Key Points
We tested the hypothesis that B-cell lymphomas arising in HCV-infected patients express B-cell receptors specific to the virus.
We analyzed the reactivity of these B-cell receptors with HCV proteins using several experimental approaches, none of which supported the hypothesis.
Abstract
Chronic hepatitis C virus (HCV) infection has been implicated in the induction and maintenance of B-cell lymphomas. The strongest evidence for this derives from clinical observations of tumor regressions upon antiviral treatments. Here we used multiple methods to test the hypothesis that the expansion of HCV-specific B cells gives rise to lymphomas. We obtained lymphoma tissues from HCV-infected lymphoma patients, including some that later regressed upon antiviral treatments. We expressed the lymphoma B-cell receptors as soluble immunoglobulin Gs and membrane IgMs, and analyzed their reactivity with HCV proteins and with HCV virions. We confirmed previous reports that HCV-associated lymphomas use a restricted immunoglobulin variable region gene repertoire. However, we found no evidence for their binding to the HCV antigens. We conclude that most lymphomas of HCV-infected patients do not arise from B cells aimed at eliminating the virus.
Introduction
Survival of B cells requires the expression of B-cell receptors (BCRs), as demonstrated in knockout mice1,2 and in some patients with non–X-linked agammaglobulinemia. Lymphoma B cells undergo somatic hypermutation in their variable region (V) genes, which would be expected to generate protein loss variants. However, in the various lymphoma types, the BCR is retained,3 suggesting immunoglobulin G (IgG)'s importance for lymphoma cell survival. Yet lymphomas’ cognate antigens are not known. B-cell proliferative diseases such as mixed cryoglobulinemia (MC) and B-cell non-Hodgkin lymphoma (B-NHL) that arise in hepatitis C virus (HCV)-infected patients represent a special opportunity to study antigenic drive in lymphomagenesis. First, both MC and B-NHL use a restricted V-gene repertoire shared by anti-HCV envelope antibodies.4,5 Second, elimination of HCV by antiviral therapies in patients with these B-cell diseases has been associated with their regression.6 Moreover, we previously identified an HCV-associated lymphoma whose BCR bound the HCV envelope protein E2.7 Normal B cells aimed at eliminating HCV would be expected to bind the virus via 2 receptors: the cognate BCR and the viral entry receptor CD81, which is a member of a costimulatory complex with CD19/CD21. Such B cells would receive dual stimulatory signals and might undergo unchecked proliferation during chronic HCV infection.
Here we tested this hypothesis by expressing BCRs from lymphomas of HCV-infected patients as soluble IgGs, suggesting their importance, and as membrane IgMs. We included patients who had tumor regressions after antiviral therapies,8 expecting that they would be more likely to express anti-HCV BCRs. We used several methods to test the reactivity of the rescued lymphoma BCRs with viral proteins and particles. However, we found no reactivity and therefore no evidence to support the hypothesis that viral antigens drive B-cell lymphomas.
Methods
Patients
Biopsy specimens of patients with B-NHL and chronic HCV infection were collected at Stanford University Medical Center, Sloan Kettering Memorial Cancer Center, and the University of Pavia Medical School. Patients’ medical record numbers were de-identified and reassigned numbers. The institutional review boards at each center approved this study, and written informed consent was obtained from all patients in accordance with the Declaration of Helsinki.
V-gene rescue
mRNA was isolated using RNeasy (Qiagen, Valencia, CA), cDNA was amplified using SMARTer RACE (Clontech, Mountain View, CA), and V-region amplification used 5′ RACE and the following constant regions primers:
IgM 5′-ggtggargcctgaggagacggtgacc-3′
IgG 5′- ggagsagggygccagggggaagac-3′
κ 5′-tgtgacgggcgagctcaggccctgat-3′
λ 5′-gcgtcaggcacagatagctgctggccgc-3′
Expression of lymphoma idiotypes (Ids)
Amplified products were inserted into an IgG1 expression vector9 and then expressed, as was done previously.4 IgGs in the supernatant of transiently transfected COS-7 cells were quantitated by enzyme-linked immunosorbent assay (ELISA). Expression of the rescued V regions in A20 cells as membrane IgM was done, as previously.10
HCV proteins
Expression of soluble envelope protein of HCV genotype 1a (E2661) and envelope protein of HCV genotype 2a (J6E2) were done as previously.7,11 HCV envelope proteins expressed intracellularly (HCV-E1E2) of various genotypes were encoded by pCR 3.1-UKN1B12.16, -UKN1B5.23, -UKN2A1.2, and -UKN2A2.4.12 The E1E2 sequences from these plasmids, and the E1E2 of the H77c strain (genotype 1a) were ligated into pCDM8 expression plasmids and transiently transfected into 293T cells. An anti-HCV ELISA kit (DIAsource, Louvain-la-Neuve, Belgium) analyzed interaction of patient IgGs with core, NS3, NS5A, and NS5B proteins.
Binding of rescued IgG and IgM
ELISA was done to detect binding to HCV-E2, as was done previously.7 Flow cytometry was used to detect rescue IgG binding to intracellular E1E2 in permeabilized 293T cells, and the binding of A20 cell surface IgM to soluble E2.
Results and discussion
The incidence of B-cell proliferative diseases, including MC and NHL, is higher in HCV-infected patients than in noninfected individuals, especially in certain geographical areas such as Italy.8,13 Moreover, the regression of B-cell diseases in response to successful antiviral therapies implies a causative link between HCV infection and B-cell proliferative diseases.6,8 Here, we aimed to validate the hypothesis that B-cell lymphomas arise from expansion of antiviral B cells in HCV-infected patients by analyzing the reactivity of their lymphoma BCRs with HCV. Patients were diagnosed in the US and in Italy, and the latter received antiviral therapy and included oncological responders and nonresponders. Analysis of V-gene usage showed a restricted repertoire, specifically usage of VH-169 and Vκ3-20 (Table 1).
Table 1.
Immunoglobulins rescued from lymphomas of HCV-infected patients do not react with HCV proteins
| Patient | Isotype | Diagnosis | Lymphoma response to antiviral therapy | VH | VL | Accession # VH/VL | HCV genotype | E2661 | J6E2 | E1E2 | NS + Core |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 101 | IgM/k | DLBCL | ND | VH1-69 | Vk3-20 | KF895775/6 | ND | Neg | ND | ND | ND |
| 102 | IgM/k | FL | ND | VH3-48 | Vk1-39 | KF895777/8 | ND | Neg | ND | ND | ND |
| 103 | Ig*/k | MZL | ND | VH4-59 | Vk3-20 | KF895779/80 | ND | Neg | ND | ND | ND |
| 104 | IgM/k | DLBCL | ND | VH1-69 | Vk1D-16 | KF895781/2 | ND | Neg | ND | ND | ND |
| 105 | IgM/k | DLBCL | ND | VH1-69 | Vk3-20 | KF895783/4 | ND | Neg | ND | ND | ND |
| 106 | IgM/k | NHL | ND | VH4-59 | Vk3-15 | KF895785/6 | ND | Neg | ND | ND | ND |
| 107 | IgM/k | NA | ND | VH3-21 | Vk1-39 | KF895787/8 | ND | Neg | ND | ND | ND |
| 108 | IgM/k | NA | ND | VH4-59 | Vk3-15 | KF895789/90 | ND | Neg | Neg | Neg | Neg |
| 109 | IgM/k | NA | ND | VH4-34 | Vk3-20 | KF895791/2 | ND | Neg | Neg | Neg | Neg |
| 110 | IgM/k | NA | ND | VH4-59 | Vk3-20 | KF895793/4 | ND | Neg | Neg | Neg | Neg |
| 111 | IgG/k | NA | ND | VH1-02 | Vk2-30 | KF895795/6 | ND | Neg | Neg | Neg | Neg |
| 112 | IgM/k | NA | ND | VH4-34 | Vk3-20 | KF895797/8 | ND | Neg | Neg | Neg | Neg |
| 121 | IgM/k | MALT MZL | Yes | VH1-69 | Vk3-20 | KF895799/800 | 2a/2c | Neg | Neg | Neg | Neg |
| 122 | IgM/k | SMZL | No | VH4-59 | Vk3D-15 | KF895801/2 | 1b | Neg | Neg | Neg | Neg |
| 123 | IgM/k | SMZL | Yes | VH1-69 | Vk3-20 | KF895803/4 | 2a/2c | Neg | Neg | Neg | Neg |
| 124 | IgM/k | MALT MZL | Yes | VH4-30 | Vk3-15 | KF895805/6 | 2a/2c | Neg | Neg | Neg | Neg |
| 125 | IgM/k | SMZL | Yes | VH3-30 | Vk1-8 | KF895807/8 | 2a/2c | Neg | Neg | Neg | Neg |
| 126 | IgM/k | MALT MZL | Yes | VH1-69 | Vk3-20 | KF895809/10 | 1b | Neg | Neg | Neg | Neg |
| 127 | IgM/k | NHL | No | VH1-69 | Vk3-20 | KF895811/2 | 1b | Neg | Neg | Neg | Neg |
DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma; Ig*, undetermined heavy chain; MALT, mucosa-associated lymphoid tissue; MZL, marginal zone lymphoma; NA, not available; ND, not done; Neg, negative; NS + Core, HCV nonstructural + core proteins; SMZL, splenic marginal zone lymphoma.
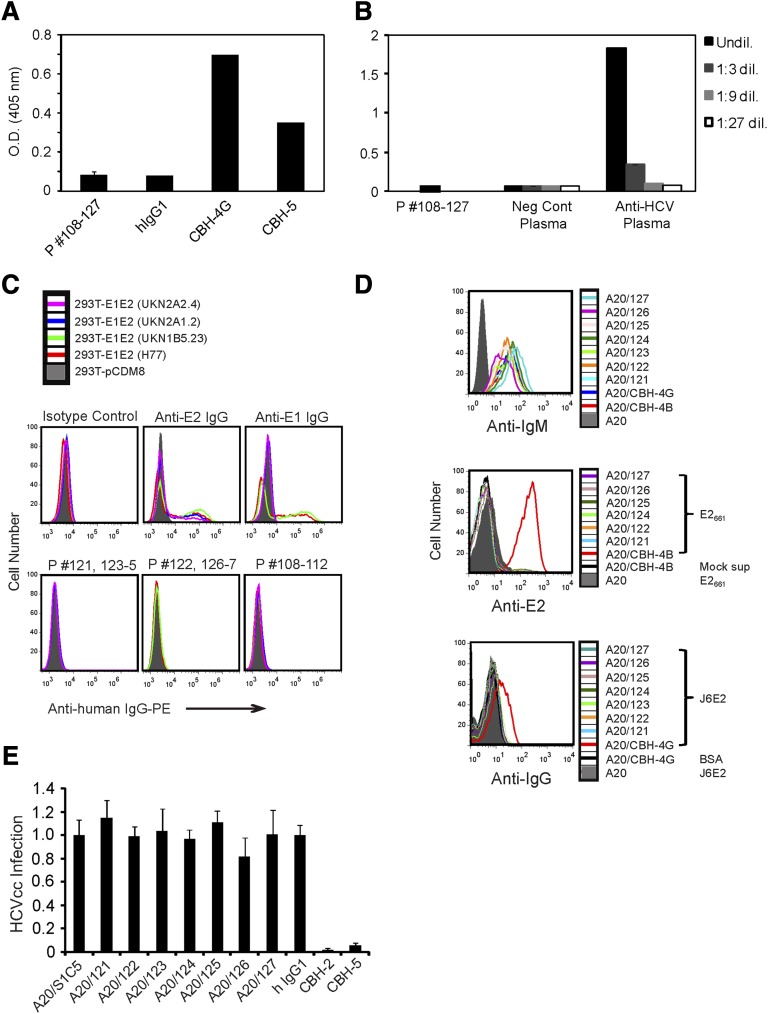
We sequenced the V-region genes and expressed them as secreted human IgG1/κ in transfected cells. We then analyzed the reactivity of all rescued IgG1 with the soluble HCV E2 ectodomain, E2661 (genotype 1a)7 or J6E2 (genotype 2a).11 However, except for the anti-E2 monoclonal antibody (mAb) controls, none of the tested IgG1 reacted with E2661 (Table 1) or with J6E2 (Figure 1A).14 Next we tested all of the rescued IgG1s with internal HCV antigens, which are included in a diagnostic kit; however, none reacted with core, NS3, NS5A, and NS5B proteins (Figure 1B).
Figure 1.
Rescued HCV-associated lymphoma Ids do not react with viral proteins when expressed as soluble IgGs or as cell surface IgMs. (A) Purified J6E2 captured on lectin-coated 96-well plates was incubated with the indicated patients’ Id IgG1s, with anti-E2 mAbs CBH-4G or CBH-5, or with a human IgG1/κ isotype control. Plate-bound IgGs were detected with an HRP-conjugated antihuman IgG. (B) 96-well plates coated with HCV core, NS3, and NS5 antigens were incubated with the indicated patients’ Id IgG1s, with anti-HCV plasma, or with negative control plasma and diluted as indicated. After washing, plate-bound Igs were detected with HRP-conjugated HCV core, NS3, and NS5 antigens. (A-B) Each bar represents the mean optical density of wells incubated with each group of Id IgGs ± the standard deviation. Representative results of 2 experiments for each assay are shown. (C) Single-cell suspensions of 293 T cells transfected with empty pCDM8 vector (filled gray) or with pCDM8 vectors encoding E1E2 of the indicated genotypes were fixed and permeabilized. The cells were then stained with the anti-E2, anti-E1, a human IgG1/κ isotype control mAb, or with a mixture of the indicated patients’ IgG1 containing 0.5 μg of each Id. Cells were then washed, stained with PE-conjugated antihuman IgG, and analyzed by flow cytometry. (D) The patients’ Ids were expressed as human IgMs on the surface of the mouse B-cell line, A20. Positive controls were A20 cells expressing CBH-4B or CBH-4G. Cells were stained with FITC conjugated anti-human IgM (top panel). Cells were incubated for 1 hour on ice with cell culture supernatant containing soluble E2661 or mock supernatant. Cells were then washed, stained with AlexaFluor 647-conjugated mouse anti-E2 mAb (H53) (middle panel). Cells were incubated for 1 hour on ice with soluble J6E2 or bovine serum albumin, washed, and further incubated for 1 hour on ice with a 1:1 mixture of the anti-E2 mAbs CBH-2 and CBH-5. After washing, cells were stained with AlexaFluor 647-conjugated antihuman IgG (bottom panel). (C-D) Cells were washed and analyzed by flow cytometry. (E) A20 cells expressing the indicated surface lymphoma patients’ Id IgM or an IgM of irrelevant specificity (SIC5); neutralizing anti-E2 mAbs (CBH2 or CBH5) or a control human IgG1 mAb were incubated with luciferase-reporter HCV produced in cell culture (HCVcc) (J6/JFH(p7-Rluc2A) HCV,14 titer: 6.3 × 105 TCID50/mL) for 1 hour at 37°C. These samples were then used to inoculate naïve huh-7.5 cells. To measure infectivity, cells were lysed at 48 hours and subjected to standard luciferase assays. Y-axis represents HCVcc infection relative to the A20 SIC5 control. Data represent means and standard deviations (error bars).
HCV is enveloped by 2 heterodimeric proteins, E1 and E2.15 We tested the possibility that the rescued IgG1s recognize the heterodimer in its native membrane-bound form by using 293T cells transfected with constructs encoding full E1E2 polypeptides.12 We specifically selected E1E2 of the HCV genotypes 1b and 2a matching the infected patients genotypes, as well as E1E2 derived from the HCV isolate H77 of genotype 1a. The heterodimers were expressed intracellularly, as detected by flow cytometry using anti-E1 and anti-E2 mAb (Figure 1C, top panels). However, none of the patients’ rescued IgG1 showed reactivity (Figure 1C, bottom panels). Chronic HCV infection is thought to have a causative role in MC, characterized by the benign proliferation of B-cell–secreting IgM with rheumatoid factor (RF) activity. Evidence also exists that implicates MC as the precursor to frank NHL. However, we did not find RF activity in any of the rescued IgG1s (data not shown).
The VH-169 gene is also repetitively used in human mAbs that react with HIV16 and with the influenza hemagglutinin (HA) protein.17 Of note, in a recent study, germline VH-169 expressed as surface IgM reacted with HA, whereas soluble versions of the mAb were unreactive. It was proposed that clustering of surface BCR on a naïve B cell increases the avidity of the otherwise low-affinity germline BCR to HA to a level sufficient to trigger B-cell activation.18 We therefore expressed the lymphoma Ids as human IgMs on A20, a mouse B-cell line (Figure 1D, top panel), and tested their reactivity with soluble E2 proteins. However, only positive control cells expressing IgM with known anti-E2 reactivity showed binding to soluble E2661 and J6E2 proteins (Figure 1D, middle and bottom panels, respectively). In addition, these cell-surface–expressed lymphoma Id IgMs did not bind HCV-core or NS proteins (data not shown).
Finally, we took a step further to explore whether lymphoma Ids interact with HCV proteins on an assembled virion. HCVcc are associated with lipids19,20 and may contain other antigens not studied in previous experiments. However, incubation of HCVcc with A20 cells expressing surface lymphoma Id IgMs did not reduce their infectivity, whereas neutralizing anti-E2 mAbs (CBH-2 and CBH-5) blocked infection (Figure 1E). Nonneutralizing anti-E2 mAbs (CBH-4B, CBH-4G) expressed as cell surface IgMs (Figure 1E), or as soluble CBH-4G mAb (data not shown), did not block infection.
This study tested the hypothesis that B cells aimed at eliminating the virus give rise to HCV-associated B-cell lymphomas. We included patients that responded to antiviral therapy expecting them to be more likely to bind the virus. However, while confirming a restricted usage of VH-169, we did not identify a single BCR that reacted with HCV (Table 1), hence there was no evidence to support the hypothesis.
Acknowledgments
The authors thank Drs William Robinson and Jeremy Sokolove for providing RF-positive and -negative plasma, and technical expertise in testing RF activity.
This study was supported by a grant from Stanford’s Institute for Immunity, Transplantation, and Infection; and the National Institutes of Health, National Institute of Allergy and Infectious Diseases (K08 AI079406) (S.E.).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: P.P.N., C.-C.K., and S.W. performed the experiments; P.P.N. and S.W. analyzed the results and created the figures; L.A., M.P., C.S.P., and R.L. provided biopsy specimens; J.M. provided the J6E2; A.T. and J.B. provided the plasmids encoding E1E2 glycoproteins; and P.P.N., S.W., S.E., R.L., and S.L. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Shoshana Levy, Stanford University Medical Center, CCSR Building, Room 1105a, Stanford, CA 94305; e-mail: slevy@stanford.edu.
References
- 1.Gu H, Kitamura D, Rajewsky K. B cell development regulated by gene rearrangement: arrest of maturation by membrane-bound D mu protein and selection of DH element reading frames. Cell. 1991;65(1):47–54. doi: 10.1016/0092-8674(91)90406-o. [DOI] [PubMed] [Google Scholar]
- 2.Kitamura D, Roes J, Kühn R, Rajewsky KA. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350(6317):423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 3.Gururajan M, Jennings CD, Bondada S. Cutting edge: constitutive B cell receptor signaling is critical for basal growth of B lymphoma. J Immunol. 2006;176(10):5715–5719. doi: 10.4049/jimmunol.176.10.5715. [DOI] [PubMed] [Google Scholar]
- 4.Chan CH, Hadlock KG, Foung SK, Levy S. V(H)1-69 gene is preferentially used by hepatitis C virus-associated B cell lymphomas and by normal B cells responding to the E2 viral antigen. Blood. 2001;97(4):1023–1026. doi: 10.1182/blood.v97.4.1023. [DOI] [PubMed] [Google Scholar]
- 5.Charles ED, Green RM, Marukian S, et al. Clonal expansion of immunoglobulin M+CD27+ B cells in HCV-associated mixed cryoglobulinemia. Blood. 2008;111(3):1344–1356. doi: 10.1182/blood-2007-07-101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermine O, Lefrère F, Bronowicki JP, et al. Regression of splenic lymphoma with villous lymphocytes after treatment of hepatitis C virus infection. N Engl J Med. 2002;347(2):89–94. doi: 10.1056/NEJMoa013376. [DOI] [PubMed] [Google Scholar]
- 7.Quinn ER, Chan CH, Hadlock KG, Foung SK, Flint M, Levy S. The B-cell receptor of a hepatitis C virus (HCV)-associated non-Hodgkin lymphoma binds the viral E2 envelope protein, implicating HCV in lymphomagenesis. Blood. 2001;98(13):3745–3749. doi: 10.1182/blood.v98.13.3745. [DOI] [PubMed] [Google Scholar]
- 8.Peveling-Oberhag J, Arcaini L, Hansmann ML, Zeuzem S. Hepatitis C-associated B-cell non-Hodgkin lymphomas. Epidemiology, molecular signature and clinical management. J Hepatol. 2013;59(1):169–177. doi: 10.1016/j.jhep.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Reff ME, Carner K, Chambers KS, et al. Depletion of B cells in vivo by a chimeric mouse human monoclonal antibody to CD20. Blood. 1994;83(2):435–445. [PubMed] [Google Scholar]
- 10.Ng PP, Jia M, Patel KG, et al. A vaccine directed to B cells and produced by cell-free protein synthesis generates potent antilymphoma immunity. Proc Natl Acad Sci USA. 2012;109(36):14526–14531. doi: 10.1073/pnas.1211018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whidby J, Mateu G, Scarborough H, Demeler B, Grakoui A, Marcotrigiano J. Blocking hepatitis C virus infection with recombinant form of envelope protein 2 ectodomain. J Virol. 2009;83(21):11078–11089. doi: 10.1128/JVI.00800-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarr AW, Urbanowicz RA, Hamed MR, et al. Hepatitis C patient-derived glycoproteins exhibit marked differences in susceptibility to serum neutralizing antibodies: genetic subtype defines antigenic but not neutralization serotype. J Virol. 2011;85(9):4246–4257. doi: 10.1128/JVI.01332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sautto G, Mancini N, Clementi M, Burioni R. Molecular signatures of hepatitis C virus (HCV)-induced type II mixed cryoglobulinemia (MCII). Viruses. 2012;4(11):2924–2944. doi: 10.3390/v4112924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moradpour D, Penin F. Hepatitis C virus proteins: from structure to function. Curr Top Microbiol Immunol. 2013;369:113–142. doi: 10.1007/978-3-642-27340-7_5. [DOI] [PubMed] [Google Scholar]
- 15.Gorny MK, Pan R, Williams C, et al. Functional and immunochemical cross-reactivity of V2-specific monoclonal antibodies from HIV-1-infected individuals. Virology. 2012;427(2):198–207. doi: 10.1016/j.virol.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohshima N, Iba Y, Kubota-Koketsu R, Asano Y, Okuno Y, Kurosawa Y. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J Virol. 2011;85(21):11048–11057. doi: 10.1128/JVI.05397-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lingwood D, McTamney PM, Yassine HM, et al. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489(7417):566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moriishi K, Matsuura Y. Exploitation of lipid components by viral and host proteins for hepatitis C virus infection. Front Microbiol. 2012;3:54. doi: 10.3389/fmicb.2012.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neveu G, Barouch-Bentov R, Ziv-Av A, Gerber D, Jacob Y, Einav S. Identification and targeting of an interaction between a tyrosine motif within hepatitis C virus core protein and AP2M1 essential for viral assembly. PLoS Pathog. 2012;8(8):e1002845. doi: 10.1371/journal.ppat.1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray CL, Jones CT, Tassello J, Rice CM. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J Virol. 2007;81(19):10220–10231. doi: 10.1128/JVI.00793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]