Abstract
Purpose.
Determine the impact of rod photoreceptor-specific expression of Cre recombinase on the kinetics of phototransduction in the mouse eye and identify changes in gene expression that underlie any observed phenotypic differences.
Methods.
Transretinal ERG and single-cell suction electrode recordings were used to measure the kinetics of phototransduction in a mouse line exhibiting rod photoreceptor–specific Cre recombinase expression, and the results were compared with those from control non–Cre-expressing littermates. Gene expression changes were evaluated using RNA sequencing transcriptome analysis. The pattern of expression of Rgs9bp was determined by mapping sequencing reads to the mouse genome and performing 3′-rapid amplification of cDNA ends (3′-RACE).
Results.
Expression of the rod-specific iCre75 transgene was accompanied by accelerated phototransduction inactivation, likely due to overexpression of the Rgs9bp gene, which encodes the Rgs9 anchor protein (R9AP). R9AP upregulation stabilized the RGS9 GAP complex, altering phototransduction kinetics. 3′-Race identified an abundant, unexpected Rgs9bp-Prm1 fusion mRNA in Cre-expressing mouse retinas, which was determined to be derived from a second transgene present in the iCre75 line.
Conclusions.
Here we report the presence of a second, R9AP-expressing transgene in the iCre75 mouse line, leading to altered kinetics of phototransduction. These results highlight an important caveat that must be considered when utilizing this mouse line for rod photoreceptor–specific gene loss of function studies.
Keywords: rod photoreceptors, phototransduction, Cre recombinase, mouse, retina, R9AP
We report that the presence of a second R9AP-expressing transgene in the iCre75 mouse line influences the rate of recovery of phototransduction.
Introduction
Engineered chromosomal deletions driven by Cre-mediated recombination of tandem locus of X-over P1 recognition sequences to generate conditional knockout (cKO) mouse models constitute a powerful tool for cell-type and tissue-specific gene loss of function studies with diverse applications. Cre recombinase–dependent conditional knockout mouse technology has been used extensively in the visual system to evaluate the functional impact of gene loss on visual system development and function.1–12 Several mouse lines have been developed that exhibit photoreceptor-specific Cre expression.13–16 Specifically, the iCre75 mouse line has emerged as an invaluable tool for rod photoreceptor specific gene loss of function studies as it exhibits robust rod photoreceptor-specific Cre recombinase expression without any of the morphological and functional defects observed in other rhodopsin promoter-driven Cre lines.15,16
While evaluating the kinetics of phototransduction in rod photoreceptor-specific Dicer conditional knockout (cKO) mice, we identified an increase in the rate of phototransduction shutdown in iCre75-expressing mice. This kinetic defect was accompanied by a dramatic increase in expression of the Rgs9 anchor protein (R9AP). R9AP anchors the regulator of G-protein signaling 9 (RGS9) GTPase-accelerating protein (GAP) complex to outer segment disk membranes, both stabilizing the GAP complex against proteolysis and enhancing its activity by directing it to the appropriate subcellular location.17–21 Loss of R9AP function in mice or human patients results in decreased temporal resolution of visual signaling evidenced by defects in motion discrimination under bright light conditions, highlighting the importance of R9AP in the regulation of phototransduction recovery kinetics.17,20,22 In transgenic mice, overexpression of R9AP has been directly demonstrated to enhance the rate of phototransduction shutdown.23 Here we report that the rate of recovery of phototransduction is altered in iCre75 mice due to dysregulation of R9AP. Moreover, this dysregulation results from an additional R9AP-encoding transgene. These results highlight a need for caution when designing studies involving rod-specific cKO mice.
Methods
Animals
All animal procedures and experiments were approved by the Case Western Reserve University and Washington University in St. Louis animal care committees and conformed to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Rod photoreceptor–specific Dicer cKO mice were generated by crossing Dicerfl/fl mice24 (obtained from The Jackson Laboratory, Bar Harbor, ME) with iCre75 mice15 (a gift from Neena Haider, Harvard Medical School, Boston, MA), which express Cre recombinase specifically in rods driven by a fragment of the opsin gene promoter. Animals used for this study resulted from crossing cKO mice (Dicerfl/fl iCre75+) with control mice (Dicerfl/fl iCre75−) yielding 50% cKO mice and 50% control littermates. iCre75 mice (Dicer+/+iCre75+) were generated by crossing with C57BL/6J wild type mice (The Jackson Laboratory) yielding 50% iCre75 mice and 50% wild-type littermates. Genotyping was performed by PCR as described previously.15,24 Mice were housed in the animal resource center at the Case Western Reserve University School of Medicine where they were maintained on a standard chow diet in a 12-hour light (∼10 lux), 12-hour dark cycle.
Transretinal ERG and Single-Cell Recording
Mice were euthanized by CO2 asphyxiation after 6 to 24 hours dark adaptation. Eyes were enucleated under dim red light and retinas were dissected under infrared (IR) light with IR converters attached to the dissection microscope. Each retina was placed photoreceptor-side upward on a dark filter paper (HABG01300; Millipore Corp., Billerica, MA) attached to a custom-built specimen holder similar to that described previously by Nymark et al.25 Retinas were perfused with Ringer's solution containing (mM): Na+, 133.9; K+, 3.3; Mg2+, 2.0; Ca2+, 1.0; Cl−, 143.2; glucose, 10.0; EDTA, 0.01; HEPES, 12.0, buffered to pH 7.5 with NaOH. The solution was supplemented with 0.72 g/L Leibovitz culture medium L-15 (Sigma-Aldrich; St. Louis, MO) to improve retinal viability and with 40 μM DL-AP4 and 100 μM BaCl2 to isolate the photoreceptor component of the ERG signal. The temperature readings from the thermocouple near the retina were adjusted to 37°C by heating the perfusion solution just before the specimen holder. Light-dependent changes in the voltage across the retina were measured by two pellet electrodes connected to a differential amplifier (DP-311, Warner Instruments LLC, Hamden, CT). ERG signals were low pass filtered at 300 Hz (8-pole Bessel filter) and sampled at 10 kHz with a digitizer and data acquisition software (Digidata1440A and pClamp 9; Molecular Devices, Sunnyvale, CA). A cyan 505-nm LED light (SR-01-E0070, Luxeon Star; Quadica Developments, Inc., Brantford, Ontario, Canada) was aligned to produce a homogenous light spot that was larger than the effective measurement area (determined by the ∅0.5 mm lower electrode channel below the retina; see Fig. 1 in Nymark et al.25) at the plane of the retina. Short 1-ms light pulses were produced by a voltage-to-current converter (LDC210C, Thorlabs, Inc., Newton, NJ) controlled with analog voltage output of the digitizer together with data acquisition software (Molecular Devices). Light power was measured with a calibrated optometer (UDT S471; Gamma Scientific, San Diego, CA) and converted into intensity by dividing it with the light spot area measured at the level of the retina. Photon flux was converted into photoisomerizations of rhodopsin molecules (R*) per rod as described previously.26 An exponential function was fitted to normalized response amplitude data,
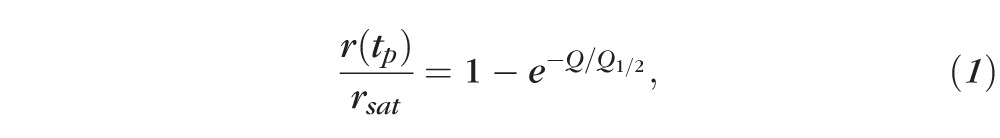 |
where r(tp) is the response amplitude measured at peak response, rsat is the maximal amplitude of the saturated photoresponse amplitude, and Q1/2 describes the flash intensity that produces 50% of the maximal response. Fractional sensitivity SF of rods to dim flashes producing less than 20% of the maximal response was calculated as
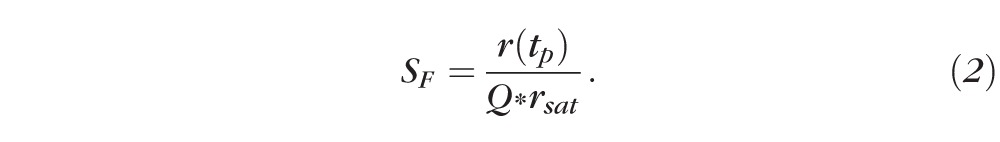 |
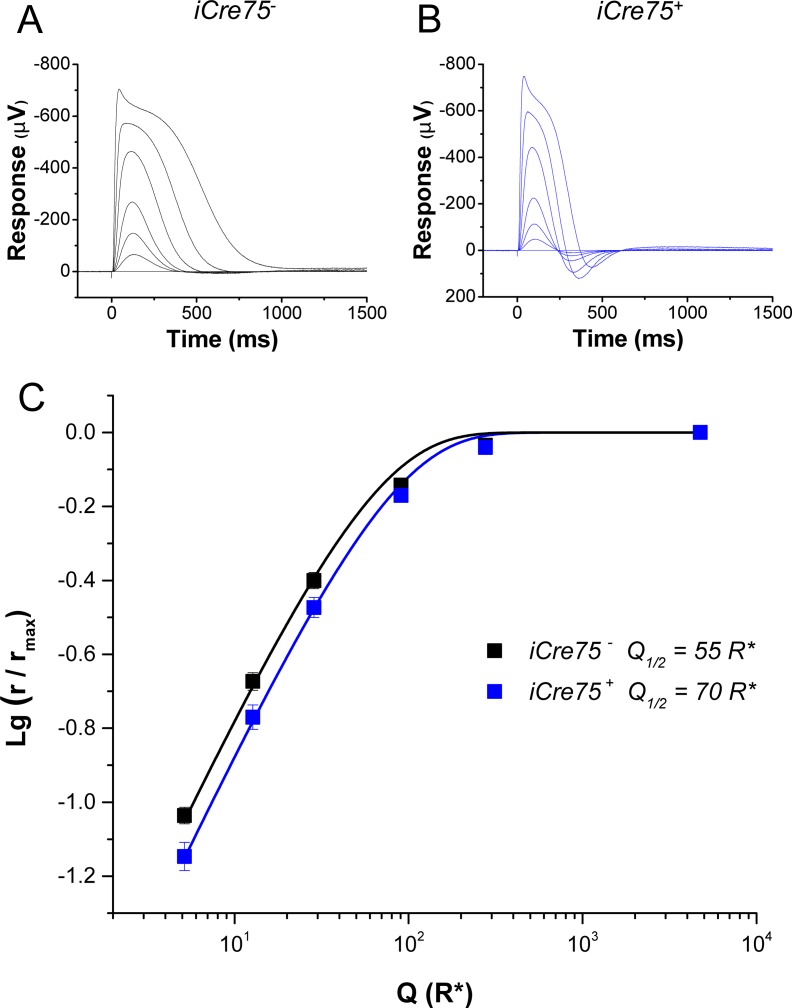
Figure 1.
Dark-adapted photoreceptor responses, recorded by transretinal ERG from isolated retinas of iCre75− (A) and iCre75+ (B) mice, to short 1-ms flashes of 505-nm light. (C) Population averaged response amplitudes (normalized to saturated response amplitude) as a function of flash strength in R* per rod for iCre75− (black squares, six retinas) and iCre75+ (blue squares, six retinas) mice. Fitting of an exponential function (Equation 1) to the r/rmax data yielded a half-saturating flash intensity, Q1/2, of 55 and 70 R* per rod for control and iCre75 mouse rods, respectively.
The estimated fractional single photon response was derived in a similar manner by dividing a response in linear range (r[tp] < 0.2*rsat) by Q and rsat. The dominant time constant of the rod photoresponse recovery, (τD), was determined by the slope of the linear fit to the (LnQ, Tsat) data, where Tsat is the time in saturation determined from saturated responses at the criterion level of 30% recovery.27
For single-cell recordings, the retina was dissected as described above and then chopped in small pieces with a razor blade. An aliquot was placed on the recording chamber, fit to the stage of an inverted microscope, and perfused with Locke solution (112 mM NaCl, 3.6 mM KCl, 2.4 mM MgCl2, 1.2 mM CaCl2, 10 mM HEPES, 20 mM NaHCO3, 3 mM Na2-succinate, 0.5 mM Na-glutamate, 10 mM glucose, pH 7.4) equilibrated with 95% O2/5% CO2 and heated from 34 to 37°C. Membrane current was recorded by drawing the outer segment of a single rod protruding from a piece of retina into a suction recording electrode. The recording electrode and the reference electrode placed in the bath adjacent to the retina were filled with a solution containing 140 mM NaCl, 3.6 mM KCl, 2.4 mM MgCl2, 1.2 mM CaCl2, 3 mM HEPES, 10 mM glucose, pH 7.4. Test flashes of 500-nm light and 20-ms duration were delivered from a calibrated light source controlled by computer-driven shutters. Dark current, Idark, was estimated from the amplitude of the saturated light response for each rod. Rod sensitivity was estimated from the flash intensity, I1/2, required to produce a half maximal response, derived by fitting the intensity-response data with the Naka-Rushton function
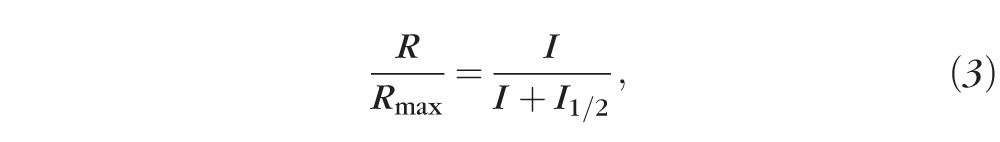 |
where R is the response amplitude, Rmax is the saturated rod response amplitude, and I is the test flash intensity. Integration time (Tintegr) was measured as the time integral of the normalized dim flash response, and the recovery time constant (τrec) was measured from the single exponential fit to the second half of the response shutoff phase. The dominant time constant (τD) was measured as for transretinal recordings. All analyses were performed with data acquisition and analysis software (ClampFit; Molecular Devices and Origin; OriginLab Corp., Northampton, MA). Statistical significance was determined using a two-tailed Student's t-test with values of P < 0.05 considered different.
RNAseq Transcriptome Analysis
Mice were euthanized by cervical dislocation between 7:30 and 9:00 AM. Eyes were enucleated and retinas were carefully dissected out and immediately placed in stabilization reagent (RNALater; Qiagen, Venlo, Limburg). Mouse retina sequencing libraries were prepared as previously described.28 Pooled total RNA samples from eight mouse retinas were used for each library preparation. Libraries were made from three independent biological replicates for each genotype. Each library was run on one lane of a sequencer (Illumina HiScan; Illumina, Inc., San Diego, CA) in the CWRU Genomics Core Facility using 50-bp single-end sequencing. Data were processed and aligned with the UCSC mouse genome assembly and transcript annotation (mm9) using the Genomic Short-read Nucleotide Alignment Program. Raw read counts for mouse RefSeq genes were extracted with the HTSeq-count program and used for calculation of reads per kilobase per million mapped reads (RPKM) values.
Antibodies and Immunoblotting
For immunoblotting analysis, retinas were dissected, put into 1× RIPA buffer (25 mM Tris, pH 7.6, 150 mM NaCl, 1% [vol/vol] NP-40, 1% [vol/vol] sodium deoxycholate, and 0.1% [wt/vol] SDS), triturated by pipetting, and lysed by sonication. Samples were normalized by protein concentration and resolved by SDS-PAGE on 4% to 12% gradient gels. Immunoblots were developed using antibodies against β-tubulin (mouse monoclonal; Sigma-Aldrich); Cre recombinase (mouse monoclonal 2D8, Millipore Corp.); R9AP (rabbit polyclonal, provided by Vadim Arshavsky, Duke University, Durham, NC); Gβ5 (goat polyclonal G4718, a gift from Theodore Wensel, Baylor College of Medicine, Houston, TX); or RGS9 (rabbit polyclonal R4433, also from Theodore Wensel).
3′-RACE and PCR Analysis
To determine the sequence of the 3′-end of Rgs9bp mRNA, we dissected retinas from iCre75 and cKO mice along with control littermates and extracted total RNA using an RNA kit (NucleoSpin RNA II kit; Macherey-Nagel, Düren, Germany) and prepared cDNA with Superscript III Reverse transcriptase (Invitrogen) using primer 1 (5′-gcctagctcgagtttttttttttttttttttt-3′). The 3′-end of Rgs9bp cDNA was then amplified by PCR using primer 1 and the Rgs9bp-specific primer 2 (5′-gcgcaaaagcttcggtagaggagcgcgcgggc-3′), which anneals 116 bp upstream of the termination codon. PCR amplification (60°C annealing temperature, 2-minute extension time) resulted in a product of around 450 bp with cDNA derived from Cre-expressing retinas. This fragment was cloned between the XhoI and HindIII sites of pBluescriptSK+ plasmid. We then purified plasmid DNA and subjected it to Sanger sequencing. Five clones each were sequenced from cKO and iCre75 mice. To determine whether a mouse Opsin promoter fragment was present upstream of the Rgs9bp coding DNA sequence (CDS) in iCre75-containing mice, PCR was performed with a forward primer (5′-gcgcaaaagcttggcagacaagatgagacaccctttc-3′) annealing upstream of the mouse Opsin gene and a reverse primer (5′-caagagtctcgaggaaggtggctcccagcgctagtg-3′) annealing within the CDS of Rgs9bp. PCR amplification, employing mouse tail–derived genomic DNA (55°C annealing temperature, 1-minute extension time) yielded a product around 650 bp for iCre75 and cKO mice. PCR products derived from 2 cKO and 2 iCre75 mice were subjected to Sanger sequencing.
Results
iCre75 Mice Exhibit Altered Light Response Kinetics
We began the physiological analysis of Cre-expressing mice by performing transretinal recordings to evaluate their rod function. Control iCre75− (C57Bl/6) mice produced robust rod-driven responses (Fig. 1A) with normal sensitivity (Fig. 1C). Producing a half-saturated light response required the isomerization of 55 rhodopsin molecules (R*) per rod (Fig. 1C, Table 1). The iCre75+ retinas also produced robust rod responses (Fig. 1B) with comparable maximal amplitudes (Table 1). However, the sensitivity of iCre75+ retinas was slightly reduced (Fig. 1C) and they required 70 R* for a half-saturated response (Table 1). Another measure of sensitivity, SF, representing the amplitude of the response per R* to a dim flash stimulus, also revealed a slight reduction in the sensitivity of rods from iCre75 retinas (Table 1). The iCre75+ responses also produced a notable overshoot in the recovery phase and appeared faster than those of control rods (Fig. 1C).
Table 1.
Rod Photoresponse Parameters From Transretinal Recordings
|
rsat, μV |
tp, ms |
τD, ms |
SF, %/R* |
Q1/2, R* |
|
| iCre75− | 594 ± 67 | 132 ± 2.6 | 120 ± 7 | 1.9 ± 0.1 | 56 ± 3 |
| iCre75+ | 663 ± 73 | 112 ± 3† | 72 ± 5† | 1.5 ± 0.1* | 71 ± 8 |
sat, saturated response amplitude measured at plateau; tp, time to peak of a dim flash response; τD, dominant time constant; SF, fractional sensitivity of a dim flash response; Q1/2, half saturating flash strength in R* per rod. Values are mean ± SEM.
P < 0.05 compared with control iCre75− rods.
P < 0.005 compared with control iCre75− rods.
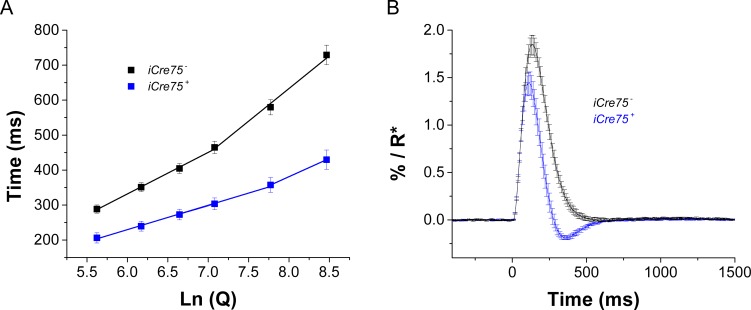
To examine the kinetics of transretinal rod responses in detail, we inspected the recovery from saturated flash responses to determine the dominant time constant of phototransduction inactivation (τD).27 We found that rod-saturated responses in iCre75+ retinas recovered faster than in the iCre75− controls (Fig. 2A). Moreover, iCre75+ retinas had a shorter rod dominant time constant (Table 1), indicating accelerated inactivation of the rod G protein, transducin (Gt)/ phosphodiesterase (PDE) complex and overall phototransduction. The waveform of the estimated rod single photon response (see “Methods” section) was also substantially accelerated in iCre75+ retinas compared with iCre75− controls (Fig. 2B). Consistent with this observation, rods from iCre75 retinas evidenced a faster time to peak than controls (Table 1). Together, these results clearly demonstrate that iCre75 transgenic mice exhibit distinct physiological properties of their rods as measured with transretinal recordings.
Figure 2.
iCre75 mice exhibit accelerated recovery of rod photoresponses in transretinal ERG recordings. (A) Saturation times measured at 30% recovery (from plateau after the peak) as a function of Ln (Q) are shown as black squares for control and blue squares for iCre75 mice (six retinas for each genotype). Linear fit between ca. 300 and 1000 R* per rod gave a τD of 119 ms and 67 ms for iCre75− and iCre75+ mice, respectively. Above approximately 2000 R* per rod, the slope increased to 186 ms and 104 ms for control and iCre75 retinas. (B) Average estimated single photon responses of WT (black, six retinas) and iCre75+ (blue, six retinas) mouse rods were derived by dividing the response to a dim flash by Q and rsat.
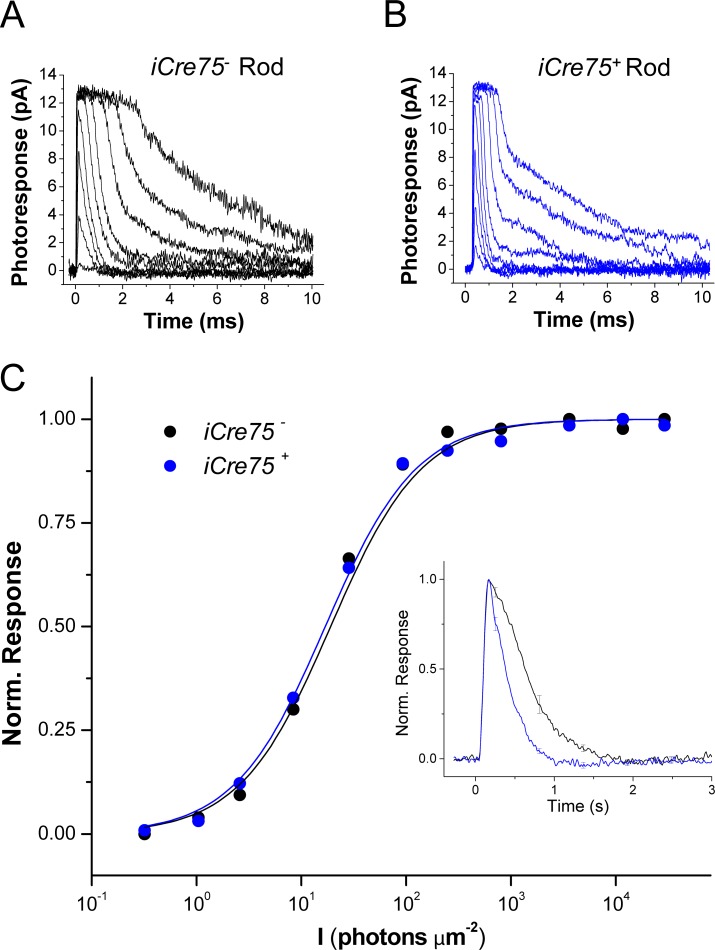
These results are consistent with a direct effect on the rod phototransduction cascade. However, as these transretinal measurements were obtained from the whole retina, it is possible that components other than rod outer segments also affect the measurements of sensitivity and kinetics. To address this issue, we next performed single-cell suction recordings from control iCre75− and transgenic iCre75+ rods. Consistent with the transretinal results above, we found that individual iCre75+ rods produce responses with normal saturated amplitudes (compare Figs. 3A, 3B), indicating that their dark current was not altered (Table 2). Their sensitivity was somewhat decreased as evidenced by the slight increase in half-saturating flash intensity, I1/2 (Table 2). Notably, the accelerated response kinetics of iCre75 mice persisted and was clearly observed in the population-averaged normalized dim flash responses (Fig. 3C, inset) and their associated parameters: time to peak (Tpeak), integration time (Tintegr), and recovery time constant (τrec; Table 2). The dominant time constant (τD), estimated from bright flash responses, was also significantly shortened in iCre75+ rods compared with iCre75− controls (Table 2). Together, these results are consistent with our findings from transretinal recordings and confirm the acceleration of phototransduction inactivation in iCre75 mice.
Figure 3.
iCre75 mice evidence accelerated recovery of rod photoresponses in single-cell suction electrode recordings. Shown are dark-adapted photoreceptor responses recorded with a suction electrode from single rods of iCre75− (A) and iCre75+ (B) mice to short 20-ms flashes of 500-nm light. (C) Normalized response amplitudes for these two rods are plotted as a function of flash strength, I, and fit with Equation 3 (see “Methods” section). Inset: population-averaged normalized dim flash responses from iCre75− (black, n = 26) and iCre75+ (blue, n = 21) mouse rods.
Table 2.
Rod Photoresponse Parameters From Single-Cell Recordings
|
Idark, pA |
I1/2, phot μm−2 |
Tpeak, ms |
Tintegr, ms |
τrec, ms |
τD, ms |
|
| iCre75− rods, n = 28 | 12.8 ± 0.7 | 21.3 ± 1.7 | 222 ± 16 | 528 ± 36 | 354 ± 24 | 273 ± 17 |
| iCre75+ rods, n = 21 | 12.4 ± 0.4 | 26.1 ± 2.0 | 171 ± 6* | 288 ± 18* | 210 ± 13* | 166 ± 10* |
dark, dark current measured from saturated responses; I1/2, half-saturating light intensity; time-to-peak (Tpeak) and integration time (Tintegr) refer to responses with amplitudes < 0.2 Idark that fell within the linear range; τrec, time constant for single-exponential decay of a dim flash response recovery phase; τD, dominant time constant of recovery after supersaturating flashes determined from the linear fit to time in saturation versus intensity semilog plots. Values are mean ± SEM.
Indicates significant difference (P < 0.05) compared with control iCre75− rods.
iCre75-Expressing Mice Also Overexpress R9AP
To gain insight into the molecular mechanism underlying the observed enhancement in the rate of phototransduction shutdown in iCre75-expressing mice, we analyzed the global retinal transcriptome of a previously developed iCre75-expressing mouse line, namely Dicer conditional knockout (cKO) mice. One of the most highly dysregulated genes in cKO animals was Rgs9bp, which encodes R9AP, the anchor protein for the photoreceptor Rgs9 GAP complex. Rgs9bp mRNA was upregulated by 1.83-fold in cKO mice (Fig. 4A, Table 3). Previous work has demonstrated that Rgs9 GAP complex activity is rate-limiting for shutdown of the phototransduction signaling cascade and that expression of R9AP limits the abundance of the GAP complex through stabilization of its other major constituents, Rgs9 and Gβ5.20,23,29 Indeed, overexpression of R9AP in transgenic mice has previously been directly demonstrated to result in accelerated shutoff of photoresponses.23 Therefore, upregulation of R9AP is sufficient to elicit more rapid phototransduction inactivation.
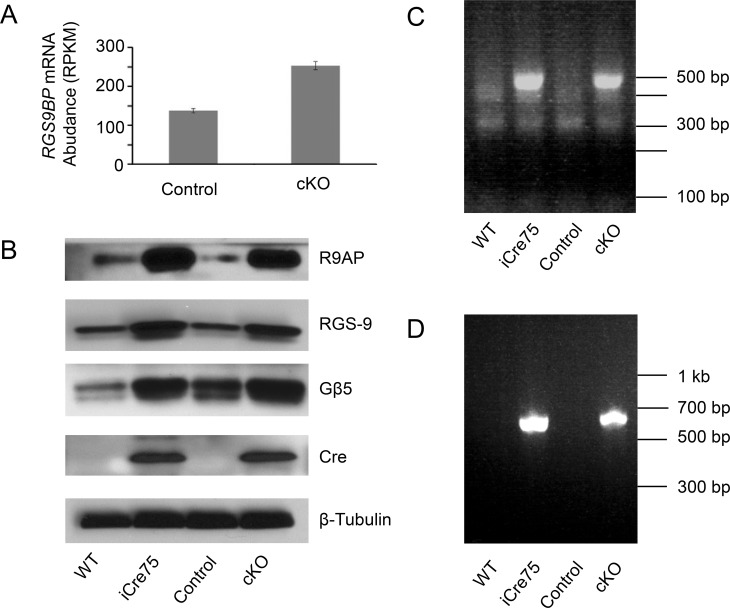
Figure 4.
Expression of the Rgs9 GAP complex in iCre75-expressing mouse retinas. (A) Bar graph showing the mean expression level of full length Rgs9bp mRNA in cKO mouse retinas and retinas from control littermates. Error bars represent standard deviations from three biological replicates. (B) Immunoblots comparing the protein levels in the retina of components of the Rgs9 GAP complex, namely Rgs9, Gβ5, and R9AP. β-tubulin was used as a loading control, and a Cre immunoblot is included to confirm Cre-recombinase expression. iCre75 and cKO mice are compared with their littermate controls. (C) PCR amplification of the 3′-end of cDNA derived from Rgs9bp transcripts in iCre75 or cKO mice and control littermates. Note that only the aberrant transcript is detected as PCR conditions did not support amplification of the >5 kb 3′-UTR of the canonical mRNA. (D) PCR amplification of genomic DNA from iCre75 or cKO animals and control littermates with primers annealing within the mouse opsin promoter and Rgs9bp CDS. iCre75, Dicer+/+ iCre75+; cKO, Dicerfl/fl iCre75+; control, Dicerfl/fl iCre75−.
Table 3.
mRNA Expression Levels of Relevant Genes and Gene Fragments Derived From RNAseq
|
Gene |
Region |
Expression in WT, RPKM |
Expression in Control, RPKM |
Expression in cKO, RPKM |
Fold Change, cKO vs. Control |
|
| Rgs9bp | Total | 166 ± 7.6 | 138 ± 5.5 | 253 ± 10.3 | 1.83 |
| Rgs9bp | CDS | 101 ± 26.1 | 90.2 ± 19.0 | 510 ± 21.1 | 5.65 |
| Rgs9bp | 3′-UTR | 76.4 ± 5.9 | 62.4 ± 3.70 | 51.7 ± 3.4 | 0.83 |
| Prm1 | Total | 0.21 ± 0.05 | 0.19 ± 0.19 | 914 ± 194 | 4810 |
| Rgs9 | Total | 139 ± 9.9 | 118 ± 2.79 | 113 ± 1.07 | 0.96 |
| Gnb5 | Total | 254 ± 38.5 | 198.7 ± 22.1 | 179.9 ± 15.2 | 0.91 |
Expression levels are given as RPKM ± SD.
Next, we evaluated the expression of the Rgs9 GAP complex components at the protein level in the retina. By immunoblotting, we observed a dramatic upregulation of R9AP in Cre-expressing animals. This effect was not dependent on Dicer, as cKO mice (Dicerfl/fl iCre75+) showed R9AP expression similar to that of iCre75 mice (Dicer+/+ iCre75+) and both were dramatically increased relative to iCre75− littermates (Fig. 4B). In addition, we observed upregulation of two other GAP complex components, Rgs9 and Gβ5 in iCre75-expressing retinas (Fig. 4B), despite little difference in mRNA levels for these two factors in cKO mice (Table 3). This result is consistent with previous reports of R9AP-dependent enhancement of Rgs9 GAP complex stability.20,23,29
iCre75-Expressing Mice Accumulate an Aberrant Rgs9bp mRNA Species
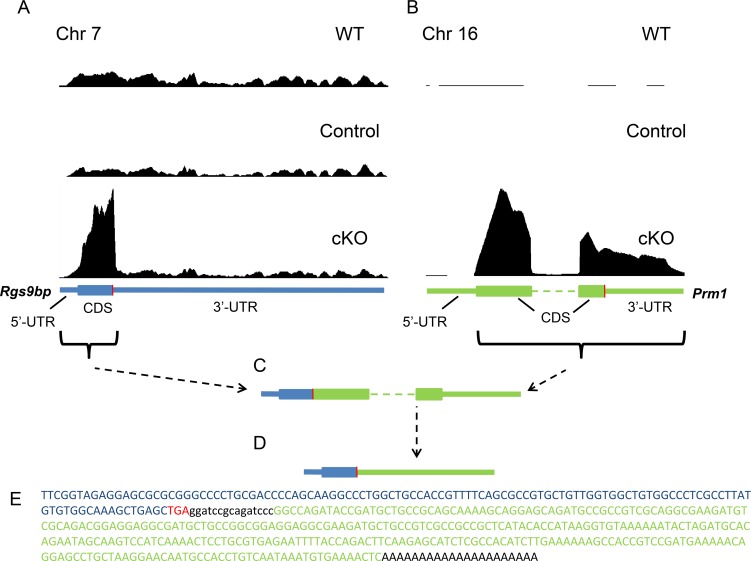
Though not quantitative, the difference in R9AP immunoblotting signals between iCre75-expressing animals and controls appeared to be greater than the 1.83-fold increase in mRNA expression observed in the RNA sequencing (RNAseq) data. To investigate this disparity, we mapped the RNAseq reads onto the mouse genome to evaluate the sequence coverage of the Rgs9bp gene in cKO mice. Surprisingly, we saw a striking difference in the pattern of expression of this mRNA (Fig. 5A). Although the 3′-untranslated region (3′-UTR) was expressed at similar levels compared with controls (reduced 1.2-fold, Table 3), the coding sequence (CDS) was selectively and dramatically upregulated in cKO mice (increased 5.65-fold). This result suggested that iCre75-expressing rods also expressed an aberrant Rgs9bp mRNA variant lacking the canonical 3′-UTR.
Figure 5.
iCre75-expressing mice also express a Rgs9bp-Prm1 fusion mRNA. (A) Pattern of RNAseq reads mapped to the Rgs9bp gene in WT (C57BL/6J) mice, cKO mice, and control (Dicerfl/fl iCre75−) mouse littermates. The sequence encoding Rgs9bp mRNA is shown below in blue with the translation termination codon indicated in red. The y-axis in (A) and (B) is normalized based on the total number of reads from each sequencing run. (B) Pattern of RNAseq reads mapped to the Prm1 gene in WT (C57BL/6J) mice, cKO mice, and control littermates. The sequence encoding Prm1 mRNA is shown below in green. (C) Schematic representation of the fusion mRNA composed of elements of the Rgs9bp (blue) and Prm1 (green) messages. This fusion mRNA was present in both cKO and iCre75 mouse retinas. Note that this schematic is not drawn to scale as the canonical Rgs9bp transcript (∼6.5 kb) is much larger than the Prm1 transcript (∼500 bp). (D) As the translation termination codon derived from the Rgs9bp mRNA was retained in the fusion transcript, the Prm1 CDS and 3′-UTR together form the 3′-UTR of the fusion mRNA. (E) Sequence of cDNA derived from the 3′-end of the aberrant Rgs9bp transcript in Cre-expressing retinas. Sequences derived from Rgs9bp and Prm1 are shown in blue and green, respectively, with the termination codon shown in red.
Such an observation could result from a number of different processes including premature transcription termination or an alternative splicing event. To differentiate between these possibilities, we used 3′-RACE to determine the sequence of the 3′-end of this novel Rgs9bp mRNA species in iCre75 and cKO mice. The forward primer for cDNA amplification was designed to anneal to a region upstream of the translation termination codon, and a poly(dT)-containing reverse primer was designed to anneal to the mRNA 3′-poly(A) tail. As expected, PCR amplification of cDNA derived from control retinas lacking Cre expression yielded no products (amplification of the >5 kb 3′-UTR of the canonical transcript was not supported by the PCR conditions employed). In contrast, amplification of cDNA derived from Cre-expressing retinas yielded a fragment of approximately 450 bp (Fig. 4C). This phenomenon was observed in both cKO mice and iCre75 mice and therefore was independent of Dicer activity. We then cloned PCR fragments derived from either cKO or iCre75 retinas and performed Sanger sequencing on five clones from each. The majority of clones (4/5 for cKO and 4/5 for iCre75) yielded a nearly identical insert sequence. Consistent with the RNAseq results, the inserts contained a sequence corresponding to the 3′-end of the Rgs9bp CDS concluding with the termination codon. Most unexpectedly, an additional ∼300-bp sequence was observed between the termination codon and the poly(A) tail (Figs. 5C–E). This sequence did not match any sequence within the Rgs9bp gene, nor anywhere else in mouse chromosome 7. To identify the source of this novel 3′-UTR, we performed a basic local alignment search tool search on the mouse genome that identified perfect homology to the CDS and 3′-UTR of the Prm1 gene on mouse chromosome 16 (Fig. 5E). Prm1 encodes a male germ line–specific DNA packaging factor which is negligibly expressed in the eye. The fragment of Prm1 represented in the fusion transcript began with the third nucleotide of the initiation codon (Fig. 5C). The fusion message is expected to encode the same protein as the canonical Rgs9bp mRNA with the sequence derived from the Prm1 CDS and 3′-UTR serving as the 3′-UTR of the novel transcript (Fig. 5D). Analysis of Prm1 expression from RNAseq data identified robust expression of the identified sequence in cKO mice, consistent with the 3′-RACE results (Fig. 5B).
iCre75 Mice Express an Rgs9bp Transgene
There were several possible explanations for this unexpected result including: (1) a Cre-dependent trans-splicing event linking the Rgs9bp and Prm1 mRNAs; (2) a Cre-dependent transposition of the Rgs9bp CDS onto chromosome 16; (3) a Cre-driven chromosomal translocation event; or (4) contamination of the iCre75 transgenic mouse line with an additional transgene encoding Rgs9bp. Krispel et al.23 previously reported the development of an Rgs9bp transgenic mouse line. Interestingly, this transgene was driven by a fragment of the mouse opsin promoter and utilized a 600-bp fragment of the mouse Prm1 gene as a polyadenylation element. Therefore, we reasoned that the iCre75 mouse line might contain this Rgs9bp transgene. To test this hypothesis, we performed PCR on genomic DNA from iCre75 mice, cKO mice, or control littermates using a forward primer designed to anneal near the 3′ end of the opsin promoter and a reverse primer annealing within the CDS of Rgs9bp. This PCR generated a ∼650-bp product for iCre75 and cKO animals, with no product observed in control littermates (Fig. 4D). Direct sequencing of PCR products confirmed the presence of the mouse opsin promoter region upstream of the Rgs9bp CDS in both iCre75 and cKO animals.
Discussion
Our results demonstrate that iCre75 mice exhibit accelerated phototransduction inactivation that can be explained by upregulation of the RGS9 GAP complex. GAP complex upregulation results from overexpression of the anchor protein R9AP, which in turn is due to the expression of an aberrant R9AP-expressing transcript. Similar to a previously described transgenic mouse line,23 this transcript is driven by a fragment of the mouse opsin promoter and a fragment of the mouse Prm1 gene lies downstream, acting as polyadenylation element. Therefore, we surmise that the iCre75 mouse line also contains this Rgs9bp transgene. As the iCre75 and Rgs9bp transgenes have faithfully cosegregated across many generations, it's likely that they are inserted very close to each other on the same chromosome. These results highlight the power of next generation sequencing to precisely identify altered gene expression patterns, helping to unambiguously explain an unexpected observation. Due to its robust Cre expression leading to highly efficient rod-specific gene excision along with the absence of any observable abnormalities in retinal morphology or cell survival, the iCre75 mouse line remains a powerful tool for dissecting gene function in rods. However, the results presented here represent an important caveat that must be considered when designing experiments using this line.
Acknowledgments
The authors thank Lesle T. Webster Jr (Case Western Reserve University, CWRU, Cleveland, Ohio) for his comments on the manuscript; Neena Haider (Harvard Medical School, Boston, Massachusetts) and Jason Chen (Virginia Commonwealth University, Richmond, Virginia) for providing mice; and Theodore Wensel (Baylor College of Medicine, Houston, Texas) and Vadim Arshavsky (Duke University, Durham, NC) for providing antibodies.
Supported by Grants R01EY0022326 (KP), R24EY021126 (KP, VJK), R01EY019312 (VJK), and EY002687 to the Department of Ophthalmology and Visual Sciences at Washington University; Grant P30EY11373 from the National Eye Institute of the National Institutes of Health; and Research to Prevent Blindness and Foundation Fighting Blindness. The authors alone are responsible for the content and writing of the paper.
Disclosure: T.R. Sundermeier, None; F. Vinberg, None; D. Mustafi, None; X. Bai, None; V.J. Kefalov, None; K. Palczewski, None
References
- 1. Maxeiner S, Dedek K, Janssen-Bienhold U, et al. Deletion of connexin45 in mouse retinal neurons disrupts the rod/cone signaling pathway between AII amacrine and ON cone bipolar cells and leads to impaired visual transmission. J Neurosci. 2005; 25: 566–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Longbottom R, Fruttiger M, Douglas RH, Martinez-Barbera JP, Greenwood J, Moss SE. Genetic ablation of retinal pigment epithelial cells reveals the adaptive response of the epithelium and impact on photoreceptors. Proc Natl Acad Sci U S A. 2009; 106: 18728–18733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ivanovic I, Allen DT, Dighe R, Le YZ, Anderson RE, Rajala RV. Phosphoinositide 3-kinase signaling in retinal rod photoreceptors. Invest Ophthalmol Vis Sci. 2011; 52: 6355–6362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ivanovic I, Anderson RE, Le YZ, Fliesler SJ, Sherry DM, Rajala RV. Deletion of the p85alpha regulatory subunit of phosphoinositide 3-kinase in cone photoreceptor cells results in cone photoreceptor degeneration. Invest Ophthalmol Vis Sci. 2011; 52: 3775–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li L, Khan N, Hurd T, et al. Ablation of the X-linked retinitis pigmentosa 2 (Rp2) gene in mice results in opsin mislocalization and photoreceptor degeneration. Invest Ophthalmol Vis Sci. 2013; 54: 4503–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wavre-Shapton ST, Tolmachova T, Lopes da Silva M, Futter CE, Seabra MC. Conditional ablation of the choroideremia gene causes age-related changes in mouse retinal pigment epithelium. PLoS One. 2013; 8: e57769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keady BT, Le YZ, Pazour GJ. IFT20 is required for opsin trafficking and photoreceptor outer segment development. Mol Biol Cell. 2011; 22: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rajala A, Tanito M, Le YZ, Kahn CR, Rajala RV. Loss of neuroprotective survival signal in mice lacking insulin receptor gene in rod photoreceptor cells. J Biol Chem. 2008; 283: 19781–19792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rajala RV, Tanito M, Neel BG, Rajala A. Enhanced retinal insulin receptor-activated neuroprotective survival signal in mice lacking the protein-tyrosine phosphatase-1B gene. J Biol Chem. 2010; 285: 8894–8904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. La Torre A, Georgi S, Reh TA. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc Natl Acad Sci U S A. 2013; 110: E2362–E2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Georgi SA, Reh TA. Dicer is required for the maintenance of notch signaling and gliogenic competence during mouse retinal development. Dev Neurobiol. 2011; 71: 1153–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Georgi SA, Reh TA. Dicer is required for the transition from early to late progenitor state in the developing mouse retina. J Neurosci. 2010; 30: 4048–4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Le YZ, Zheng L, Zheng W, et al. Mouse opsin promoter-directed Cre recombinase expression in transgenic mice. Mol Vis. 2006; 12: 389–398 [PubMed] [Google Scholar]
- 14. Le YZ, Ash JD, Al-Ubaidi MR, Chen Y, Ma JX, Anderson RE. Targeted expression of Cre recombinase to cone photoreceptors in transgenic mice. Mol Vis. 2004; 10: 1011–1018 [PubMed] [Google Scholar]
- 15. Li S, Chen D, Sauve Y, McCandless J, Chen YJ, Chen CK. Rhodopsin-iCre transgenic mouse line for Cre-mediated rod-specific gene targeting. Genesis. 2005; 41: 73–80 [DOI] [PubMed] [Google Scholar]
- 16. Jimeno D, Feiner L, Lillo C, et al. Analysis of kinesin-2 function in photoreceptor cells using synchronous Cre-loxP knockout of Kif3a with RHO-Cre. Invest Ophthalmol Vis Sci. 2006; 47: 5039–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Umino Y, Herrmann R, Chen CK, Barlow RB, Arshavsky VY, Solessio E. The relationship between slow photoresponse recovery rate and temporal resolution of vision. J Neurosci. 2012; 32: 14364–14373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu G, Zhang Z, Wensel TG. Activation of RGS9-1GTPase acceleration by its membrane anchor, R9AP. J Biol Chem. 2003; 278: 14550–14554 [DOI] [PubMed] [Google Scholar]
- 19. Hu G, Wensel TG. R9AP, a membrane anchor for the photoreceptor GTPase accelerating protein, RGS9-1. Proc Natl Acad Sci U S A. 2002; 99: 9755–9760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Keresztes G, Martemyanov KA, Krispel CM, et al. Absence of the RGS9.Gbeta5 GTPase-activating complex in photoreceptors of the R9AP knockout mouse. J Biol Chem. 2004; 279: 1581–1584 [DOI] [PubMed] [Google Scholar]
- 21. Baker SA, Martemyanov KA, Shavkunov AS, Arshavsky VY. Kinetic mechanism of RGS9-1 potentiation by R9AP. Biochemistry. 2006; 45: 10690–10697 [DOI] [PubMed] [Google Scholar]
- 22. Nishiguchi KM, Sandberg MA, Kooijman AC, et al. Defects in RGS9 or its anchor protein R9AP in patients with slow photoreceptor deactivation. Nature. 2004; 427: 75–78 [DOI] [PubMed] [Google Scholar]
- 23. Krispel CM, Chen D, Melling N, et al. RGS expression rate-limits recovery of rod photoresponses. Neuron. 2006; 51: 409–416 [DOI] [PubMed] [Google Scholar]
- 24. Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005; 102: 10898–10903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nymark S, Haldin C, Tenhu H, Koskelainen A. A new method for measuring free drug concentration: retinal tissue as a biosensor. Invest Ophthamol Vis Sci. 2006; 47: 2583–2588 [DOI] [PubMed] [Google Scholar]
- 26. Heikkinen H, Nymark S, Koskelainen A. Mouse cone photoresponses obtained with electroretinogram from the isolated retina. Vision Res. 2008; 48: 264–272 [DOI] [PubMed] [Google Scholar]
- 27. Pepperberg DR, Cornwall MC, Kahlert M, et al. Light-dependent delay in the falling phase of the retinal rod photoresponse. Vis Neurosci. 1992; 8: 9–18 [DOI] [PubMed] [Google Scholar]
- 28. Mustafi D, Kevany BM, Genoud C, et al. Defective photoreceptor phagocytosis in a mouse model of enhanced S-cone syndrome causes progressive retinal degeneration. FASEB J. 2011; 25: 3157–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gospe SM III, Baker SA, Kessler C, et al. Membrane attachment is key to protecting transducin GTPase-activating complex from intracellular proteolysis in photoreceptors. J Neurosci. 2011; 31: 14660–14668 [DOI] [PMC free article] [PubMed] [Google Scholar]