Abstract
The eye lens protein γD-crystallin contributes to cataract formation in the lens. In vitro experiments show that γD-crystallin has a high propensity to form amyloid fibers when denatured, and that denaturation by acid or UV-B photodamage results in its C-terminal domain forming the β-sheet core of amyloid fibers. Here, we show that thermal denaturation results in sheet-like aggregates that contain cross-linked oligomers of the protein, according to transmission electron microscopy and SDS-PAGE. We use two-dimensional infrared spectroscopy to show that these aggregates have an amyloid-like secondary structure with extended β-sheets, and use isotope dilution experiments to show that each protein contributes approximately one β-strand to each β-sheet in the aggregates. Using segmental 13C labeling, we show that the organization of the protein's two domains in thermally induced aggregates results in a previously unobserved structure in which both the N-terminal and C-terminal domains contribute to β-sheets. We propose a model for the structural organization of the aggregates and attribute the recruitment of the N-terminal domain into the fiber structure to intermolecular cross linking.
Keywords: amyloid, crystallin, cataract, cross-linking, aggregate, infrared, isotope labeling, 2D IR spectroscopy
Introduction
Cataracts are a common protein aggregation disease of the eye lens, resulting from the formation of insoluble deposits of lens crystallin proteins that blur vision.1,2 Aggregation may result from mutations3 as well as accumulated damage from sources such as UV radiation,4–8 oxidative stress,5,9 and a variety of post-translational modifications1,10 that induce precipitation or misfolding of lens crystallins. Although the causes of damage are well known, the resulting structures of aggregated lens crystallin proteins are poorly understood. In vitro denaturation studies have shown that crystallins can form a variety of different kinds of aggregates, but that they have a high propensity for the formation of amyloid fibers.7,11–15 In fact, acidic conditions,11–13 chemical denaturants, UV radiation,7 and thermal denaturation16 have been shown to result in amyloid fiber formation.
Human γD-crystallin is a structural eye lens protein containing two domains with similar folds17 but very different stabilities and behaviors during aggregation.7,8,11–13,18–21 In the native state, each of the domains of γD-crystallin is made up of a β-sandwich composed of two Greek key subdomains.17 Equilibrium unfolding and fluorescence studies have shown that the protein's N-terminal domain is less stable than its C-terminal domain, and that a folding intermediate with a disordered N-terminal domain and native-like C-terminal domain exists.19 The hydrophobic interface between the domains contains specific residue contacts that stabilize the native fold, and the presence of this interface allows the C-terminal domain to act as a template for the folding of the N-terminal domain.21 The specific association between N-and C-terminal domains can also result in the formation of dimers or polymers through a domain-swapping mechanism that has been hypothesized as a possible route to precipitation and cataract formation.22,23 In addition to domain swapping, amorphous aggregation and amyloid fiber formation have been observed.7,11–13,24 Amyloid fiber formation is the dominant in vitro mode of aggregation for γD-crystallin and a number of other crystallins.7,11–16,25 Both the isolated N-and C-terminal domains of γD-crystallin, as well as the full-length protein, can form amyloid fibers in vitro upon denaturation with acid.11–13 Previously, we found that the acid-induced fibers formed by the full-length protein behave differently than might be expected from examination of the isolated domains.11,12 Full-length, acid-induced γD-crystallin aggregates only contain core β-strands from the C-terminal domain, while the N-terminal domain becomes highly disordered.12 In fact, nearly the entire C-terminal domain forms amyloid β-sheets.11 We also observed similar domain behaviors in UV-B-induced aggregates of γD-crystallin, in which amyloid fiber formation by the C-terminal domain is accompanied by extensive side chain oxidation, peptide bond cleavage, and covalent protein cross-linking.7 The results of these studies suggest that the C-terminal domain has a higher intrinsic amyloid propensity than the N-terminal domain, independent of the mode of denaturation.
In this paper, we show that γD-crystallin can form alternative aggregate structures upon thermal denaturation. Using transmission electron microscopy and SDS-PAGE, we show that the aggregates formed by thermal denaturation have sheet-like morphology and may contain covalently cross-linked proteins. To study the structure of these aggregates, we use a combination of two-dimensional infrared (2D IR) spectroscopy26 and segmental 13C labeling, which we implement through expressed protein ligation.7,11,12 2D IR spectroscopy is a useful technique for determining protein structures in complex systems such as aggregates or membranes because spectra can be collected straightforwardly in heterogeneous samples.7,11,12,27–36 Like FTIR spectroscopy, 2D IR spectroscopy is sensitive to the secondary structure, solvent exposure, and dynamics of a protein based on IR frequencies and line shapes.26 Further structural resolution can be obtained using isotope labeling of a single amino acid,28–35,37–40 labeling of an entire sequence segment,7,11,12,41 or through the incorporation of non-natural vibrational probes.27,42–47 2D IR spectroscopy has a number of important advantages over FTIR spectroscopy. First, it allows the resolution of cross-peaks, which reveal coupling between different vibrational modes.26,31,36,48 Second, 2D IR signal strengths scale with the fourth power of the transition dipole moment, which results in increased resolution of the various peaks in a 2D IR spectrum.26,38 Third, the relative contributions of inhomogeneous and homogeneous broadening can be resolved from 2D line shapes, revealing important information about structural order and dynamics.12,29,32,45,49,50 Finally, all of these advantages can be obtained in kinetics studies that take advantage of rapid-scan technology based on mid-IR pulse shaping.38,51 Here, we use 2D IR spectroscopy, segmental 13C labeling, and isotope dilution to show that the aggregates formed from thermal denaturation of γD-crystallin contain β-strands from both the N-terminal and C-terminal domains, a result we tentatively attribute to intermolecular cross-linking. These results are strikingly different from those obtained from acid11,12 and UV-B7-induced denaturation, and demonstrate that different structural isoforms may be produced depending on the mode of denaturation that induces aggregation. With this information, and comparisons to acid-induced amyloid fibers of γD-crystallin,12 we propose a model for the thermally induced aggregates of the protein. Finally, we discuss the implications of this model for the possible role of amyloid-like crystallin aggregates in cataracts.
Results and Discussion
In this section, we present the results associated with the aggregation of human γD-crystallin (S84C) as induced by thermal denaturation. We also present a comparison of the thermally induced aggregates to acid-induced amyloid fibers of γD-crystallin, for which we have developed a structural model in previous publications using 2D IR spectroscopy,12 and which is consistent with mass spectrometry.11
Thermal denaturation of human γD-crystallin (S84C) resulted in visible turbidity of the samples within 30 min, followed by continued formation of visible precipitates over the course of 3 h of incubation at 80°C. Transmission electron micrographs showed aggregates as either twisted [Fig. 1(A)] or flat sheets [Fig. 1(B)]. Analysis of the samples by SDS-PAGE [Fig. 1(D)] reveals that the aggregates are composed not only of monomers (∼20 kDa), but also dimers and a small amount of trimers and higher-order oligomers. Based on integration of the gel band intensities and the presence of some high molecular weight species that did not enter the gel, we estimate that a minimum of 25% of the total protein is cross-linked. Because SDS-PAGE samples were prepared by incubating the aggregates at 95°C in the presence of detergent and a thiol reductant, non-covalent and disulfide-based oligomers are unlikely to be present. Aggregates prepared in the presence of 2-mercaptoethanol [Fig. 1(D), Lane 4] also showed the formation of oligomeric or cross-linked species. Thus, we tentatively conclude that the oligomers are produced by covalent, non-disulfide cross-links between protein molecules. However, exceptionally stable disulfide-based oligomers52 and stable non-covalent assemblies53 cannot be ruled out. Wild type protein was analyzed in the same manner and resulted again in identical band patterns, showing that the S84C mutation, which is required for expressed protein ligation in segmentally labeled proteins, does not influence the product distribution. For the purpose of comparison, we present a typical TEM image of acid-induced aggregates [Fig. 1(C)], which form uniform suspensions of fibers with diameters of ∼5–10 nm.11 UV-B-induced aggregates also form fibers with similar diameters.7 The amyloid fibers formed by acid and UV-B-induced denaturation are much narrower than the sheets formed from thermal denaturation, which have irregular dimensions that range from ∼50 to 1,000 nm across. Previous SDS-PAGE analysis of acid-induced aggregates showed neither cross-linking nor polypeptide cleavage,11 while UV-B-induced aggregates showed both kinds of protein modifications.7
Figure 1.
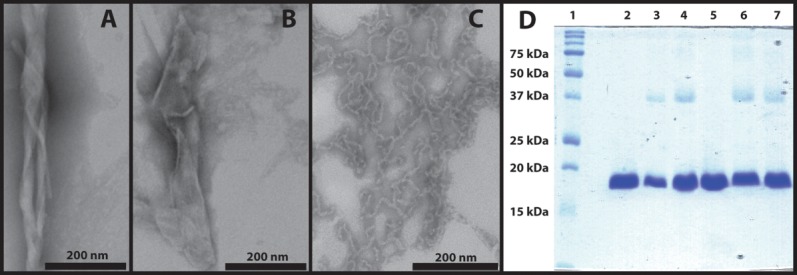
Characterization of thermally induced γD-crystallin aggregates. A. TEM image of thermally induced aggregates with twisted sheet morphology. B. TEM image of thermally induced aggregates with flat sheet morphology. C. Acid-induced amyloid fibers. D. SDS-PAGE analysis of thermally induced γD-crystallin aggregates. Lanes from left: (1) Molecular weight marker; (2) native γD-crystallin (S84C); (3) thermally induced aggregates of γD-crystallin (S84C); (4) thermally induced aggregates of γD-crystallin (S84C) with 2-mercaptoethanol; (5) native wild type γD-crystallin; (6) thermally induced aggregates of wild type γD-crystallin; (7) thermally induced aggregates of wild type γD-crystallin with 2-mercaptoethanol. Image C is reproduced from data described in Ref.11.
To characterize the secondary structure of the thermally induced aggregates, we turn to 2D IR spectroscopy. The secondary structure sensitivity of infrared spectroscopy is largely a result of the differences in vibrational coupling between peptide bonds in α-helices, β-sheets, and random coil structures.26,54,55 The relative orientations of the vibrational transition dipole moments and their respective coupling constants associated with each secondary structure cause delocalization of vibrational motions that results in frequency shifts and intensity changes.26 The 2D IR spectrum of unlabeled, thermally induced human γD-crystallin [Fig. 2(A)] shows features that are typical of amyloid fibers, including a narrow, low-frequency peak pair (ωpump = 1614 cm−1) assigned to β-sheets and a broader, high-frequency peak pair (ωpump = 1625–1700 cm−1) assigned to disordered structures.12 In 2D IR spectra, peaks come in pairs because the pulse sequence probes both the (0–1) and (1–2) vibrational transitions.26 The characteristic amyloid β-sheet peak at 1614–1620 cm−1 appears at low frequency because coupling constants between adjacent residues in β-sheets are strongly negative,39,54,55 resulting in a red shift of the amide band that scales with the size of the β-sheet.54,55 The β-sheet amide I band is also very narrow compared to that of the native protein and the disordered signal in the aggregates, reflecting a relatively small inhomogeneous contribution to the line width that reveals a highly uniform environment for these β-sheet modes.12,26 The 13C-labeled aggregates [Fig. 2(B)] contain similar sets of peak pairs at 1570 cm−1 and 1580–1660 cm−1, respectively, consistent with the ∼40 cm−1 shift in frequency that occurs upon replacement of 12C with 13C.12
Figure 2.
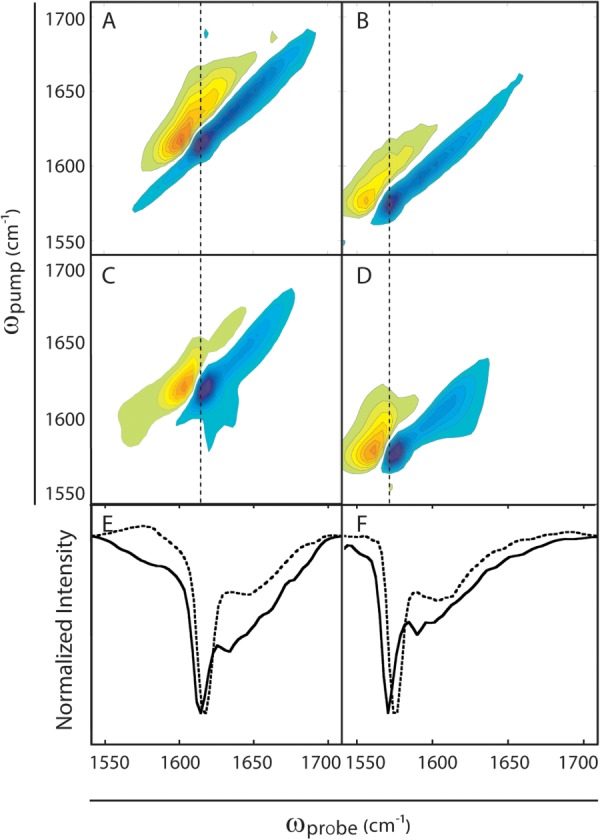
2D IR spectra of γD-crystallin amyloid aggregates. A. Thermally induced 12C γD-crystallin aggregates. B. Thermally induced 13C γD-crystallin aggregates. C. Acid-induced 12C γD-crystallin amyloid fibers. D. Acid-induced 13C γD-crystallin amyloid fibers. E. Diagonal slices of spectra in A (solid lines) and C (dashed lines). F. Diagonal slices of spectra in B (solid lines) and D (dashed lines). Dotted lines though the maxima of the β-sheet peaks reveal frequency differences between thermally induced and acid-induced β-sheets. Spectra of the acid-induced aggregates are reproduced from data described in Ref.12.
For comparison, the 2D IR spectra of unlabeled (12C) and labeled (13C) γD-crystallin fibers prepared by acid denaturation are reproduced in Figure 2(C,D) from data reported previously.12 Slices through the diagonals of the acid-and thermally denatured 2D IR spectra [Fig. 2(E,F)], which reflect the same modes that would be observed in FTIR spectra, aid in their comparison. The spectra are similar but the frequency of the β-sheet feature near 1620 cm−1 in the thermally induced aggregates is lower than that in the acid-induced fibers. This frequency difference, though small, reflects structural differences between the thermally induced and acid-induced fibers. The frequency of this mode depends on the size and structural order of the β-sheets, which indicates that the thermally induced fibers are larger and more ordered than those formed from acid denaturation.
Another difference between the thermally induced and acid-induced aggregates is apparent in the broad peak pairs that are observed between ωpump ≈ 1625–1700 cm−1 in unlabeled aggregates and ωpump ≈ 1585–1660 cm−1 in the labeled aggregates. These signals are due to disordered regions of the proteins in the aggregates.11,12 It is clear from the diagonal slices in Figure 2(E,F) that the disordered structures in the thermally induced aggregates have greater relative intensities than their acid-induced counterparts, and also appear to contain multiple discrete peaks. Because the samples were cooled to room temperature for analysis, some refolding of denatured structures is possible, and may explain the presence of multiple features in the disordered region of the spectrum. Thus, the unlabeled spectra in Figure 2 provide an overview of the secondary structure of the proteins, but do not reveal which residues contribute to the β-sheets or the disordered regions. To locate these regions within the proteins, we turn to isotope labeling.
To obtain additional information about the architecture of the aggregates, we use isotope dilution. The frequency of the amide I β-sheet signal is related to the extent of vibrational delocalization in the aggregates.12,55 The vibrational coupling in amyloid β-sheets is very strong and so if all strands are composed of the same isotopes, the vibrational modes will extend over many β-strands in the β-sheets. The larger the delocalization of the vibrational mode, the lower the β-sheet frequency.26,54,55 If the strands are not all composed of the same isotopes, then the couplings do not delocalize vibrational modes as effectively, giving rise to a higher frequency.12,28,55 We can use this fact to provide an estimate of the number of β-strands each protein molecule contributes to each β-sheet.12 By mixing 12C-and 13C-labeled proteins in various proportions prior to aggregation, we observe a 13C β-sheet frequency trend in the aggregates that reflects the coupling between 13C-labeled β-strands (Fig. 3). Thus, the frequency of the 13C amide I band provides a measure of the number of β-strands each protein contributes to the β-sheets. We quantify the frequency as a function of the number of contributed β-strands using transition dipole coupling simulations,55 described in detail in a previous publication.12 Shown in Figure 3 are the frequency trends for 13C amide I maxima for amyloid β-sheets in which each individual protein contributes between one and four β-strands to each β-sheet. We find that the frequencies are consistent with approximately one β-strand per β-sheet for each protein molecule in the aggregates. We previously found a similar result for the acid-induced fibers.12
Figure 3.
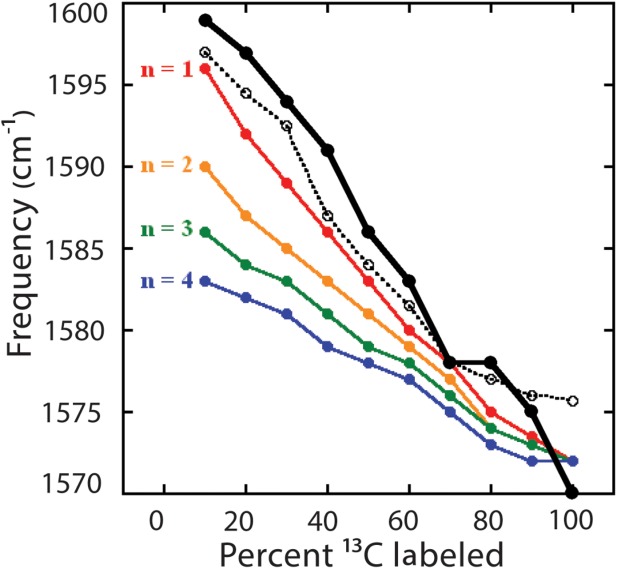
Isotope dilution study of thermally induced γD-crystallin aggregates (solid black line, closed circles) compared to acid-induced amyloid fibers (dashed black line, open circles) and transition dipole coupling simulations (colored lines, closed circles) for aggregate models with n = 1–4 strands per β-sheet per protein. Acid-induced fiber data and simulation results are reproduced from Ref.12.
We have also learned that each of the two domains contribute β-strands to the amyloid β-sheets of the thermally induced aggregates. Using expressed protein ligation, we generated a variant of γD-crystallin (S84C) in which the N-terminal domain is 13C-labeled and the C-terminal domain is unlabeled (12C).7,11,12 The spectrum of segmentally labeled, thermally induced aggregates of γD-crystallin is shown in Figure 4(A). First, we consider the signal from the unlabeled C-terminal domain. In the thermally induced aggregates, this signal appears at ωpump = 1624 cm−1, a much lower frequency than the native 12C C-terminal domain [which appears at ωpump ≈ 1640 cm−1, Fig. 4(B)] and within the typical frequency range for amyloid fibers.7 Thus, we conclude that the C-terminal domain still forms amyloid β-sheets. Turning to the 13C-labeled N-terminal domain, we see a peak with a maximum at ωpump ≈ 1600 cm−1, similar to the native frequency.12 However, we also see a low-frequency shoulder at ωpump = 1591 cm−1. To better visualize the appearance of this shoulder, we subtracted the native protein spectrum [Fig. 4(B)] from the spectrum of the thermally induced aggregates [Fig. 4(A)], each of which was normalized to the most intense diagonal amide I signal. This difference spectrum [Fig. 4(C)], which is useful as a qualitative indicator of frequency shifts, contains three difference peak pairs along the diagonal. The lowest frequency pair, centered at ωpump = 1591 cm−1, results from the structural transition of the N-terminal domain. The two other peak pairs, at ωpump = 1625 cm−1 and with opposite sign at ωpump = 1638 cm−1, are the signal from the amyloid-like C-terminal β-sheets and the loss of the native β-sandwich fold of the C-terminal domain, respectively. Thus, it is clear that both the N-and C-terminal domains form β-sheets upon thermal denaturation, and that both domains undergo conformational changes from the native state to the aggregated state.
Figure 4.
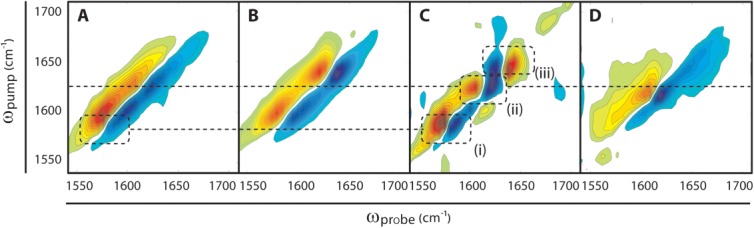
2D IR spectra of segmentally labeled γD-crystallin aggregates. A. Thermally induced amyloid aggregates of N-terminally 13C-labeled γD-crystallin. B. Native N-terminally 13C-labeled γD-crystallin. C. Difference spectrum of thermally induced aggregates (A) minus native (B) N-terminally 13C-labeled γD-crystallin. Features in the difference spectrum clarify the amide I peak shifts associated with thermally induced aggregation: (i) additional low frequency intensity in the 13C-labeled (N-terminal domain) region; (ii) additional low-frequency intensity in the unlabeled (C-terminal domain) region; (iii) loss of intensity in the region of the native, unlabeled C-terminal domain indicated by a difference peak with opposite sign. D. Acid-induced amyloid fibers of 13C N-terminally labeled γD-crystallin. Spectra B and D are reproduced from data described in Ref.12.
Also shown in Figure 4 is the spectrum of acid-induced amyloid fibers with the 13C N-terminally labeled [Fig. 4(D)].12 The acid-induced spectrum indicates that only the C-terminal domain is involved in amyloid formation, since sharp peaks only appear at ωpump = 1617 cm−1 and are identical to that in the unlabeled acid-induced amyloid fibers [Fig. 2(C)], as previously reported and confirmed with mass spectrometry.11,12 Thus, it is immediately apparent that both domains contribute to the amyloid β-sheets of the thermally induced aggregates whereas in acid denaturation only the C-terminal domain forms amyloid β-sheets. Moreover, in the thermally induced aggregates, there is a ∼10 cm−1 difference between the unlabeled amyloid β-sheet peak in Figure 2A and the amyloid signal from the unlabeled C-terminal domain in Figure 4A. Because the frequencies do not match but the protein structures are identical, we must conclude that it is the 13C labeling of the N-terminal domain that causes the frequency shift in the 12C C-terminal domain. In other words, the 13C labels of the N-terminal domain disrupt the vibrational coupling between the 12C residues of the C-terminal domain. In order for one domain to alter the frequency of the other domain, both domains must contribute to the same amyloid β-sheets, as we describe below.
Three hypothetical structural models for thermally-induced aggregates of full-length γD-crystallin (and its cross-linked products) are postulated in Figure 5 assuming parallel β-sheets, which are common in amyloid aggregates.56 Corresponding models with antiparallel strands could also be constructed, but because β-sheets containing parallel and antiparallel β-strands have similar amide I frequencies, our conclusions (described below) would be the same. We consider simple models in which the N-terminal domain and C-terminal domain form interacting amyloid β-sheets, consistent with our data above. These models, shown in Figure 5, are: (A) amyloid aggregates with the N-terminal and C-terminal domains forming non-interspersed β-sheets; (B) amyloid aggregates in which the N-terminal and C-terminal domains of the same protein are hydrogen-bonded to one another in the same β-sheet; and (C) amyloid aggregates in which the N-terminal and C-terminal domains of different proteins are hydrogen-bonded to one another in the same β-sheet. Models like these have been proposed for other amyloid-forming proteins, and so might be thought of as representative of the canonical set of plausible amyloid aggregate structures.52,56–58 Based on the frequencies of both the N-and C-terminal domains in Figure 4(A), we exclude model (A), because although β-strands may interact at the boundary of labeled and unlabeled regions the intra-strand coupling constants are too weak to cause a significant shift in β-sheet frequency.39,54,55 Additionally, the frequency of the shoulder on the labeled N-terminal domain signal [Figure 4(A,C)] is too high for extensively coupled N-terminal domains. We can also exclude model (B) because the isotope dilution data obtained from uniformly labeled proteins shows a single strand per β-sheet per protein, whereas a lower frequency would be expected in dilute 13C aggregates with more than one consecutive β-strand from each protein.12 Based on the exclusion of models (A) and (B), we are thus left with model (C) as the most likely explanation for our data. Model (C) is the only model that is consistent with the low-frequency β-sheet peaks in the 2D IR spectra of uniformly labeled samples, the β-sheet frequency trend observed upon isotope dilution of uniformly labeled aggregates, and the frequency shifts of β-sheet peaks that occur when the N-terminal domain of the aggregates protein is 13C labeled.
Figure 5.
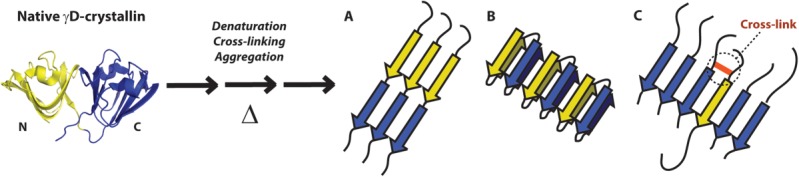
Structural models for the thermally induced aggregation of γD-crystallin. Native γD-crystallin (left) is denatured by heat, and undergoes both cross-linking and aggregation into amyloid-like β-sheets. Three general aggregate structures are possible. A. N-terminal (yellow) and C-terminal (blue) domains form consecutive, non-hydrogen-bonded β-strands. B. Both the N-terminal and C-terminal domains of a single protein form β-strands in the same β-sheet. C. N-terminal domains of a minority of proteins are recruited into a C-terminal domain β-sheet, possibly as a result of intermolecular cross-linking (red).
It is known that both the N-terminal domain and C-terminal domain can individually form amyloid structures after denaturation with acid,13 which may not be surprising given the sequence and native structure similarity of the N-and C-terminal domains.17 However, in our previous studies of full-length γD-crystallin, only the C-terminal domain formed amyloid β-sheets.11,12 In fact, even when the polypeptide backbone is cleaved by UV-B radiation, thereby separating N-and C-terminal sequences, no amyloid structures containing N-terminal domain residues were observed.7 Moreover, it is clear from our SDS-PAGE results that polypeptide cleavage does not occur upon heating, so domain separation cannot explain the inclusion of both domains in the aggregate β-sheets. Additionally, because the N-terminal domain is less stable than the C-terminal domain under our experimental conditions,19 the ratio of N-terminal and C-terminal strands cannot be explained by the relative proportions of unfolded domains at 80°C. Thus, we hypothesize that the observed cross-linking drives the formation of mixed-domain amyloid structures. Similar recruitment of peripheral β-strands into the amyloid fiber core, driven by cross-linking, has been observed before.52 Also, amyloid cross-seeding and co-polymerization experiments have shown that amyloid proteins with different sequences or modifications can co-localize in the same fibers.59–62 Thus, it is possible that the N-and C-terminal domains can both contribute to the same β-sheets, even though they have different amino acid sequences. Our data supports that conclusion.
Based on the relative shifts of the signals from the N-terminal and C-terminal domains, we conclude that the amide I vibrations of the unlabeled C-terminal domain are more delocalized in the aggregates (14 cm−1 red shift vs. native Greek key) than those of the N-terminal domain (7 cm−1 red shift vs. native Greek key). This observation indicates that the proportion of C-terminal domains in the aggregate β-sheets is larger than the proportion of N-terminal domains (Fig. 3). Because vibrational coupling constants are invariant with 13C labeling,12,63 it is reasonable to assume that the 13C β-sheet frequency trend observed upon isotope dilution is identical to that of 12C β-sheets. Referring to this trend in Figure 3, we estimate that the β-sheets contain approximately 30–40% N-terminal strands and 60–70% C-terminal strands. This result is similar to the proportion of putatively cross-linked products observed in our SDS-PAGE results [Fig. 1(D)], and thus corroborates our cross-linking hypothesis. Although this difference in domain content may seem small, we note that the frequency shift and intensity increase due to vibrational coupling is dependent on the proximity of multiple β-strands of the same carbon isotopes. This may lead to large differences in the 2D IR spectra based on small differences in the number of labeled N-terminal and unlabeled C-terminal strands. For example, in a randomly formed aggregate containing 65% unlabeled (12C) β-strands, the probability of producing three consecutive 12C strands is (0.65)3 = 0.275 while the probability of producing three consecutive 13C strands is (0.35)3 = 0.043. Of course, cross-linked samples are not random; in our model [Fig. 5(C)] labeled and unlabeled strands come in pairs. However, because we observe a greater shift in unlabeled C-terminal amide I frequency, we know that the aggregate β-sheets contain more C-terminal strands than N-terminal strands. This fact may be explained by the exclusion of some cross-linked N-terminal β-strands from the aggregate β-sheets, or from the incorporation of C-terminal strands from the non-cross linked portion of the sample. Both of these possibilities can be explained by the greater intrinsic amyloid propensity of the C-terminal domain, which we have observed in our previous studies.7,11,12 Thus, in model (C) of Figure 5, we draw a higher proportion of C-terminal domain than N-terminal domain in the β-sheets.
Conclusions
In this paper, we demonstrate the formation of sheet-like aggregates from thermally denatured human γD-crystallin with extended, ordered β-sheets that are similar to those that occur in amyloid fibers.56 Although the morphologies of these aggregates are very different from amyloid fibers formed by acid11,13 and UV-B7-induced denaturation of the protein, one might conclude that the aggregate structures are the same based on the similarity of their FTIR (not shown) or 2D IR spectra. Aside from small differences in amide I frequency and line shape, 2D IR spectra of uniformly labeled aggregates produced by thermal and acid denaturation are indistinguishable. However, that is far from correct. Segmental isotope labeling of the N-terminal domain shows that that the amyloid-like β-sheets of the thermally induced aggregates are actually very different from those in the acid-induced fibers. In the thermally induced aggregates, both the N-and C-terminal domains contribute to the same β-sheets. The C-terminal domain constitutes the majority of the β-strands in these aggregates, consistent with it being the more amyloidogenic of the two domains.7,11,12 The identification of this alternative aggregate structure highlights the importance of isotope labeling in structural studies of protein aggregates; without the resolution of domains provided by segmental 13C labeling, the participation of the N-terminal domain in the aggregate β-sheets could not have been observed. Our results also prove that ordered amyloid fibers can accommodate β-strands with sequences that are not identical, which is an important observation because sequence mismatches between β-strands are relevant to amyloid polymorphism,64 amyloid cross-seeding,65 co-aggregation62 of amyloid proteins, and some proposed amyloid structures of proteins with repeat sequences.66 The 2D IR and isotope labeling methods presented here provide a way to understand β-strand organization in these and other complex amyloid aggregates.
These results indicate that amyloid-like aggregates of γD-crystallin can occur in a variety of different isoforms depending on the mode of denaturation. In previous studies, we showed that only the C-terminal domain forms amyloid β-sheets after acid-or UV-B-induced denaturation. Here, we attribute the recruitment of N-terminal domain sequences into the β-sheets to the formation of highly stable associations between protein molecules we observed in the SDS-PAGE analysis of the thermally induced aggregates, a phenomenon that is not observed in acid-induced aggregates12 but has been identified in other systems.52 We tentatively attribute these associations to covalent cross-linking, which can result from the incubation of proteins at elevated temperatures.67 However, some amyloid aggregates, such as those formed by Aβ, also form non-covalent SDS-stable dimers upon disruption,53 suggesting that strong non-covalent interactions between γD-crystallin molecules may account for the observed dimers and oligomers. Although some cross-linking was also observed in UV-B-induced fibers, the presence of other modifications such as side-chain oxidation and peptide bond cleavage may prevent the recruitment of the N-terminal domain into the UV-B-induced fiber β-sheets.
The differences between amyloid fibers formed from thermal and acid denaturation highlight the complexity of amyloid aggregation processes. The final structures of amyloid aggregates likely depend on the details of the misfolding pathway, including the structural characteristics of the initial denatured state, barrier heights between folding intermediates, and the stability of the final aggregates. In γD-crystallin, the N-terminal domain is less stable to chemical and thermal denaturation than the C-terminal domain,68,69 but it is the C-terminal domain that forms the majority of the amyloid β-sheet core in aggregates prepared by a variety of denaturation methods.11,70,71 Additionally, Brubaker et al.72 showed that the thermal stabilities of mutants of the closely related protein γS-crystallin do not predict their aggregation propensities. Because the final γD-crystallin amyloid aggregates prepared by thermal and acid denaturation are different at both the morphological and molecular structure level, it is clear that a detailed analysis of the entire aggregation pathway is required to understand how the different structural isoforms are favored under different conditions. Rapid-scan 2D IR spectroscopy38 provides a possible means of deciphering the structural evolution of lens crystallin proteins during amyloid aggregate formation.
The results presented here also have important implications for the study of γ-crystallin aggregation in cataracts. To date, amyloid fibers have not been observed in protein aggregates taken from cataractous lenses, despite the apparent high propensity for crystallins to form amyloid fibers in vitro.1,7,11–16,20,25,73 However, in a few cases where the infrared spectra of such aggregates have been measured, low-frequency absorption near 1620 cm−1, which is characteristic of extended amyloid β-sheets, have been reported.74 The fact that such spectral features are observed for aggregates that form sheets instead of fibers indicates that amyloid-like aggregation may occur even where fibers are not observed. In fact, amyloid-like infrared spectra of γ-crystallins, without the presence of fiber morphology in TEM images, has been observed in vitro.6 In addition, the presence of intermolecular cross-links in thermally induced γD-crystallin aggregates mirrors the cross-linking observed in heavily damaged, aged lenses.2,9,75,76 Although the nature of our in vitro cross-links remains to be determined, our results suggest that the presence of intermolecular cross-links may influence morphologies and molecular structures of lens crystallin aggregates.
Materials and Methods
All chemicals were purchased from Sigma-Aldrich and used as received, unless otherwise specified. Human γD-crystallin, and its segmentally labeled variants, were expressed in Escherichia coli and purified as described previously.12
Samples of human γD-crystallin were dissolved to ∼20 µM in 20 mM sodium phosphate buffer (pH 7.0) with 100 mM NaCl. For samples used in 2D IR experiments, buffers were prepared with D2O instead of H2O. All samples were thermally denatured by placing them in microcentrifuge tubes on a heating block set to 80°C, and incubated for 3 h. The resulting precipitates were gently resuspended by pipetting. Transmission electron micrographs of methylamine tungstate stained samples were collected at the University of Wisconsin Medical School Electron Microscope Facility, using a Philips CM 120 transmission electron microscope. SDS-PAGE samples were prepared by immediately dissolving the aggregates in Laemmli sample buffer (containing 20 mM 2-mercaptoethanol), and were heated for 10 min at 95°C to disperse the aggregates. Band intensities on the SDS gel were integrated using ImageJ software, which is available free of charge from http://rsbweb.nih.gov/ij/.
Samples of γD-crystallin were cooled to room temperature and placed between CaF2 windows separated by a 56 µM Teflon spacer. 2D IR spectra were collected and analyzed as described previously.12,38 In addition to uniformly labeled and segmentally labeled samples, samples were prepared with mixtures of 12C and 13C proteins for an isotope dilution study. Proteins were mixed prior to denaturation, with compositions ranging from 10 to 90% 13C-labeled γD-crystallin. Frequencies of the 13C-labeled amyloid β-sheet peaks were obtained from the spectra directly or through the subtraction of the unlabeled (12C) aggregate spectrum from dilute (10–20%) 13C aggregates.
Acknowledgments
The authors would like to thank Randall Massey of the University of Wisconsin Medical Sciences Electron Microscopy Facility for his help in the collection of TEM images, and would also like to thank Sean M. Decatur for useful discussion.
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- 1.Wang Y, King JA. Protein misfolding diseases. Hoboken: John Wiley & Sons; 2010. Cataract as a protein-aggregation disease; pp. 487–515. In Marina Ramirez-Alvarado, Jeffery W. Kelly, and Christopher M. Dobson, editor. [Google Scholar]
- 2.Bloemendal H, de Jong W, Jaenicke R, Lubsen NH, Slingsby C, Tardieu A. Ageing and vision: structure, stability and function of lens crystallins. Prog Biophys Mol Biol. 2004;86:407–485. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Graw J. Congenital hereditary cataracts. Int J Dev Biol. 2004;48:1031–1044. doi: 10.1387/ijdb.041854jg. [DOI] [PubMed] [Google Scholar]
- 4.Taylor HR. Ultraviolet radiation and the eye: an epidemiologic study. Trans Am Opthalmol Soc. 1989;87:802–853. [PMC free article] [PubMed] [Google Scholar]
- 5.Davies MJ, Truscott RJW. Photo-oxidation of proteins and its role in cataractogenesis. J Photochem Photobiol B. 2001;63:114–125. doi: 10.1016/s1011-1344(01)00208-1. [DOI] [PubMed] [Google Scholar]
- 6.Fatima U, Sharma S, Guptasarma P. Structures of differently aggregated and precipitated forms of γB crystallin: an FTIR spectroscopic and EM study. Protein Pept Lett. 2010;17:1155–1162. doi: 10.2174/092986610791760414. [DOI] [PubMed] [Google Scholar]
- 7.Moran SD, Zhang TO, Decatur SM, Zanni MT. Amyloid fiber formation in human γD-crystallin induced by UV-B photodamage. Biochemistry. 2013 doi: 10.1021/bi4008353. doi: 10.1021/bi4008353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schafheimer N, King J. Tryptophan cluster protects human γD-crystallin from ultraviolet radiation-induced photoaggregation in vitro. Photochem Photobiol. 2013;89:1106–1115. doi: 10.1111/php.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Truscott RJW. Age-related nuclear cataract—oxidation is the key. Exp Eye Res. 2005;80:709–725. doi: 10.1016/j.exer.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Hains PG, Truscott RJ. Post-translational modifications in the nuclear region of young, aged, and cataract human lenses. J Proteome Res. 2007;6:3935–3943. doi: 10.1021/pr070138h. [DOI] [PubMed] [Google Scholar]
- 11.Moran SD, Decatur SM, Zanni MT. Structural and sequence analysis of the human γD-crystallin amyloid fibril core using 2D IR spectroscopy, segmental 13C labeling, and mass spectrometry. J Am Chem Soc. 2012;134:18410–18416. doi: 10.1021/ja307898g. [DOI] [PubMed] [Google Scholar]
- 12.Moran SD, Woys AM, Buchanan LE, Bixby E, Decatur SM, Zanni MT. Two-dimensional IR spectroscopy and segmental 13C labeling reveals the domain structure of human γD-crystallin amyloid fibrils. Proc Natl Acad Sci USA. 2012;109:3329–3334. doi: 10.1073/pnas.1117704109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papanikolopoulou K, Mills-Henry I, Tho SL, Wang YT, Gross AAR, Kirschner DA, Decatur SM, King J. Formation of amyloid fibrils in vitro by human γD-crystallin and its isolated domains. Mol Vis. 2008;14:81–89. [PMC free article] [PubMed] [Google Scholar]
- 14.Ecroyd H, Carver JA. Crystallin proteins and amyloid fibrils. Cell Mol Life Sci. 2009;66:62–81. doi: 10.1007/s00018-008-8327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meehan S, Knowles TPJ, Baldwin AJ, Smith JF, Squires AM, Clements P, Treweek TM, Ecroyd H, Tartaglia GG, Vendruscolo M, MacPhee CE, Dobson CM, Carver JA. Characterisation of amyloid fibril formation by small heat-shock chaperone proteins human αA-, αB-and R120G αB-crystallins. J Mol Biol. 2007;372:470–484. doi: 10.1016/j.jmb.2007.06.060. [DOI] [PubMed] [Google Scholar]
- 16.Meehan S, Berry Y, Luisi B, Dobson CM, Carver JA, MacPhee CE. Amyloid fibril formation by lens crystallin proteins and its implications for cataract formation. J Biol Chem. 2004;279:3413–3419. doi: 10.1074/jbc.M308203200. [DOI] [PubMed] [Google Scholar]
- 17.Basak A, Bateman O, Slingsby C, Pande A, Asherie N, Ogun O, Benedek GB, Pande J. High-resolution X-ray crystal structures of human γD crystallin (1.25 A) and the R58H mutant (1.15 A) associated with aculeiform cataract. J Mol Biol. 2003;328:1137–1147. doi: 10.1016/s0022-2836(03)00375-9. [DOI] [PubMed] [Google Scholar]
- 18.Mills IA, Flaugh SL, Kosinski-Collins MS, King JA. Folding and stability of the isolated Greek key domains of the long-lived human lens proteins γD-crystallin and γS-crystallin. Protein Sci. 2007;16:2427–2444. doi: 10.1110/ps.072970207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosinski-Collins MS, Flaugh SL, King J. Probing folding and fluorescence quenching in human γD crystallin Greek key domains using triple tryptophan mutant proteins. Protein Sci. 2004;13:2223–2235. doi: 10.1110/ps.04627004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosinski-Collins MS, King J. In vitro unfolding, refolding, and polymerization of human γD crystallin, a protein involved in cataract formation. Protein Sci. 2003;12:480–490. doi: 10.1110/ps.0225503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flaugh SL, Kosinski-Collins MS, King J. Interdomain side-chain interactions in human γD crystallin influencing folding and stability. Protein Sci. 2005;14:2030–2043. doi: 10.1110/ps.051460505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das P, King JA, Zhou R. β-strand interactions at the domain interface critical for the stability of human lens γD-crystallin. Protein Sci. 2010;19:131–140. doi: 10.1002/pro.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das P, King JA, Zhou R. Aggregation of gamma-crystallins associated with human cataracts via domain swapping at the C-terminal beta-strands. Proc Natl Acad Sci USA. 2011;108:10514–10519. doi: 10.1073/pnas.1019152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreau KL, King JA. Cataract-causing defect of a mutant γ-crystallin proceeds through an aggregation pathway which bypasses recognition by the α-crystallin chaperone. PLoS One. 2012;7:e37256. doi: 10.1371/journal.pone.0037256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Petty S, Trojanowski A, Knee K, Goulet D, Mukerji I, King J. Formation of amyloid fibrils in vitro from partially unfolded intermediates of human γC-crystallin. Invest Ophthalmol Vis Sci. 2010;51:672–678. doi: 10.1167/iovs.09-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamm P, Zanni MT. Concepts and methods of 2D infrared spectroscopy. New York: Cambridge University Press; 2011. [Google Scholar]
- 27.Woys AM, Mukherjee SS, Skoff DR, Moran SD, Zanni MT. A strongly absorbing class of non-natural labels for probing protein electrostatics and solvation with FTIR and 2D IR spectroscopies. J Phys Chem B. 2013;117:5009–5018. doi: 10.1021/jp402946c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. Two-dimensional infrared spectra of isotopically diluted amyloid fibrils from Aβ40. Proc Natl Acad Sci USA. 2008;105:7720–7725. doi: 10.1073/pnas.0802993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee P, Kass I, Arkin IT, Zanni MT. Picosecond dynamics of a membrane protein revealed by 2D IR. Proc Natl Acad Sci USA. 2006;103:3528–3533. doi: 10.1073/pnas.0508833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Middleton CT, Marek P, Cao P, Chiu CC, Singh S, Woys AM, de Pablo JJ, Raleigh DP, Zanni MT. Two-dimensional infrared spectroscopy reveals the complex behaviour of an amyloid fibril inhibitor. Nat Chem. 2012;4:355–360. doi: 10.1038/nchem.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunkelberger EB, Woys AM, Zanni MT. 2D IR cross peaks reveal hydrogen–deuterium exchange with single residue specificity. J Phys Chem B. 2013 doi: 10.1021/jp402942s. doi: 10.1021/jp402942s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woys AM, Lin Y-S, Reddy AS, Xiong W, de Pablo JJ, Skinner JL, Zanni MT. 2D IR line shapes probe ovispirin peptide conformation and depth in lipid bilayers. J Am Chem Soc. 2010;132:2832–2838. doi: 10.1021/ja9101776. [DOI] [PubMed] [Google Scholar]
- 33.Remorino A, Korendovych IV, Wu Y, DeGrado WF, Hochstrasser RM. Residue-specific vibrational echoes yield 3D structures of a transmembrane helix dimer. Science. 2011;332:1206–1209. doi: 10.1126/science.1202997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Middleton CT, Singh S, Reddy AS, Woys AM, Strasfeld DB, Marek P, Raleigh DP, de Pablo JJ, Zanni MT, Skinner JL. 2D IR spectroscopy of human amylin fibrils reflects stable β-sheet structure. J Am Chem Soc. 2011;133:16062–16071. doi: 10.1021/ja204035k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunkelberger EB, Buchanan LE, Marek P, Cao P, Raleigh DP, Zanni MT. Deamidation accelerates amyloid formation and alters amylin fiber structure. J Am Chem Soc. 2012;134:12658–12667. doi: 10.1021/ja3039486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shim S-H, Gupta R, Ling YL, Strasfeld DB, Raleigh DP, Zanni MT. Two-dimensional IR spectroscopy and isotope labeling defines the pathway of amyloid formation with residue-specific resolution. Proc Natl Acad Sci USA. 2009;106:6614–6619. doi: 10.1073/pnas.0805957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith AW, Lessing J, Ganim Z, Peng CS, Tokmakoff A, Roy S, Jansen TLC, Knoester J. Melting of a β-hairpin peptide using isotope-edited 2D IR spectroscopy and simulations. J Phys Chem B. 2010;114:10913–10924. doi: 10.1021/jp104017h. [DOI] [PubMed] [Google Scholar]
- 38.Middleton CT, Woys AM, Mukherjee SS, Zanni MT. Residue-specific structural kinetics of proteins through the union of isotope labeling, mid-IR pulse shaping, and coherent 2D IR spectroscopy. Methods. 2010;52:12–22. doi: 10.1016/j.ymeth.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woys AM, Almeida AM, Wang L, Chiu C-C, McGovern M, de Pablo JJ, Skinner JL, Gellman SH, Zanni MT. Parallel β-sheet vibrational couplings revealed by 2D IR spectroscopy of an isotopically labeled macrocycle: quantitative benchmark for the interpretation of amyloid and protein infrared spectra. J Am Chem Soc. 2012;134:19118–19128. doi: 10.1021/ja3074962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Backus EHG, Bloem R, Donaldson PM, Ihalainen JA, Pfister R, Paoli B, Caflisch A, Hamm P. 2D-IR study of a photoswitchable isotope-labeled α-helix. J Phys Chem B. 2010;114:3735–3740. doi: 10.1021/jp911849n. [DOI] [PubMed] [Google Scholar]
- 41.Liang C, Louhivuori M, Marrink SJ, Jansen TLC, Knoester J. Vibrational spectra of a mechanosensitive channel. J Phys Chem Lett. 2013;4:448–452. doi: 10.1021/jz3019258. [DOI] [PubMed] [Google Scholar]
- 42.King JT, Kubarych KJ. Site-specific coupling of hydration water and protein flexibility studied in solution with ultrafast 2D-IR spectroscopy. J Am Chem Soc. 2012;134:18705–18712. doi: 10.1021/ja307401r. [DOI] [PubMed] [Google Scholar]
- 43.Thielges MC, Axup JY, Wong D, Lee HS, Chung JK, Schultz PG, Fayer MD. Two-dimensional IR spectroscopy of protein dynamics using two vibrational labels: a site-specific genetically encoded unnatural amino acid and an active site ligand. J Phys Chem B. 2011;115:11294–11304. doi: 10.1021/jp206986v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi J-H, Raleigh D, Cho M. Azido homoalanine is a useful infrared probe for monitoring local electrostatistics and side-chain solvation in proteins. J Phys Chem Lett. 2011;2:2158–2162. doi: 10.1021/jz200980g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dutta S, Li Y-L, Rock W, Houtman JCD, Kohen A, Cheatum CM. 3-Picolyl azide adenine dinucleotide as a probe of femtosecond to picosecond enzyme dynamics. J Phys Chem B. 2011;116:542–548. doi: 10.1021/jp208677u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jo H, Culik RM, Korendovych IV, Degrado WF, Gai F. Selective incorporation of nitrile-based infrared probes into proteins via cysteine alkylation. Biochemistry. 2010;49:10354–10356. doi: 10.1021/bi101711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rubtsov IV. Relaxation-assisted two-dimensional infrared (RA 2DIR) method: accessing distances over 10 A and measuring bond connectivity patterns. Acc Chem Res. 2009;42:1385–1394. doi: 10.1021/ar900008p. [DOI] [PubMed] [Google Scholar]
- 48.Maekawa H, Poli MD, Moretto A, Toniolo C, Ge N-H. Toward detecting the formation of a single helical turn by 2D IR cross peaks between the amide-I and-II modes. J Phys Chem B. 2009;113:11775–11786. doi: 10.1021/jp9045879. [DOI] [PubMed] [Google Scholar]
- 49.Kwak K, Park S, Finkelstein IJ, Fayer MD. Frequency-frequency correlation functions and apodization in two-dimensional infrared vibrational echo spectroscopy: a new approach. J Chem Phys. 2007;127:124503–124517. doi: 10.1063/1.2772269. [DOI] [PubMed] [Google Scholar]
- 50.Roberts ST, Loparo JJ, Tokmakoff A. Characterization of spectral diffusion from two-dimensional line shapes. J Chem Phys. 2006;125:084502. doi: 10.1063/1.2232271. [DOI] [PubMed] [Google Scholar]
- 51.Skoff DR, Laaser JE, Mukherjee SS, Middleton C, Zanni MT. Simplified and economical 2D IR spectrometer design using a dual acousto-optic modulator. Chem Phys. 2012;422:8–15. doi: 10.1016/j.chemphys.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Louis JM, Byeon I-JL, Baxa U, Gronenborn AM. The GB1 amyloid fibril: Recruitment of the peripheral β-strands of the domain swapped dimer into the polymeric interface. J Mol Biol. 2005;348:687–698. doi: 10.1016/j.jmb.2005.02.071. [DOI] [PubMed] [Google Scholar]
- 53.O'Nuallain B, Freir DB, Nicoll AJ, Risse E, Ferguson N, Herron CE, Collinge J, Walsh DM. Amyloid beta-protein dimers rapidly form stable synaptotoxic protofibrils. J Neurosci. 2010;30:14411–14419. doi: 10.1523/JNEUROSCI.3537-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hahn S, Kim S-S, Lee C, Cho M. Characteristic two-dimensional IR spectroscopic features of antiparallel and parallel β-sheet polypeptides: simulation studies. J Chem Phys. 2005;123:084905. doi: 10.1063/1.1997151. [DOI] [PubMed] [Google Scholar]
- 55.Strasfeld DB, Ling YL, Gupta R, Raleigh DP, Zanni MT. Strategies for extracting structural information from 2D IR spectroscopy of amyloid: application to islet amyloid polypeptide. J Phys Chem B. 2009;113:15679–15691. doi: 10.1021/jp9072203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jahn TR, Makin OS, Morris KL, Marshall KE, Tian P, Sikorski P, Serpell LC. The common architecture of cross-β amyloid. J Mol Biol. 2010;395:717–727. doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 57.Tycko R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Q Rev Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 58.Teoh CL, Pham CLL, Todorova N, Hung A, Lincoln CN, Lees E, Lam YH, Binger KJ, Thomson NH, Radford SE, Smith TA, Müller SA, Engel A, Griffin MDW, Yarovsky I, Gooley PR, Howlett GJ. A structural model for apolipoprotein C-II amyloid fibrils: experimental characterization and molecular dynamics simulations. J Mol Biol. 2011;405:1246–1266. doi: 10.1016/j.jmb.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 59.Gal N, Morag A, Kolusheva S, Winter R, Landau M, Jelinek R. Lipid bilayers significantly modulate cross-fibrillation of two distinct amyloidogenic peptides. J Am Chem Soc. 2013;135:13582–13589. doi: 10.1021/ja4070427. [DOI] [PubMed] [Google Scholar]
- 60.Seeliger J, Evers F, Jeworrek C, Kapoor S, Weise K, Andreetto E, Tolan M, Kapurniotu A, Winter R. Cross-amyloid interaction of Aβ and IAPP at lipid membranes. Angew Chem Intl Ed. 2012;51:679–683. doi: 10.1002/anie.201105877. [DOI] [PubMed] [Google Scholar]
- 61.Sarell CJ, Stockley PG, Radford SE. Assessing the causes and consequences of co-polymerization in amyloid formation. Prion. 2013;7:0–1. doi: 10.4161/pri.26415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sarell CJ, Woods LA, Su Y, Debelouchina GT, Ashcroft AE, Griffin RG, Stockley PG, Radford SE. Expanding the repertoire of amyloid polymorphs by co-polymerization of related protein precursors. J Biol Chem. 2013;288:7327–7337. doi: 10.1074/jbc.M112.447524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lam AR, Moran SD, Preketes NK, Zhang TO, Zanni MT, Mukamel S. Study of the γD-crystallin protein using two-dimensional infrared (2DIR) spectroscopy: experiment and simulation. J Phys Chem B. 2013;117:15436–15443. doi: 10.1021/jp405159v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma B, Nussinov R. Selective molecular recognition in amyloid growth and transmission and cross-species barriers. J Mol Biol. 2012;421:172–184. doi: 10.1016/j.jmb.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McGlinchey RP, Yap TL, Lee JC. The yin and yang of amyloid: insights from [small alpha]-synuclein and repeat domain of Pmel17. Phys Chem Chem Phys. 2011;13:20066–20075. doi: 10.1039/c1cp21376h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luppi B, Bigucci F, Corace G, Delucca A, Cerchiara T, Sorrenti M, Catenacci L, Di Pietra AM, Zecchi V. Albumin nanoparticles carrying cyclodextrins for nasal delivery of the anti-Alzheimer drug tacrine. Eur J Pharm Sci. 2011;44:559–565. doi: 10.1016/j.ejps.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 68.Kosinski-Collins MS, Flaugh SL, King J. Probing folding and fluorescence quenching in human γD crystallin Greek key domains using triple tryptophan mutant proteins. Protein Sci. 2004;13:2223–2235. doi: 10.1110/ps.04627004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mills IA, Flaugh SL, Kosinski-Collins MS, King JA. Folding and stability of the isolated Greek key domains of the long-lived human lens proteins γD-crystallin and γS-crystallin. Protein Sci. 2007;16:2427–2444. doi: 10.1110/ps.072970207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moran SD, Decatur SM, Zanni MT. Structural and sequence analysis of the human gammaD-crystallin amyloid fibril core using 2D IR spectroscopy, segmental (13)C labeling, and mass spectrometry. J Am Chem Soc. 2012;134:18410–18416. doi: 10.1021/ja307898g. [DOI] [PubMed] [Google Scholar]
- 71.Moran SD, Zhang TO, Decatur SM, Zanni MT. Amyloid fiber formation in human γD-crystallin induced by UV-B photodamage. Biochemistry. 2013;52:6169–6181. doi: 10.1021/bi4008353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brubaker WD, Freites JA, Golchert KJ, Shapiro RA, Morikis V, Tobias DJ, Martin RW. Separating instability from aggregation propensity in γS-crystallin variants. Biophys J. 2011;100:498–506. doi: 10.1016/j.bpj.2010.12.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang W, Cai H-C, Li F-F, Xi Y-B, Ma X, Yan Y-B. The congenital cataract-linked G61C mutation destabilizes γD-crystallin and promotes non-native aggregation. PLoS One. 2011;6:e20564. doi: 10.1371/journal.pone.0020564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee S, Lin S, Li M, Liang R. Possible mechanism of exacerbating cataract formation in cataractous human lens capsules induced by systemic hypertension or glaucoma. Ophthalmic Res. 1997;29:83–90. doi: 10.1159/000268001. [DOI] [PubMed] [Google Scholar]
- 75.Hanson SRA, Hasan A, Smith DL, Smith JB. The major in vivo modifications of the human water-insoluble lens crystallins are disulfide bonds, deamidation, methionine oxidation and backbone cleavage. Exp Eye Res. 2000;71:195–207. doi: 10.1006/exer.2000.0868. [DOI] [PubMed] [Google Scholar]
- 76.Takemoto L, Sorensen CM. Protein–protein interactions and lens transparency. Exp Eye Res. 2008;87:496–501. doi: 10.1016/j.exer.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


