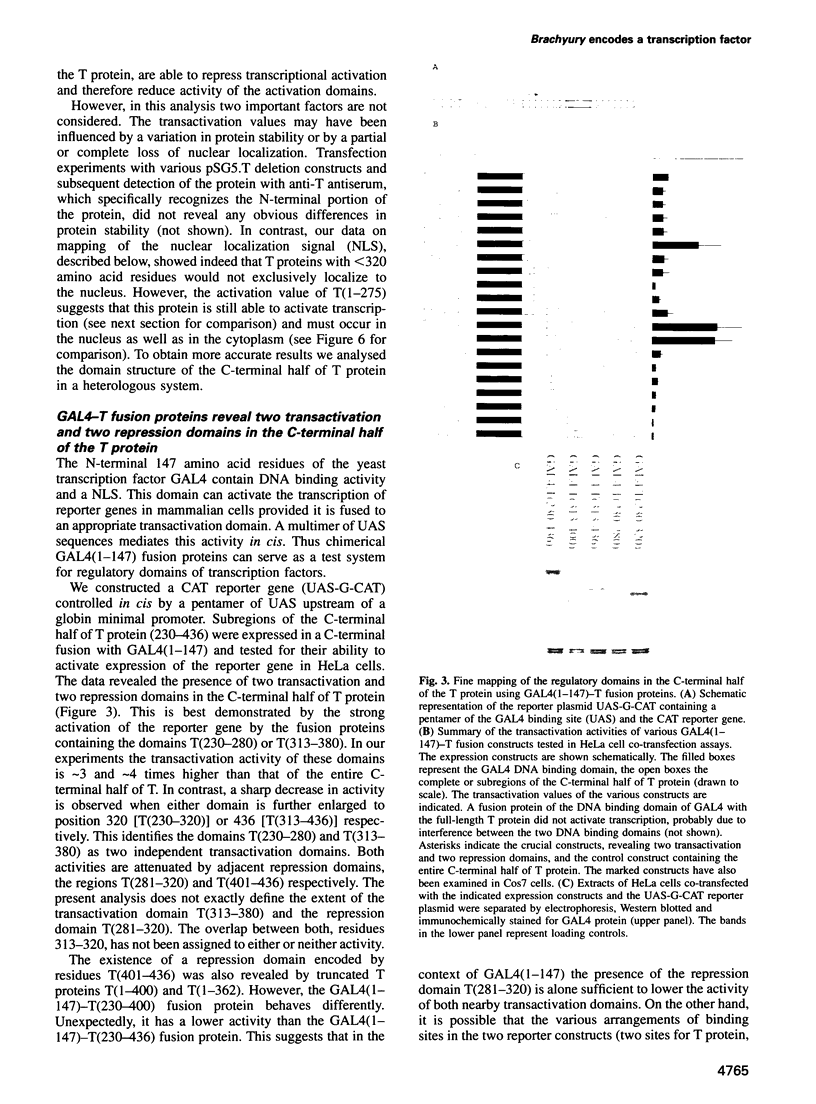
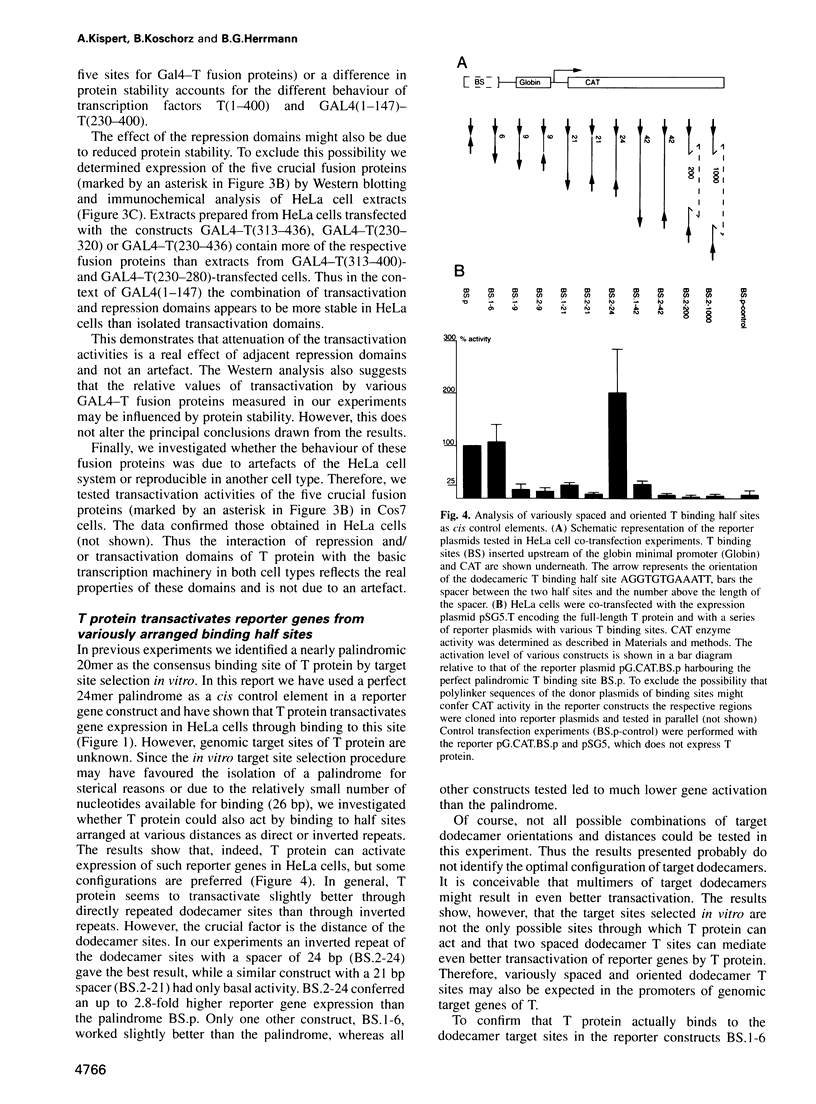
Abstract
The mouse Brachyury (T) gene is required for differentiation of the notochord and formation of mesoderm during posterior development. Homozygous embryos lacking T activity do not develop a trunk and tail and die in utero. The T gene is specifically expressed in notochord and early mesoderm cells in the embryo. recent data have demonstrated that the T protein is localized in the cell nucleus and specifically binds to a palindrome of 20 bp (the T site) in vitro. We show that the T protein activates expression of a reporter gene in HeLa cells through binding to the T site. Thus T is a novel tissue-specific transcription factor. It consists of a large N-terminal DNA binding domain (amino acids 1-229) and two pairs of transactivation and repression domains in the C-terminal protein half. T can also transactivate transcription through variously oriented and spaced T sites, a fact that may be relevant in the search for genes controlled by T protein and important in mesoderm development.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agulnik S. I., Bollag R. J., Silver L. M. Conservation of the T-box gene family from Mus musculus to Caenorhabditis elegans. Genomics. 1995 Jan 1;25(1):214–219. doi: 10.1016/0888-7543(95)80128-9. [DOI] [PubMed] [Google Scholar]
- Bollag R. J., Siegfried Z., Cebra-Thomas J. A., Garvey N., Davison E. M., Silver L. M. An ancient family of embryonically expressed mouse genes sharing a conserved protein motif with the T locus. Nat Genet. 1994 Jul;7(3):383–389. doi: 10.1038/ng0794-383. [DOI] [PubMed] [Google Scholar]
- Carey M. Mechanistic advances in eukaryotic gene activation. Curr Opin Cell Biol. 1991 Jun;3(3):452–460. doi: 10.1016/0955-0674(91)90073-8. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988 Dec 2;55(5):887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Cunliffe V., Smith J. C. Specification of mesodermal pattern in Xenopus laevis by interactions between Brachyury, noggin and Xwnt-8. EMBO J. 1994 Jan 15;13(2):349–359. doi: 10.1002/j.1460-2075.1994.tb06268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Laskey R. A. Nuclear targeting sequences--a consensus? Trends Biochem Sci. 1991 Dec;16(12):478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Frankel A. D., Kim P. S. Modular structure of transcription factors: implications for gene regulation. Cell. 1991 May 31;65(5):717–719. doi: 10.1016/0092-8674(91)90378-c. [DOI] [PubMed] [Google Scholar]
- Garcia-Bustos J., Heitman J., Hall M. N. Nuclear protein localization. Biochim Biophys Acta. 1991 Mar 7;1071(1):83–101. doi: 10.1016/0304-4157(91)90013-m. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hall C. V., Jacob P. E., Ringold G. M., Lee F. Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells. J Mol Appl Genet. 1983;2(1):101–109. [PubMed] [Google Scholar]
- Harrison S. C. A structural taxonomy of DNA-binding domains. Nature. 1991 Oct 24;353(6346):715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Herrmann B. G. Expression pattern of the Brachyury gene in whole-mount TWis/TWis mutant embryos. Development. 1991 Nov;113(3):913–917. doi: 10.1242/dev.113.3.913. [DOI] [PubMed] [Google Scholar]
- Herrmann B. G., Kispert A. The T genes in embryogenesis. Trends Genet. 1994 Aug;10(8):280–286. doi: 10.1016/0168-9525(90)90011-t. [DOI] [PubMed] [Google Scholar]
- Herrmann B. G., Labeit S., Poustka A., King T. R., Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990 Feb 15;343(6259):617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Hwang J. J., Chambon P., Davidson I. Characterization of the transcription activation function and the DNA binding domain of transcriptional enhancer factor-1. EMBO J. 1993 Jun;12(6):2337–2348. doi: 10.1002/j.1460-2075.1993.tb05888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Richardson W. D., Markham A. F., Smith A. E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984 Sep 6;311(5981):33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Kispert A., Herrmann B. G. Immunohistochemical analysis of the Brachyury protein in wild-type and mutant mouse embryos. Dev Biol. 1994 Jan;161(1):179–193. doi: 10.1006/dbio.1994.1019. [DOI] [PubMed] [Google Scholar]
- Kispert A., Herrmann B. G., Leptin M., Reuter R. Homologs of the mouse Brachyury gene are involved in the specification of posterior terminal structures in Drosophila, Tribolium, and Locusta. Genes Dev. 1994 Sep 15;8(18):2137–2150. doi: 10.1101/gad.8.18.2137. [DOI] [PubMed] [Google Scholar]
- Kispert A., Herrmann B. G. The Brachyury gene encodes a novel DNA binding protein. EMBO J. 1993 Aug;12(8):3211–3220. doi: 10.1002/j.1460-2075.1993.tb05990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispert A., Ortner H., Cooke J., Herrmann B. G. The chick Brachyury gene: developmental expression pattern and response to axial induction by localized activin. Dev Biol. 1995 Apr;168(2):406–415. doi: 10.1006/dbio.1995.1090. [DOI] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Klein-Hitpass L., Schorpp M., Wagner U., Ryffel G. U. An estrogen-responsive element derived from the 5' flanking region of the Xenopus vitellogenin A2 gene functions in transfected human cells. Cell. 1986 Sep 26;46(7):1053–1061. doi: 10.1016/0092-8674(86)90705-1. [DOI] [PubMed] [Google Scholar]
- Lai E., Prezioso V. R., Tao W. F., Chen W. S., Darnell J. E., Jr Hepatocyte nuclear factor 3 alpha belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 1991 Mar;5(3):416–427. doi: 10.1101/gad.5.3.416. [DOI] [PubMed] [Google Scholar]
- MacGregor G. R., Caskey C. T. Construction of plasmids that express E. coli beta-galactosidase in mammalian cells. Nucleic Acids Res. 1989 Mar 25;17(6):2365–2365. doi: 10.1093/nar/17.6.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermod N., O'Neill E. A., Kelly T. J., Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989 Aug 25;58(4):741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Nagpal S., Friant S., Nakshatri H., Chambon P. RARs and RXRs: evidence for two autonomous transactivation functions (AF-1 and AF-2) and heterodimerization in vivo. EMBO J. 1993 Jun;12(6):2349–2360. doi: 10.1002/j.1460-2075.1993.tb05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye J. A., Petersen J. M., Gunther C. V., Jonsen M. D., Graves B. J. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992 Jun;6(6):975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- Pani L., Overdier D. G., Porcella A., Qian X., Lai E., Costa R. H. Hepatocyte nuclear factor 3 beta contains two transcriptional activation domains, one of which is novel and conserved with the Drosophila fork head protein. Mol Cell Biol. 1992 Sep;12(9):3723–3732. doi: 10.1128/mcb.12.9.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugfelder G. O., Roth H., Poeck B. A homology domain shared between Drosophila optomotor-blind and mouse Brachyury is involved in DNA binding. Biochem Biophys Res Commun. 1992 Jul 31;186(2):918–925. doi: 10.1016/0006-291x(92)90833-7. [DOI] [PubMed] [Google Scholar]
- Qian Y. Q., Billeter M., Otting G., Müller M., Gehring W. J., Wüthrich K. The structure of the Antennapedia homeodomain determined by NMR spectroscopy in solution: comparison with prokaryotic repressors. Cell. 1989 Nov 3;59(3):573–580. doi: 10.1016/0092-8674(89)90040-8. [DOI] [PubMed] [Google Scholar]
- Rao Y. Conversion of a mesodermalizing molecule, the Xenopus Brachyury gene, into a neuralizing factor. Genes Dev. 1994 Apr 15;8(8):939–947. doi: 10.1101/gad.8.8.939. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S., Ho R. K., Herrmann B. G., Nüsslein-Volhard C. The protein product of the zebrafish homologue of the mouse T gene is expressed in nuclei of the germ ring and the notochord of the early embryo. Development. 1992 Dec;116(4):1021–1032. doi: 10.1242/dev.116.4.1021. [DOI] [PubMed] [Google Scholar]
- Smith J. C., Price B. M., Green J. B., Weigel D., Herrmann B. G. Expression of a Xenopus homolog of Brachyury (T) is an immediate-early response to mesoderm induction. Cell. 1991 Oct 4;67(1):79–87. doi: 10.1016/0092-8674(91)90573-h. [DOI] [PubMed] [Google Scholar]
- Stunnenberg H. G. Mechanisms of transactivation by retinoic acid receptors. Bioessays. 1993 May;15(5):309–315. doi: 10.1002/bies.950150504. [DOI] [PubMed] [Google Scholar]
- Treisman J., Harris E., Desplan C. The paired box encodes a second DNA-binding domain in the paired homeo domain protein. Genes Dev. 1991 Apr;5(4):594–604. doi: 10.1101/gad.5.4.594. [DOI] [PubMed] [Google Scholar]
- Treisman R., Ammerer G. The SRF and MCM1 transcription factors. Curr Opin Genet Dev. 1992 Apr;2(2):221–226. doi: 10.1016/s0959-437x(05)80277-1. [DOI] [PubMed] [Google Scholar]
- Wasylyk C., Kerckaert J. P., Wasylyk B. A novel modulator domain of Ets transcription factors. Genes Dev. 1992 Jun;6(6):965–974. doi: 10.1101/gad.6.6.965. [DOI] [PubMed] [Google Scholar]
- Webster N. J., Green S., Jin J. R., Chambon P. The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell. 1988 Jul 15;54(2):199–207. doi: 10.1016/0092-8674(88)90552-1. [DOI] [PubMed] [Google Scholar]
- Weigel D., Jäckle H. The fork head domain: a novel DNA binding motif of eukaryotic transcription factors? Cell. 1990 Nov 2;63(3):455–456. doi: 10.1016/0092-8674(90)90439-l. [DOI] [PubMed] [Google Scholar]
- Wilkinson D. G., Bhatt S., Herrmann B. G. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990 Feb 15;343(6259):657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- Xiao J. H., Davidson I., Matthes H., Garnier J. M., Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991 May 17;65(4):551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- Xu W., Rould M. A., Jun S., Desplan C., Pabo C. O. Crystal structure of a paired domain-DNA complex at 2.5 A resolution reveals structural basis for Pax developmental mutations. Cell. 1995 Feb 24;80(4):639–650. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]
- Yasuo H., Satoh N. Function of vertebrate T gene. Nature. 1993 Aug 12;364(6438):582–583. doi: 10.1038/364582b0. [DOI] [PubMed] [Google Scholar]