Abstract
N-formylated sugars have been observed on the O-antigens of such pathogenic Gram-negative bacteria as Campylobacter jejuni and Francisella tularensis. Until recently, however, little was known regarding the overall molecular architectures of the N-formyltransferases that are required for the biosynthesis of these unusual sugars. Here we demonstrate that the protein encoded by the wbtj gene from F. tularensis is an N-formyltransferase that functions on dTDP-4-amino-4,6-dideoxy-d-glucose as its substrate. The enzyme, hereafter referred to as WbtJ, demonstrates a strict requirement for N10-formyltetrahydrofolate as its carbon source. In addition to the kinetic analysis, the three-dimensional structure of the enzyme was solved in the presence of dTDP-sugar ligands to a nominal resolution of 2.1 Å. Each subunit of the dimeric enzyme is dominated by a “core” domain defined by Met 1 to Ser 185. This core motif harbors the active site residues. Following the core domain, the last 56 residues fold into two α-helices and a β-hairpin motif. The hairpin motif is responsible primarily for the subunit:subunit interface, which is characterized by a rather hydrophobic pocket. From the study presented here, it is now known that WbtJ functions on C-4′ amino sugars. Another enzyme recently investigated in the laboratory, WlaRD, formylates only C-3′ amino sugars. Strikingly, the quaternary structures of WbtJ and WlaRD are remarkably different. In addition, there are several significant variations in the side chains that line their active site pockets, which may be important for substrate specificity. Details concerning the kinetic and structural properties of WbtJ are presented.
Keywords: N-formyltransferase; lipopolysaccharide; 4,6-dideoxy-4-formamido-d-glucose; O-antigen; protein structure; bacterial sugar biosynthesis; formylated sugars
Introduction
Since the devastating attacks on September 11, 2001, concern over the use of biological weapons has grown considerably. In reality, however, the utilization of bioweapons is not new. Indeed, the first documented case of biological warfare occurred during the Anatolian war (1320–1318 BC) where the Neshites and the Arzawans sent infected rams towards one another.1 Most likely these animals were carriers of the pathogenic bacterium, Francisella tularensis, a virulent Gram-negative organism that is the causative agent of tularemia or “rabbit fever.” More than two hundred species of vertebrates and invertebrates have now been shown to be infected by the bacterium.2 F. tularensis is transmitted to humans via deer flies, mosquitoes, or ticks, for example, or by the inappropriate handling of infected animals. Because of its highly infectious nature, the ease in which it can be produced as an aerosol, and its ability to cause severe respiratory illnesses and systemic infections, F. tularensis is classified as a select agent by the Centers for Disease Control and requires, at a minimum, biosafety level 2 handling practices.
As observed in most Gram-negative bacteria, the outer membranes of F. tularensis contain complex glycoconjugates referred to as lipopolysaccharides.2 Each lipopolysaccharide entity is composed of a lipid A molecule, a core oligosaccharide, and an O-antigen. Unlike the core oligosaccharides, the O-antigens of Gram-negative bacteria often contain unusual dideoxysugars such as 4,6-dideoxy-4-formamido-d-glucose or Qui4NFo as in the case of F. tularensis (Scheme 1). These sugars are added to the lipopolysaccharide via the action of glycosyltransferases, which employ nucleotide-linked derivatives such as dTDP-Qui4NFo as their substrates.
Scheme 1.
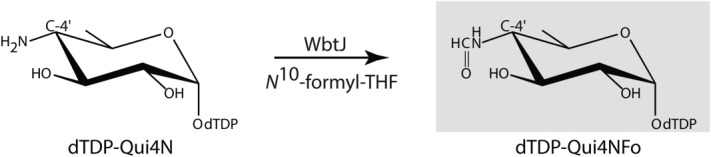
Reaction catalyzed by WbtJ.
The amino acid sequence for the hypothetical protein from F. tularensis subsp. tularensis that is thought to catalyze the last step in the formation of dTDP-Qui4NFo (Scheme 1) has been deposited in GenBank (AAS60274.1) and is referred to as WbtJ. Here we report the first structural and biochemical characterization of this putative N-formyltransferase. Our results demonstrate that, indeed, WbtJ catalyzes the conversion of dTDP-4,6-dideoxy-4-amino-d-glucose (dTDP-Qui4N) to dTDP-4,6-dideoxy-4-formamido-d-glucose (dTDP-Qui4NFo) using N10-formyltetrahydrofolate as the carbon donor. Details concerning the overall molecular architecture of WbtJ and its catalytic properties are presented.
Results and Discussion
Enzymatic activity of WbtJ
Given that the assignment of WbtJ as an N-formyltransferase was based simply on amino acid sequence homologies, we began our investigation by determining its enzymatic activity. Kinetic parameters were determined at 23°C via a discontinuous assay using HPLC as described in Materials and Methods. The enzyme demonstrated an absolute requirement for N10-formyltetrahydrofolate. A plot of substrate concentration (dTDP-Qui4N) versus initial rate was analyzed and fitted to the equation vo = (Vmax[S])/(KM + [S]) (Fig. 1). The Km for dTDP-Qui4N was determined to be 0.73 ± 0.08 mM and the kcat was 12.3 ± 0.5 s−1. The overall catalytic efficiency of WbtJ (1.7 × 104 M−1 s−1) is comparable to those values observed for other sugar N-formyltransferases.3,4 The enzyme demonstrated no catalytic activity with GDP-perosamine, dTDP-3,6-dideoxy-3-amino-d-glucose, or dTDP-3,6-dideoxy-3-amino-d-galactose as a substrate.
Figure 1.
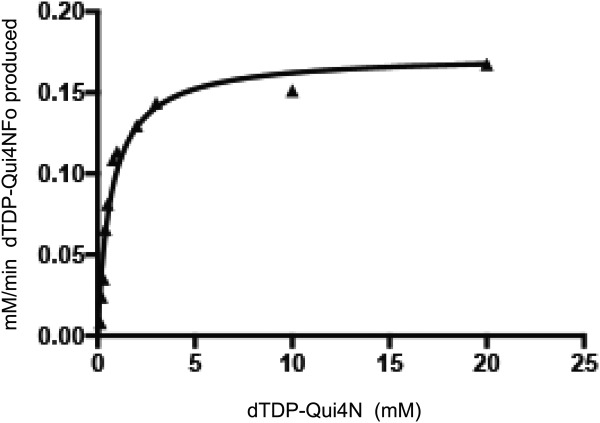
Plot for the determination of kcat and Km.
Overall structure of WbtJ
Crystallization conditions were initially surveyed for WbtJ in the absence of ligands or in the presence of dTDP and N5-formyltetrahydrofolate. The catalytically competent N10-formyltetrahydrofolate cofactor is unstable. The best crystals were observed growing from solutions containing 5 mM dTDP and 5 mM N5-formyltetrahydrofolate. These crystals belonged to the space group P21 and contained four subunits in the asymmetric unit. The structure of WbtJ was solved via SAD phasing as described in Materials and Methods. Unfortunately, no electron density for the N5-formyltetrahydrofolate cofactor was observed in the initial map. Subsequently, crystals of WbtJ, in complex with N5-formyltetrahydrofolate and either the substrate (dTDP-Qui4N) or the product (dTDP-Qui4NFo) were also grown. None of the electron density maps based on X-ray data collected from these crystals revealed bound N5-formyltetrahydrofolate. The electron density maps did show, however, that dTDP-sugars were bound in the active sites, but the pyranosyl moieties of the ligands were somewhat disordered.
A second crystal form, cocrystallized in the presence of N5-formyltetrahydrofolate and dTDP-Qui4NFo, was subsequently obtained that belonged to the space group P1 with eight subunits in the asymmetric unit. An electron density map using X-ray data collected from this crystal form revealed the position of a dTDP-Qui4NFo ligand in one subunit. In addition, four other subunits in the asymmetric unit contained dTDP-Qui4N, most likely a contaminant in the dTDP-Qui4NFo solutions. The remaining three subunits contained bound ligands, but the electron densities were ordered only for the dTDP moieties. Again, no bound tetrahydrofolate derivatives were observed in the electron density map. The model was refined to a nominal resolution of 2.1 Å (Roverall and Rfree to 19.5 and 27.4%) and is the focus of the following discussion. Given that there were eight subunits in the asymmetric unit, only subunits A and B, which form a functional dimer, are described below. The electron densities for the dTDP-Qui4N and dTDP-Qui4NFo ligands, when bound to subunits A and B, respectively, are displayed in Figure 2(a,b).
Figure 2.
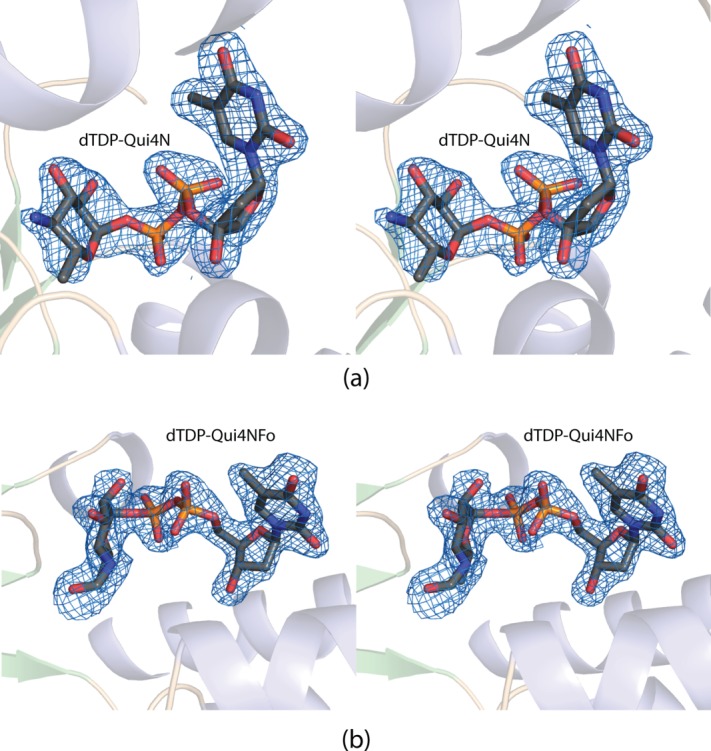
Electron density corresponding to the dTDP-ligands. The substrate for WbtJ, dTDP-Qui4N, was observed binding in the active site of subunit A. The electron density map shown in (a) was calculated with coefficients of the form Fo − Fc, where Fo was the native structure factor amplitude and Fc was the calculated structure factor amplitude. The map was contoured at 3σ. The ligand coordinates were not included in the map calculation or in the initial model refinements. Shown in (b) is the electron density corresponding to the dTDP-Qui4NFo ligand bound to subunit B. The map was calculated as described above. All figures were prepared with the software package PyMOL.5
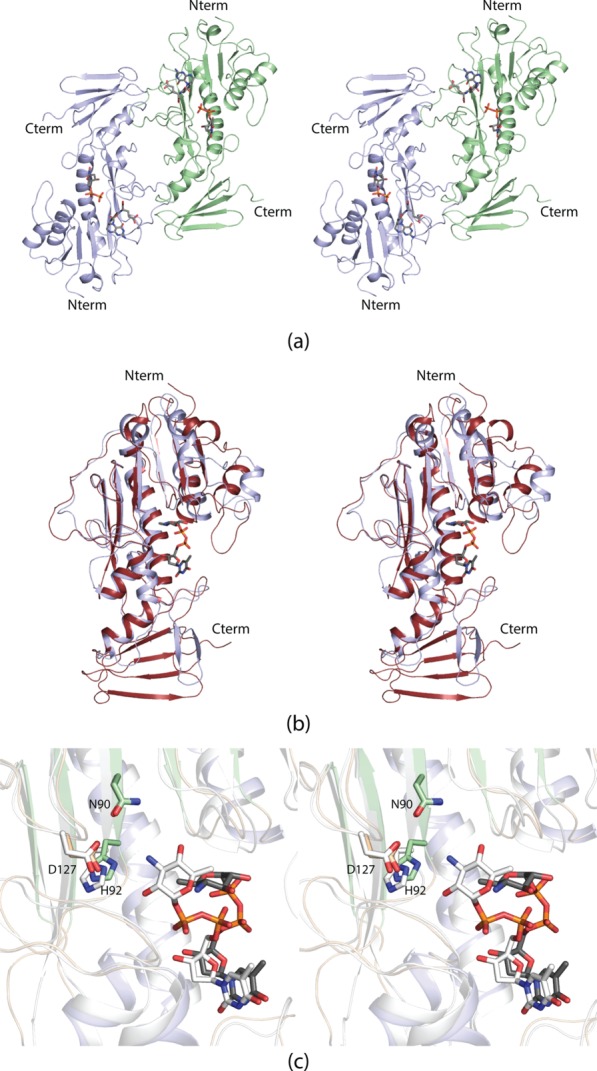
A ribbon representation of WbtJ is shown in Figure 3(a). The dimer has overall dimensions of ∼92 Å × 74 Å × 66 Å and a buried surface area of ∼1700 Å2 per subunit. The active sites, separated by ∼50 Å, are located in relatively shallow clefts at the surface of the enzyme. The pyrophosphoryl groups of the dTDP-sugar ligands are solvent exposed. Each monomer is dominated by a globular “core” formed by Met 1 to Ser 185. This N-terminal domain consists of a seven-stranded mixed β-sheet flanked on one side by three α-helices and on the other side by an α-helix and a helical turn [Fig. 3(b)].
Figure 3.
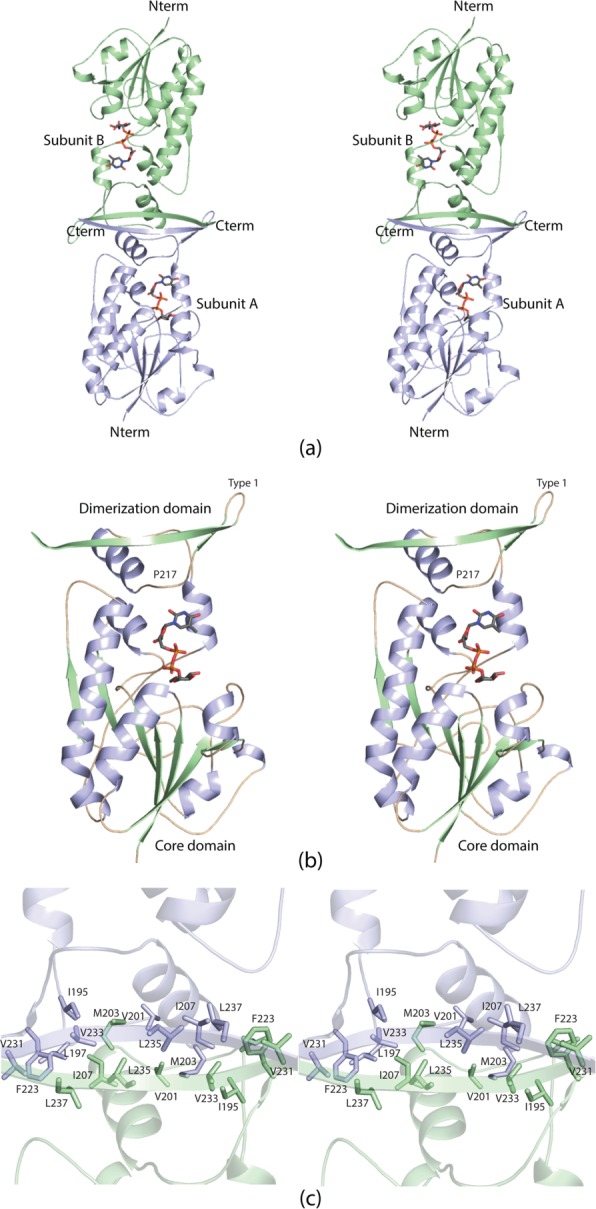
The structure of WbtJ. A ribbon representation of the WbtJ dimer is displayed in (a). The subunit:subunit interface is formed by a four stranded β-sheet. The dTDP-sugar ligands are displayed as sticks. Shown in (b) is a ribbon representation of one subunit. The overall architecture of the subunit can be envisioned as a globular “core” domain that harbors the active site region and a dimerization domain that extends away from the core motif. A close-up view of the subunit:subunit interface is presented in (c). Subunits A and B are colored in light blue and green, respectively.
Following the N-terminal core, the polypeptide chain, from Ile 186 to Ser 241, folds into two α-helices and terminates with a β-hairpin motif. Pro 217, situated in a turn connecting the second α-helix to the β-hairpin, adopts a cis conformation. The β-hairpin contains a classical Type I turn between Asp 225 and Gly 228. This portion of the polypeptide chain, extending from the core domain, is responsible for the subunit:subunit interactions required for dimerization. Specifically, the β-hairpins in each subunit pack against one another to ultimately form a four-stranded anti-parallel β-sheet. The dimeric interface is characterized by a hydrophobic patch formed by Ile 195, Leu 197, Val 201, Met 203, Ile 207, Phe 223, Val 231, Val 233, Leu 235, and Leu 237 from both monomers [Fig. 3(c)]. The two subunits of the dimer are virtually identical such that their α-carbons superimpose with a root-mean-square deviation of 0.53 Å.
A close-up view of the active site with bound dTDP-Qui4N in subunit A is displayed in Figure 4(a). The thymine ring of the dTDP-sugar forms parallel stacking interactions with the aromatic side chains of Phe 103 and Tyr 218 and participates in hydrogen-bonding interactions with the carboxamide group of Asn 220 and a water molecule. The ribosyl moiety is anchored to the protein via the side chain of Gln 105. Two water molecules and the side chains of His 73, Tyr 149, and Tyr 218 interact with the phosphoryl oxygens of the substrate. There are no interactions between the protein and the functional groups on the hexose ring of the substrate. A close-up view of the active site with bound dTDP-Qui4NFo in subunit B is presented in Figure 4(b). Binding of the product results in a few minor rotations of some side chains, but overall the active site geometries of subunits A and B are nearly identical within experimental error. The formyl group of the dTDP-Qui4NFo ligand, directed out of the active site, does not lie within hydrogen bonding distance to any protein side chains.
Figure 4.
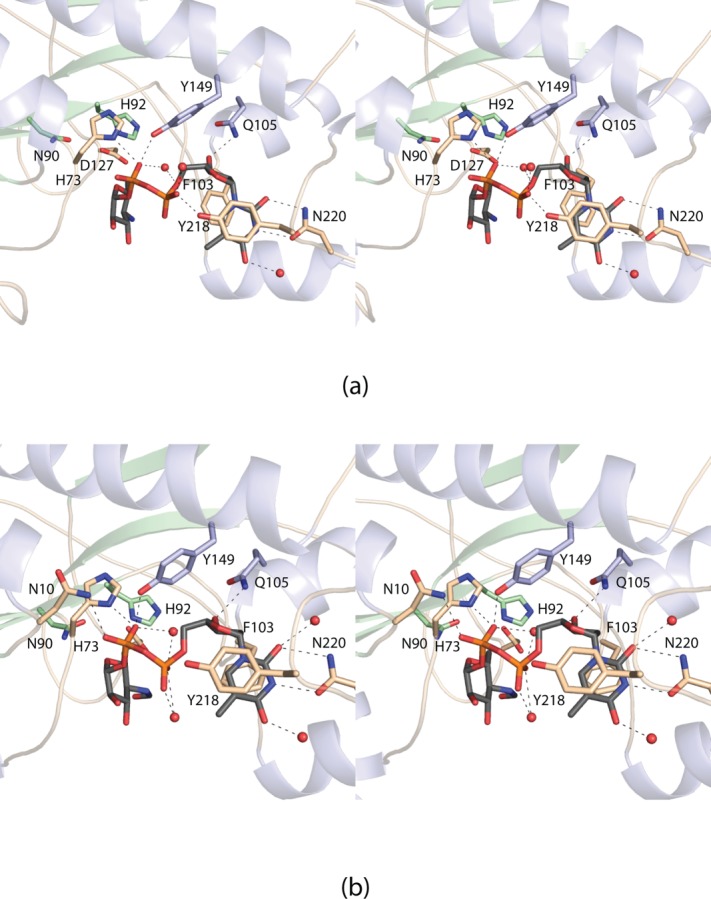
Close-up views of the active sites. The active site in subunit A contained bound dTDP-Qui4N. Those residues surrounding the ligand are shown in (a). Possible hydrogen bonding interactions are indicated by the dashed lines. The active site region for subunit B of the dimer is displayed in (b). This subunit contained the product, dTDP-Qui4NFo.
Comparison of WbtJ to WlaRD
N-formylated sugars were first observed in 1985 on the lipopolysaccharides of certain strains of Pseudomonas aeruginosa.6 Until recently, however, little had been reported on the biochemical and/or structural aspects of the enzymes required for their biosynthesis.3,6,7 The first three-dimensional structure of a sugar N-formyltransferase, that of ArnA from Escherichia coli, was reported in 2005.7 ArnA is a bifunctional enzyme whose N-terminal domain converts UDP-4-amino-4-deoxy-l-arabinose to UDP-4-formamido-4-deoxy-l-arabinose. The structure was determined in the presence of N5-formyltetrahydrofolate and UMP. The second report on a sugar N-formyltransferase, appearing in 2012, described a biochemical characterization of VioF from Providencia alcalifaciens O30.3 Like WbtJ, this enzyme catalyzes the N-formylation of dTDP-Qui4N to yield dTDP-Qui4NFo (Scheme 1) and requires N10-formyltetrahydrofolate for activity. Very recently, we reported a detailed structural and functional characterization of an enzyme referred to as WlaRD from Campylobacteri jejuni 81116. WlaRD catalyzes the N-formylation of dTDP-3,6-dideoxy-3-amino-d-glucose to yield dTDP-3,6-dideoxy-3-formamido-d-glucose.4 For this analysis, seven crystal structures were determined in the presence of various cofactors and dTDP-linked sugars. One of the models provided, for the first time, a detailed understanding of an N-formyltransferase complexed to its catalytically competent cofactor, N10-formyltetrahydrofolate, rather than the more stable N5-formyltetrahydrofolate derivative.
Structural alignments of WbtJ, WlaRD, and ArnA reveal three conserved residues located within their active sites. These correspond to Asn 90, His 92, and Asp 127 in WbtJ [Fig. 4(a)]. Mutation of any of these conserved residues in WlaRD resulted in an inactive enzyme under the assay conditions employed.4 The conserved histidine in WlaRD was predicted to function as the active site base to deprotonate the C-3′ amino group of the dTDP-sugar substrate. In WbtJ, the imidazole side chain of His 92 is located at 7.8 Å from the C-4′ amino group of the substrate. This longer than expected distance is most likely a result of the dTDP-sugar substrate adopting a nonproductive binding conformation in the active site as discussed below.
Both WlaRD and WbtJ function as dimers, but the manner in which their subunits associate is different as can be seen by comparing the ribbon representation of WlaRD shown in Figure 5(a) to that of WbtJ displayed in Figure 3(a). The WlaRD polypeptide chain, extending for an additional 33 amino acid residues at the C-terminus, folds into a four-stranded antiparallel β-sheet. As a consequence, the subunit:subunit interface in WlaRD is markedly different to that observed for WbtJ. A superposition of the WlaRD and WbtJ subunits is presented in Figure 5(b). Whereas the quaternary structures are different between the two enzymes, the overall folds of their “core” domains superimpose well with a root-mean-square deviation 1.3 Å for 172 α-carbons. The amino acid sequence identity between these core domains is 29%.
Figure 5.
Comparison of the WlaRD and WbtJ structures. A ribbon representation of the WlaRD dimer is presented in (a). Note the difference in quaternary structures between WlaRD and WbtJ [Fig. 3(a)]. A superposition of the ribbon drawings for WlaRD (red) and WbtJ (light blue) is shown in (b). Whereas the core domains are similar between the two enzymes, the C-terminal regions are completely different. A comparison of the WlaRD and WbtJ active sites is provided in (c). The positions of the three absolutely conserved residues, Asn 90, His 92, and Asp 127 are shown for WbtJ (green and wheat bonds) and WlaRD (white bonds). The dTDP-Qui4N ligand bound to WbtJ is highlighted in gray and orange bonds whereas the dTDP-Qui3N substrate bound to WlaRD is depicted in white bonds. The amino acid numbering refers to those residues in WbtJ.
A comparison of the active sites for WlaRD and WbtJ is shown in Figure 5(c). The thymine rings of the substrates are accommodated in the active sites of these enzymes in similar manners. Specifically, in both enzymes, the thymine ring of the substrate is surrounded by a phenylalanine residue (Phe 103 in WbtJ and Phe 107 in WlaRD) and a tyrosine residue (Tyr 218 in WbtJ and Tyr 222 in WlaRD). In addition, the thymine rings lie within hydrogen bonding distance to either Asn 220 in WbtJ or Gln 223 in WlaRD. The cis-proline (Pro 217) in WbtJ that lies near the thymine ring of the dTDP-sugar is not conserved in WlaRD, however.
The major difference in substrate binding between these two enzymes arises primarily through the dihedral angles of the pyrophosphoryl groups. The dTDP-sugar in WlaRD adopts a more extended conformation such that its C′-3 amino group projects towards the conserved asparagine, histidine, and aspartate residues. Importantly, all of the structures of WlaRD were solved in the presence of either N5-formyltetrahydrofolate or N10-formyltetrahydrofolate. Most likely the dTDP-sugar in WbtJ adopts a non-productive conformation due to the absence of a bound tetrahydrofolate derivative. In the presence of the appropriate cofactor, the WbtJ substrate probably adopts an extended conformation that places the hexose C′-4 amino group within hydrogen bonding distance to His 92.
There are also several notable differences in the amino acid residues lining the active sites of WlaRD versus WbtJ. In WlaRD, the phosphoryl moiety of the dTDP-sugar substrate interacts with Lys 9. Strikingly, in WbtJ, the corresponding residue is Asp 9 whose side chain projects away from the active site pocket. Likewise, the positively charged Arg 192 in WlaRD, which is directed toward the active site, is replaced with Ile 186 in WbtJ. Finally, the indole side chain of Trp 102 in WbtJ, that abuts one side of the pyranosyl ring of the dTDP-sugar substrate, is replaced with Val 106 in WlaRD. These variations in the active site pockets make it difficult to predict with any certainty the manner in which WbtJ accommodates the dTDP-sugar substrate in a catalytically competent mode.
In summary, the kinetic experiments described herein demonstrate for the first time that WbtJ functions as a sugar N-formyltransferase using dTDP-Qui4N as its substrate and N10-formyltetrahydrofolate as its cofactor. The overall catalytic efficiency of WbtJ is, indeed, comparable to that observed for other sugar N-formyltransferases. The structural analysis of WbtJ shows that it adopts a decidedly different quaternary structure as compared to WlaRD. Experiments designed to obtain a model for the ternary complex of WbtJ are presently underway.
Materials and Methods
Cloning of wbtj
A synthetic gene encoding for a protein with a C-terminal His-tag was originally synthesized by DNA2.0 using E. coli optimized codons. This C-terminally tagged construct did not yield soluble protein, however. Thus, the synthesized gene was subsequently employed as a template for further cloning. A new construct was prepared via PCR using Platinum Pfx DNA polymerase (Invitrogen) and primers that incorporated NdeI, 5′-CATATG AAGAAGATTTTTGTTGTGACTGATAACCGTACC-3′, and XhoI, 5′-CTCGAG TTAGCTGATCTTTTCCAGTTCCAATGCGACGAAAACTTTGTTACC-3′ restriction sites. The PCR product was digested with NdeI and XhoI and ligated into pET28t, a lab pET28b(+) vector that had been previously modified to incorporate a TEV protease recognition site after the N-terminal polyhistidine tag.8
Protein expression and purification
The pET28t-wbtj plasmid was utilized to transform Rosetta2(DE3) E. coli cells (Novagen). The cultures were grown in lysogeny broth supplemented with kanamycin and chloramphenicol at 37°C with shaking until an optical density of 0.8 was reached at 600 nm. The flasks were cooled in an ice bath, and the cells were induced with 1 mM IPTG and allowed to express protein at 16°C for 24 h.
The cells were harvested by centrifugation and disrupted by sonication on ice. The lysate was cleared by centrifugation, and WbtJ was purified with Ni-NTA resin (Qiagen) according to the manufacturer′s instructions. The protein was dialyzed against 10 mM Tris-HCl (pH 8.0) and 200 mM NaCl and concentrated to 14 mg mL−1 based on an extinction coefficient of 1.33 (mg mL−1)−1 cm−1. For removal of the N-terminal His6-tag, a solution containing a 30:1 molar ratio (WbtJ:TEV protease) was allowed to digest at 4°C for 48 h. Uncleaved protein and the TEV protease were removed by passage over Ni-NTA resin. Cleaved enzyme was dialyzed and concentrated to 13.5 mg mL−1 in the same manner as described above for the N-terminally tagged version.
Selenomethionine-labeled protein was prepared via standard methods used in the laboratory.9 The E. coli cells were grown in minimal media, and prior to the addition of IPTG, methionine biosynthesis was suppressed by the addition of lysine, threonine, phenylalanine, leucine, isoleucine, valine, and selenomethionine. The selenomethionine labeled protein was purified in the same manner as the wild-type enzyme, the His6-tag was removed, and the protein was concentrated to 12.5 mg mL−1.
Synthesis and purification of the dTDP-sugar ligands
dTDP-Qui4N was prepared by modifications of a previously reported procedure.10 Solutions (50 mL) containing 3.5 mM dTDP-glucose, 0.25 mg mL−1 E. coli RmlB (a 4,6-dehydratase), 0.4 mg mL−1 DesI (an aminotransferase from Streptomyces venezuelae), 40 mM sodium glutamate, and 50 mM HEPPS (pH 8.5) were allowed to react at room temperature for 12 h to generate dTDP-Qui4N. All enzymes were then removed by filtration through a 10,000 MW cutoff Amicon filter. The filtered reaction mixture was diluted 20-fold with water and loaded onto a 50 mL HiLoad 26/10 Q-Sepharose HP column. The dTDP-Qui4N was purified with a 12-column volume gradient of 0–0.9 M ammonium acetate (pH 4.0) and eluted at a concentration of 0.250 M ammonium acetate. All enzymes required for the dTDP-sugar synthesis were cloned and purified in the laboratory.
N10-formyltetrahydrofolate was synthesized as previously described.11 Specifically, N5-formyltetrahydrofolate was first converted to 5,10-methylenetetrahydrofolate by mixing 20 mg of N5-formyltetrahydrofolate and 50 µL β-mercaptoethanol in 4 mL water and adjusting the pH to 1.9 with 0.1 M HCl. The solution was incubated at room temperature for 4 h. The mixture was then converted to N10-formyltetrahydrofolate by raising the pH to 8.5 with 1 M KOH followed by incubation at 37°C for 60 min.
For the preparation of the dTDP-Qui4NFo sugar used in this investigation, an 18 mL N10-formyltetrahydrofolate mixture was combined with 100 mg of dTDP-Qui4N, 30 mg WbtJ, and 5 mL of 500 mM HEPPS (pH 8.5) and incubated overnight at 37°C. The reaction mixture was passed through a 10,000 MW cutoff Amicon filter to remove enzymes, diluted to 700 mL with water, loaded onto a 50 mL HiLoad 26/10 Q-Sepharose HP column, and eluted with a 15-column volume gradient of 0–1.2 M ammonium acetate (pH 4.0). The dTDP-Qui4NFo eluted at a concentration of 750 mM ammonium acetate. The material was diluted, the pH adjusted to 8.5, and the sample loaded onto a second 50 mL 26/10 column for further purification. A 10-column volume gradient of 0–600 mM ammonium bicarbonate (pH 8.5) was used for this step, with the dTDP-Qui4NFo eluting at a concentration of 200 mM ammonium bicarbonate. ESI mass spectrometry of the product in negative ion ionization mode gave an M-1 peak mass of 574, which corresponds to the calculated mass for dTDP-Qui4NFo.
Crystallization of WbtJ complexes
Crystallization conditions were surveyed by the hanging drop method of vapor diffusion using a laboratory-based sparse matrix screen. The enzyme was initially tested in the absence of ligands or in the presence of 5 mM dTDP and 5 mM N5-formyltetrahydrofolate at both room temperature and 4°C. The untagged version of the enzyme, at room temperature, yielded the best crystals from the initial screens. X-ray diffraction quality crystals of the protein in complex with 5 mM dTDP and 5 mM N5-formyltetrahydrofolate were subsequently grown from precipitant solutions composed of 27–31% 2-methyl-2,4-pentanediol (MPD) and 100 mM MOPS (pH 7.0). The crystals belonged to the monoclinic space group P21 with unit cell dimensions of ∼a = 80 Å, b = 72 Å, c = 107 Å and β = 107° and four subunits in the asymmetric unit. Crystals of the selenomethionine-labeled protein were also grown under the same conditions and belonged to the P21 space group.
A second crystal form was subsequently obtained from 12 to 15% poly(ethylene glycol) 8000 and 100 mM HEPPS (pH 8.0) in the presence of 5 mM dTDP and 5 mM N5-formyltetrahydrofolate. These crystals belonged to the P1 space group with unit cell dimensions of ∼a = 72 Å, b = 80 Å, c = 110 Å, α = 72°, β = 88°, and γ = 90° and eight subunits in the asymmetric unit. Crystals of WbtJ complexed with N5-formyltetrahydrofolate and either dTDP-Qui4N or dTDP-Qui4NFo were grown under similar conditions.
Structural analysis of WbtJ complexes
The crystals grown from MPD were transferred into a cryo-protectant solution containing 35% MPD, 300 mM NaCl, 5% ethylene glycol, and ligands in the same concentrations used for the crystallization trials. The crystals grown from the poly(ethylene glycol) 8000 solutions were first transferred to a solution containing 19% poly(ethylene glycol) 8000, 200 mM NaCl, and ligands in the same concentrations used for the crystallization experiments. They were then transferred to a final solution containing 25% poly(ethylene glycol) 8000, 400 mM NaCl, 16% ethylene glycol, and ligands in the same concentrations used for crystallization trials.
All X-ray data were collected at the Structural Biology Center Beamline 19-BM (Advanced Photon Source). The data were processed and scaled with HKL3000.12 Relevant X-ray data collection statistics are listed in Table 1.
Table I.
X-ray Data Collection Statistics
| Selenomethionine-labeled protein | Wild-type enzyme | |
|---|---|---|
| Wavelength (Å) | 0.9794 | 0.9794 |
| Resolution limits (Å) | 30.0–2.24 (2.30–2.24)a | 30.0–2.10 (2.18–2.10)a |
| Unit cell dimensions (Å, deg) | a = 80.0, b = 72.0 c = 107.3, β = 107.3 | a = 71.9, b = 80.1, c = 109.9, α = 71.7, β = 88.6,γ = 89.8 |
| Number of independent reflections | 56,279 (4288) | 121,975 (11,250) |
| Completeness (%) | 99.9 (99.9) | 90.2 (82.9) |
| Redundancy | 5.2 (5.0) | 2.1 (1.7) |
| avg I/avg σ(I) | 37.4 (9.0) | 24.0 (3.9) |
| Rsym (%)b | 5.1 (22.8) | 7.0 (23.1) |
Statistics for the highest resolution bin.
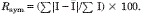
The first structure of WbtJ was solved via a SAD experiment (selenium peak wavelength 0.9794 Å) using selenomethionine labeled crystals obtained in the presence of 5 mM dTDP, 5 mM N5-formyltetrahydrofolate and MPD and belonging to the P21 space group. Analysis of the X-ray data with SHELXD located 16 of the expected 24 sites.13 Initial phases were calculated with SOLVE, and further improved with fourfold averaging and solvent flattening using RESOLVE.14,15 The resulting electron density map allowed for an almost complete tracing of the polypeptide chain. The model was then used as a search probe to solve the structures of the proteins in complex with various substrate analogs, substrates, or products via molecular replacement. Unfortunately, no crystals that were examined contained bound tetrahydrofolate derivatives.
An electron density map using X-ray data collected from a crystal belonging to the P1 space group did reveal the position of a dTDP-Qui4NFo ligand in one subunit. In addition, four other subunits within the asymmetric unit contained dTDP-Qui4N. The remaining three subunits also contained bound ligands, but the electron density was sufficiently ordered only for the dTDP moiety of the ligand. Iterative cycles of model building with COOT and refinement with REFMAC reduced the Rwork and Rfree to 19.1 and 27.4%, respectively, from 30 to 2.1 Å resolution.16,17 Model refinement statistics are listed in Table 2.
Table II.
Refinement Statistics
| Resolution limits (Å) | 30–2.1 |
| R-factor (overall)%/no. reflectionsa | 19.5/121975 |
| R-factor (working)%/no. reflections | 19.1/115826 |
| R-factor (free)%/no. reflections | 27.4/6149 |
| Number of protein atoms | 15686 |
| Number of heteroatoms | 1012 |
| Average B values | |
| Protein atoms (Å2) | 39.9 |
| Ligand (Å2) | 41.3 |
| Solvent (Å2) | 38.8 |
| Weighted RMS deviations from ideality | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 2.29 |
| Planar groups (Å) | 0.010 |
| Ramachandran regions (%)b | |
| Most favored | 89.1 |
| Additionally allowed | 10.3 |
| Generously allowed | 0.6 |
R-factor = (Σ|Fo − Fc|/Σ|Fo|) × 100 where Fo is the observed structure-factor amplitude and Fc. is the calculated structure-factor amplitude.
Distribution of Ramachandran angles according to PROCHECK.18
Kinetic analyses
Kinetic parameters for WbtJ were obtained via a discontinuous assay using an ÄKTA HPLC. The reaction rates were determined by calculating the amount of dTDP-Qui4NFo produced on the basis of the peak area of the HPLC trace as measured at 267 nm. The area was correlated to concentration via a calibration curve created with standard samples that had been treated in the same manner as the reaction aliquots.
Specifically, for these experiments, 1.1 mL reactions were set up that contained 7 mM N10-formyltetrahydrofolate, 50 mM HEPPS (pH 8.5), 0.002 mg mL−1 enzyme, and substrate concentrations ranging from 0.15 to 20 mM dTDP-Qui4N. 250-µL aliquots were taken at various time points over 4 min and quenched by the addition of 12 µL of 6 M HCL. Afterwards, 200 µL of carbon tetrachloride were added, the samples were vigorously mixed, spun at 14,000g for 1 min, and a 200-µL aliquot of the aqueous phase taken for subsequent analysis via HPLC. The samples were diluted with 2 mL water and loaded onto a 1 mL ResQ column, and the products quantified after elution with an eight-column volume gradient of 0–400 mM LiCl (pH 4.0, HCl). Plots of concentrations versus initial rates were analyzed using PRISM (GraphPad Software) and were fitted to the equation vo = (Vmax [S])/(KM + [S]). The kinetic parameters determined were as follows: Km = 0.73 ± 0.08 mM and kcat = 12.3 ± 0.5 s−1.
Acknowledgments
The authors thank Professor Grover L. Waldrop for helpful comments. A portion of the research described in this article was performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. The authors acknowledge Dr. Norma E. C. Duke for assistance during the X-ray data collection at Argonne.
Glossary
- dTDP
thymidine diphosphate
- dTDP-Qui4N
dTDP-4-amino-4,6-dideoxy-D-glucose
- dTDP-Qui4NFo
dTDP-4,6-dideoxy-4-formamido-D-glucose
- ESI
electrospray ionization
- HEPPS
N-2-hydroxyethylpiperazine-N′-3-propanesulfonic acid
- HPLC
high-performance liquid chromatography
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- MOPS
3-(N-morpholino)-propanesulfonic acid
- MPD
2-methyl-2,4-pentanediol
- Ni-NTA
Ni nitrilotriacetic acid
- PCR
polymerase chain reaction
- SAD
single anomalous dispersion
- TEV
tobacco etch virus
- Tris
tris-(hydroxymethyl)aminomethane
References
- 1.Trevisanato SI. The “Hittite plague,” an epidemic of tularemia and the first record of biological warfare. Med Hypotheses. 2007;69:1371–1374. doi: 10.1016/j.mehy.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Okan NA, Kasper DL. The atypical lipopolysaccharide of Francisella. Carbohydr Res. 2013;378:79–83. doi: 10.1016/j.carres.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu B, Chen M, Perepelov AV, Liu J, Ovchinnikova OG, Zhou D, Feng L, Rozalski A, Knirel YA, Wang L. Genetic analysis of the O-antigen of Providencia alcalifaciens O30 and biochemical characterization of a formyltransferase involved in the synthesis of a Qui4N derivative. Glycobiology. 2012;22:1236–1244. doi: 10.1093/glycob/cws089. [DOI] [PubMed] [Google Scholar]
- 4.Thoden JB, Goneau MF, Gilbert M, Holden HM. Structure of a sugar N-formyltransferase from Campylobacter jejuni. Biochemistry. 2013;52:8374–8385. doi: 10.1021/bi4009006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeLano WL. The PyMOL molecular graphics system. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 6.Knirel YA, Vinogradov EV, Shashkov AS, Dmitriev BA, Kochetkov NK, Stanislavsky ES, Mashilova GM. Somatic antigens of Pseudomonas aeruginosa. The structure of the O-specific polysaccharide chains of lipopolysaccharides of P. aeruginosa serogroup O4 (Lanyi) and related serotype O6 (Habs) and immunotype 1 (Fisher) Eur J Biochem. 1985;150:541–550. doi: 10.1111/j.1432-1033.1985.tb09055.x. [DOI] [PubMed] [Google Scholar]
- 7.Williams GJ, Breazeale SD, Raetz CR, Naismith JH. Structure and function of both domains of ArnA, a dual function decarboxylase and a formyltransferase, involved in 4-amino-4-deoxy-L-arabinose biosynthesis. J Biol Chem. 2005;280:23000–23008. doi: 10.1074/jbc.M501534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thoden JB, Holden HM. The molecular architecture of human N-acetylgalactosamine kinase. J Biol Chem. 2005;280:32784–32791. doi: 10.1074/jbc.M505730200. [DOI] [PubMed] [Google Scholar]
- 9.Thoden JB, Holden HM. Molecular structure of galactokinase. J Biol Chem. 2003;278:33305–33311. doi: 10.1074/jbc.M304789200. [DOI] [PubMed] [Google Scholar]
- 10.Burgie ES, Holden HM. Molecular architecture of DesI: a key enzyme in the biosynthesis of desosamine. Biochemistry. 2007;46:8999–9006. doi: 10.1021/bi700751d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breazeale SD, Ribeiro AA, Raetz CR. Oxidative decarboxylation of UDP-glucuronic acid in extracts of polymyxin-resistant Escherichia coli. Origin of lipid a species modified with 4-amino-4-deoxy-L-arabinose. J Biol Chem. 2002;277:2886–2896. doi: 10.1074/jbc.M109377200. [DOI] [PubMed] [Google Scholar]
- 12.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution-from diffraction images to an initial model in minutes. Acta Cryst. 2006;D62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 13.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Cryst. 1999;D55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terwilliger TC. Maximum-likelihood density modification. Acta Cryst. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Cryst. 2003;D59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 17.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 18.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]


