Abstract
The TaqMan real-time polymerase chain reaction (PCR) assay was evaluated systematically with respect to the standard curve, linear range, and used for detecting Mycobacterium leprae DNA in paraffin-embedded skin biopsy specimens from 60 confirmed leprosy patients and three healthy individuals and 29 other dermatoses and bacterial DNA from 21 different species. The test was further evaluated with 51 paucibacillary (PB) patients. The results showed that the test had good sensitivity (8 fg) and good specificity with no cross-reactivity with 21 other bacterial species and the control specimens, except one with Xanthomatosis. The real-time PCR detection rate for the 51 PB specimens was 74.5% (38 of 51). We conclude that the real-time PCR test is a useful adjunct test for diagnosing early stage or PB leprosy cases.
Introduction
Leprosy is an infectious disease caused by Mycobacterium leprae, which is an acid-fast, rod-shaped bacterium that cannot be cultured in vitro. Mycobacterium leprae shows preference for certain body sites, such as the skin and peripheral nerves. Leprosy diagnosis often relies on microscopic detection of acid-fast bacilli (AFB) in tissue smears and clinical evaluations.1 Early stage leprosy is difficult to diagnose by clinical criteria alone as the sensitivity of AFB staining is quite low.2 Serological tests based on specific M. leprae antigens do not detect all clinical cases because most of the patients at the paucibacillary (PB) stage of infection do not develop significant levels of antibody response.3,4 This group potentially consists of patients with diverse clinical, bacteriological, and histopathological features. Frequently, M. leprae cannot be detected in the tissues of early lesions, histopathology can be non-specific, clinical findings are inconclusive, and patient histories can be unreliable, making diagnosis of early stage leprosy very difficult.5
Molecular tests have a great potential for detection and identification of M. leprae in tissues because they are more sensitive than the conventional methods.6–8 The M. leprae-specific repetitive element (RLEP) was found to be capable of detecting M. leprae DNA in 73% of patients with a bacterial index (BI) of 0.9,10 Furthermore, real-time polymerase chain reaction (PCR) is more sensitive and quantitative for detecting DNA sequences from specimens than conventional PCR. It can also be used as a robust method for detection of bacteria in clinical situations,11 and for quantification in footpad mouse tissues.12 The PCR detecting the presence of M. leprae DNA in fixed tissues coupled with histopathology may help clinicians arrive at a more rapid and definitive diagnosis of the disease.13 Recently, we have found that a nested PCR method that detects M. leprae DNA in patients' whole blood can be used for rapid and early diagnosis of leprosy.14 In this study, we evaluated the use of a real-time PCR test targeting RLEP DNA sequence for adjunct diagnosis of PB leprosy in comparison to the nested PCR test14 and also conventional histopathology for diagnosis of leprosy.
Materials and Methods
Study populations.
This study was conducted within a hyper-endemic site in Yunnan Province of Southwest China. The study group was composed of paraffin specimens from 60 confirmed leprosy patients and three healthy control individuals and twenty-nine other dermatoses. The 60 cases were classified within the spectrum of leprosy according to Ridley-Jopling's criteria: 8 tuberculoid (TT), 8 borderline tuberculoid (BT), 10 borderline-borderline (BB), 10 borderline-lepromatous (BL), and 9 lepromatous leprosy (LL) patients. To compare the effect of storage time on the result of the experiment, we collected the paraffin specimens of BL 7 in 2008 and BL 8 in 2013, respectively. The 51 possible PB biopsy specimens from patients having only clinical symptoms were collected as part of a normal diagnostic routine. All 143 specimens were paraffin-embedded skin biopsy specimens.
Ethics statement.
This study was approved by the Ethical Committee of Beijing Friendship Hospital Institutional Committee. All patients provided informed written consent.
Pathological examination.
A small piece of the skin lesion was excised from each patient and processed for routine histopathological diagnosis. The tissue was fixed in 10% formalin and processed for paraffin embedding. Skin biopsies were performed on the most representative lesions in all patients. The sections were stained with hematoxylin and eosin staining (H&E) and AFB staining and then examined by pathologists. The type and characteristic of granuloma lesions and the presence of AFB were noted. Ridley-Jopling classification of leprosy into lepromatous leprosy (LL), borderline lepromatous (BL), borderline tuberculoid (BT), tuberculoid (TT), and indeterminate leprosy (I), was performed as described.15,16
Bacterial DNA.
Mycobacterium leprae DNA was obtained from Colorado State University, Fort Collins, Co. Seventeen other mycobacterial species (M. tuberculosis, M. lufu, M. avium, M. marinum, M. bovis BCG-Pasteur, M. chelonae, M. bovis [Ravenel], M. flavescens, M. smegmatis, M. gordonae, M. ulcerans, M. intracellulare, M. simiae, M. bovis [AFZ/ZZ/97], M. lepraemurium, M. kansasii, and M. phlei) and four other non-mycobacterial species (Streptococcus pyogenes, Clostridium perfringens, Escherichia coli, and Staphylococcus epidermidis) were also included as controls for this study. These were kindly provided by Dr. Thomas Gillis from National Hansen's Disease Program (NHOP), Baton Rouge, LA, and the DNA samples were adjusted to 10 μg/mL before use in the PCR.
DNA extraction from specimens.
From each paraffin-embedded tissue block, 10 μm-thick sections of paraffin-embedded tissue were cut with a microtome blade. To prevent carryover tissue contamination of the samples, the microtome blade was cleaned with 100% ethanol after sectioning each sample. All specimens were extracted and processed with the DNeasy Blood and Tissue kit (Qiagen, Inc., Valencia, CA) for DNA isolation according to the manufacturer's recommendations.
Real-time PCR analysis.
Polymerase chain reaction and data analyses were performed in a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA). Primers and probes for the RLEP9 (RLEP-372 base pair [bp]) TaqMan PCR were selected from a common region of the RLEP family of dispersed repeats. The RLEP primers and fluorescent probes
5′-GTGCATGTCATGGCCTTGAGgtgtcggcgtggtcaatgtggccgcacctgaacaggcacgtccccgtgcacggtatAACTATTCGCACCTGATGTTATCCC-3′.
were designed and synthesized by the ABI manufacturer (Applied Biosystems), based on the criteria established for TaqMan PCR reactions. The real-time PCR amplified a 101 bp sequence of the RLEP element, which is present at 37 copies/cell.17 All reagents used in the TaqMan assay were those recommended by the manufacturer (Applied Biosystems). The PCR amplifications were performed in duplicate wells under the following conditions: 10 min at 95°C, followed by 42 temperature cycles (15 s at 95°C and 1 min at 60°C).
Specificity and sensitivity of the real-time PCR assay.
The specificity of the real-time PCR TaqMan assay was determined by analyzing purified DNA from 60 leprosy patients and three healthy control individuals and 29 other dermatoses, also from 21 other bacterial species. The sensitivity or lower limit of detection was determined by analyzing 2-fold serial dilutions of M. leprae DNA from 500 to 4 fg DNA and amplified as described previously (threshold cycle [CT] values of ≤ 35).
Bacillary load determination.
We constructed DNA plasmid standards, consisting of purified plasmid DNA, specific for the 101 bp sequence of the RLEP element target. By including a serial dilution of a standard in each PCR run, with known amounts of input copy number, the target gene could be quantified in the unknown samples. The 101 bp RLEP fragment was amplified from the DNA of M. leprae strain NHDP63 using the same primers used for the real-time PCR assay and cloned in pGEMT vector (Promega, Madison, WI). The recombinant clones were confirmed by restriction digestion and DNA sequencing (Beijing Dingguo Biotechnology Co., Ltd). Copy numbers of the recombinant plasmids pGEM-T101 were calculated. Ten to 107 copies of the plasmid DNA were used for the standard curve preparation.
Nested PCR assay.
The levels of M. leprae DNA in the same set of the formalin-fixed paraffin-embedded specimens were estimated by a nested PCR assay as we previously described in Reference 14. Briefly, a 372 bp fragment of M. leprae-specific repetitive sequence was performed using the outer set of primers sense 5′- GCACGTAAGCCTGTCGGTGG-3′ (ML1) and antisense 5′- CGGCCGGATCCTCGATGCAC-3′ (ML2). An inner nested set of primers was designed to amplify a 131 bp fragment using 5′-GTGAGGGTAGTTGTT-3′ (LP1) and 5′-GGTGCGAATAGTT-3′ (LP2). The PCR amplification of template DNA was carried out using a thermal cycler PTC 200 (MJ Research, Munich, Germany). Cycling parameters were as follows: initial denaturation at 95°C, 15 min, followed by 40 cycles of denaturation at 94°C, 30 s; annealing at 60°C for 1 min 30 s; extension at 72°C for 1 min 30 s; and final extension at 72°C, 10 min. The PCR was performed in a 25 μL reaction mix consisting of QIAGEN Multiplex PCR, 2 μL of DNA, and 200 ng of each primer. The LP1 and LP2 primers were used for the second round PCR amplification as described previously, except the annealing temperature was lowered to 40°C and 1 μL of the first round PCR was used as the DNA template. Amplified PCR products were analyzed by DNA agarose gel electrophoresis and visualized in a Gel Doc XR System (Bio-Rad, Hercules, CA).
Statistical analysis.
Data are presented as means ± SD. P values < 0.05 were considered statistically significant, and Kappa value > 0.7 was considered to have good correlation. All statistical analyses were performed using IBM SPSS Version 10 (Armonk, NY).
Results
Standard curve for the RLEP real-time PCR assay.
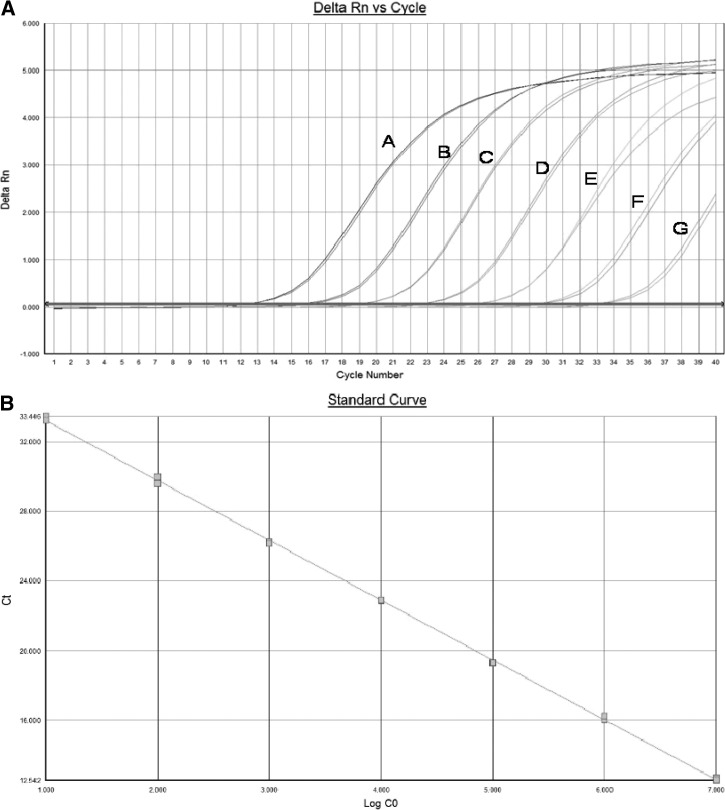
Recombinant plasmids carrying the cloned amplicon were prepared using the same primer set as the real-time PCR analysis and used for the preparation of a standard curve. The standard curve obtained was linear over seven logs with a correlation coefficient of 0.99 and amplification efficiency of 95% (Figure 1A and B).
Figure 1.
Standardization of the real-time polymerase chain reaction (PCR) assay. Ten to 107 copies of the plasmid DNA were used for standard curve preparation. Ct = −3.44 log + 36.65; R2 = 0.9995.
Sensitivity and specificity of the real-time PCR assay.
To evaluate the sensitivity of the real-time PCR assay, serial dilutions of M. leprae DNA from 500 to 4 fg were amplified during 40 cycles. The detection limit of M. leprae DNA is shown in Table 1. A titration of M. leprae DNA in the TaqMan PCR using RLEP primers and probes gave a lower limit of detection of 8 fg, equaling approximately three organisms based on the M. leprae chromosome size of ∼3.27 Mb.
Table 1.
Results of sensitivity of detection of Mycobacterium leprae DNA by the real-time PCR assay
| M. leprae DNA | Average Ct | Average copy |
|---|---|---|
| 1 pg | 29 | 5.48 × 1,000 |
| 500 fg | 30 | 2.2 × 1,000 |
| 250 fg | 31 | 932 |
| 125 fg | 32 | 792 |
| 62 fg | 33 | 515 |
| 31 fg | 34 | 202 |
| 16 fg | 35 | 35 |
| 8 fg | 35 | 24 |
| 4 fg | 36 | − |
To determine the specificity of the real-time PCR assay, 17 other mycobacterial species and four other bacterial species and control samples were tested in the real-time PCR. Apart from one Xanthomatosis the 101 bp fragment was only amplified by the real-time PCR with M. leprae DNA but not from DNA of other mycobacterial species or bacteria belonging to other genera and controls.
Based on the clinical and the histopathological features, the confirmed patients were classified within the spectrum of leprosy according to Ridley-Jopling's criteria: LL:9;BL:10;BB:10;BT:8;TT:8. Except for 8 cases of TT, the real-time PCR, the nested PCR, and AFB staining all showed positive results for 52 of the 60 confirmed leprosy patient tissue specimens. It is worth noting that for the 8 cases of TT, the positive rates of detection by the real-time PCR test and nested PCR were 50% and 25%, respectively, whereas AFB staining was negative for all 8 TT cases. The real-time PCR test improved the M. leprae DNA detection rate from the specimens of TT patients. To compare the effects of the storage time of paraffin specimens for PCR, specimens from 7 cases in 2008 and 8 cases in 2013 were tested at the same time. The results showed that the real-time PCR test was 100% positive (Table 2). Thus, these findings suggest that the real-time PCR test is highly sensitive and specific.
Table 2.
Diagnosis of paucibacillary (PB) patients by real-time polymerase chain reaction (PCR) and nested PCR tests
| Classification | No. | Real-time PCR | BI | PCR vs. BI | Nested PCR | Two kinds of PCR | ||
|---|---|---|---|---|---|---|---|---|
| Ct number* | Copy number | Positive (%) | Positive (%) | P value | Positive (%) | P value | ||
| LL | 9 | 24.9 ± 3.89 | 1.2 × 106 ± 3.3 × 106 | 9/9 (100) | 9/9 (100) | P = 1 | 9/9 (100) | P = 1 |
| BL | 10 | 27 ± 4.62 | 2.6 × 105 ± 4.1 × 105 | 10/10 (100) | 10/10 (100) | P = 1 | 10/10 (100) | P = 1 |
| BB | 10 | 30.8 ± 5.05 | 3.4 × 105 ± 1.0 × 106 | 10/10 (100) | 10/10 (100) | P = 1 | 10/10 (100) | P = 1 |
| BT | 8 | 32.6 ± 2.33 | 1.2 × 103 ± 1.8 × 103 | 8/8 (100) | 8/8 (100) | P = 1 | 8/8 (100) | P = 1 |
| TT | 8 | 34.6 ± 1.82 | 2.3 × 102 ± 3.2 × 102 | 4/8 (50) | 0/8 (−) | P = 0.04 | 2/8 (25) | P = 0.24 |
| BL 2008 year | 7 | 31.0 ± 5.16 | 4.7 × 105 ± 1.2 × 106 | 8/8 (100) | 8/8 (100) | P = 1 | 7/8 (88) | P = 0.5 |
| BL 2013 year | 8 | 27.4 ± 4.98 | 9.5 × 105 ± 2.3 × 106 | 10/10 (100) | 10/10 (100) | P = 1 | 10/10 (100) | P = 1 |
| Tinea versicolor | 5 | − | − | 0/5 | 0/5 | P = 1 | ||
| Psoriasis | 5 | − | − | 0/5 | 0/5 | P = 1 | ||
| Pityriasis rosea | 5 | − | − | 0/5 | 0/5 | P = 1 | ||
| Tinea corporis | 5 | − | − | 0/5 | 1/5 (20) | P = 0.5 | ||
| Multiple neurofibromatosis | 2 | − | − | 0/2 | 0/2 | P = 1 | ||
| Xanthomatosis | 2 | 34.6 ± 1.82 | 101 | 1/2 (50) | 0/2 | P = 0.5 | ||
| Scleroderma | 5 | − | − | 0/5 | 1/5 (20) | P = 0.5 | ||
| Normal person | 3 | − | − | 0/3 | 0/3 | P = 1 | ||
Threshold cycle [CT] values of ≤ 35. CT values < 35 were considered positive.
BI = bacterial index; LL = lepromatous leprosy; BL = borderline-lepromatous; BB = borderline-borderline; BT = borderline tuberculoid; TT = tuberculoid.
Real-time PCR analysis of clinical samples.
We evaluated the use of the real-time PCR assay developed previously on 51 clinically diagnosed PB patients. Out of the 51 cases, 34 were male and 17 female with a male/female ratio of 2:1. The age range was from 7 to 66 years of age. Of the 51 PB cases, the clinical diagnosis was BT in 39 (76.5%), TT in 10 (19.6%), and indeterminate in 2 (3.9%) cases. Of the 51 cases in paraffin-embedded skin biopsy specimens, the positive rates with the real-time PCR showed a much better correlation with clinical classification for BT AFB-positive cases (93.3%), BT AFB-negative cases (70.8%), and TT AFB-negative cases (55.6%). However, the results of histopathological classification showed 66.67%, 58.3%, and 11.1% concordance with clinical classification for BT AFB-positive cases, BT AFB-negative cases, and TT AFB- negative cases, respectively. The overall positive rates of the real-time PCR test and histopathological classification were 74.5% and 52.9%, respectively (P < 0.05). It is worth noting for AFB-negative BT and TT, the positive real-time PCR rates were 70.8% and 55.6%, respectively (Table 3).
Table 3.
Results of polymerase chain reaction (PCR) tests and pathological examination of 51 confirmed PB samples
| Clinical | No. | Real-time PCR | Nested PCR | Two kinds of PCR | Histopath. | Real-time PCR vs. Histopath. | ||
|---|---|---|---|---|---|---|---|---|
| Positive (%) | Ct | Copy | Positive (%) | P value | Agreement with clinical | P value | ||
| BT AFB+ | 15 | 14/15 (93.3) | 30.7 ± 2 | 2.3 × 103 ± 5.5 × 103 | 15/15 (100) | P > 0.05 | 10/15 (66.7) | P > 0.05 |
| BT AFB– | 24 | 17/24 (70.8) | 33.4 ± 1.8 | 6.2 × 102 ± 8.9 × 102 | 15/24 (62.5) | P > 0.05 | 14/24 (58.3) | P > 0.05 |
| TT AFB+ | 1 | 1/1 (100) | 34 | 1.7 × 102 | 1/1 (100) | − | 1/1 (100) | − |
| TT AFB– | 9 | 5/9 (55.6) | 32 ± 1.8 | 9.5 × 102 ± 1.4 × 103 | 5/9 (55.6) | − | 1/9 (11.1) | P > 0.05 |
| I | 2 | 1/2 (50) | 35 | 1.3 × 102 | 1/2 (50) | − | 1/2 (50) | − |
| Total | 51 | 38/51 (74.5) | 37/51 (72.5) | P > 0.05 | 27/51 (52.9) | P < 0.05 | ||
Threshold cycle [CT] values of ≤ 35. CT values < 35 were considered positive.
BT AFB = borderline tuberculoid acid-fast bacilli; TT AFB = tuberculoid acid-fast bacilli; I = indeterminate leprosy.
Comparison of the real-time PCR assay and a nested PCR test for diagnosis of PB patients.
We have recently shown that a nested PCR test based on detecting the same M. leprae repeat DNA sequences as the current study is a useful technique for sensitive detection of leprosy from the whole blood of leprosy patients.14 Therefore, we wanted to compare the real-time PCR developed here with the nested PCR test for detecting M. leprae DNA using the formalin-fixed paraffin-embedded samples. The results are shown in Table 3. The first round of the nested PCR was positive in 30 of the 51 samples and through the second round PCR 7 was positive from 21 first-round negative samples. Among all the 51 samples, 36 were positive by both nested PCR and real-time PCR, 12 were negative by both methods. There were two samples positive by the real-time PCR but negative by the nested PCR, whereas 1 sample was positive by the nested PCR but negative by the real-time PCR. There was no statistical difference between the two methods in detecting PB patients (P > 0.05, Kappa value = 0.97).
Discussion
Leprosy is a chronic granulomatous disease caused by M. leprae. Because M. leprae cannot be grown in vitro the disease cannot be diagnosed by culture techniques, the diagnosis of leprosy relies on clinical and histopathological findings and AFB staining. Paucibacillary and early stage leprosy are difficult to diagnose because of the small number of bacilli available for detection by AFB staining. In general the threshold limit of detection for direct microscopic counting is ∼1 × 104 bacilli12; even when it is diagnosed, detection of bacteria is difficult and histopathological findings can be non-specific. For patients with negative AFB, the sensitivity and the specificity of the diagnosis can be potentially improved significantly by molecular tests. Fortunately, in recent years, several molecular techniques based on PCR have become available that are helpful in the diagnosis of leprosy based on detection of M. leprae DNA in clinical samples.10,14,18,19 In this study, we developed and validated a real-time PCR assay and showed that this test could rapidly detect M. leprae DNA from formalin-fixed paraffin-embedded specimens.
We set out to investigate the feasibility of using real-time PCR to accurately estimate the number of bacilli in specimens using predetermined standards. This study describes a simple and sensitive real-time PCR with fluorescent probe for quantitative detection of M. leprae. When DNA extraction was used in conjunction with the real-time PCR, the results could be obtained rapidly in 2 hours. The time needed to complete the real-time PCR (RT-PCR) tests for the whole blood and slit skin smear specimens and the paraffin-embedded samples is similar. However, the biopsy specimens required a slightly longer time to allow for tissue homogenization and digestion, the results of real-time PCR test could be obtained within two and a one-half hours. The sensitivity of the real-time PCR test could detect as low as 8 fg of M. leprae DNA, or ∼240 bacteria in infected tissues.12 Quantification of DNA from microorganisms using real-time PCR can be conducted based on the crossing point for each sample, which allows the construction of a standard curve for determining the corresponding concentrations of unknown samples. The specificity of the real-time PCR test was found to be highly specific with 60 confirmed patients and showed no cross-reactivity with other mycobacterial or bacterial DNA. Only one Xanthomatosis had cross-reactivity but cycle threshold (CT) value was near to the negative threshold. The real-time PCR test is especially valuable for detection of low numbers of bacilli with BI negative samples, with the positive rate reaching 50% for such patients in this study. The excellent sensitivity and specificity of the real-time PCR test suggest the test may be useful not only for early diagnosis of leprosy but also for excluding some skin lesions not caused by leprosy.
Leprosy is a spectral disease. The clinical manifestations are dependent on the host immune response.20 The Ridley-Jopling classification based on clinical, histopathological, and immunological features is widely accepted. However, the discrepancy between clinical and histopathological diagnosis is significant, especially for PB and patients in early stages of the disease because the clinical diagnoses are based on tissue lesions, even when a histopathological examination has been done and there are usually no specific changes in the pathological analysis.16,21 In particular, patients with a single lesion are often confused with other skin diseases. The high proportion of PB cases may reflect an improvement on clinical detection of new cases in high endemic areas. Martinez and others22 showed identification of 79.2% of leprosy patients with no detectable bacilli by PCR tests. In this study, we obtained similar results (70.8%) with the real-time PCR test in paraffin-embedded skin biopsy for AFB-negative specimens. It was found that the overall positive rate of AFB was 31.4% (16 of 51), compared with positive real-time PCR and histopathological results of 74.5% and 52.9% (P < 0.05), respectively. It is of interest to note that for negative AFB samples belonging to BT and TT the real-time PCR positive rates were 70.8% and 55.6%, respectively, indicating the real-time PCR test is more sensitive in detecting AFB-negative leprosy cases. Among 51 cases with clinical symptoms, BI was positive in 16, through pathological examination the other 11 with leprosy pathological changes were confirmed to be leprosy, a total of 27 patients were diagnosed with leprosy, the remaining 24 were increased by 11, 10 positive cases, respectively, by real-time PCR and nested PCR, the total diagnosed cases went up to 38 by real-time PCR. The greater sensitivity and specificity of the RLEP TaqMan PCR can be an especially useful tool for the rapid detection of M. lepare DNA in clinical specimens in which no AFB are detectable microscopically, and should be used in difficult to diagnose cases such as the specimens with no leprosy pathological changes. The ability of real-time PCR to detect M. leprae DNA on regular bacteriological negative samples would be helpful in differentiating leprosy from diseases that cause similar symptoms ensuring a correct diagnosis.
We also compared the amplification of M. leprae DNA using the real-time PCR with the nested PCR test for the same specimens. Through correlation Kappa analysis, there was no statistical difference between the two methods (P > 0.05), indicating that both the real-time PCR and the nested PCR behaved similarly in sensitivity for detection of PB cases. The PCR-based tests can improve sensitivity of the PB diagnosis by AFB, clinical evaluation, and histopathology. However, real-time PCR detection of M. leprae DNA could be superior to nested PCR, which is prone to contamination with PCR products in the second round of amplification but can be used as a rapid, sensitive, and specific confirmatory test to identify the presence of M. leprae in tissue specimens for diagnosis of PB leprosy. Although PCR is a useful tool for the detection of early clinical infection, it remains to be seen if all PCR-positive cases will develop into active disease.23 We will continue to follow up the 13 of 51 undiagnosed cases. Further research is needed to determine the relationship between the amount of bacterial DNA or bacterial load, as assessed by the real-time PCR test in this study, and the disease development in future studies.
ACKNOWLEDGMENTS
We are grateful to P. J. Brennan and T. P. Gillis for providing bacterial DNA samples.
Footnotes
Authors' addresses: Wen Yan, Yan Xing, Lian Chao Yuan, and Huan-Ying Li, Capital University of Medicine Affiliated Beijing Friendship Hospital, Beijing, China, E-mails: weny8@163.com, hellen19801@163.com, yuanlc@126.com, and lhybj@163.com. Rong De Yang and Fu Yue Tan, Yunnan Wenshan Skin Disease Dispensary, China, E-mails: yangrd@qq.com and tfyyyy@qq.com Ying Zhang, Department of Molecular Microbiology and Immunology, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD, E-mail: yzhang@jhsph.edu.
References
- 1.Foss NT, Motta ACF. Leprosy, a neglected disease that causes a wide variety of clinical conditions in tropical countries. Mem Inst Oswaldo Cruz, Rio de Janeiro. (2012);107((Suppl. 1)):28–33. doi: 10.1590/s0074-02762012000900006. [DOI] [PubMed] [Google Scholar]
- 2.Shepard CC, McRae DH. A method for counting acid-fast bacteria. Int J Lepr. 1968;36:78–82. [PubMed] [Google Scholar]
- 3.Buhrer-Sekula S, Smits HL, Gussenhoven GC, van Leeuwen J, Amador S, Fujiwara T, Klatser PR, Oskam L. Simple and fast lateral flow test for classification of leprosy patients and identification of contacts with high risk of developing leprosy. J Clin Microbiol. 2003;41:1991–1995. doi: 10.1128/JCM.41.5.1991-1995.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douglas JT, Steven LM, Fajardo T, Cellona RV, Madarang MG, Abalos RM, Steenbergen GJ. The effects of chemotherapy on antibody levels in lepromatous patients. Lepr Rev. 1988;59:127–135. doi: 10.5935/0305-7518.19880017. [DOI] [PubMed] [Google Scholar]
- 5.Santos VS, de Mendonça Neto PT, Falcão Raposo OF, Fakhouri R, Reis FP, Feitosa VL. Evaluation of agreement between clinical and histopathological data for classifying leprosy. Int J Infect Dis. 2013;17:e189–e192. doi: 10.1016/j.ijid.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Clark-Curtiss JE, Docherty MA. A species-specific repetitive sequence in Mycobacterium leprae DNA. J Infect Dis. 1989;159:7–15. doi: 10.1093/infdis/159.1.7. [DOI] [PubMed] [Google Scholar]
- 7.Katoch VM. Newer diagnostic techniques for tuberculosis. Indian J Med Res. 2001;120:418–428. [PubMed] [Google Scholar]
- 8.Hampson SJ, Portaels F, Thompson J, Green EP, Moss MT, Hermon-Taylor J, McFadden JJ. DNA probes demonstrate a single highly conserved strain of Mycobacterium avium infecting AIDS patients. Lancet. 1989;1:65–68. doi: 10.1016/s0140-6736(89)91427-x. [DOI] [PubMed] [Google Scholar]
- 9.Yoon D-H, Cho S-N, Lee MK, Abalos RM, Cellona RV, Fajardo Jr TT, Guido LS, Dela Cruz EC, Walsh GP, Kim JD. Evaluation of polymerase chain reaction amplification of Mycobacterium leprae-specific repetitive sequence in biopsy specimens from leprosy patients. J Clin Microbiol. 1993;31:895–899. doi: 10.1128/jcm.31.4.895-899.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez AN, Ribeiro-Alves M, Sarno EN, Moraes MO. Evaluation of qPCR-based assays for leprosy diagnosis directly in clinical specimens. PLoS Negl Trop Dis. 2011;5:e1354. doi: 10.1371/journal.pntd.0001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kramme S, Bretzel G, Panning M, Kawuma J, Drosten C. Detection and quantification of Mycobacterium leprae in tissue samples by real-time PCR. Med Microbiol Immunol (Berl) 2004;193:189–193. doi: 10.1007/s00430-003-0188-8. [DOI] [PubMed] [Google Scholar]
- 12.Truman RW, Andrews PK, Robbins NY, Adams LB, Krahenbuhl JL, Gillis TP. Enumeration of Mycobacterium leprae using real-time PCR. PLoS Negl Trop Dis. 2008;2:e328. doi: 10.1371/journal.pntd.0000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardman WJ, Benian GM, Howard T, McGowan JE, Jr, Metchock B, Murtagh JJ. Rapid detection of mycobacteria in inflammatory necrotizing granulomas from formalin fixed, paraffin-embedded tissue by PCR in clinically high-risk patients with acid-fast stain and culture-negative tissue biopsies. Am J Clin Pathol. 1996;106:384–389. doi: 10.1093/ajcp/106.3.384. [DOI] [PubMed] [Google Scholar]
- 14.Wen Y, Xing Y, Yuan LC, Liu J, Zhang Y, Li HY. Whole-blood nested-PCR amplification of M. leprae-specific DNA for early diagnosis of leprosy. Am J Trop Med Hyg. 2013;88:918–922. doi: 10.4269/ajtmh.11-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ridley DS, Jopling WH. Classification of leprosy according to immunity – a five group system. Int J Lepr. 1966;34:255–273. [PubMed] [Google Scholar]
- 16.Lockwood DN, Nicholls P, Smith WC, Das L, Barkataki P, van Brakel W, Suneetha S. Comparing the clinical and histological diagnosis of leprosy and leprosy reactions in the INFIR cohort of Indian patients with multibacillary leprosy. PLoS Negl Trop Dis. 2012;6:e1702. doi: 10.1371/journal.pntd.0001702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole ST, Supply P, Honoré N. Repetitive sequences in Mycobacterium leprae and their impact on genome plasticity. Lepr Rev. 2001;72:449–461. [PubMed] [Google Scholar]
- 18.Rodrigues LC, Lockwood DN. Leprosy now: epidemiology, progress, challenges, and research gaps. Lancet Infect Dis. 2011;11:464–470. doi: 10.1016/S1473-3099(11)70006-8. [DOI] [PubMed] [Google Scholar]
- 19.Lini N, Shankernarayan NP, Dharmalingam K. Quantitative real-time PCR analysis of Mycobacterium leprae DNA and mRNA in human biopsy material from leprosy and reactional cases. J Med Microbiol. 2009;58:753–759. doi: 10.1099/jmm.0.007252-0. [DOI] [PubMed] [Google Scholar]
- 20.Scollard DM, Adams LB, Gillis TP, Kranenbuthl JL, Truman RW, Williams DL. The continuing challenges of leprosy. Clin Microbiol Rev. 2006;19:338–381. doi: 10.1128/CMR.19.2.338-381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhatia AS, Katoch K, Narayanan RB, Ramu G, Mukherjee A, Lavania RK. Clinical and histopathological correlation in the classification of leprosy. Int J Lepr. 1993;61:433–438. [PubMed] [Google Scholar]
- 22.Martinez AN, Britto CF, Nery JA, Sampaio EP, Jardim MR, Sarno EN, Moraes MO. Evaluation of real-time and conventional PCR targeting complex 85 genes for detection of Mycobacterium leprae DNA in skin biopsy samples from patients diagnosed with leprosy. J Clin Microbiol. 2006;44:3154–3159. doi: 10.1128/JCM.02250-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patrocínio LG, Goulart IM, Goulart LR, Patrocínio JA, Ferreira FR, Fleury RN. Detection of Mycobacterium leprae in nasal mucosa biopsies by the polymerase chain reaction. FEMS Immunol Med Microbiol. 2005;44:311–316. doi: 10.1016/j.femsim.2005.01.002. [DOI] [PubMed] [Google Scholar]