the sialome of the adult female mosquito Aedes aegypti. Insect Biochem Mol Biol. 2002;32:1101–1122. doi: 10.1016/s0965-1748(02)00047-4. [DOI] [PubMed] [Google Scholar]
- 17.Ribeiro JM, Arca B, Lombardo F, Calvo E, Phan VM, Chandra PK, Wikel SK. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics. 2007;8:6. doi: 10.1186/1471-2164-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smartt CT, Kim AP, Grossman GL, James AA. The Apyrase gene of the vector mosquito, Aedes aegypti, is expressed specifically in the adult female salivary glands. Exp Parasitol. 1995;81:239–248. doi: 10.1006/expr.1995.1114. [DOI] [PubMed] [Google Scholar]
- 19.Sim S, Ramirez JL, Dimopoulos G. Dengue virus infection of the Aedes aegypti salivary gland and chemosensory apparatus induces genes that modulate infection and blood-feeding behavior. PLoS Pathog. 2012;8:e1002631. doi: 10.1371/journal.ppat.1002631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almeras L, Fontaine A, Belghazi M, Bourdon S, Boucomont-Chapeaublanc E, Orlandi-Pradines E, Baragatti M, Corre-Catelin N, Reiter P, Pradines B, Fusai T, Rogier C. Salivary gland protein repertoire from Aedes aegypti mosquitoes. Vector Borne Zoonotic Dis. 2010;10:391–402. doi: 10.1089/vbz.2009.0042. [DOI] [PubMed] [Google Scholar]
- 21.Almeras L, Orlandi-Pradines E, Fontaine A, Villard C, Boucomont E, de Senneville LD, Baragatti M, Pascual A, Pradines B, Corre-Catelin N, Pages F, Reiter P, Rogier C, Fusai T. Sialome individuality between Aedes aegypti colonies. Vector Borne Zoonotic Dis. 2009;9:531–541. doi: 10.1089/vbz.2008.0056. [DOI] [PubMed] [Google Scholar]
- 22.Ribeiro JM, Francischetti IM. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- 23.Ribeiro JM. Blood-feeding arthropods: live syringes or invertebrate pharmacologists? Infect Agents Dis. 1995;4:143–152. [PubMed] [Google Scholar]
- 24.Guo X, Xu Y, Bian G, Pike AD, Xie Y, Xi Z. Response of the mosquito protein interaction network to dengue infection. BMC Genomics. 2010;11:380. doi: 10.1186/1471-2164-11-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wasinpiyamongkol L, Patramool S, Thongrungkiat S, Maneekan P, Sangmukdanan S, Misse D, Luplertlop N. Protein expression in the salivary glands of dengue-infected Aedes aegypti mosquitoes and blood-feeding success. Southeast Asian J Trop Med Public Health. 2012;43:1346–1357. [PubMed] [Google Scholar]
- 26.Tchankouo-Nguetcheu S, Bourguet E, Lenormand P, Rousselle JC, Namane A, Choumet V. Infection by chikungunya virus modulates the expression of several proteins in Aedes aegypti salivary glands. Parasit Vectors. 2012;5:264. doi: 10.1186/1756-3305-5-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chisenhall DM, Mores CN. Diversification of West Nile virus in a subtropical region. Virol J. 2009;6:106. doi: 10.1186/1743-422X-6-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Megy K, Emrich SJ, Lawson D, Campbell D, Dialynas E, Hughes DS, Koscielny G, Louis C, Maccallum RM, Redmond SN, Sheehan A, Topalis P, Wilson D. VectorBase: improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic Acids Res. 2012;40:D729–D734. doi: 10.1093/nar/gkr1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonizzoni M, Dunn WA, Campbell CL, Olson KE, Marinotti O, James AA. Complex modulation of the Aedes aegypti transcriptome in response to dengue virus infection. PLoS One. 2012;7:e50512. doi: 10.1371/journal.pone.0050512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sim S, Dimopoulos G. Dengue virus inhibits immune responses in Aedes aegypti cells. PLoS One. 2010;5:e10678. doi: 10.1371/journal.pone.0010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, Hoffmann JA, Imler JL. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol. 2005;6:946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- 32.Sabatier L, Jouanguy E, Dostert C, Zachary D, Dimarcq JL, Bulet P, Imler JL. Pherokine-2 and -3. Eur J Biochem. 2003;270:3398–3407. doi: 10.1046/j.1432-1033.2003.03725.x. [DOI] [PubMed] [Google Scholar]
- 33.Mounsey A, Bauer P, Hope IA. Evidence suggesting that a fifth of annotated Caenorhabditis elegans genes may be pseudogenes. Genome Res. 2002;12:770–775. doi: 10.1101/gr208802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Platt KB, Linthicum KJ, Myint KS, Innis BL, Lerdthusnee K, Vaughn DW. Impact of dengue virus infection on feeding behavior of Aedes aegypti. Am J Trop Med Hyg. 1997;57:119–125. doi: 10.4269/ajtmh.1997.57.119. [DOI] [PubMed] [Google Scholar]
- 35.Salazar MI, Richardson JH, Sanchez-Vargas I, Olson KE, Beaty BJ. Dengue virus type 2: replication and tropisms in orally infected Aedes aegypti mosquitoes. BMC Microbiol. 2007;7:9. doi: 10.1186/1471-2180-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialynas E, Hammond M, Hill CA, Konopinski N, Lobo NF, MacCallum RM, Madey G, Megy K, Meyer J, Redmond S, Severson DW, Stinson EO, Topalis P, Birney E, Gelbart WM, Kafatos FC, Louis C, Collins FH. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 2009;37:D583–D587. doi: 10.1093/nar/gkn857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calvo E, Tokumasu F, Marinotti O, Villeval JL, Ribeiro JM, Francischetti IM. Aegyptin, a novel mosquito salivary gland protein, specifically binds to collagen and prevents its interaction with platelet glycoprotein VI, integrin alpha2beta1, and von Willebrand factor. J Biol Chem. 2007;282:26928–26938. doi: 10.1074/jbc.M705669200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rossingol PA, Spielman A. Fluid transport across the ducts of a mosquito. J Insect Physiol. 1982;28:579–583. [Google Scholar]
- 39.Peng Z, Simons FE. Advances in mosquito allergy. Curr Opin Allergy Clin Immunol. 2007;7:350–354. doi: 10.1097/ACI.0b013e328259c313. [DOI] [PubMed] [Google Scholar]
- 40.Simons FE, Peng Z. Mosquito allergy: recombinant mosquito salivary antigens for new diagnostic tests. Int Arch Allergy Immunol. 2001;124:403–405. doi: 10.1159/000053771. [DOI] [PubMed] [Google Scholar]
- 41.Sylvestre G, Gandini M, Maciel-de-Freitas R. Age-dependent effects of oral infection with dengue virus on Aedes aegypti (Diptera: Culicidae) feeding behavior, survival, oviposition success and fecundity. PLoS One. 2013;8:e59933. doi: 10.1371/journal.pone.0059933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramirez JL, Dimopoulos G. The Toll immune signaling pathway control conserved anti-dengue defenses across diverse Ae. aegypti strains and against multiple dengue virus serotypes. Dev Comp Immunol. 2010;34:625–629. doi: 10.1016/j.dci.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linthicum KJ, Platt K, Myint KS, Lerdthusnee K, Innis BL, Vaughn DW. Dengue 3 virus distribution in the mosquito Aedes aegypti: an immunocytochemical study. Med Vet Entomol. 1996;10:87–92. doi: 10.1111/j.1365-2915.1996.tb00086.x. [DOI] [PubMed] [Google Scholar]
- 44.Mackenzie J. Wrapping things up about virus RNA replication. Traffic. 2005;6:967–977. doi: 10.1111/j.1600-0854.2005.00339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandez-Garcia MD, Mazzon M, Jacobs M, Amara A. Pathogenesis of flavivirus infections: using and abusing the host cell. Cell Host Microbe. 2009;5:318–328. doi: 10.1016/j.chom.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 46.Jin JP, Chong SM. Localization of the two tropomyosin-binding sites of troponin T. Arch Biochem Biophys. 2010;500:144–150. doi: 10.1016/j.abb.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng ML, Hong SS. Flavivirus infection: essential ultrastructural changes and association of Kunjin virus NS3 protein with microtubules. Arch Virol. 1989;106:103–120. doi: 10.1007/BF01311042. [DOI] [PubMed] [Google Scholar]
- 48.Lyles DS. Cytopathogenesis and inhibition of host gene expression by RNA viruses. Microbiol Mol Biol Rev. 2000;64:709–724. doi: 10.1128/mmbr.64.4.709-724.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 50.Sheehan D, Meade G, Foley VM, Dowd CA. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem J. 2001;360:1–16. doi: 10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gui Z, Hou C, Liu T, Qin G, Li M, Jin B. Effects of insect viruses and pesticides on glutathione S-transferase activity and gene expression in Bombyx mori. J Econ Entomol. 2009;102:1591–1598. doi: 10.1603/029.102.0425. [DOI] [PubMed] [Google Scholar]
- 52.Drakesmith H, Prentice A. Viral infection and iron metabolism. Nat Rev Microbiol. 2008;6:541–552. doi: 10.1038/nrmicro1930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.