Abstract
Purpose
Pax6 is a key regulatory gene for eye, brain, and pancreas development. It acts as a transcriptional activator and repressor. Loss-of-function of Pax6 results in down- and upregulation of a comparable number of genes, although many are secondary targets. Recently, we found a prototype of a Pax6-binding site that acts as a transcriptional repressor. We also identified the Trpm3 gene as a Pax6-direct target containing the miR-204 gene located in intron 6. Thus, there are multiple Pax6-dependent mechanisms of transcriptional repression in the cell. More than 50 Pax6 missense mutations have been identified in humans and mice. Two of these mutations, N50K (Leca4) and R128C (Leca2), were analyzed in depth resulting in different numbers of regulated genes and different ratios of down- and upregulated targets. Thus, additional studies of these mutants are warranted to better understand the molecular mechanisms of the mutants’ action.
Methods
Mutations in PAX6 and PAX6(5a), including G18W, R26G, N50K, G64V, R128C, and R242T, were generated with site-directed mutagenesis. A panel of ten luciferase reporters driven by six copies of Pax6-binding sites representing a spectrum of sites that act as repressors, moderate activators, and strong activators were used. Two additional reporters, including the Pax6-regulated enhancer from mouse Trpm3 and six copies of its individual Pax6-binding site, were also tested in P19 cells.
Results
PAX6 (N50K) acted either as a loss-of-function or neutral mutation. In contrast, PAX6 (R128C) and (R242T) acted as loss-, neutral, and gain-of-function mutations. With three distinct reporters, the PAX6 (N50K) mutation broke the pattern of effects produced by substitutions in the surrounding helices of the N-terminal region of the paired domain. All six mutations tested acted as loss-of-function using the Trpm3 Pax6-binding site.
Conclusions
These studies highlight the complexity of Pax6-dependent transcriptional activation and repression mechanisms, and identify the N50K and R128C substitutions as valuable tools for testing interactions between Pax6, Pax6 (N50K), and Pax6 (R128C) with other regulatory proteins, including chromatin remodelers.
Introduction
Pax6 functions as a key regulatory gene for vertebrate eye development [1-4]. Outside the eye, Pax6 regulates neurogenesis, as well as olfactory and pancreatic development [5-7]. To achieve these diverse roles, Pax6 proteins regulate common and distinct sets of genes in different cell types, and Pax6’s function clearly depends on the cell-type context [8-15]. Pax6 directly regulates genes encoding a wide range of regulatory, signaling, and structural proteins required for eye morphogenesis [16-21]. Comprehensive understanding of Pax6 function in individual cell types in conjunction with elucidation of molecular processes disrupted in the presence of disease-causing mutations in human PAX6, as well as developmental abnormalities linked to rodent Pax6 mutant alleles, are essential for understanding human organogenesis and tissue maintenance.
At the structural level, Pax6 contains two DNA-binding domains, a paired domain (PD) and an internal homeodomain (HD). The PD comprises two independent helix-turn-helix DNA-binding subdomains, called PAI and RED [22]. The amino acid sequences of the human and mouse PAX6/Pax6 gene are identical. Alternate splicing produces two main Pax6 proteins, Pax6/p46 and Pax6(5a)/p48 [23,24], which differ by an insertion of 14 amino acids into the PAI subdomain, encoded by exon 5a, that disrupts its DNA-binding activity [25]. DNA-binding studies identified consensus sequences recognized by isolated Pax6 and Pax6(5a) PDs [25,26] followed by identification of binding sites recognized by Pax6 PD/HD and PD(5a)/HD [21]. These studies lead to the identification of a continuum of eight Pax6-binding sequences that show high, moderate, and weak transcriptional activation. The ninth sequence variant (motif 1–2) functions as a transcriptional repressor of Pax6 and Pax6(5a) [21].
Human genetic studies on aniridia and related ocular syndromes have led to the identification of more than 400 PAX6 mutations, including more than 50 missense mutations. These include a mutation R128C in the RED subdomain that caused foveal hypoplasia in two unrelated patients [27,28]. In contrast, a much smaller number of mouse missense mutations were isolated, including Pax6Leca1 (V256G), Pax6Leca2 (R128C), Pax6Leca4 (N50G) [29,30], and Pax64Neu (S259P) [31]. Externally, the Leca allelic series are displayed as a lens-corneal adhesion: Leca2 displays central corneal and lens opacities, and Leca4 is characterized by cataract in the presence of a relatively normal iris [29]. Recent analysis of Leca2 and Leca4 homozygous mutations in the mouse forebrain using high-density oligonucleotide array hybridizations revealed 94 and 416 disregulated genes, respectively [32]. Interestingly, a higher proportion of downregulated genes were found in the Leca2 (59/94, 63%) compared to the Leca4 (179/416, 43%) model. The majority of these genes are unique suggesting that different types of Pax6-binding sites respond to these mutations.
The molecular mechanisms of transcriptional activation and repression by Pax6 remain to be elucidated [2]. Pax6 has been shown to bind the Brg1 and Brm catalytical subunits of the SWI/SNF complex [33-35]. In mouse lens, Brg1 and Pax6 regulate expression of a joint set of genes [36]. Pax6 also binds p300 histone acetyltransferase [37]. Use of different chromatin remodeling complexes recruited by Pax6 to specific genes is thought to play a pivotal role in Pax6’s capacity to control critical steps in embryonic development [34]. However, it is not known if Pax6 missense mutations modulate these protein–protein interactions. Transcriptional repression by Pax6 is even less understood. Apart from the site-specific repression described above, two additional mechanisms address this Pax6 function. First, Pax6 activates expression of the Trpm3 gene [15], which harbors miR-204, a key regulatory microRNA (miRNA) of mammalian eye development [38]. Thus, Pax6 indirectly controls expression of genes in the specific miR-204 pathway. Second, complex formation between Pax6 and pRb can also reduce gene expression by preventing Pax6 from binding DNA [39,40].
To gain additional insights into Pax6-mediated gene activation and repression, we examined the functional properties of a representative group of six missense PAX6 and PAX6(5a) mutants. In particular, Pax6 mutations included two mouse missense mutations, N50K (Leca4) and R128C (Leca2), as both were analyzed in depth resulting in different numbers of regulated genes and different ratios of down- and upregulated targets (see above). Notably, mouse Leca2 is identical to a human PAX6 mutation identified in multiple patients as described above. The R242T mutation represents one of the few mutations known within the PAX6 homeodomain [41]. The remaining PAX6 mutations, including G18W, R26G, and G64V, were examined elsewhere using P6CON/luciferase reporters [40,42,43]. Herein, we used a panel of 12 distinct luciferase reporters, with a primary focus on N50K and R128C, as genetic and molecular studies of these Pax6-mutated genes are now available for comparisons [32]. The reporter systems used here included a panel of ten Pax6-binding sites recognized by different combinations of the PAI, RED, and HD subdomains [21], coupled with two reporters based on a recently identified Pax6-dependent enhancer in the Trpm3/miR-204 locus [15]. We found that the N50K and R128C mutations in PAX6 have unexpected properties relative to other mutations that impair structure and function of either the PAI or RED subdomain that can be used for future mechanistic studies of protein–protein complexes recruited by PAX6 to chromatin DNA.
Methods
Reporter gene construction
A 1.5 kb region (mm9: chr19:22524101–22525510) of the mouse Trpm3 gene that includes a previously identified Pax6-binding site [15] was synthesized by GenScript and cloned into a pGL3-Basic vector, which contains a firefly luciferase reporter gene. Six copies of Pax6-binding site 3.4 (5′-ACT TTA AAG GAT GAC AGA ATT-3′, Pax6-binding nucleotides underlined) and motifs P6CON, 1–1, 1–2, 1–3, 2–1, 2–2, 3–1, 3–2, 3–3, and 4–1 were also made as described above and cloned into the E4-TATA-firefly luciferase reporter as described earlier [21,42].
Cell cultures, transfections, and reporter assays
The transient transfections were conducted in embryonic carcinoma P19 cells that lack endogenous Pax6 expression as described earlier [21]. PAX6 and PAX6(5a) cDNAs were generated in CMV-containing vector pKW10 as described earlier [32,42]. Briefly, the cells were cultured on 10 cm dishes in DMEM (Life Technologies, Carlsbad, CA, Cat # 11965) medium supplemented with 10% fetal bovine serum (FBS). Sixteen hours before transfection, 5 × 104 cells were seeded into each well of the 24-well microplate in the medium without antibiotics. The cells were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, Cat # 11668027) with 0.6 µg of firefly luciferase reporter plasmids, and 50 ng of empty vector (pKW10) or expression plasmids encoding PAX6, PAX6(5a), and their mutants. As an internal control, 20 ng of Renilla-TK luciferase plasmids was also included in the transfection mixtures. After 4 h, fresh medium was added, and the cells were incubated for another 26–30 h. The dual luciferase reporter assay system (Promega, Fitchburg, WI, Cat# E1960) was used to measure promoter activity. The experiments were performed in triplicate with two independent repeats. For statistical evaluation of the results, p values were calculated from paired Student t tests between reactions with cDNA vector and wild-type Pax6/Pax6(5a) cDNA for each motif. Paired Student t tests were also performed between wild-type Pax6/Pax6(5a) and each Pax6 mutant.
Homology model of human PAX6 bound to DNA
PAI and RED domains in complex with cognate DNA were obtained from the Protein Data Bank (PDB) coordinate file of 6PAX. Separately, a homology model of the human HD domain was generated using the Drosophila melanogaster HD domain template structure (PDB entry 3A01). A set of seven protein–nucleotide atom–atom distances were extracted from this complex. The combined complex structure containing PAI, RED, and HD domains with the cognate DNA was constructed using these structures, models, and distance restraints. The crystal structure of the human TBP domain in complex with DNA (PDB entry 1CDW) and the structure of the mouse bZIP bound to DNA (PDB entry 4AUW) were also included in the PAX6 model. The homology model was produced using Modeler 9v2. The figure was prepared with Pymol 1.1r1 (PyMOL Molecular Graphics System; Schrödinger, LLC, Portland, OR).
Results and Discussion
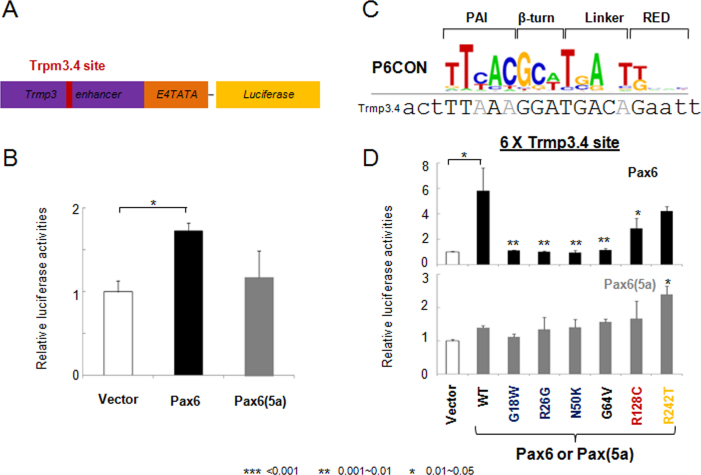
The PAX6 mutations studied here are shown in Figure 1A. In the first set of experiments, we used a panel of ten reporter genes driven by Pax6-binding sites of different structure. They represent a continuum of sequences that provide high and moderate levels of Pax6 activation as well as transcriptional repression [21]. The 6xP6CON reporter is derived from the Pax6 consensus binding site identified by Epstein et al. [26] (see Figure 1B,C). This reporter was activated by wild-type PAX6 approximately ten times in the P19 cells. Five PAX6 mutants, including G18W, R26G, N50K G64V, and R242T, resulted in reduced activity. In contrast, the R128C mutation elicited nearly 2.5-fold activation of the P6CON. As expected, no activation of P6CON was found with PAX6(5a) as this splice variant does not bind P6CON [25,44]. These results are similar to our earlier studies of G18W, R26G, G64V, and R128C in P19 cells [43] and the N50K mutation in HEK293 cells [32]. The R242T mutation was evaluated for the first time and showed reduced activation of P6CON in P19 cells.
Figure 1.
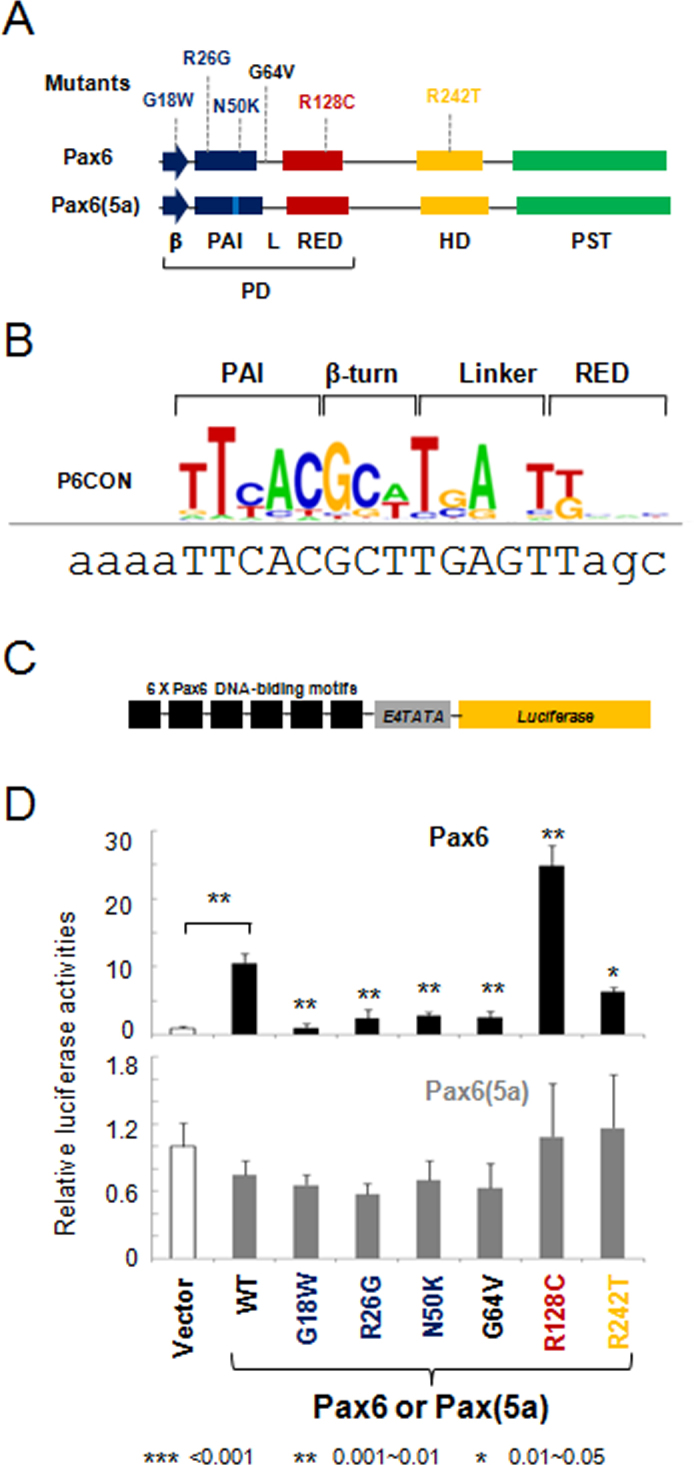
Transcriptional regulation by PAX6 and PAX6(5a) on P6CON luciferase reporters. A: A schematic diagram of PAX6 and PAX6(5a) proteins including six missense mutations G18W, R26G, N50K, G64V, R128C, and R242T. Paired domain, PD; homeodomain, HD, proline/serine/threonine-rich domain, PST; β-turn, β; linker between the PAI and RED subdomains, L. B: Distribution of nucleotides in the P6CON Pax6-binding site. The DNA sequence for gene synthesis is aligned with the P6CON motif. The binding sites and flanking sequences are in upper and lower case, respectively. C: A schematic representation of the 6x Pax6 DNA-binding motif/E4TATA-luciferase reporter. D: Evaluation of PAX6 (black bars) and PAX6(5a) (gray bars) series in cotransfected P19 cells. The data are expressed as relative fold changes elicited in the presence of PAX6 or PAX6(5a) compared with the changes found with the empty vector, pKW10. The sample size n=6 from two independent triplicates. The error bars represent the standard deviation. The significant fold-changes are indicated by the asterisks. The range of the p values are: *** <0.001, ** 0.001~0.01, * 0.01~0.05. The p values between empty vector and wild-type Pax6/Pax6(5a) are calculated with paired Student t tests. The p value for each mutant is calculated by comparing it with the wild-type.
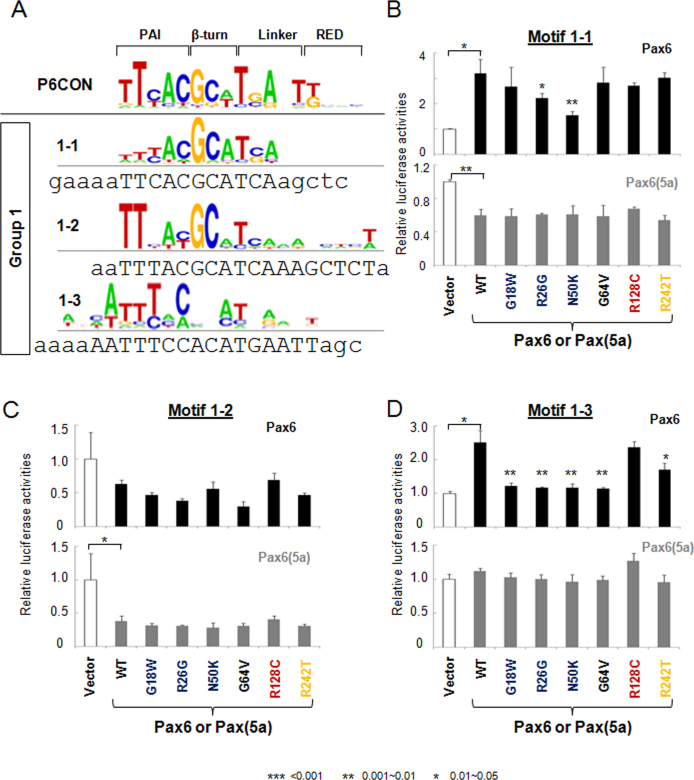
A group of three Pax6-binding site variants, motifs 1–1, 1–2, and 1–3, are related to the P6CON sequence (Figure 2A); however, motifs 1–1 and 1–3 activate the reporter gene (range of activation two- to four-fold) while motif 1–2 acts as a transcriptional repressor (Figure 2B-D). The PAX6 N50K showed the lowest activation on the motif 1–1 reporter, while the other mutants mostly showed no change (Figure 2B). PAX6 N50K and R128C showed no significant repression on the motif 1–2 reporter. Both these activities were similar to the wild type PAX6 (Figure 2C). In contrast, the R26G and G64V PAX6 proteins were more potent repressors compared to the wild-type PAX6 (Figure 2C). Motif 1–3 was not activated by PAX6 carrying mutations G18W, R26G, N50K, and G64V (Figure 2D). The R128C mutation apparently did not abrogate PAX6 function on motif 1–3 (Figure 2D). Although PAX6(5a) can bind motifs 1–1, 1–2, and 1–3 [21], none of the mutants studied here showed any significant difference compared to the wild-type PAX6(5a; Figure 2B-D).
Figure 2.
Transcriptional regulation by PAX6 and PAX6(5a) on motif 1–1, 1–2, and 1–3 luciferase reporters. A: Distribution of the nucleotides in the Pax6-binding motifs 1–1, 1–2, and 1–3. The DNA sequences for gene synthesis are aligned with the corresponding motifs. The binding sites and flanking sequences are in upper and lower case, respectively. Evaluation of PAX6 (black bars) and PAX6(5a) (gray bars) series with 1–1, 1–2, and 1–3 luciferase reporters in cotransfected P19 cells is shown in panel (B), (C), and (D) respectively. The sample size n=6 from two independent triplicates. The error bars represent the standard deviation. The significant fold-changes are indicated by the asterisks. The range of the p values are: *** <0.001, ** 0.001~0.01, * 0.01~0.05.
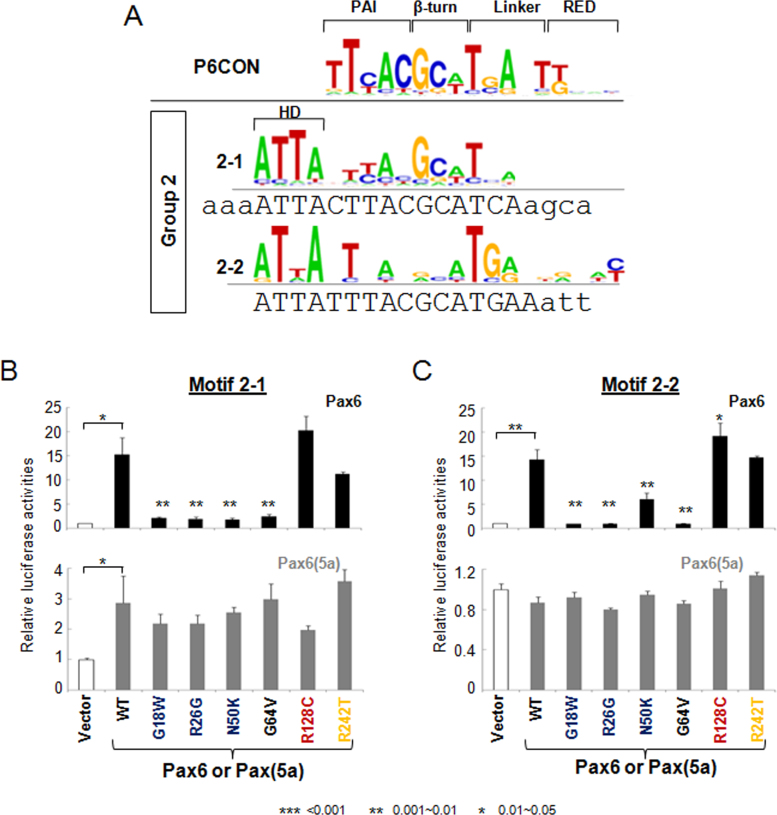
A group of two Pax6-binding site variants (2–1 and 2–2) is characterized by a presumptive HD-binding site, 5′-ATTA’-3 (Figure 3A). This sequence is localized upstream from the target sequence recognized by the PAI, β-turn, and linker regions and is similar to P6CON (Figure 3A). The 2–1 and 2–2 motifs were activated approximately 15-fold by PAX6 (Figure 3B,C). For motif 2–1, the majority of missense mutants showed approximately 10% of the wild-type PAX6 activity (Figure 3B), and the overall pattern of their properties was similar to motif 1–3 (compare Figure 2D and Figure 3B). On motif 2–2, this pattern was “broken” by mutant N50K, which had about one third of the wild-type PAX6 activity (Figure 3C). Although motifs 2–1 and 2–2 differed only at the G residue in the linker region, motif 2–1 (Figure 3B), but not motif 2–2 (Figure 3C), was nearly three-fold activated by PAX6(5a). No change in reporter gene activity on motif 2–1 was found for PAX6(5a) carrying mutations G18W, R26G, N50K, and G64V (Figure 3B). In contrast, PAX6(5a) R128C (R242T) showed moderate loss (gain) of activity, respectively (Figure 3B). Interestingly, no change in reporter gene activity was found on motif 2–2 (Figure 3C).
Figure 3.
Transcriptional regulation by PAX6 and PAX6(5a) on motif 2–1 and 2–2 luciferase reporters. A: Distribution of the nucleotides in Pax6-binding motifs 2–1 and 2–2. The DNA sequences for gene synthesis are aligned with the corresponding motifs. The binding sites and flanking sequences are in upper and lower case, respectively. Evaluation of PAX6 (black bars) and PAX6(5a) (gray bars) series with 2–1 (B) and 2–2 (C) luciferase reporter in cotransfected P19 cells. The sample size n=6 from two independent triplicates. The error bars represent the standard deviation. The significant fold-changes are indicated by the asterisks. The range of the p values are: *** <0.001, ** 0.001~0.01, * 0.01~0.05.
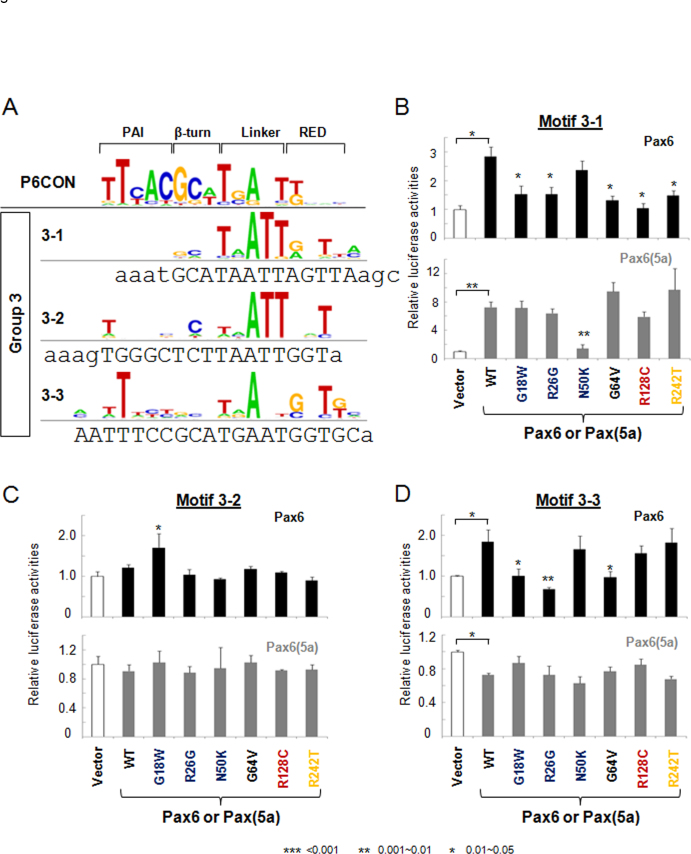
The third group of Pax6-binding sites, including motifs 3–1, 3–2 and 3–3, were characterized by nucleotides recognized by the linker region and RED subdomain (Figure 4A). Sites 3–1 and 3–2 preferred PAX6(5a) over PAX6, while site 3–3 was bound and activated only by PAX6 [21]. On motif 3–1, five Pax6 mutants, including G18W, R26G, G64V, R128C, and R242T, showed at least 50% loss of their activation potential (Figure 4B). In contrast, PAX6 N50K retained more than 80% of the wild-type PAX6 activity (Figure 4B). On motif 3–2, wild-type PAX6 acted as a weak activator, and the G18W mutation produced a moderate approximate 1.6-fold activation (Figure 4C). Three mutants, including G18W, R26G, and G64V, evaluated with motif 3–3 lost their activation potential (Figure 4D). The replacement of the T to A nucleotide in the PAI-RED linker binding region (ATT to AAT) had only a marginal effect in PAX6 R128C (Figure 4D). The most notable finding in the PAX6(5a) series (Figure 4B–D) was strongly reduced activation by PAX6(5a) N50K using the motif 3–1 reporters (Figure 4B).
Figure 4.
Transcriptional regulation by PAX6 and PAX6(5a) on motif 3–1, 3–2, and 3–3 luciferase reporters. A: Distribution of the nucleotides in Pax6-binding motifs 3–1, 3–2, and 3–3. The DNA sequences for gene synthesis are aligned with the corresponding motifs. The binding sites and flanking sequences are in upper and lower case, respectively. Evaluation of PAX6 (black bars) and PAX6(5a; gray bars) series with 3–1 (B), 3–2 (C), and 3–3 (D) luciferase reporters in cotransfected P19 cells. The sample size n=6 from two independent triplicates. The error bars represent the standard deviation. The significant fold-changes are indicated by the asterisks. The range of the p values are: *** <0.001, ** 0.001~0.01, * 0.01~0.05.
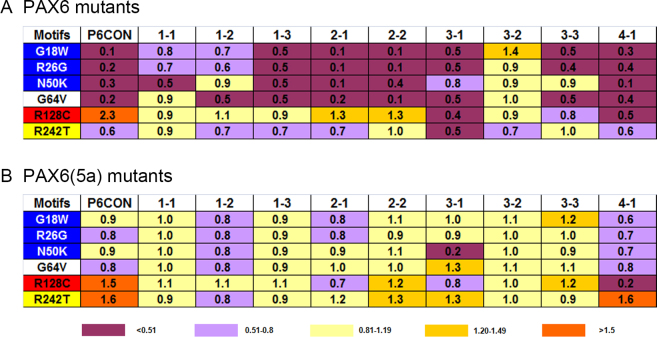
Motif 4–1 was comprised from a presumptive HD-binding sequence 5′-TAAT-3′ followed by nucleotides preferentially recognized by PAX6(5a) (Figure 5A) though PAX6 activation was higher compared to PAX6(5a) (Figure 5B). All PAX6 mutants evaluated lost their activation potential with the highest effect found for PAX6 N50K (Figure 5B). In the PAX6(5a) series, the highest decrease in promoter activity was found for R128C. In contrast, the activity of mutant PAX6(5a) R242T nearly doubled compared to wild-type PAX6(5a). To visualize similar and distinct properties of individual PAX6 mutations examined here, Figure 6 provides a color-coded summary of data from Figure 1-Figure 5.
Figure 5.
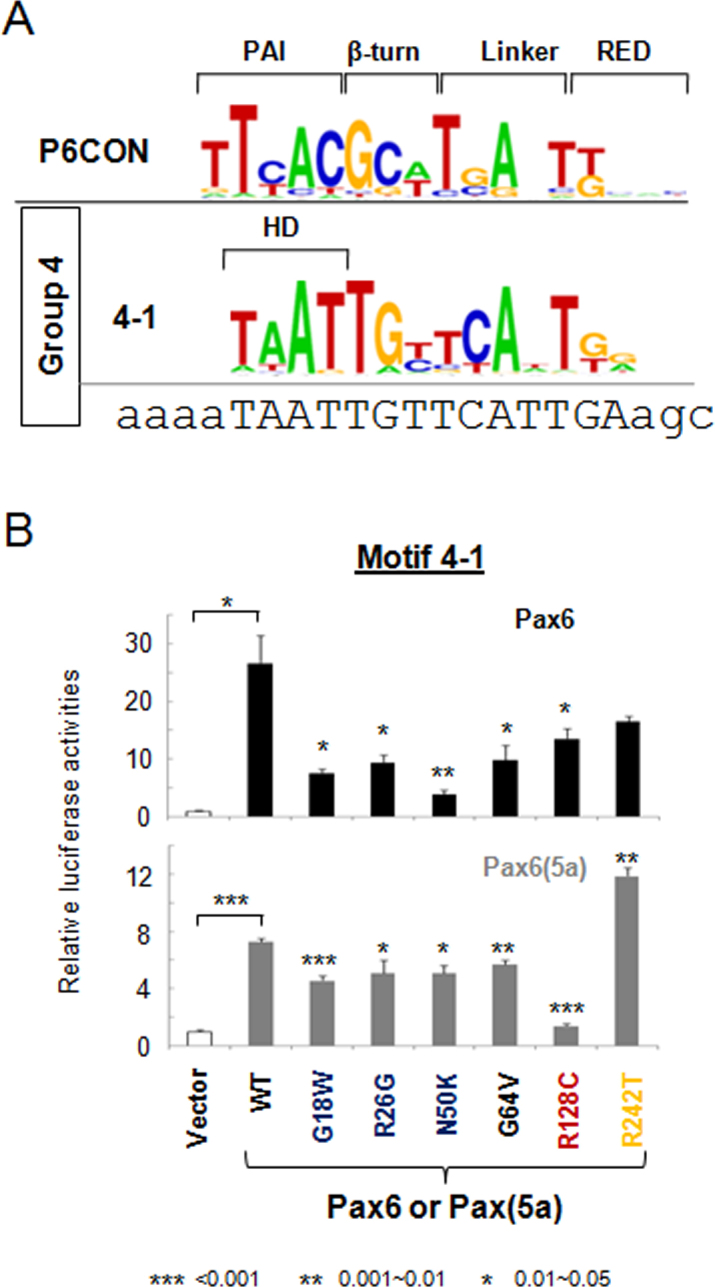
Transcriptional regulation by PAX6 and PAX6(5a) on motif 4–1 luciferase reporters. A: Distribution of the nucleotides in Pax6-binding motif 4–1. The DNA sequences for gene synthesis are aligned with the corresponding motifs. The binding sites and flanking sequences are in upper and lower case, respectively. Evaluation of PAX6 (black bars) and PAX6(5a; gray bars) series with 4–1 motif luciferase reporters in cotransfected P19 cells. The sample size n=6 from two independent triplicates. The error bars represent the standard deviation. The significant fold-changes are indicated by the asterisks. The range of the p values are: *** <0.001, ** 0.001~0.01, * 0.01~0.05.
Figure 6.
A summary of relative transactivation of six PAX6 and corresponding PAX6(5a) missense mutants compared to wild-type PAX6 and PAX6(5a) proteins.
The Trpm3/miR-204 model is a novel Pax6-regulated system [15]. We first tested a 1.5 kb genomic fragment from intron 6 of the mouse Trmp3 locus. We found that this fragment was activated by PAX6 by a factor of 1.8 in cotransfected P19 cells (Figure 7A,B). This result was nearly identical to the activity of the zebrafish homologous genomic fragment tested in HeLa cells [15]. We next generated a reporter plasmid driven by six copies of the Pax6-binding site (Figure 7C) identified by a combination of electrophoretic mobility shift assays (EMSAs) and chromatin immunoprecipitation (ChIP) assays [15]. With PAX6, this reporter yielded approximately six-fold activation that was reduced in the presence of all PAX6 mutants (Figure 7D). PAX6(5a) did not activate this promoter although the R242T mutant showed almost two-fold activation (Figure 7D).
Figure 7.
Transcriptional regulation Trpm3 luciferase reporters by PAX6 and PAX6(5a). A: A schematic representation of the Trpm3 enhancer and 6x Pax6 DNA-binding motif/E4TATA-luciferase reporter. B: Cotransfection of Trpm3/E4TATA/luc with PAX6 or PAX6(5a). C: A comparison of Trpm3.4 Pax6-binding site with P6CON. The DNA sequence for gene synthesis is aligned with the P6CON motif. The binding sites and flanking sequences are in upper and lower case, respectively. D: Evaluation of PAX6 (black bars) and PAX6(5a; gray bars) series with the 6xTrpm3.4 luciferase reporter in cotransfected P19 cells. The sample size n=6 from two independent triplicates. The error bars represent the standard deviation. The significant fold-changes are indicated by the asterisks. The range of the p values are: *** <0.001, ** 0.001~0.01, * 0.01~0.05.
Previous systematic studies of PAX6 missense mutants primarily focused on the correlation between their transactivation and DNA-binding in in vitro assays [43,45,46]. For mutations G18W, R26G, G64V, and R128C, we did not find any correlation between their DNA-binding and reporter gene activity [43]. Similarly, no correlation between in vitro DNA binding and transactivation/repression was found using the current panel of reporters used here [21]. Although no data are available to explain this apparent discrepancy, it is obvious that a typical missense mutation does not work as a DNA-binding but rather as a functional loss-of-function mutant in the context of the execution of the transcription process. More recently, extensive analyses of 50 PAX6 missense mutations predicted that the majority (77%) of these mutations influence intrinsic protein stability while the minority (30%) are thought to impair DNA binding [45]. The HD mutation R242T gave higher levels of activation using the P3 HD-dimeric reporter in HeLa cells [41]. Reduced sensitivity to trypsin digestion was found for the mutated protein concomitant with increased amounts of the nuclear proteins raising the possibility that the increased activation of the reporter gene is a protein dosage effect. In the current studies, we found that PAX6 R242T mutants had either decreased or unchanged activation (Figure 1, Figure 2, Figure 3, Figure 4, Figure 5, Figure 7). Only PAX6(5a) R242T produced more activity using motifs 3–1 and 4–1 and Trpm3.4 (Figure 4, Figure 5, Figure 7). Thus, multiple mechanisms can be used to explain the pleiotropic effects of this HD mutation.
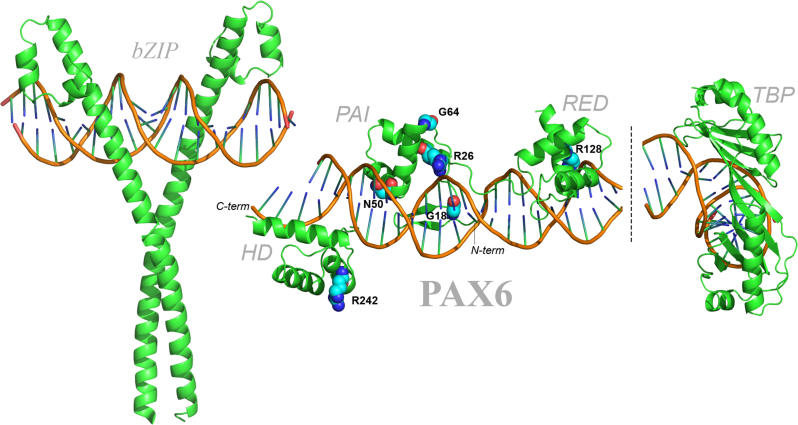
To put in a structural context of the studied mutations, a PAX6 protein–DNA model was built (Figure 8). In this model, we used a DNA-binding site recognized by all five DNA-binding subdomains (β-turn, PAI, linker, RED and HD; e.g., motifs 2–1 and 2–2) with the HD-binding region upstream of the paired domain. The PAX6 PD–DNA interaction is based on a crystallographic model determined with P6CON [47]. The PAX6 HD interaction with DNA was based on similar data [48]. The structure of the approximate 80 amino acid region that connects the PD and HD is not known, and, thus, was not included in Figure 8. The present data show that the N50K and R128C mutations differ from each other (see Figure 6) and that they represent “outliers” from the panel of mutations investigated here and those tested in our earlier studies, including A33P, S43P, I87R, and V126D [43]. Both mutations are at the interface of protein–DNA interaction. The N50 residue is located in the beginning of the third α-helix comprising the PAI subdomain [47]. A substitution of polar uncharged asparagine for the charged lysine residue established a new type of interaction with the DNA, though not experimentally examined by Alibes et al. [45]. The outlier property of this substitution is documented on motifs 2–2 and 3–3 [31,41] activated by PAX6 [PAX6(5a)], respectively. The R128 residue is located in the central part of the third α-helix comprising the RED subdomain [47] (Figure 8). In addition to the change from a positively charged residue to a neutral one, the nonsynonymous arginine to cysteine substitution is known to disrupt helical conformation in many proteins [49]. The odd property of this substitution is documented on motifs P6CON (hyperactivation) and a larger group, including 1–1, 1–3, 2–1, 2–2, 3–2, and 3–3 (no change), by PAX6. It was argued earlier that the R128C mutation does not cause any significant conformational change in the mutated PAX6 proteins bound to the Pax6-binding site evaluated with FoldX [45]. However, the most likely explanation is that the major impact is on PAX6(5a) type-binding sites (i.e., motifs 3–1 and 4–1), recognized by PAX6 and PAX6(5a) proteins, an idea that remains to be experimentally tested with FoldX and potentially via direct crystallographic studies taking advantage of choosing from the DNA-binding motifs evaluated here. We conclude that the N50K and R128C PAX6 missense mutations represent useful systems for further dissecting molecular mechanisms of Pax6 in gene activation and repression.
Figure 8.
Homology model of the human paired box protein PAX6 bound to DNA. Interaction of PAX6 with DNA and localization of missense disease-causing mutations within the protein–DNA complex. The protein and DNA molecules are shown using cartoon representation. Residues with observed mutations in patients are shown using sphere representation. The color coding is atom specific. Red, blue, and light blue correspond to oxygen, nitrogen, and carbon atoms, respectively. The long flexible linker between the RED and homeodomain (HD) domains [54] was not included in the model. Binding of the bZIP domain transcription factor (e.g., c-Maf) and TBP in the vicinity of PAX6, e.g., identified in αA- and αB-crystallin promoters [40], is also shown.
Indeed, recent functional studies of the Leca4 (N50K) and Leca2 (R128C) Pax6 alleles revealed similarities and differences in cellular responses controlled by Pax6 [32]. Pax6 target genes, disrupted by the N50K mutation, include genes that control forebrain patterning, neurogenesis, and decreasing cell proliferation. In contrast, Pax6-dependent genes, affected by the R128C mutation, include genes that increase cell proliferation, without any apparent impact on neurogenesis. An attractive hypothesis is that these mutations impair interactions with chromatin remodeling enzymes, including Brg1 and Brm [33-35]. The availability of specific antibodies for ChIP-seq studies in conjunction with mouse genetic studies of Brg1, Brm, BAF155, and BAF170 alleles crossed with the Pax6Leca2/Leca4 alleles allows direct testing of this model.
To further illustrate how PAX6 mutations can disrupt physical interactions with other proteins bound, we included a model of bZIP factor-bound to DNA (Figure 8, left) and TATA-box binding protein (TBP, Figure 8, right). It has been shown earlier that c-Maf, a bZIP transcription factor, binds to Pax6 [50], and local configurations of Pax6- and Maf-binding sites support different levels of transcriptional synergism [42]. Pax6 and c-Maf are known to bind p300 histone acetyltransferase [51]. Pax6 and CBP/p300 regulate multiple common genes during the lens placode formation [52] and crystallins in differentiating lens fibers [53]. Binding of TBP to the TATA box induces large DNA bending (Figure 8). The Pax6/TBP interaction has been established elsewhere [39]. Taken together, the present models show that major and minor conformational changes in mutated PAX6 proteins can have large impacts on the protein–protein interactions with other regulatory proteins, including c-Maf, TBP, and their associated chromatin remodeling complexes.
Some PAX6 missense mutations display pleiotropic activity in different cell types examined, such P19, 293T, CHO-K1, and N/N1003A [43]. The P19 cells were used here as the panel of ten luciferase reporters displayed a wide range of transcriptional activation and repression [21] and mutations N50K and R128C gave similar results in P19 and HEK293 cells [32]. In conclusion, the present studies emphasize the complexity of Pax6-dependent transcriptional activation and repression mechanisms, and identify the N50K and R128C substitutions as valuable tools for testing interactions between Pax6, Pax6N50K, and Pax6R128C with other regulatory proteins, including chromatin remodelers and their impact on eye and brain development.
Acknowledgments
We thank Dr. Rebecca McGreal for a critical reading of the manuscript. This work was supported by NIH grant R01 EY012200 (AC), and an unrestricted grant from Research to Prevent Blindness to the Department of Ophthalmology and Visual Sciences. DU is a student in the Magistère de Génétique Graduate Program at Université Paris Diderot, Sorbonne Paris Cité.
References
- 1.Cvekl A, Piatigorsky J. Lens development and crystallin gene expression: many roles for Pax-6. Bioessays. 1996;18:621–30. doi: 10.1002/bies.950180805. [DOI] [PubMed] [Google Scholar]
- 2.Shaham O, Menuchin Y, Farhy C, Ashery-Padan R. Pax6: a multi-level regulator of ocular development. Prog Retin Eye Res. 2012;31:351–76. doi: 10.1016/j.preteyeres.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Medina-Martinez O, Jamrich M. Foxe view of lens development and disease. Development. 2007;134:1455–63. doi: 10.1242/dev.000117. [DOI] [PubMed] [Google Scholar]
- 4.Swamynathan SK. Ocular surface development and gene expression. J Ophthalmol. 2013;2013:103947. doi: 10.1155/2013/103947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osumi N, Shinohara H, Numayama-Tsuruta K, Maekawa M. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells. 2008;26:1663–72. doi: 10.1634/stemcells.2007-0884. [DOI] [PubMed] [Google Scholar]
- 6.Dohrmann C, Gruss P, Lemaire L. Pax genes and the differentiation of hormone-producing endocrine cells in the pancreas. Mech Dev. 2000;92:47–54. doi: 10.1016/s0925-4773(99)00324-x. [DOI] [PubMed] [Google Scholar]
- 7.Nomura T, Haba H, Osumi N. Role of a transcription factor Pax6 in the developing vertebrate olfactory system. Dev Growth Differ. 2007;49:683–90. doi: 10.1111/j.1440-169X.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- 8.Duparc RH, Boutemmine D, Champagne MP, Tetreault N, Bernier G. Pax6 is required for delta-catenin/neurojugin expression during retinal, cerebellar and cortical development in mice. Dev Biol. 2006;300:647–55. doi: 10.1016/j.ydbio.2006.07.045. [DOI] [PubMed] [Google Scholar]
- 9.Holm PC, Mader MT, Haubst N, Wizenmann A, Sigvardsson M, Gotz M. Loss- and gain-of-function analyses reveal targets of Pax6 in the developing mouse telencephalon. Mol Cell Neurosci. 2007;34:99–119. doi: 10.1016/j.mcn.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Wolf LV, Yang Y, Wang J, Xie Q, Braunger B, Tamm ER, Zavadil J, Cvekl A. Identification of pax6-dependent gene regulatory networks in the mouse lens. PLoS ONE. 2009;4:e4159. doi: 10.1371/journal.pone.0004159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Numayama-Tsuruta K, Arai Y, Takahashi M, Sasaki-Hoshino M, Funatsu N, Nakamura S, Osumi N. Downstream genes of Pax6 revealed by comprehensive transcriptome profiling in the developing rat hindbrain. BMC Dev Biol. 2010;10:6. doi: 10.1186/1471-213X-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, Rajagopal R, Liu Y, Dattilo LK, Shaham O, Ashery-Padan R, Beebe DC. The mechanism of lens placode formation: a case of matrix-mediated morphogenesis. Dev Biol. 2011;355:32–42. doi: 10.1016/j.ydbio.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Q, Yang Y, Huang J, Ninkovic J, Walcher T, Wolf L, Vitenzon A, Zheng D, Gotz M, Beebe DC, Zavadil J, Cvekl A. Pax6 interactions with chromatin and identification of its novel direct target genes in lens and forebrain. PLoS ONE. 2013;8:e54507. doi: 10.1371/journal.pone.0054507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaham O, Gueta K, Mor E, Oren-Giladi P, Grinberg D, Xie Q, Cvekl A, Shomron N, Davis N, Keydar-Prizant M, Raviv S, Pasmanik-Chor M, Bell RE, Levy C, Avellino R, Banfi S, Conte I, Ashery-Padan R. Pax6 regulates gene expression in the vertebrate lens through miR-204. PLoS Genet. 2013;9:e1003357. doi: 10.1371/journal.pgen.1003357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grinchuk O, Kozmik Z, Wu X, Tomarev S. The Optimedin gene is a downstream target of Pax6. J Biol Chem. 2005;280:35228–37. doi: 10.1074/jbc.M506195200. [DOI] [PubMed] [Google Scholar]
- 17.Liu W, Lagutin OV, Mende M, Streit A, Oliver G. Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J. 2006;25:5383–95. doi: 10.1038/sj.emboj.7601398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaniolo K, Leclerc S, Cvekl A, Vallieres L, Bazin R, Larouche K, Guerin SL. Expression of the alpha4 integrin subunit gene promoter is modulated by the transcription factor Pax-6 in corneal epithelial cells. Invest Ophthalmol Vis Sci. 2004;45:1692–704. doi: 10.1167/iovs.03-0908. [DOI] [PubMed] [Google Scholar]
- 19.Machon O, Kreslova J, Ruzickova J, Vacik T, Klimova L, Fujimura N, Lachova J, Kozmik Z. Lens morphogenesis is dependent on Pax6-mediated inhibition of the canonical Wnt/beta-catenin signaling in the lens surface ectoderm. Genesis. 2010;48:86–95. doi: 10.1002/dvg.20583. [DOI] [PubMed] [Google Scholar]
- 20.Oron-Karni V, Farhy C, Elgart M, Marquardt T, Remizova L, Yaron O, Xie Q, Cvekl A, Ashery-Padan R. Dual requirement for Pax6 in retinal progenitor cells. Development. 2008;135:4037–47. doi: 10.1242/dev.028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Q, Cvekl A. The orchestration of mammalian tissue morphogenesis through a series of coherent feed-forward loops. J Biol Chem. 2011;286:43259–71. doi: 10.1074/jbc.M111.264580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jun S, Desplan C. Cooperative interactions between paired domain and homeodomain. Development. 1996;122:2639–50. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- 23.Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–49. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 24.Carriere C, Plaza S, Martin P, Quatannens B, Bailly M, Stehelin D, Saule S. Characterization of quail Pax-6 (Pax-QNR) proteins expressed in the neuroretina. Mol Cell Biol. 1993;13:7257–66. doi: 10.1128/mcb.13.12.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Epstein JA, Glaser T, Cai J, Jepeal L, Walton DS, Maas RL. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–34. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 26.Epstein J, Cai J, Glaser T, Jepeal L, Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 1994;269:8355–61. [PubMed] [Google Scholar]
- 27.Azuma N, Nishina S, Yanagisawa H, Okuyama T, Yamada M. PAX6 missense mutation in isolated foveal hypoplasia. Nat Genet. 1996;13:141–2. doi: 10.1038/ng0696-141. [DOI] [PubMed] [Google Scholar]
- 28.van Heyningen V, Williamson KA. PAX6 in sensory development. Hum Mol Genet. 2002;11:1161–7. doi: 10.1093/hmg/11.10.1161. [DOI] [PubMed] [Google Scholar]
- 29.Thaung C, West K, Clark BJ, McKie L, Morgan JE, Arnold K, Nolan PM, Peters J, Hunter AJ, Brown SD, Jackson IJ, Cross SH. Novel ENU-induced eye mutations in the mouse: models for human eye disease. Hum Mol Genet. 2002;11:755–67. doi: 10.1093/hmg/11.7.755. [DOI] [PubMed] [Google Scholar]
- 30.Ramaesh T, Williams SE, Paul C, Ramaesh K, Dhillon B, West JD. Histopathological characterisation of effects of the mouse Pax6(Leca4) missense mutation on eye development. Exp Eye Res. 2009;89:263–73. doi: 10.1016/j.exer.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 31.Favor J, Peters H, Hermann T, Schmahl W, Chatterjee B, Neuhauser-Klaus A, Sandulache R. Molecular characterization of Pax6(2Neu) through Pax6(10Neu): an extension of the Pax6 allelic series and the identification of two possible hypomorph alleles in the mouse Mus musculus. Genetics. 2001;159:1689–700. doi: 10.1093/genetics/159.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walcher T, Xie Q, Sun J, Irmler M, Beckers J, Ozturk T, Niessing D, Stoykova A, Cvekl A, Ninkovic J, Gotz M. Functional dissection of the paired domain of Pax6 reveals molecular mechanisms of coordinating neurogenesis and proliferation. Development. 2013;140:1123–36. doi: 10.1242/dev.082875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ninkovic J, Steiner-Mezzadri A, Jawerka M, Akinci U, Masserdotti G, Petricca S, Fischer J, von Holst A, Beckers J, Lie CD, Petrik D, Miller E, Tang J, Wu J, Lefebvre V, Demmers J, Eisch A, Metzger D, Crabtree G, Irmler M, Poot R, Gotz M. The BAF Complex Interacts with Pax6 in Adult Neural Progenitors to Establish a Neurogenic Cross-Regulatory Transcriptional Network. Cell Stem Cell. 2013;13:403–18. doi: 10.1016/j.stem.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuoc TC, Boretius S, Sansom SN, Pitulescu ME, Frahm J, Livesey FJ, Stoykova A. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev Cell. 2013;25:256–69. doi: 10.1016/j.devcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y, Stopka T, Golestaneh N, Wang Y, Wu K, Li A, Chauhan BK, Gao CY, Cveklova K, Duncan MK, Pestell RG, Chepelinsky AB, Skoultchi AI, Cvekl A. Regulation of alphaA-crystallin via Pax6, c-Maf, CREB and a broad domain of lens-specific chromatin. EMBO J. 2006;25:2107–18. doi: 10.1038/sj.emboj.7601114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He S, Pirity MK, Wang WL, Wolf L, Chauhan BK, Cveklova K, Tamm ER, Ashery-Padan R, Metzger D, Nakai A, Chambon P, Zavadil J, Cvekl A. Chromatin remodeling enzyme Brg1 is required for mouse lens fiber cell terminal differentiation and its denucleation. Epigenetics Chromatin. 2010;3:21. doi: 10.1186/1756-8935-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hussain MA, Habener JF. Glucagon gene transcription activation mediated by synergistic interactions of pax-6 and cdx-2 with the p300 co-activator. J Biol Chem. 1999;274:28950–7. doi: 10.1074/jbc.274.41.28950. [DOI] [PubMed] [Google Scholar]
- 38.Conte I, Carrella S, Avellino R, Karali M, Marco-Ferreres R, Bovolenta P, Banfi S. miR-204 is required for lens and retinal development via Meis2 targeting. Proc Natl Acad Sci USA. 2010;107:15491–6. doi: 10.1073/pnas.0914785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cvekl A, Kashanchi F, Brady JN, Piatigorsky J. Pax-6 interactions with TATA-box-binding protein and retinoblastoma protein. Invest Ophthalmol Vis Sci. 1999;40:1343–50. [PubMed] [Google Scholar]
- 40.Cvekl A, Yang Y, Chauhan BK, Cveklova K. Regulation of gene expression by Pax6 in ocular cells: a case of tissue-preferred expression of crystallins in lens. Int J Dev Biol. 2004;48:829–44. doi: 10.1387/ijdb.041866ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D'Elia AV, Puppin C, Pellizzari L, Pianta A, Bregant E, Lonigro R, Tell G, Fogolari F, van Heyningen V, Damante G. Molecular analysis of a human PAX6 homeobox mutant. Eur J Hum Genet. 2006;14:744–51. doi: 10.1038/sj.ejhg.5201579. [DOI] [PubMed] [Google Scholar]
- 42.Chauhan BK, Yang Y, Cveklova K, Cvekl A. Functional interactions between alternatively spliced forms of Pax6 in crystallin gene regulation and in haploinsufficiency. Nucleic Acids Res. 2004;32:1696–709. doi: 10.1093/nar/gkh334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chauhan BK, Yang Y, Cveklova K, Cvekl A. Functional properties of natural human PAX6 and PAX6(5a) mutants. Invest Ophthalmol Vis Sci. 2004;45:385–92. doi: 10.1167/iovs.03-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duncan MK, Kozmik Z, Cveklova K, Piatigorsky J, Cvekl A. Overexpression of PAX6(5a) in lens fiber cells results in cataract and upregulation of (alpha)5(beta)1 integrin expression. J Cell Sci. 2000;113:3173–85. doi: 10.1242/jcs.113.18.3173. [DOI] [PubMed] [Google Scholar]
- 45.Alibés A, Nadra AD, De Masi F, Bulyk ML, Serrano L, Stricher F. Using protein design algorithms to understand the molecular basis of disease caused by protein-DNA interactions: the Pax6 example. Nucleic Acids Res. 2010;38:7422–31. doi: 10.1093/nar/gkq683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shukla S, Mishra R. Predictions on impact of missense mutations on structure function relationship of PAX6 and its alternatively spliced isoform PAX6(5a). Interdiscip Sci. 2012;4:54–73. doi: 10.1007/s12539-012-0114-0. [DOI] [PubMed] [Google Scholar]
- 47.Xu HE, Rould MA, Xu W, Epstein JA, Maas RL, Pabo CO. Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and C-terminal subdomain in DNA binding. Genes Dev. 1999;13:1263–75. doi: 10.1101/gad.13.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson DS, Guenther B, Desplan C, Kuriyan J. High resolution crystal structure of a paired (Pax) class cooperative homeodomain dimer on DNA. Cell. 1995;82:709–19. doi: 10.1016/0092-8674(95)90468-9. [DOI] [PubMed] [Google Scholar]
- 49.Lund A, Joensen F, Christensen E, Duno M, Skovby F, Schwartz M. A novel arginine-to-cysteine substitution in the triple helical region of the alpha1(I) collagen chain in a family with an osteogenesis imperfecta/Ehlers-Danlos phenotype. Clin Genet. 2008;73:97–101. doi: 10.1111/j.1399-0004.2007.00926.x. [DOI] [PubMed] [Google Scholar]
- 50.Planque N, Leconte L, Coquelle FM, Benkhelifa S, Martin P, Felder-Schmittbuhl MP, Saule S. Interaction of Maf transcription factors with Pax-6 results in synergistic activation of the glucagon promoter. J Biol Chem. 2001;276:35751–60. doi: 10.1074/jbc.M104523200. [DOI] [PubMed] [Google Scholar]
- 51.Cvekl A, Mitton KP. Epigenetic regulatory mechanisms in vertebrate eye development and disease. Heredity (Edinb) 2010;105:135–51. doi: 10.1038/hdy.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolf L, Harrison W, Huang J, Xie Q, Xiao N, Sun J, Kong L, Lachke SA, Kuracha MR, Govindarajan V, Brindle PK, Ashery-Padan R, Beebe DC, Overbeek PA, Cvekl A. Histone posttranslational modifications and cell fate determination: lens induction requires the lysine acetyltransferases CBP and p300. Nucleic Acids Res. 2013;41:10199–10214. doi: 10.1093/nar/gkt824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Q, Dowhan DH, Liang D, Moore DD, Overbeek PA. CREB-binding protein/p300 co-activation of crystallin gene expression. J Biol Chem. 2002;277:24081–9. doi: 10.1074/jbc.M201821200. [DOI] [PubMed] [Google Scholar]
- 54.Vuzman D, Polonsky M, Levy Y. Facilitated DNA search by multidomain transcription factors: cross talk via a flexible linker. Biophys J. 2010;99:1202–11. doi: 10.1016/j.bpj.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]