Abstract
The transcription factor NF-κB plays a pivotal role in innate immunity in response to a variety of stimuli, and the coordinated regulation of this pathway determines the proper host responses to extracellular signals. In this study, we identified RACK1 as a novel negative regulator of NF-κB signaling, NF-κB-mediated cytokine induction and inflammatory reactions. RACK1 physically associates with the IKK complex in a TNF-triggered manner. This interaction interferes with the recruitment of the IKK complex to TRAF2, which is a critical step for IKK phosphorylation and subsequent activation triggered by TNF. By modulating the interaction between TRAF2 and IKK, RACK1 regulates the levels of NF-κB activation in response to different intensities of stimuli. Our findings suggest that RACK1 plays an important role in controlling the sensitivity of TNF-triggered NF-κB signaling by regulating IKK activation and provide new insight into the negative regulation of inflammatory reactions.
Keywords: IKK complex, inflammatory reactions, NF-κB, RACK1, sensitivity
Introduction
The transcription factor NF-κB plays a pivotal role in innate immune responses to a variety of stimuli and has a key function in cytokine-mediated inflammation1,2. In most cells, the IκB family of inhibitory proteins sequesters NF-κB in the cytoplasm, which holds the NF-κB pathway in an inactive status. Many extracellular stimuli, such as TNF and interleukin 1 (IL-1), lead to the activation of the IκB kinase (IKK) complex, which phosphorylates IκB proteins and promotes their ubiquitination and subsequent degradation3. This process leads to the translocation of NF-κB from the cytoplasm to the nucleus, where it initiates the transcription of a series of target genes4.
Many studies have noted that a key event that controls NF-κB activation in response to TNF stimulation is the formation of the TNF receptor 1 (TNFR1) signaling complex5,6. The process of accurate assembly and subsequent dissociation of the signaling proteins in this complex determines the initiation and duration of NF-κB activation, respectively7,8. After TNF stimulation, TRAF2 rapidly recruits TAK1 to TNFR1. Meanwhile, the IKK complex is directly recruited by TRAF2/5 and is further stabilized at the signaling platform by recognizing the K63-modified ubiquitination chain of RIP18,9,10. After the assembly of all of these signaling proteins, the TAK1 complex mediates phosphorylation and activation of the IKK complex, and the IKK complex dissociates from the signaling platform and subsequently transmits the signal to downstream factors11,12. It is widely thought that the effective regulation of signal transduction is always controlled by opposing activities, such as phosphorylation and dephosphorylation and ubiquitination and deubiquitination, which are mediated by various effector proteins around TNFR1 signaling complex. Currently, several negative regulators have been reported: Smad7 disturbs the recruitment of the TAK1 complex by TRAF210; A20 edits the K63-modified ubiquitination chain of RIP1, which destroys the interaction between IKK and RIP113; and CUEDC2, a newly identified negative regulator that mediates the dephosphorylation of the IKK complex14. Despite extensive studies on the mechanisms underlying the recruitment of the IKK complex to TNFR1, the molecular mechanisms of the negative regulation of this recruitment process remain unknown. As the recruitment of the IKK complex to TNFR1 is a key step in NF-κB activation, it would be interesting to determine whether negative regulation of IKK recruitment exists as another opposing regulatory mechanism.
The receptor of activated protein kinase C 1 (RACK1, GNB2L1) is a 36-kDa, cytosolic protein containing seven Trp-Asp 40 (WD40) repeats and is ubiquitously expressed in a diverse variety of species with high conservation15,16. Due to its interaction with a range of signaling proteins, such as PKC, Src and IFNα receptor, RACK1 has been widely perceived as an indispensable hub for signaling transduction in multiple signaling pathways17,18,19,20,21,22. Previous studies have demonstrated that RACK1 plays an important role in regulating cell growth, apoptosis and mobility23,24. RACK1 is also involved in mediating extracellular stress-induced cell signaling, such as JNK and p38 signaling25,26. All of these reports indicate that RACK1 has important functions in the response to extracellular stresses. However, whether and how RACK1 is involved in innate immune responses in mammals is still unknown.
In this study, we identified that RACK1 negatively regulates NF-κB activation by interacting with IKKα and IKKβ. By associating with IKK, RACK1 hinders TRAF2-mediated recruitment and the subsequent phosphorylation of IKK triggered by TNF. More importantly, by delaying the formation of the signaling complex of TRAF2 and the IKK complex, RACK1 controls the rate and intensity of NF-κB activation in response to TNF. Our results suggest that RACK1 acts as a novel negative regulator in controlling the sensitivity of NF-κB signaling in response to TNF.
Results
RACK1 represses the activity of NF-κB and the transcription of its target genes
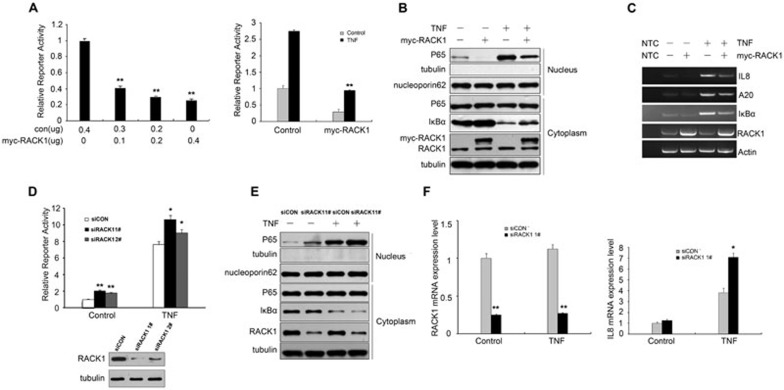
RACK1 is recognized as a scaffold protein that is involved in various biological responses. Recently, it was reported that RACK1 might function to modulate rice innate immune responses27. Considering the conservation of RACK1 in a diverse range of species, we wondered whether mammalian RACK1 is also involved in the regulation of innate immunity. Given the key role of NF-κB signaling in innate immunity, we chose this pathway to test our hypothesis, using NF-κB-responsive promoter reporter assays. As shown in Figure 1A, overexpression of RACK1 significantly repressed the activity of an NF-κB-responsive promoter in HEK293T cells in a dose-dependent manner. In addition to the basal activity of the reporter, we examined the effect of RACK1 on TNF-induced NF-κB-responsive promoter activity. Similarly, we found that the elevation of the reporter activity that was induced by TNF was more dramatic in control cells than in RACK1-overexpressing cells (Figure 1A), and this phenomenon was also observed in HEK293 and Hela cells (Supplementary information, Figure S1A). As translocation of the transcription factor p65 into the nucleus is critical for activating the NF-κB-responsive promoter, we then examined the effects of RACK1 on the cytoplasmic-nuclear translocation of p65. Overexpression of RACK1 impaired p65 accumulation in the nucleus at basal levels as well as after TNF stimulation (Figure 1B), suggesting that RACK1 might be a novel negative regulator of the NF-κB signaling pathway by impairing the nuclear translocation of p65. To further confirm this hypothesis, we examined the effects of RACK1 on the transcription of NF-κB target genes in either TNF-treated 293T or resting cells. The results of semiquantitative RT-PCR demonstrated that overexpression of RACK1 indeed inhibited TNF-induced transcription of NF-κB target genes, including IL8, A20 and IκBα (Figure 1C).
Figure 1.
RACK1 repressed the activity of NF-κB and the transcription of its target genes. (A) Overexpression of RACK1 inhibited an NF-κB-responsive luciferase reporter activity. RACK1 repressed the basal level activity of the reporter in a dose-dependent manner (left). The reporter response of 293T cells to TNF (10 ng/ml) was impaired by RACK1 overexpression (right). (B) Overexpression of RACK1 inhibited basal level and TNF-induced unclear translocation of p65. (C) RACK1 repressed TNF-induced transcription of endogenous IL8, A20 and IκBα genes. 293T cells were transfected with the indicated expression plasmids for 24 h, and then the cells were treated with TNF or left untreated for 45 min before semiquantitative RT-PCR. (D) Knockdown of RACK1 potentiated the activity of the NF-κB-responsive luciferase reporter. (E) Silencing of RACK1 expression promoted the translocation of p65 into the nucleus at the basal level and after TNF treatment (5 ng/ml for 15 min). (F) Knockdown of RACK1 enhanced TNF-induced transcription of NF-κB target genes. The indicated 293T cells were treated with TNF or left untreated for 45 min before quantitative RT-PCR. Graphs show the mean ± SD, n = 3. *P < 0.05; **P< 0.01.
Next, we determined whether endogenous RACK1 was required for TNF-induced signaling. We knocked down endogenous RACK1 by lentiviral-based RNAi in 293T cells and examined the NF-κB reporter activity. As shown in Figure 1D, knockdown of RACK1 increased the activity of the NF-κB reporter compared to the control cells. Unlike other negative regulators of this pathway, knockdown of RACK1 increased the reporter activity more dramatically in the resting state (∼2.5-fold) than after TNF stimulation (∼1.5-fold), which suggested that RACK1 might function as a buffering factor for the basal activity of the NF-κB signaling pathway. Consistently, knockdown of RACK1 increased p65 accumulation in the nucleus in the resting state as well as after TNF stimulation (Figure 1E). Additionally, knockdown of RACK1 promoted TNF-triggered transcription of IL8, A20 and IκBα (Figure 1F and Supplementary information, Figure S1B). Taken together, these results indicated that RACK1 acted as a negative regulator of NF-κB signaling.
RACK1 regulates TNF-induced NF-κB activation through the IKK complex
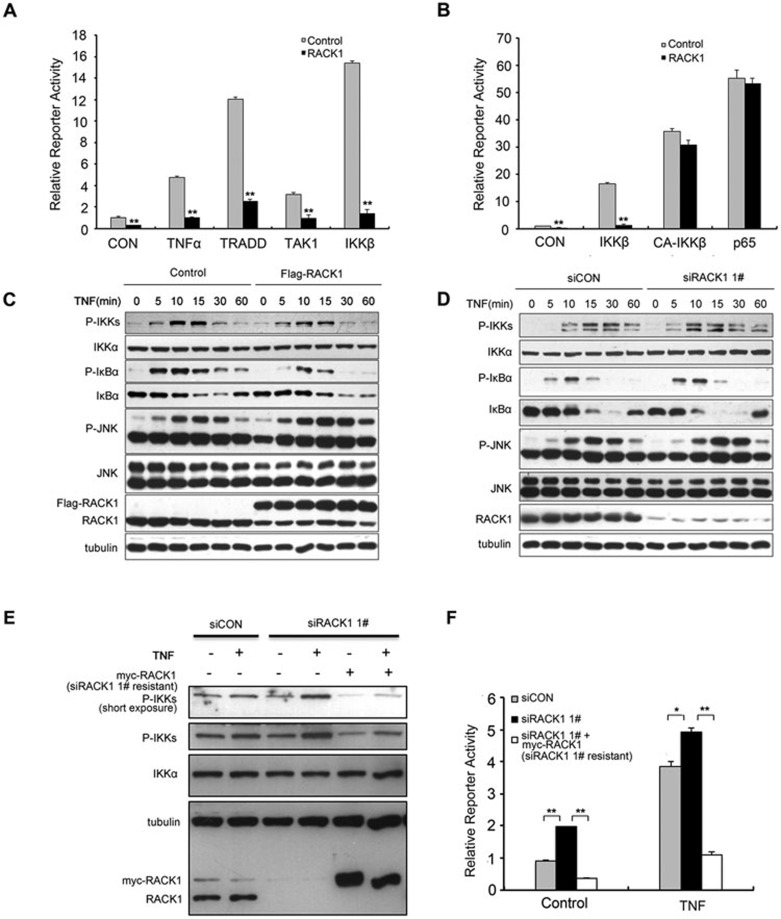
Several proteins are involved in the regulation of TNF-induced NF-κB signaling, such as TRADD and the TAK1 and IKK complexes, which could transmit extracellular signals to downstream transcription factors. To determine the molecular position of RACK1 in this signaling pathway, we examined the effects of RACK1 on NF-κB activation induced by these mediators via reporter assays. Overexpression of RACK1 nearly completely inhibited NF-κB activation induced by TRADD, TAK1 and IKKβ (Figure 2A and Supplementary information, Figure S2A). However, unlike the inhibitory effects on NF-κB activation that were induced by the overexpression of wild-type IKKβ, RACK1 showed little effect on NF-κB activation induced by either a constitutively active mutant of IKKβ or the NF-κB subunit p65 (Figure 2B and Supplementary information, Figure S2B). More importantly, overexpression of RACK1 inhibited IκBα degradation mediated by IKKβ (Supplementary information, Figure S2C). These results suggested that the inhibitory effects of RACK1 depend on the activity of IKK during NF-κB activation.
Figure 2.
RACK1 inhibited NF-κB activation by impairing IKK phosphorylation. (A) Overexpression of RACK1 inhibited the NF-κB reporter activity induced by the components of the TNF pathway. 293T cells were transfected with the indicated plasmids. Twenty-four hours later, the cells were left untreated or treated with TNF (10 ng/ml) for 6 h before luciferase assays were performed. (B) RACK1 only blocked IKK-dependent NF-κB activation. RACK1 but did not affect the NF-κB activation induced by a constitutively active mutant of IKKβ or p65. CA-IKKβ, constitutively active IKKβ mutant. (C) Overexpression of RACK1 impaired TNF-induced phosphorylation of the IKK complex and delayed degradation of IκBα. 293T cells were infected with Flag-RACK1-expressing or control lentivirus. After puromycin selection, the stable cell lines were treated with TNF (5 ng/ml) before lysis. (D) Knockdown of RACK1 accelerated the TNF-triggered phosphorylation of the IKK complex and degradation of IκBα. (E, F) 293T cells were infected with siRACK1 1# or control siRNA lentivirus and, 48 h later, were transfected with NF-κB-responsive luciferase reporter plasmids and the siRACK1 1#-resistant RACK1 overexpression or control vector. Ninety-six hours after viral infection, the cells were treated with TNF for immunoblotting and luciferase reporter assays. Graphs show the mean ± SD, n = 3. *P< 0.05; **P< 0.01.
Then, we investigated whether RACK1 affects IKK activation triggered by TNF. We first infected 293T cells with RACK1-overexpressing lentivirus or control virus. After drug selection, we treated RACK1-overexpressing stable cells and control cells with TNF, and examined the effects of RACK1 on IKK phosphorylation with antibodies specific for phospho-IKKα (Ser180) and phospho-IKKβ (Ser181). Upon TNF treatment, the phosphorylation levels of these proteins increased more significantly in the control cells than in RACK1-overexpressing cells (Figure 2C); however, the total amount of IKK protein remained unchanged during the treatment. In addition, overexpression of RACK1 weakened the TNF-induced phosphorylation of IκBα, thus delaying the subsequent degradation of IκBα (Figure 2C); whereas overexpression of RACK1 did not affect the activation of JNK induced by TNF treatment, which is another important signaling pathway controlled by TNFR1 (Figure 2C). These results indicated that overexpression of RACK1 could effectively and specifically retard the activation of IKK induced by TNF.
We then examined the effects of RACK1 knockdown on IKK activation induced by TNF. Phosphorylation of IKKα and IKKβ, but not JNK, occurred more promptly in RACK1-knockdown cells than in control cells, especially at the early stages (from 5 to 10 min) of TNF treatment (Figure 2D). This observation suggested that downregulation of RACK1 expression facilitated TNF-induced activation of the IKK complex. Consistently, knockdown of RACK1 potentiated the IKK complex-mediated phosphorylation of IκBα and accelerated the subsequent degradation of IκBα (Figure 2D). To exclude the off-target effects of RACK1 knockdown experiments, we constructed a RACK1-overexpression plasmid with a sequence that was resistant to siRACK1 1#. Restoring the expression of RACK1 in RACK1-knockdown cells effectively inhibited TNF-induced phosphorylation of the IKK complex and NF-κB reporter activity (Figure 2E and 2F), which confirmed the specificity of RACK1's function in the NF-κB signaling pathway. IL1β functions as a key regulator of inflammation and innate immunity and is another activator of NF-κB signaling. Thus, we examined whether RACK1 could modulate IL1-induced NF-κB signaling. Overexpression of RACK1 in HeLa cells influenced neither the phosphorylation levels of the IKK complex and IκBα nor the subsequent degradation of IκBα after IL1β induction, suggesting that RACK1 exerted inhibitory effects mainly on TNF-mediated NF-κB activation via interfering with TNF-induced phosphorylation of IKKs (Suppelmentary information, Figure S2D).
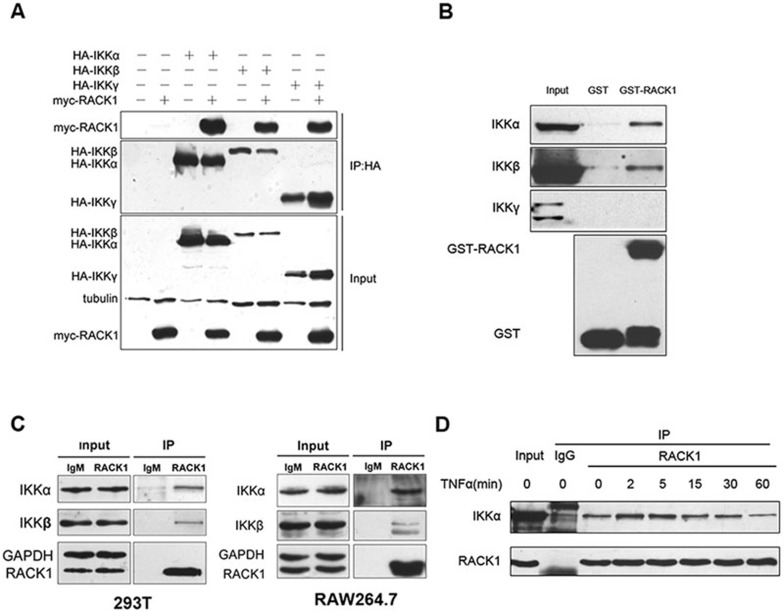
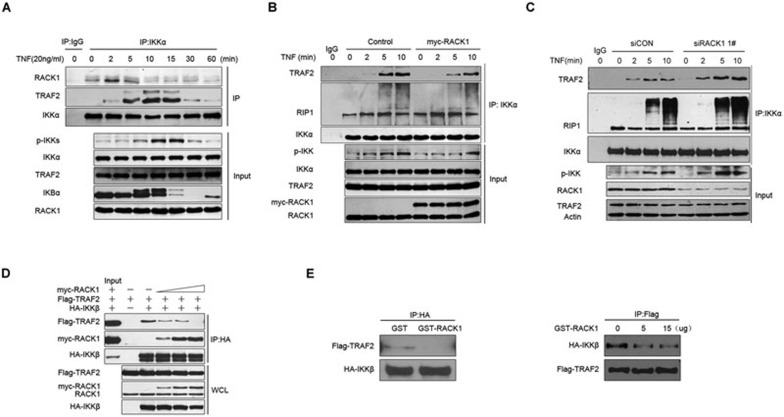
RACK1 directly interacts with IKKα and IKKβ
As a scaffold protein, RACK1 interacts with various proteins, including signaling transduction complexes. We further investigated whether RACK1 negatively regulated NF-κB signaling through a direct interaction with the IKK complex. To test this hypothesis, we first examined whether RACK1 could interact with the three subunits of the IKK complex. The results of exogenous, co-immunoprecipitation assays showed each subunit of the IKK complex was able to interact with RACK1 (Figure 3A), although all of the molecules were introduced by overexpression. We also examined the interaction between RACK1 and IKKs via an in vitro binding assay. Endogenous IKKα and IKKβ could be pulled down by GST-RACK1, whereas the regulator subunit, IKKγ, showed no detectable interaction with GST-RACK1 (Figure 3B). This observation indicated that RACK1 associated with the IKK complex via direct binding to IKKα and IKKβ. To exclude the possibility that abundant monomer IKKα and IKKβ in the cell lysate prevented access of GST-RACK1 to IKKγ, we purified the GST-IKKγ fusion protein and examined whether it could interact with RACK1 via an in vitro binding assay. As shown in Supplementary information, Figure S3A, neither GST nor GST-IKKγ could bind to the His-RACK1 protein, which further confirmed our observation. To verify whether the interaction between RACK1 and IKKs occurred under physiological conditions, we conducted endogenous coimmunoprecipitation assays using 293T and RAW264.7 cells (Figure 3C). In addition, we monitored the dynamics of the interaction between RACK1 and IKKs upon TNF treatment. The association of these proteins was enhanced 2 to 5 min after TNF stimulation, and began to decrease to the basal level after 1 h (Figure 3D). These results identified a direct interaction between RACK1 and the IKK complex under physiological conditions, with dynamic alteration after TNF stimulation.
Figure 3.
RACK1 interacted with IKKα and IKKβ. (A) Overexpressed RACK1 interacted with the overexpressed IKK complex. Myc-RACK1-293T stable cell lines were transfected with the indicated plasmids. Twenty-four hours later, the cells were harvested, and immunoprecipitated with the anti-HA antibody and analyzed by immunoblotting with anti-Flag antibody. (B) GST-RACK1 pulled down endogenous IKKα and IKKβ, but not IKKγ. 293T cell extracts were incubated with GST alone or GST-RACK1 and then immunoprecipitated with Sepharose beads. Immunoblots of the immunoprecipitates were analyzed with immunoprecipitated IKK antibodies. (C) RACK1 physically interacted with IKKα and IKKβ. 293T or RAW264.7 cell extracts were immunoprecipitated with mouse IgM or anti-RACK1 antibody and then subjected to immunoblot analysis of the endogenous IKK proteins. (D) RACK1 interacted with IKK in a TNF-triggered manner. 293T cells were treated with TNF (10 ng/ml) for the indicated time, and the cells were then harvested for immunoprecipitation with mouse IgM or anti-RACK1 antibody. The interacting IKK proteins were analyzed by immunoblotting.
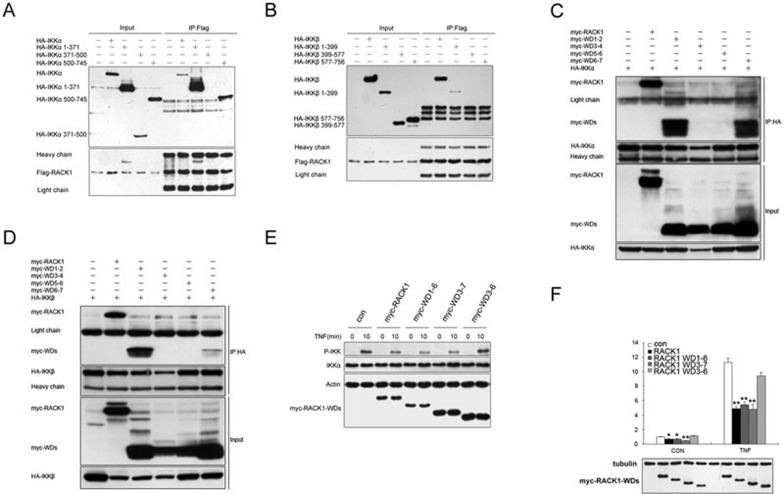
Mapping the RACK1-IKK interaction domains
As reported previously, IKKα and IKKβ have three functional domains: the amino-terminal kinase, leucine zipper and carboxy-terminal helix-loop-helix (HLH) domains. To identify the docking sites within the IKKs for RACK1, we performed co-immunoprecipitation assays using several deletion mutants of IKKα and IKKβ (Supplementary information, Figure S4). As shown in Figure 4A, like the full-length IKKα, the kinase (amino acids 1-371) and helix-loop-helix domains (amino acids 500-745) of the IKKα mutants immunoprecipitated with Flag-RACK1. However, only the kinase domain (amino acids 1-399) of the IKKβ mutant immunoprecipitated with Flag-RACK1 (Figure 4B). These results indicated that the kinase domains of IKKα and IKKβ were critical for their interaction with RACK1, and the HLH domain of IKKα was also required.
Figure 4.
RACK1 inhibited IKK phosphorylation and NF-κB activation by interacting with IKK. (A, B) The Flag-RACK1 293T stable cell line was transfected with HA-tagged IKKα or IKKβ or various IKKα or IKKβ mutants; lysates were immunoprecipitated with anti-Flag antibody (mouse IgG) and analyzed by immunoblotting with anti-HA antibody (Rabbit IgG). (C, D) 293T cells were transfected with HA-tagged IKKα or IKKβ and myc-tagged wild-type RACK1 or RACK1 WD deletion mutants; lysates were immunoprecipitated with anti-HA antibody and analyzed by immunoblotting with anti-myc antibody. (E) Immunoblot analysis of the effects of overexpression of RACK1 and RACK1 WD deletion mutants on endogenous IKK phosphorylation after treatment with TNF (5 ng/ml) for 0 to 10 min. (F) Luciferase reporter assay of the inhibition of NF-κB activation in 293T cells transfected with RACK1 or RACK1 WD deletion mutants then untreated or treated with TNF (10 ng/ml) for 6 h. Graphs show the mean ± SD, n = 3. *P< 0.05; **P< 0.01.
To determine which WD40 domains of RACK1 are responsible for its interaction with IKKs, we generated several deletion mutants of RACK1 consisting of WD1-2, WD3-4, WD5-6 and WD6-7. The result of the co-immunoprecipitation experiments indicated that WD1-2 and WD6-7 of RACK1 interacted with IKKα, and only WD1-2 of RACK1 could interact with IKKβ (Figure 4C and 4D). Next, we tested whether the inhibitory effects of RACK1 on NF-κB activation depended on its interaction with IKKs. We constructed several deletion mutants of RACK1 lacking the docking sites for IKKs, including WD1-6, WD3-7 and WD3-6. Of the three mutants, due to the lack of docking sites for both IKKs, WD3-6 could not interact with either IKKα or IKKβ (Data not shown). We overexpressed RACK1 and its mutants in 293T cells and treated the cells with TNF. Similar to the wild-type RACK1, the WD1-6 and WD3-7 RACK1 mutants repressed TNF-induced IKK phosphorylation, while the WD3-6 RACK1 mutant had no effect on IKK phosphorylation (Figure 4E). Consistent with these results, the WD1-6 and WD3-7 RACK1 mutants inhibited the reporter activity of the NF-κB-responsive promoter, while the WD3-6 RACK1 mutant showed no influence on NF-κB-responsive reporter activity under basal or TNF-stimulation conditions (Figure 4F). Thus, the direct interaction between RACK1 and IKKs is crucial for the inhibitory effects of RACK1 on IKK phosphorylation and TNF-mediated NF-κB activation.
RACK1 interferes with the TNF-induced recruitment of IKK complex to TRAF2
As previously reported, the IKK complex is rapidly phosphorylated at the signaling transduction platform around TNFR1 upon TNF stimulation8. Therefore, we sought to determine whether RACK1 was simultaneously recruited. As shown by the result of immunoprecipitation assays in TNFR1-overexpressing 293T cells, IKK began to be recruited to Flag-TNFR1 after 2 min upon TNF treatment and disassociated from the platform after 10 min, while RACK1 persistently associated with TNFR1 (Supplementary information, Figure S5). This observation was in agreement with previous studies showing that RACK1 associated with TNFR128,29. These results suggested that RACK1 might modulate the IKK phosphorylation process at the TNFR1 signaling complex. As reported previously, after being triggered by TNF, the IKK complex was rapidly recruited to TNFR1 by TRAF2 within 5 to 10 min. The strongest interaction between RACK1 and the IKK complex occurred at 2 to 5 min after TNF stimulation (Figure 3D), which prompted us to determine whether RACK1 played an inhibitory role at this early stage of TNF treatment. To explore this, we performed immunoprecipitation experiments with endogenous protein using an IKKα-specific antibody. Upon TNF stimulation, enhanced interaction of RACK1 and the IKK complex could be detected as early as 2 min and was sustained to 5 min. Meanwhile, a large amount of IKK complex began to be recruited by TRAF2 at 5 min, which led to strong phosphorylation of IKK at 10 min (Figure 5A). These results suggested that RACK1 might play an important role in the early process of IKK complex recruitment. Furthermore, overexpression of RACK1 impaired the association between IKK and TRAF2 but did not affect the interaction between IKK and RIP1, which is another important mediator that stabilizes the IKK complex around the TNFR1 signaling complex (Figure 5B). In contrast, during signaling complex assembly around the TNFR1, in RACK1-knockdown cells TRAF2 but not RIP1 promptly recruited more IKK complexes, which were subsequently phosphorylated (Figure 5C). All of these observations suggested that RACK1 exerted its inhibitory effect on NF-κB activation by interfering with the interaction between the IKK complex and TRAF2. To further address this point, we performed coimmunoprecipitation assays in 293T cells co-expressing HA-tagged IKKβ and Flag-tagged TRAF2 as well as different amounts of myc-tagged RACK1. As shown in Figure 5D, the amount of myc-RACK1 that precipitated with IKKβ increased when exogenous myc-RACK1 increased, and immunoprecipitation of TRAF2 with IKKβ gradually decreased. Moreover, results of the GST pull-down assays also showed that GST-RACK1 could directly interfere with the interaction between IKKβ and TRAF2 in vitro (Figure 5E). All of these data supported the notion that RACK1 represses NF-κB activation by directly interfering with TRAF2-mediated recruitment of the IKK complex during TNF-induced signaling complex assembly.
Figure 5.
RACK1 interfered with TRAF2-mediated recruitment of the IKK complex. (A) 293T cells were treated with TNF (10 ng/ml) for 0 to 60 min; the lysate was immunoprecipitated with mouse anti-IKKα antibody. (B) 293T cells were transfected with control vectors or myc-RACK1 vectors, and 24 h later, the cells were treated with TNF (10 ng/ml) for the indicated time. The lysates were immunoprecipitated with mouse anti-IKKα antibody. (C) Knockdown of RACK1 enhanced the TRAF2-mediated recruitment of the IKK complex. (D) 293T cells were transfected with the indicated vectors, and 24 h later, the lysates were immunoprecipitated with mouse anti-HA antibody. (E) 293T cells were transfected with HA-IKKβ and Flag-TRAF2 vectors, and 24 h later, the lysates were incubated with purified GST protein (10 μg) or GST-RACK1 protein (10 μg). After 4 h of incubation, half of the lysate was immunoprecipitated with mouse anti-HA antibody, and the other half was immunoprecipitated with mouse anti-Flag antibody.
RACK1 regulates the sensitivity of NF-κB activation in response to TNF
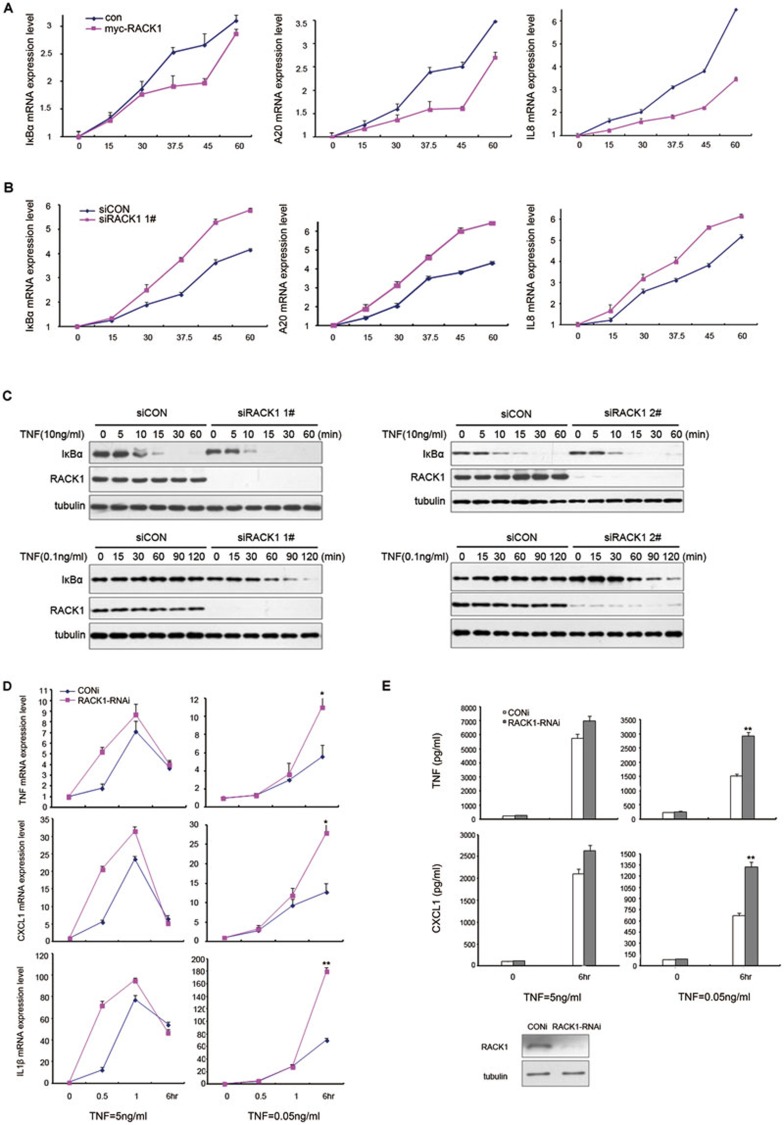
The above data showed that knockdown of RACK1 accelerated the TNF-triggered phosphorylation process of IKK (Figure 2D), but it did not extend the duration of this process, which was controlled by other negative feedback regulators such as A20 and CUEDC2. These observations suggested that RACK1 might control the response rate of TNF-induced NF-κB activation. To address this possibility, we manipulated the protein level of RACK1 in 293T cells and then treated the cells with TNF. By assessing the mRNA level of NF-κB target genes, we found that overexpression of RACK1 retarded the induction of all NF-κB-responsive genes, including A20, IκBα, TNFα and IL8 (Figure 6A and Supplementary information, Figure S6A). Consistent with these results, knockdown of RACK1 promoted the expression of TNF-induced target genes (Figure 6B and Supplementary information, Figure S6B). Considering the more dramatic elevation of NF-κB-responsive promoter transcriptional activity by knockdown of RACK1 in the resting state compared to after TNF treatment (Figure 1D), we hypothesized that the inhibitory effects of RACK1 on NF-κB activation may be more prominent under conditions of low levels of TNF. To test this hypothesis, we treated RACK1-knockdown cells with different concentrations of TNF and examined the rate of IκBα degradation triggered by TNF, which was the index of the activation rate of NF-κB. Consistent with previous data, when cells were treated with a commonly used concentration of TNF (10 ng/ml), knockdown of RACK1 accelerated the degradation of IκBα, and most of the IκBα protein had been degraded in RACK1 knockdown cells by 15 min (Figure 6C). When stimulating the RACK1-knockdown cells with a low concentration of TNF (0.1 ng/ml), IκBα gradually degraded from 60 to 120 min. By contrast, the protein level of IκBα remained unchanged during the same treatment in control cells (Figure 6C). These data indicated that RACK1 might be more essential for the regulation of NF-κB signaling in response to low concentrations of TNF.
Figure 6.
RACK1 regulates the sensitivity of NF-κB activation and inflammatory reactions induced by TNF. (A) Overexpression of RACK1 postponed NF-κB target gene expression. Human 293T cells were transfected with control vectors or myc-RACK1 vectors and, 24 h later, were treated with TNF (10 ng/ml) for 0 to 60 min. The mRNA expression level of A20, IκBα, and IL8 was analyzed by RT-PCR. (B) Knockdown of RACK1 accelerated NF-κB-target gene expression. Human 293T cells were infected with control siRNA or RACK1-specific siRNA lentivirus and, 72 h later, were treated with TNF (10 ng/ml) for 0 to 60 min. The mRNA level of IκBα, A20 and IL8 was analyzed by RT-PCR. (C) Knockdown of RACK1 promoted the IκBα degradation induced by different concentrations of TNF. Control cells or RACK1-knockdown cells were treated with TNF (10 ng/ml or 0.1 ng/ml) for the indicated time. (D) Knockdown of RACK1 promoted the inflammatory gene expression induced by different concentrations of TNF. RAW264.7 cells were transfected with control or RACK1-specific siRNAs and, 48 h later, were treated with different concentrations of TNF. (E) Knockdown of RACK1 increased the TNF-induced production of TNFα and CXCL1 in primary mouse macrophages. The primary macrophages were transfected with control or RACK1-specific siRNAs, and 48 h later, were treated with different concentrations of TNF. Graphs show the mean ± SD, n = 3. *P< 0.05; **P< 0.01.
TNF is a well-known inducer of inflammatory reactions that triggers the expression of a series of cytokines. Challenging the cells with a high concentration of TNF results in rapid expression of many cytokines. However, when confronting a low level of TNF, cells must respond properly to this “noise” stimulation. To explore this, we examined the expression of NF-κB target genes in response to stimulation with different doses of TNF in RAW264.7 cells and primary mouse peritoneal macrophages via RT-PCR and enzyme-linked immunoassays (ELISAs). The results of the RT-PCR assays showed that the expression of NF-κB-targeted inflammatory genes (including TNFα, IL1β, CXCL1 and CXCL2) and negative feedback genes (IκBα and A20) was induced more quickly within the first hour of exposure to a high concentration TNF (5 ng/ml) in RACK1-knockdown cells (Figure 6D and Supplementary information, Figure S6C and S6D), which was consistent with the previous observation in 293T cells. When treated with a relatively low concentration of TNF (0.05 ng/ml), RACK1-knockdown cells finally transcribed more NF-κB target genes after 6 h in response to this weak stimulation compared to control cells (Figure 6D and Supplementary information, Figure S6D). These observations indicated that RACK1 controlled the activation rate and intensity of the NF-κB signaling pathway when faced with high and low concentrations of TNF, respectively. To further clarify the physiological significance of RACK1 in the regulation of the inflammatory response, we analyzed TNF-induced production of TNFα and CXCL1 in peritoneal macrophages. The primary macrophages were transfected with control siRNA or RACK1-specific siRNA, and 48 h later, the cells were challenged with TNF at different concentrations (5 ng/ml or 0.05 ng/ml). Consistent with the above observation, inhibition of RACK1 expression led to a modest increase in TNFα and CXCL1 production induced by high concentrations of TNF (Figure 6E). By contrast, knockdown of RACK1 resulted in a significant increase in both TNFα and CXCL1 production when induced by a low concentration of TNF (Figure 6E). Collectively, these results demonstrated that RACK1 regulated the inflammatory response to different extents of TNF stimulation, and raised the possibility that RACK1 functions as a rheostat of TNFα stimulation.
Discussion
As a number of studies have reported, the accurate regulation of IKK activity is a key step in the activation of the NF-κB pathway by extracellular stimulators such as TNF and IL1β30,31,32. After stimulation, IKK is recruited immediately to the newly assembled signaling complex around membrane receptors, where a platform is provided for phosphorylation and subsequent activation of the IKK complex33,34,35. Thus, the regulation of IKK complex recruitment is critical for an extracellular stimulation-induced NF-κB activation. As previously reported, the process of IKK recruitment is executed by TRAF2/5 and RIP1 around the TNFR136. Upon TNF challenge, TRADD first responds to the stimulation and initiates the assembly of a signaling complex mainly containing TRAFs and RIP1. After their respective modifications, the TRAFs directly recruit the IKK complex, whereas the newly modified K63 ubiquitination chain of RIP1 is recognized by IKKγ. These two types of protein-protein interactions fix the IKK complex at the signaling transduction platform for its subsequent phosphorylation. Although many previous studies have reported mechanisms of IKK activation, the negative regulators responsible for IKK activation have not been identified until recently. For example, A20, which edits the ubiquitin chain of RIP1, terminates IKK activation by interfering with the interaction between the IKK complex and RIP113. Another negative regulator, CUEDC2, recruits PP1c to dephosphorylate the IKK complex and maintains IKK in an inactive state14. Both of these negative regulators exert their inhibitory effects through opposing actions to those activating IKK in a feedback loop model. Here, we have reported that RACK1, a scaffold protein with little known immunological function, responds to TNF treatment immediately and represses IKK activity by interfering with the recruitment of the IKK complex by TRAF2. We have shown that RACK1 executes the opposing action of IKK recruitment via direct interaction with IKKα and IKKβ. This discovery extends our knowledge of the deactivation mechanisms of IKKs.
RACK1 was first recognized as a scaffold protein of PKC18 and has been demonstrated to interact with a range of signaling transduction complexes. By in vivo and in vitro protein binding assays, we have identified RACK1 as a novel IKK signalosome component. Through an interaction with the IKK complex, RACK1 represses IKK activity and the subsequent NF-κB activation triggered by TNF. More notably, the interaction between RACK1 and IKK is regulated in a TNF-triggered manner. At the early stages of TNF challenge (from 2 to 5 min), the interaction between RACK1 and the IKK complex rapidly increases and begins to decrease to basal level. The dynamic interaction between RACK1 and IKK regulates the process of IKK activation, which is critical for ensuring precise cellular responses to extracellular stimuli. Unlike the other two negative regulators, A20 and CUEDC2, which begin to downregulate IKK activity after full activation of the IKK complex and control the activation period of NF-κB signaling, RACK1 determines the sensitivity of NF-κB activation by modulating the rate and extent of IKK activation at the beginning of the response to TNF stimulation. However, how RACK1 responds to TNF treatment to change its affinity for the IKK complex is a question that remains to be addressed. As previous studies reported, RACK1 could become phosphorylated at different amino acid residues through extracellular signaling21,22. Thus, we hypothesized that TNF treatment might induce RACK1 phosphorylation at specific sites that would modulate RACK1's function in this signaling pathway. Therefore, we constructed 11 RACK1 phosphorylation mutants, including all of the tyrosine mutants and several of the serine mutants that were previously reported or predicted using software. Unfortunately, all of the RACK1 mutants showed no defect in inhibiting TNF-induced NF-κB reporter activity or IKK phosphorylation, and all of the mutants could bind HA-IKKα as strongly as wild-type RACK1 (Supplementary information, Figure S7A, S7B and S7C). Thus, we speculate that other phosphorylation sites or modifications of RACK1 could exist for regulating its modulation of the IKK complex.
The immune system is constantly challenged with different spatiotemporal stimuli and immune cells must respond to extracellular signals appropriately to effectively execute biological functions. Therefore, interruption by noise signals should be avoided to economize energy for cells. A previous study reported that the responses of cells treated with low doses of TNF depended on pre-existing variation in cell sensitivities37, which implies the existence of regulations modulating the sensitivity in response to TNF challenge. In the present study, we have demonstrated that RACK1 acts as such a regulator to modulate the sensitivity of the TNF-triggered NF-κB activation. When challenged by a low-dose TNF stimulus, normal cells would recognize this stimulus as a noise signal and respond faintly; however, once the expression of RACK1 was repressed, the cells would become more sensitive to such signals, which could lead to an unnecessary waste of cellular energy.
In conclusion, we revealed a novel mechanism that modulates the sensitivity of the NF-κB signaling pathway. We found that RACK1, by acting as a component of the IKK signalosome, plays a key role in inhibiting IKK activity by interfering with TRAF2-mediated recruitment of the IKK complex. Moreover, our results suggest that RACK1 can effectively attenuate the NF-κB activation induced by a low concentration of TNF and functions as a buffering factor for TNF-triggered NF-κB activation.
Materials and Methods
Cell culture, transfection and luciferase reporter assays
The details of cell culture, transfection and reporter gene assay were available in Supplementary information, Data S1.
Plasmid construction
The open reading frames of human RACK1, TNFR1 and TRAF2 cDNAs were cloned into the eukaryotic expression vector pCMV-myc and pCMV-Tag2B (Invitrogen), respectively. The expression vectors of IKKα, IKKβ and IKKγ were kindly gifted by Prof. Xue-Min Zhang (National Center of Biomedical Analysis, China).
RNA extraction and real-time PCR (RT-PCR) analysis
RNA extraction and RT-PCR analysis were performed as described previously22. The details of procedures were in Supplementary information, Data S1.
Nuclear protein extraction
Nuclear extracts were prepared by the mini-extraction method as described previously22. The procedures were detailed in Supplementary information, Data S1.
RNAi-mediated knockdown of RACK1
In our studies, we knocked down human RACK1 by using lentivirus (FG12 lentiviral vector) to produce small double-stranded RNA (siRNA) as described previously22. Two sequences (1# and 2#) targeting the human mRNA of RACK1 were available in Supplementary information, Data S1. Further, to knock down mouse RACK1, we used siRNA to transfect RAW264.7 cells and primary macrophages by Lipofectamine RNAi MAX (Invitrogen). The sequence targeting the mouse mRNA of RACK1 was available in Supplementary information, Data S1.
Western blot analysis and immunoprecipitation
Western blot analysis and immunoprecipitation were performed as described previously17. Details were available in Supplementary information, Data S1. The proteins of human TNFα and IL1β were purchased from R&D Systems. Antibodies against myc, p65, nucleporin62, tubulin and IKKγ (Rabbit IgG) were purchased from Santa Cruz Biotechnology. Antibodies against HA and Flag (both Rabbit IgG) were purchased from Sigma. Antibodies to RACK1, JNK, phosphor-JNK (pT183/Py185), IKKα and IKKγ were purchased from BD Transduction Laboratories, antibodies to phosphor-IKKα (Ser180)-IKKβ (Ser181), phosphor-IκBα, IκBα, IKKα, IKKβ, TRAF2 and RIP1 were purchased from Cell Signaling Technology.
GST precipitation assay
The fusion protein of GST, GST-RACK1 and GST-IKKγ were prepared as described21. His-RACK1 protein was purchased from Abcam. Approximately 5 μg GST fusion protein was added to each total lysate from 293T cells, followed by incubation for 4 h with gentle rotation, and then beads were added and incubated for another 2h. Beads were washed three times with cell lysis buffer, and proteins were eluted with 2× SDS sample buffer and analyzed by immunoblot. For in vitro GST pull-down, approximately 1 μg His-RACK1 protein was incubated with 1 μg GST fusion protein for 4 h with gentle rotation, and then the beads were added and incubated for another 2h. Beads were washed three times with GST pull-down buffer and the beads were eluted three times with Glutathione.
Macrophage preparation
Primary macrophage cells from male C57/BJ mice were plated in 6-well plates with 10% (vol/vol) FBS in DMEM medium. On day 2, adherent cells were transfected with control siRNA or RACK1-specific siRNA, and after RACK1 knockdown, the cells were used for various experiments. All studies involving mice were done according to national and institutional guidelines. The details of primary macrophage purification were available in Supplementary information, Data S1.
ELISA
Mouse primary macrophages were used for measurement of cytokine production. The cells were treated with TNF (5 ng/ml or 0.05 ng/ml) for 6 h and then supernatants were collected. The concentrations of TNF and CXCL1 were measured by ELISA according to manufacturer's instructions (Sunny ELISA Systems). Values were calculated on the basis of a standard curve constructed for each assay.
Abbreviations
RACK1 (receptor for activated C kinase 1); NF-κB (nuclear factor kappa-B); TNF-α (tumor necrosis factor-alpha); IKK (inhibitor of nuclear factor kappa-B kinase); IκBα (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha); TRAF2 (TNF receptor-associated factor 2); JNK (JUN N-terminal kinase); RIP1 (receptor (TNFRSF)-interacting serine-threonine kinase 1)
Acknowledgments
We are grateful to Dr Xue-Min Zhang (Academy of military medical sciences, Beijing) for IKK constructs, and to Dr Xiao-Fan Wang (Duke university, Durham) and Dr Dang-Sheng Li for their constructive suggestions on our work. This work was supported by the National Basic Research Program of China (2010CB912102, Ministry of Science and Technology Key Program (2012ZX10002009-017), the National Natural Science Foundation of China (81230058, 30930023, 31100551, 31201046, 81021002), CAS/SAFEA International Partnership Program for Creative Research Teams, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (SIBS2012004), and Technology Commission of Shanghai Municipality (12XD1405600).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
RACK1 repressed the activity of the NF-κB luciferase reporter and the expression of NF-κB target genes.
RACK1 inhibited the TNF signaling pathway by influencing the IκBα degradation induced by IKKβ.
Binding assays between RACK1 and the IKK complex.
Truncated mutants of IKKα and IKKβ that were used in mapping experiments.
RACK1 modulated IKK activity through the TNFR1 complex.
The expression of RACK1 in 293T and macrophages.
Various RACK1 mutants inhibited the TNF-induced NF-κB signaling pathway.
References
- Dixit V, Mak TW. NF-kappaB signaling: many roads lead to madrid. Cell. 2002;111:615–619. doi: 10.1016/s0092-8674(02)01166-2. [DOI] [PubMed] [Google Scholar]
- Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-(kappa)B activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, et al. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan P, Baltimore D. Sam68 is required for both NF-kappaB activation and apoptosis signaling by the TNF receptor. Mol Cell. 2011;43:167–179. doi: 10.1016/j.molcel.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang L, Dorf ME. PKC phosphorylation of TRAF2 mediates IKKalpha/beta recruitment and K63-linked polyubiquitination. Mol Cell. 2009;33:30–42. doi: 10.1016/j.molcel.2008.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Austenaa LM. A birthday gift for TRADD. Nat Immunol. 2008;9:1015–1016. doi: 10.1038/ni0908-1015. [DOI] [PubMed] [Google Scholar]
- Hong S, Lim S, Li AG, et al. Smad7 binds to the adaptors TAB2 and TAB3 to block recruitment of the kinase TAK1 to the adaptor TRAF2. Nat Immunol. 2007;8:504–513. doi: 10.1038/ni1451. [DOI] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- Shim JH, Xiao C, Paschal AE, et al. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 2005;19:2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shembade N, Harhaj NS, Parvatiyar K, et al. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat Immunol. 2008;9:254–262. doi: 10.1038/ni1563. [DOI] [PubMed] [Google Scholar]
- Li HY, Liu H, Wang CH, et al. Deactivation of the kinase IKK by CUEDC2 through recruitment of the phosphatase PP1. Nat Immunol. 2008;9:533–541. doi: 10.1038/ni.1600. [DOI] [PubMed] [Google Scholar]
- McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol Pharmacol. 2002;62:1261–1273. doi: 10.1124/mol.62.6.1261. [DOI] [PubMed] [Google Scholar]
- Adams DR, Ron D, Kiely PA. RACK1, a multifaceted scaffolding protein: structure and function. Cell Commun Signal. 2011;9:22. doi: 10.1186/1478-811X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Deng YZ, Zhao JS, et al. RACK1 promotes non-small-cell lung cancer tumorigenicity through activating sonic hedgehog signaling pathway. J Biol Chem. 2012;287:7845–7858. doi: 10.1074/jbc.M111.315416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Chen CH, Caldwell J, Jamieson L, Orr E, Mochly-Rosen D. Cloning of an intracellular receptor for protein kinase C: a homolog of the beta subunit of G proteins. Proc Natl Acad Sci USA. 1994;91:839–843. doi: 10.1073/pnas.91.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang BY, Conroy KB, Machleder EM, Cartwright CA. RACK1, a receptor for activated C kinase and a homolog of the beta subunit of G proteins, inhibits activity of src tyrosine kinases and growth of NIH 3T3 cells. Mol Cell Biol. 1998;18:3245–3256. doi: 10.1128/mcb.18.6.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usacheva A, Smith R, Minshall R, et al. The WD motif-containing protein receptor for activated protein kinase C (RACK1) is required for recruitment and activation of signal transducer and activator of transcription 1 through the type I interferon receptor. J Biol Chem. 2001;276:22948–22953. doi: 10.1074/jbc.M100087200. [DOI] [PubMed] [Google Scholar]
- Deng YZ, Yao F, Li JJ, et al. RACK1 suppresses gastric tumorigenesis by stabilizing the beta-catenin destruction complex. Gastroenterology. 2012;142:812–246. doi: 10.1053/j.gastro.2011.12.046. [DOI] [PubMed] [Google Scholar]
- Li G, Ji XD, Gao H, et al. EphB3 suppresses non-small-cell lung cancer metastasis via a PP2A/RACK1/Akt signalling complex. Nat Commun. 2012;3:667. doi: 10.1038/ncomms1675. [DOI] [PubMed] [Google Scholar]
- Kiely PA, Baillie GS, Lynch MJ, Houslay MD, O'Connor R. Tyrosine 302 in RACK1 is essential for insulin-like growth factor-I-mediated competitive binding of PP2A and beta1 integrin, and for tumor cell proliferation and migration. J Biol Chem. 2008;283:22952–22961. doi: 10.1074/jbc.M800802200. [DOI] [PubMed] [Google Scholar]
- Zhang W, Cheng GZ, Gong J, et al. RACK1 and CIS mediate the degradation of BimEL in cancer cells. J Biol Chem. 2008;283:16416–16426. doi: 10.1074/jbc.M802360200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bergami P, Habelhah H, Bhoumik A, Zhang W, Wang LH, Ronai Z. RACK1 mediates activation of JNK by protein kinase C (corrected) Mol Cell. 2005;19:309–320. doi: 10.1016/j.molcel.2005.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol. 2008;10:1324–1332. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- Nakashima A, Chen L, Thao NP, et al. RACK1 functions in rice innate immunity by interacting with the RAC1 immune complex. Plant Cell. 2008;20:2265–2279. doi: 10.1105/tpc.107.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipp S, Puchert M, Adam-Klages S, et al. The Polycomb group protein EED couples TNFR1 to neutral sphingomyelinase. Proc Natl Acad Sci USA. 2010;107:1112–1117. doi: 10.1073/pnas.0908486107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkasowa AE, Adam-Klages S, Kruse ML, et al. Interaction with factor associated with neutral sphingomyelinase activation, a WD motif-containing protein, identifies receptor for activated C-kinase 1 as a novel component of the signaling pathways of the p55 TNF receptor. J Immunol. 2002;169:5161–5170. doi: 10.4049/jimmunol.169.9.5161. [DOI] [PubMed] [Google Scholar]
- Hacker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006;357:re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109 Suppl:S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Koseki T, Lin J, et al. An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J Biol Chem. 2000;275:27823–27831. doi: 10.1074/jbc.M003415200. [DOI] [PubMed] [Google Scholar]
- Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- Micheau O, Lens S, Gaide O, Alevizopoulos K, Tschopp J. NF-kappaB signals induce the expression of c-FLIP. Mol Cell Biol. 2001;21:5299–5305. doi: 10.1128/MCB.21.16.5299-5305.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devin A, Cook A, Lin Y, Rodriguez Y, Kelliher M, Liu Z. The distinct roles of TRAF2 and RIP in IKK activation by TNF-R1: TRAF2 recruits IKK to TNF-R1 while RIP mediates IKK activation. Immunity. 2000;12:419–429. doi: 10.1016/s1074-7613(00)80194-6. [DOI] [PubMed] [Google Scholar]
- Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RACK1 repressed the activity of the NF-κB luciferase reporter and the expression of NF-κB target genes.
RACK1 inhibited the TNF signaling pathway by influencing the IκBα degradation induced by IKKβ.
Binding assays between RACK1 and the IKK complex.
Truncated mutants of IKKα and IKKβ that were used in mapping experiments.
RACK1 modulated IKK activity through the TNFR1 complex.
The expression of RACK1 in 293T and macrophages.
Various RACK1 mutants inhibited the TNF-induced NF-κB signaling pathway.