Abstract
Melioidosis is an endemic disease caused by the bacterium Burkholderia pseudomallei. Concerns exist regarding B. pseudomallei use as a potential bio-threat agent causing persistent infections and typically manifesting as severe pneumonia capable of causing fatal bacteremia. Development of suitable therapeutics against melioidosis is complicated due to high degree of genetic and phenotypic variability among B. pseudomallei isolates and lack of data establishing commonly accepted strains for comparative studies. Further, the impact of strain variation on virulence, disease presentation, and mortality is not well understood. Therefore, this study evaluate and compare the virulence and disease progression of B. pseudomallei strains K96243 and HBPUB10303a, following aerosol challenge in a standardized BALB/c mouse model of infection. The natural history analysis of disease progression monitored conditions such as weight, body temperature, appearance, activity, bacteremia, organ and tissue colonization (pathological and histological analysis) and immunological responses. This study provides a detailed, direct comparison of infection with different B. pseudomallei strains and set up the basis for a standardized model useful to test different medical countermeasures against Burkholderia species. Further, this protocol serves as a guideline to standardize other bacterial aerosol models of infection or to define biomarkers of infectious processes caused by other intracellular pathogens.
Burkholderia pseudomallei are soil dwelling organisms that are responsible for melioidosis, a systemic disease of humans and animals1,2,3,4. Despite the widespread geographic occurrence of naturally acquired melioidosis in Southeast Asia, Northern Australia and several regions around the world5 there are currently no licensed vaccines and antibiotic therapies suggest intensive, 12–20 week treatment regimens. However, the epidemiology of melioidosis in endemic areas would not allow adequately sized, well controlled clinical trials to evaluate drugs for post-exposure prophylactics or treatment of disease in healthy adults resulting from inhalational exposure to aerosolized B. pseudomallei1,6. Furthermore, human melioidosis presents with a variety of symptoms and is often confused with other diseases. Disparate clinical definitions of melioidosis exist, depending on route of infection, geography, and patient health status7. Variability in what clinicians observe extends to symptom progression, bacteremia, seroconversion, lesion development, organ and tissue colonization, and mortality3. Complicating matters further, B. pseudomallei can establish a latent infection and subsequently re-emerge (up to decades later) causing a secondary persistent infection8. Typical infections are correlated to ingestion, subcutaneous exposure and inhalation in individuals with a pre-existing health condition (e.g., diabetes, alcohol abuse)3,9,10. Overall, this information supports the fact that clinical trials in endemic populations would only provide supportive data but would not directly or robustly evaluate drugs to treat inhalational exposure.
In addition to naturally occurring infections, B. pseudomallei strains are classified as a CDC category B select agents due to their biothreat potential. While no evidence exists demonstrating weaponization of B. pseudomallei, the closely related organism B. mallei has been used as a bioweapon11,12,13,14. In this scenario, successful treatment and resolution of exposure to B. pseudomallei and the resultant melioidosis cases is complicated by the bacteria intrinsic resistance to multiple antibiotics3,9,15,16. Because of the prevalence in endemic regions, the existing concerns about the possibility to aerosolize B. pseudomallei, and the multidrug resistant nature of the bacteria, continued research efforts are needed to establish new therapeutics and medical countermeasures (MCMs) for melioidosis.
Given the inability to perform human clinical trials, the development of therapeutics or MCMs for inhalational melioidosis will require approval under the Food and Drug Administration's (FDA) “Animal Rule”17. The most commonly used models are the murine models, although studies have also been conducted using the Syrian golden hamster model18,19. C57Bl/6 mice have been shown to be approximately 10-fold more resistant to disease than BALB/c mice20,21 and more likely to develop a chronic form of the disease. The hamster model has been shown to be highly susceptible to disease and does not mimic a human melioidosis infection22. With respect to the alternative infection routes used to study melioidosis such as intranasal and intraperitoneal21,23,24,25,26,27,28, aerosol infection is still believed to be the most effective route in case of intentional dissemination of the pathogen. In order to develop MCMs for inhalational melioidosis, an improved understanding of inhalational acquired melioidosis and suitable animal models for human melioidosis are needed.
Currently, the lack of a well-characterized challenge strain(s) and standardized animal models of inhalational melioidosis prevents the rigorous testing of prospective MCMs. While there are several prototypic strains of B. pseudomallei and several studies have been conducted utilizing aerosol delivery29,30,31,32,33, comparative data to establish commonly accepted strain(s) to study virulence and disease progression has not been done. Primarily, B. pseudomallei strains K96243 and 1026b have been utilized in aerosol studies. Strain K96243 was isolated from a 34-year old diabetic female in Thailand in 1996 and most aerosol studies report the LD50 value to be 5–10 cfu24. Strain 1026b was isolated from a 29-year-old diabetic rice farmer in Thailand in 1993 and the LD50 is widely variable as there are different strains commonly reported in the literature. One strain of 1026b (BEI catalog number NR-4074) has a reported LD50 value of around 2000 cfu30; whereas we have determined the LD50 of a second strain (BEI catalog number NR-9910) to have an LD50 of approximately 20 cfu (unpublished data). This briefly exemplifies the impact of strain variation on virulence, disease presentation, and mortality, which is a roadblock to perform rigorous testing necessary to develop new MCMs.
Here, we describe a detailed study characterizing the pathogenesis of B. pseudomallei aerosol infection in a murine model by comparing clinical markers from a prototypical strain (K96243) and a previously undocumented, highly virulent strain (HBPUB10303a), isolated from a 40 year old patient in Thailand during 2011. In addition we show that the variability of clinical presentations is dependent on strain selection, which has a significant impact in future testing of MCMs against Burkholderia species or other respiratory pathogens.
Results
Survival and clinical observations
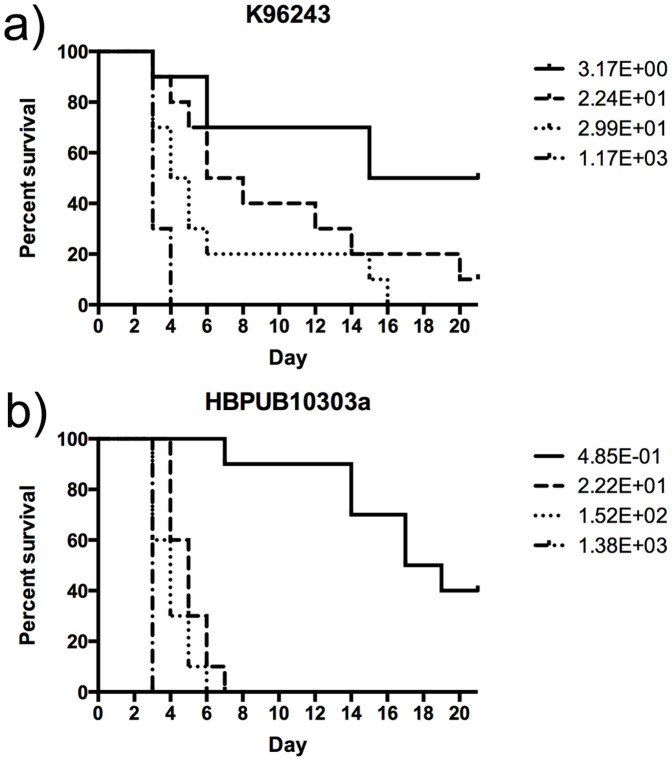
We determined the mean lethal dose (LD50) of the two strains (K96243 and HBPUB10303a) by the method or Reed and Meunch34. Groups of mice (n = 10/group) were exposed via nose-only restraint to an aerosol with an input dose of 1 × 105, 1 × 106, 1 × 107 or 1 × 108 cfu/mL generated by the Biaera aerosol system, and were monitored for clinical signs of illness and morbidity for a period of 21 days. Based on the calculated doses presented (Dp) in the survival curve shown in Figure 1, the LD50 values were determined to be approximately 1 and 4 cfu for mice exposed to HBPUB10303a and K962343, respectively. B. pseudomallei K96243 is a prototypical strain that has routinely been used during evaluation of inhalational melioidosis. The calculated LD50 value is consistent with previously reported LD50 values ranging from 5 to 10 cfu24. Values for strain HBPUB10303a have not been previously described, since this is the first report that uses this strain in a murine model of inhalational melioidosis.
Figure 1. LD50 analysis of B. pseudomallei strains.
BALB/c mice (n = 10) were infected, via aerosol delivery, with the indicated number of CFU for B. pseudomallei strains K96243 (a) and HBPUB10303a (b). Mice were monitored daily for signs and symptoms of disease. Survival data were analyzed by the Kaplan-Maier method and LD50 values were calculated by the method of Reed and Muench.
For analysis of the clinical progression of melioidosis, mice were weighed and randomized into groups prior to infection. Sixty mice were exposed via aerosol to strain HBPUB10303a, 60 mice were exposed to strain K96243 and 24 mice (12 for each strain) were uninfected control animals. Aerosolization of both strains resulted in average presented doses of 5.4 and 3.8 cfu for HBPUB10303a and K96243, respectively. On days 1, 3, 7, 10 and 14, organs of ten pre-determined animals were extracted. The lung, liver and spleen were analyzed for either bacterial burden or histopathology. Brain and mesenteric lymph nodes of infected mice were also excised and processed (data not shown). Blood and serum collected were used to analyze the blood chemistry, hematology and chemokine/cytokine levels in all animals.
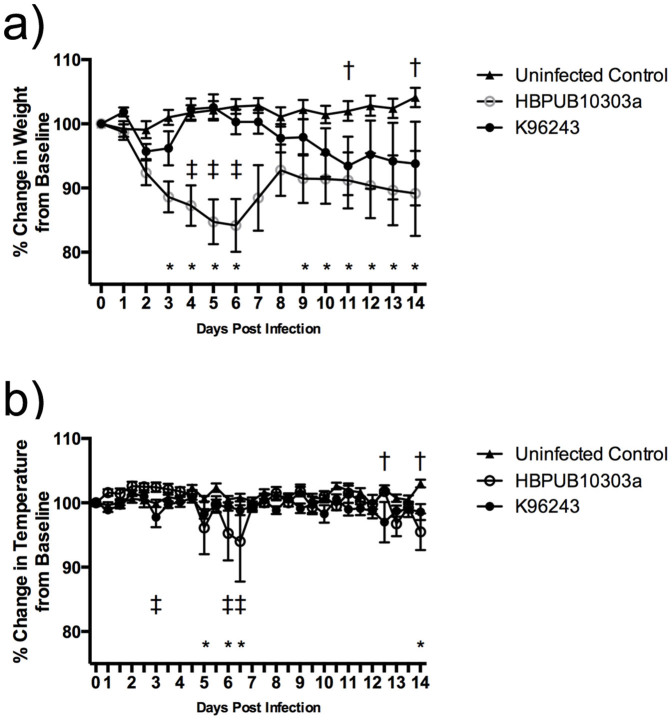
Mice exposed to both strains began to lose body weight as early as day 2 post infection (Figure 2a). The average body weight of animals infected with K96243 increased back to baseline levels on day 4 before steadily decreasing again on day 6 although, there were no significant differences in body weight when comparing these animals to uninfected controls (except on days 11 and 14). Mice exposed to strain HBPUB10303a began to lose significantly more weight on day 3 post infection when compared to uninfected control animals, and on day 4 when compared to animals infected with K96243 (Figure 2a). This trend continued until day 7 when the average body weight of animals infected with HBPUB10303a increased due to deaths. Significant differences between HBPUB10303a infected animals and the control animals were seen again at day 9 and continued to day 14 although, no further significant differences were seen between K96234 and HBPUB10303a infected animals. In general, no differences were observed in body temperatures when comparing control vs. K96243, control vs. HBPUB10303a or HBPUB10303a vs. K96243. The only significant differences could be seen on days 3, 5, 6, 12 and 14 (Figure 2b) when a rapid drop in body temperature precedes mouse death.
Figure 2. Average daily body weight and temperature.
Percent change in body weight (a) and temperature (b) of mice exposed to B. pseudomallei HBPUB10303a (open circles) or K96243 (closed circles). The symbols represent the mean value and the error bars represent the standard error of the mean. The asterisk (*) indicates significantly different values when comparing HBPUB10303a to the uninfected control animals (closed triangles), p < 0.05. The single cross (†) represents significantly different values when comparing K96243 to the uninfected controls, p < 0.05. The double cross (‡) indicates significantly different values when comparing HBPUB10303a to K96243, p < 0.05.
The clinical presentation of the disease was found to be consistent amongst both strains with signs and symptoms generally occurring earlier during HBPUB10303a infections. As previously reported, the disease progression can be divided into three stages35. The first stage of the disease progression is characterized by a lack of grooming although the behavior and responses to external stimuli remain unchanged. During this stage bacteria are generally found only in the lungs; however, all the lung, liver and spleen appeared normal (Supplemental Figure 1). A rough coat with possible nasal and/or ocular discharge characterizes stage two along with a decrease in mobility and decreased responses to external stimuli. During this stage bacteria could now be isolated from the lung, liver, spleen, mesenteric lymph nodes and blood of infected animals. Analysis of the organs during stage 2 revealed nodular lesions on the lungs as well as a mottled appearance indicative of granuloma formation (Supplemental Figure 1). Hepatomegaly and splenomegaly were commonly seen with nodules or abscesses found on the latter. Stage 3 can be identified by the cessation of grooming as well as an abnormal posture along with nasal and/or ocular discharge. Bacteria were isolated from all organs during this stage. These animals had multiple nodules and/or abscesses on the lung, liver and spleen with enlargement of the organs present in liver and spleen (Supplemental Figure 1).
Bacterial burden
The bacterial burden within the lung, liver and spleen was determined at 1, 3, 7, 10 and 14 days post-infection. Bacterial organ loads increased over the 14-day time period for both the K96243 and HBPUB10303a strains (Figure 3). There were no significant differences between the two strains. Bacteria were first detected in the spleen on day 3 for both strains. At day 7 post-infection, we cultured much more HBPUB10303a bacteria in the spleen than K96243 and this difference became significant by day 10. By day 14 post-infection, the bacterial burden in the spleen of infected animals was equivalent. Bacteria were first detected in the liver on day 3 post-infection and increased over the course of the infection for both strains. There were no significant differences in bacterial burden in the liver between the two strains.
Figure 3. Bacterial burden.
Bacterial burden in the lung (a), liver (b) and spleen (c) of mice infected with HBPUB10303a (closed circles) or K96243 (closed triangles). The symbols represent individual animals. The bars indicate the mean CFU/organ for each group. The double cross (‡) indicates significantly different values when comparing HBPUB10303a to K96243, p < 0.05.
Blood chemistry and hematology
The hematology and blood chemistry values were determined on uninfected control and B. pseudomallei infected mice, using the HemaVet and VetScan complete diagnostic systems, respectively. Infected animal samples were analyzed on days 1, 3, 7, 10 and 14 post-infection and representative markers are depicted in Figure 4. Differences between uninfected control mice and the HBPUB10303a or K96243 infected mice were typically seen on day 3 post-infection. The liver enzyme alkaline phosphatase (ALP, Figure 4a) dropped significantly in both strains when compared to uninfected controls on day 3. Although the differences were not significant, ALP levels for K96243-infected animals dropped to 47 U/L on day 3 and remained consistent at days 7, 10 and 14, where as HBPUB10303a infected animals dropped to approximately 17 U/L by day 7. Albumin (ALB), makes up approximately 50% of the total protein in the serum, and like ALP is synthesized in the liver. Significant differences between uninfected controls and the B. pseudomallei infected animals occurred at day 3 (HBPUB10303a) and day 7 (K96243). ALB levels of HBPUB10303a infected animals were significantly less at days 3, 7 and 10 post-infection compared to K96243 infected animals (Figure 4b). At day 14, the ALB levels appeared to be equivalent in both strains. Globulin (GLOB) makes up the remaining total protein in the blood serum. Unlike ALB, the GLOB levels significantly increase in both the HBPUB10303a and K96243 infected animals compared to the controls at all-time points. GLOB levels of animals infected with HBPUB10303a were significantly higher at days 3 and 7 compared to the K96423-infected animals (Figure 4c). Finally, the total white blood cell count was analyzed. Significant differences between K96243 and the uninfected control were seen on days 1, 7 and 10, while differences between HBPUB10303a are seen on days 3, 7 and 10 (Figure 4d). A significant difference was seen between K96243 infected animals and HBPUB10303a infected animals on day 1, but this difference was abolished by day 3. The VetScan and HemaVet system was used to analyze numerous other parameters however; there were less significant differences between strains for these markers (Supplemental Table 1).
Figure 4. Blood chemistry and hematology.
Analysis of the alkaline phosphatase (a), albumin (b), globulin (c) and white blood cell count (d) in mice infected with HBPUB10303a (closed circles) or K96243 (closed triangles) compared to the uninfected control animals (dashed line). The symbols represent values of individual animals. The bars indicate the mean for each group and the dashed line represents mean for uninfected control animals. The asterisk (*) indicates significantly different values when comparing HBPUB10303a to the uninfected control animals, p < 0.05. The single cross (†) represents significantly different values when comparing K96243 to the uninfected controls, p < 0.05. The double cross (‡) indicates significantly different values when comparing HBPUB10303a to K96243, p < 0.05.
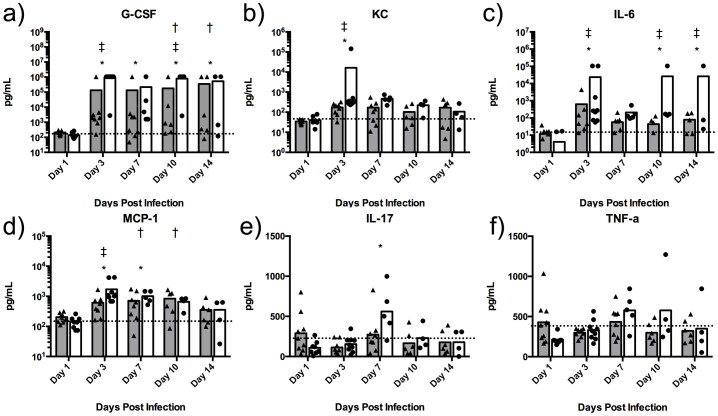
Quantitation of cytokine and chemokine levels in serum
Following 1 day of infection, there were no significant alterations in cytokine/chemokine levels (for the 23 analytes tested) in the serum of animals infected with B. pseudomallei HPUB10303A (Figure 5). By day 3, however, dramatic increases in G-CSF were observed in serum from animals infected with B. pseudomallei HPUB10303A compared to non-infected controls (Figure 5a). The values for serum G-CSF were well in excess of the highest standard and out of range (OOR high) for extrapolation; values were thus set conservatively to the highest extrapolated value. Large increases in the inflammation-associated chemokines KC (Figure 5b) and MCP-1 (Figure 5d) were also observed 3 days post-infection and were significant. IL-10, a regulatory cytokine associated with suppression of inflammation, was also significantly increased at day 3 (Supplemental Table 2). Levels of IFN-γ (Supplemental Table 2) and IL-6 (Figure 5c) also increased in several animals at day 3 but due to variability in the data set did not reach significance.
Figure 5. Blood serum chemokine/cytokine analysis.
Levels of individual chemokines/cytokines in plasma (pg/ml) of animals infected with HBPUB10303a (closed circles) or K96243 (closed triangles) compared to the uninfected control animals (dashed line). The symbols represent values of individual animals. The bars indicate the mean for each group and the dashed line represents the mean for uninfected control animals. The asterisk (*) indicates significantly different values when comparing HBPUB10303a to the uninfected control animals, p < 0.05. The single cross (†) represents significantly different values when comparing K96243 to the uninfected controls, p < 0.05. The double cross (‡) indicates significantly different values when comparing HBPUB10303a to K96243, p < 0.05.
By day 7 post-infection, levels of G-CSF, KC, and IL-10 continued to be significantly increased compared to non-infected animals. Also, at this time point, significant increases in levels of IL-17, TNF-α, IL-2, IL-3, and IL-13 were observed (Figure 5e and f, Supplemental Table 2). At day 10, KC and G-CSF continue to be significantly elevated, while TFN-α remains elevated similar to day 7, where MIP1-α (Supplemental Table 2) levels were increased. By day 14, the levels of cytokines/chemokines in the surviving animals did not differ from that of the non-infected control animals.
Compared to animals infected with B. pseudomallei HPUB10303A, the serum cytokine and chemokine responses of animals infected with B. pseudomallei K96243 were reduced. At day 3 post-infection, levels of KC, MCP-1 and G-CSF were increased as observed in response to HPUB10303A but did not reach significance (Figure 5). Several other cytokines and chemokines appear to be moderately expressed in several individual animals but did not reach significance as a group compared to control animals (Supplemental Table 2).
Quantitation of cytokine and chemokine levels in lung
Similar to the serum following 1 day of infection, the cytokine and chemokine levels in lung of animals infected with either B. pseudomallei K96243 or HPUB10303A were generally not different from baseline at day 1 post infection (Supplemental Figure 2 and Supplemental Table 3). The exception was KC, which in the lung was significantly increased at day 1 following infection with HPUB10303A (Supplemental Figure 2b).
Consistent with serum measurements, by day 3 post-infection, a pronounced increase (OOR high) in G-CSF and KC was observed in lung from animals infected with B. pseudomallei HPUB10303A compared to non-infected animals (Supplemental Figure 2a and b). MCP-1 levels were also very high at day 3 and slowly declined from days 3 to 10 but were still significantly increased compared to non-infected animals at all 3 time points (Supplemental Figure 2d). The large increases in these three chemokines (KC, G-CSF, and MCP-1) within the lung indicate a microenvironment conducive for neutrophil recruitment and activation during inflammatory responses at the site of infection.
Also at day 3, MIP1-α was dramatically increased, along with significant increases in GM-CSF, MIP1-β, TNF-α, IL-1α, IL-1β, IFN-γ, and IL-12p40 (Supplemental Figure 2 and Supplemental Table 3). In contrast to the serum, though, IL-10 levels only increased marginally at day 3 and did not reach significance. By day 7, increased levels of KC, MCP-1, G-CSF, MIP1-α, IFN-γ, IL-6, IL-1α, IL-1β were still observed. Similar to the serum observations, a significant increase in IL-17 is first seen at day 7 and, in contrast, levels of IL-2, IL-3, and IL-13 were not significantly increased in the lung during infection at day 7 or beyond. At day 10, KC, G-CSF, MCP-1, MIP1-α, IL-6, IL-17, IL-1α and IL-1β continue to be elevated. KC, G-CSF, and IL-1β continued to be elevated through day 14. These observations differed from the serum, where most cytokine/chemokine expression was observed to be decreasing by day 10 and by day 14, and no differences were noted compared to non-infected animals.
Similar to the serum cytokine/chemokine response, the expression in the lungs of animals infected with B. pseudomallei K96243 was moderate compared to animals infected with HPUB10303A (Supplemental Figure 2 and Supplemental Table 3). Similar increases in KC, G-CSF, IL-6, IL-1α were observed especially in days 3–7. Increased levels of MCP-1, GM-CSF, MIP1-α, MIP1-β, TNF-α, IFN-γ, and IL-17 were similarly noted, but in contrast to HPUB10303A-infected animals, most often did not reach statistical significance. For both strains of B. pseudomallei, no significant differences in the lung were observed in levels of Eotaxin, IL-3, IL-4, IL-5, IL-9, IL-2p70, IL-13, or RANTES (Supplemental Table 3).
Histopathology
Histological examinations of tissues (lung, liver and spleen) from mice exposed to B. pseudomallei strain K96243 and strain HBPUB10303a were conducted at 1, 3, 7, 10 and 14 days post-infection. Representative images can be seen in Figure 6a for lung, liver and spleen tissue sections from infected animals on days 7 and 14 as well as tissue sections from uninfected controls. All animals designated for the histopathology groups were analyzed and assigned and grade of 0 (no lesions, 0% of the tissue affected), 1 (minimal change, 10% and less of the tissue affected), 2 (mild change, 10–25% of the tissue affected), 3 (moderate change, 25–50% of the tissue affected), 4 (marked change, 50–75% of the tissue affected) or 5 (severe change, >75% of the tissue affected). The average grade for each of the 5 time points was assessed and presented in Figure 6b.
Figure 6. Histopathology and scoring.
Representative images from (a) hematoxylin and eosin stained lungs, liver and spleen from mice infected with K96243 and HBPUB10303a at day 7 and uninfected controls. (b) Scores assigned for lung, liver and spleen tissue sections after microscopic examination. Bars represent mean ± SEM of animals infected with HBPUB10303a (white bars) or K96243 (gray bars) and the asterisk (*) indicates significantly different values when comparing HBPUB10303a to the uninfected control animals, p < 0.05. The single cross (†) represents significantly different values when comparing K96243 to the uninfected controls p < 0.05. The double cross (‡) indicates significantly different values when comparing HBPUB10303a to K96243 p < 0.05.
On day 1 post-infection with B. pseudomallei strains HBPUB10303A or K96243, the majority of tissue sections showed minimal changes or nothing remarkable. Rare sections of lung from both groups showed one or more isolated small clusters of inflammatory cells, mostly composed of neutrophils and macrophages, and not affecting the surrounding tissue; however, there were no significant differences in either strain when compared to the uninfected control animals (Figure 6b).
On day 3 post-infection, lung sections from animals exposed to both strains of B. pseudomallei showed diffuse inflammatory infiltrates composed mainly of neutrophils and macrophages, in several cases extending to a complete lobe, and associated with hyaline membrane and cellular debris. Accumulation of proteinaceous fluid, cell debris, and inflammatory cells was observed in both small and large airways, in some cases occupying the majority of the bronchial lumen. Large clusters of inflammatory cells associated with local destruction of the normal lung parenchyma were observed, although they appeared to be at an early stage. Spleen sections collected at this time showed minimal, non-significant changes in animals infected with strain K96243 (Figure 6b), while sections collected from animals exposed to the HBPUB10303a strain showed proliferation of large foamy splenic macrophages, in many cases containing engulfed cellular debris and associated with karyorrhexis, and also scattered granulomatous formations at the very early stage. The liver sections of animals from the K96243-infected group showed minimal to no changes as well. Those sections from animals infected with HBPUB1030a were significantly different from K96243 sections (Figure 6b), showing scattered clusters of inflammatory cells, associated with necro-apoptotic hepatocytes and in some cases larger necrotic areas.
Sections of lung collected on day 7 from animals infected with the K96243 strain showed large granulomas extending over 500 μM in diameter, and in some cases 1000 μM. Sections from the animals exposed to the HBPUB10303a strain showed both large granulomatous formations and in several cases lobar pneumonia (Figure 6a). The spleen sections collected on day 7, showed large granulomas, which were extending over 500 μM in diameter in animals infected with either K96243 of HBPUB10303a. Liver sections showed scattered small clusters of inflammatory cells, and large necrotic areas and abscesses in animals infected with either strain; however, these were more frequently observed (and significantly different) in the animals exposed to the HBPUB1303a strain compared to those exposed to K96243 (Figure 6b).
On day 10, lung sections from the animals exposed to strain K96243 showed large granulomas and lobar pneumonia. The same observation, although in a larger number of animals and more severe, were seen in the sections collected from the animals exposed to the HBPUB13030a strain resulting in a significant difference between the strains (Figure 6b). Spleen sections from the HBPUB10303a infected animals showed multiple large granulomas, while sections from the K96243 animals showed minimal changes, again representing significant differences between the strains (Figure 6b). Sections from livers of animals infected with strain K96243 showed scattered small clusters of inflammatory cells, while those in the HBPUB10303a group showed both scattered inflammatory foci and large necrotic areas.
Lung sections collected on day 14 showed large granulomas and lobar pneumonia, with similar changes in the animals exposed to either strain K96243 or HBPUB10303a. However, those observations were uniform in the HBPUB10303a group, while some animals in the K96243 group showed nothing remarkable or minimal pathologic damage (data not shown). In the spleen sections, pathologic changes were mainly observed in the animals exposed to the HBPUB10303a strain and consisted of large granulomas. Liver sections showed numerous inflammatory foci and large necrotic areas and abscesses in the HBPUB10303a group, while the changes observed in the B. pseudomallei K96243 group were less remarkable and consisting mainly of small inflammatory foci scattered throughout the lobules.
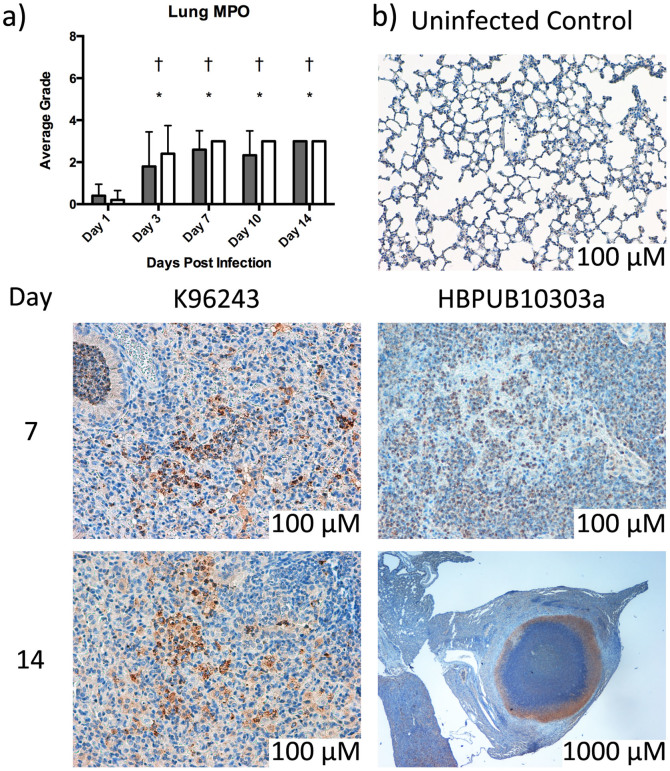
Finally, lung sections were also processed by immunohistochemical-labeling myeloperoxidase (MPO) to identify neutrophils and, to a minor extent, macrophages. The average grade for the MPO staining was calculated and reported in Figure 7a. Representative images from days 7 and 14 are shown in Figure 7b. A grade of 0 (within normal limits), 1 (focal positive staining), 2 (multifocal positive staining) or 3 (diffuse positive staining) was assigned to each tissue analyzed. Positive staining was observed in both groups beginning on day 1 after infection, and remained at day 14. Positive staining was rare and focal on day 1, and became multifocal to diffuse on days 3, 7, 10 and 14. While both strains show significant differences from the uninfected controls starting at day 3 post-infection, there appear to be no differences between the strains for the MPO staining (Figure 7b).
Figure 7. Immunohistochemistry.
(a) Immunohistochemistry scoring assigned for lung sections after microscopic examination. Bars represent mean ± SEM of animals infected with HBPUB10303a (white bars) or K96243 (gray bars). The asterisk (*) indicates significantly different values when comparing HBPUB10303a to the uninfected control animals, p < 0.05. The single cross (†) represents significantly different values when comparing K96243 to the uninfected controls p < 0.05. The double cross (‡) indicates significantly different values when comparing HBPUB10303a to K96243 p < 0.05. (b) Representative images from MPO stained lungs from mice infected with K96243 and HBPUB10303a at days 7 and 14 post-infection.
Discussion
Melioidosis, the disease caused by B. pseudomallei is an important infectious disease endemic to Southeast Asia and Northern Australia where mortality rates are approximately 50% and 20% respectively. The disease is also becoming increasingly global, as cases have been reported in Brazil and Puerto Rico2,36,37,38,39,40,41. Due to the ease of acquisition from the environment and the concern about mortality/morbidity associated with natural or human-caused large-scale illness, this pathogen has been identified as a potential biothreat agent. Therefore, an effective treatment/prevention is desired. A major challenge for development and acquisition of new MCMs is proving effectiveness since these cannot be tested on humans. The FDA establishment of the Animal Rule allows limited approval for MCMs based on adequate and well-controlled animal studies. As such, animal infections with B. pseudomallei strains to study the disease progression and/or efficacy of treatments have been extensively documented23. Our study, however, is the first to report a well-controlled comparison of two B. pseudomallei strains over the course of an infection for the purposes of establishing a consistent model to be used when evaluating potential MCMs.
Initially we examined the virulence of four strains (data not shown) in order to down-select two B. pseudomallei strains that would be ideal for further use in investigating the natural history of melioidosis. Several factors were considered when selecting the two strains, including consistency of aerosol exposure (including bacterial recovery and spray factor) and post-challenge endpoints (i.e. changes in body weight, clinical observations, gross pathology, etc.). The majority of previously published B. pseudomallei aerosol experiments have been conducted using the “prototypical” strains K96243 or 1026b23,28,29,30,31,33,35,42,43,44. B. pseudomallei 1026b, although considered a prototypical strain was concluded to behave inconsistently based on our bioaerosol parameters. Concerns over strain uniformity have also been raised as recent publications report the aerosolization of strain 1026b with drastically different dose presented LD50 values. Gelhaus et al reported the LD50 of strain 1026b to be 2200 cfu30, while Jeddeloh et. al. reported an LD50 of approximately 10 cfu31. Aerosolization of K96243 and HBPUB10303a appeared to be consistent throughout our studies as the spray factors and doses presented were similar in both the virulence study and the disease progression study.
The pathogenic differences in murine melioidosis following infection with K96243 or HBPUB10303a offer important insights for development of future MCMs. Mice challenged with the same number of bacteria (approximately 4–5 cfu) resulted in significantly different presentations of melioidosis signs and symptoms. Although infection with both strains resulted in decreases in average body weights, significant differences from uninfected control animals were seen as early as day 3 for HBPUB10303a infected mice but not until day 11 for K96243-infected mice. The significant differences between the two strains were observed on days 4–6 post-infection. Animals infected with HBPUB10303a lost 10% body weight by day 4 whereas mice challenged with K96243 lose 10% body weight around day 8. However, the bacterial burden in the lung and liver remained consistent over the course of the infection. While it appears that the bacterial counts for HBPUB10303a infected animals were higher in the spleen at day 3, 7 and 10, these differences were only statistically significant on day 10. Based on the data, it can be proposed that faster dissemination to the spleen could be an explanation for the increased virulence of B. pseudomallei strain HBPUB10303a as compared to K96243. This is supported by the qualitative observation at day 3 of bacteria in the blood of 80% of the HBPUB10303a-infected animals compared to only 22% in the K96243-infected animals (data not shown). Infection in these animals was confirmed by either bacterial cfu in other organs or histopathological observation.
With respect to hematological parameters, we observed an increase in neutrophils and white blood cells during infection with both strains when compared to uninfected control animals. Neutrophil levels rose approximately 250% and 350% by day 3 post-infection when infected with K96243 and HBPUB10303a, respectively. Increases in neutrophils are a result of the inflammatory response and subsequent granuloma formations seen in the lung, liver and spleen of infected subjects45. We also noted decreases in the liver-synthesized protein ALP and ALB. Decreases in the ALP and ALB levels are indicative of liver damage and/or malnutrition and protein deficiency. ALB levels were significantly different at day 3 between the HBPUB10303a and K96243 infected animals. This difference correlates to the increased weight loss associated with HBPUB10303a infection. GLOB makes up approximately 50% of the protein in the blood and increases in cases of acute infection, often due to increased antibody production. Significant increases in the GLOB levels between the two strains at day 3 could be a result of fast dissemination of the HBPUB10303a strain throughout the body.
Identifying serum biomarkers that are used for prognosis of disease state is very important for treatment of patients suspected of having meliodosis9,46. Here, we observed that levels of cytokines and chemokines in the serum correlated with significant inflammation in the lung and sites of dissemination. In particular, the dramatic increases in KC, G-CSF, and MCP-1 were very consistent with observed leukocyte infiltration and pathology at sites of infection. This is important given the role of these cytokines for recruitment and activation of neutrophils and monocyte/macrophage populations47,48. IL-3 (mast cell growth factor) and IL-13 cytokines contribute to pathogenesis of allergic lung disease such as asthma and the increases observed for these cytokines further suggests a highly inflammatory environment49,50. The observed increases in TNF-α and IL-6 are also important indicators associated with fever and the acute phase responses during infection and support the observed alterations in liver enzymes. The dramatic increase in IL-17 is especially of note as this cytokine is the major mechanism whereby pathogenic Th17 cells contribute to host damage via prolonged neutrophil recruitment that causes tissue damage51.
Overall, the serum cytokine and chemokines alterations mirror the histopathological changes occurring in the lung. The magnitude of the response was much greater in both the serum and lung following infection with B. pseudomallei HBPUB10303a infection than with strain K96243. Thus, serum cytokine/chemokine profiles may be predictive of disease severity due to strain virulence or exposure dose. These findings along with the hematology and blood chemistry data contribute to the understanding of the host innate immune responses during B. pseudomallei infections.
Finally, the histopathological observations presented are consistent with a severe bacterial infection resulting in multifocal and diffuse inflammatory infiltrates affecting major organ systems and correlated with several prior murine aerosol studies23. The most striking findings in our study; however, were found in the lung, where multiple large granulomas, collection of inflammatory cells, proteinaceous material, and lobar pneumonia were observed. Similar observations were made for the spleen and the liver, although changes in these organs were less frequent and severe. Overall, animals infected with either strain experienced an acute bacterial infection producing a disease similar in nature and with comparable pathologic features. However, tissue sections from animals infected with HBPUB10303a displayed more frequent and severe pathologic changes, compared to those obtained from animals infected with K96243.
Overall, this study described a well-controlled B. pseudomallei infection model using two different strains, and while aerosol infection resulted in a similar pathology, strain HBPUB10303a resulted in a more rapid progression of the disease when compared with strain K96243. Therefore, we propose that our standardized murine aerosol model should be used in future evaluations of MCMs during Burkholderia or other respiratory pathogens infections, because it provides a series of biomarkers that are easy to evaluate and can provide valuable information about the effectiveness of the treatment.
Methods
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The Animal Care and Use Committee of the University of Texas Medical Branch approved protocol 1209054.
Bacterial strains
In this study, the lethal dose at 50% (LD50) of the B. pseudomallei strains was determined in BALB/c mice challenged via aerosolization. To determine the LD50, four groups of mice (n = 10 mice per group) were exposed to nebulizer concentrations of 1 × 105, 1 × 106, 1 × 107 or 1 × 108 CFU/ml and monitored for survival. For each strain, the LD50 was calculated using methods described by Reed and Muench34.
Animals
Seven- to eight-week-old, female, BALB/c mice were obtained from Harlan Laboratories. Animals were housed in standard microisolator cages (n = 5 mice per cage) and were provided water and chow ad libitum. Mice were acclimated to study housing for at least seven days prior to bacterial infection.
Aerosol exposure
Groups of twenty un-anesthetized mice were exposed to aerosolized B. pseudomallei created by a three-jet collison nebulizer for 15 minutes at a constant flow rate of 30 L/min in a Biaera plastic aerosol rodent exposure box housed within a Class III biological safety cabinet in a biosafety level-3 suite using an automated aerosol exposure system. Animals were placed inside plastic restraint cones purchased from In-Tox products L.L.C. (Moriarty, NM). Only the noses of the animal were exposed and the plastic restraints were arranged inside perforated stainless steel aerosol boxes purchased from Biaera Technologies L.L.C. (Frederick, MD). Shortly, prior to exposure, the perforated aerosol boxes were placed inside the Biaera plastic aerosol rodent exposure box. The particle sizes generated with this system have an average of 1–2 μm mass mean aerodynamic diameter. Relative humidity was a maintained within a range of 65–68% and temperature was ambient (approximately 20–22°C). Doses presented (Dp) to each group of animals were determined by performing standard colony forming unit (CFU) counts on the samples collected from an all-glass impinger (SKC BioSampler; SKC Inc., Eighty Four, PA). Luria-Bertani broth with 4% glycerol (LBG) containing approximately 20 μL of antifoam 204 (Sigma Aldrich) was used as the collection medium in the impinger for CFU determination. The Dp was calculated using the following formulas: Dp (CFU) = CAero(CFU/mL) × exposure time (min) × minute volume (mL); minute volume = 2.1(weight [g])0.75.
Pathogenesis study design
Mice were exposed to a nebulizer concentration of 1 × 105 CFU of B. pseudomallei strain K96243 or HBPUB10303a by aerosolization as described above. Two groups of mice were used for the pathogenesis study. One group of mice (n = 10) were used to monitor survival. The second group of mice (n = 50) was used to sample mice (randomly selected) on days 0, 1, 3, 7, 10 and 14. All mice were implanted with IPTT-300 temperature chips (Biomedic Data System, Seaford, DE) to identify individual animals and monitor body temperature throughout the studies. Temperature, body weight and signs of clinical disease were observed daily. Moribund animals, defined as those demonstrating severe body weight loss, lethargy, paralysis and/or respiratory distress were humanely euthanized. One group of mice (n = 5) was selected at each time point for CFU determination. Tissues collected for CFU determination were homogenized in 1 mL of PBS using a tissue grinder (Covidien, Mansfield, MA) and then enumerated by standard plate count onto Ashdown agar plates. A second group of mice (n = 5) was selected at each time point for histopathological analysis. Blood was collected from all animals at the time of necropsy for chemokine/cytokine, blood chemistry and hematological analysis.
Histopathological evaluation
Mice were euthanized by cardiac perfusion under anesthesia and a complete necropsy was performed to collect the lung, liver, spleen and brain for histopathology. Tissues were fixed in 10% formalin and then removed from bio-containment and processed for histopathology. The tissues were paraffin-embedded, sectioned and then stained with Hematoxylin and Eosin (H&E). Duplicate sections were stained with myeloperoxidase (MPO). All sections were examined microscopically for tissue abnormalities.
Hematology and blood chemistry
Whole blood collected during necropsy was added either to and EDTA tube for complete blood count determination using a Hemavet (Drew Scientific, Dallas, TX), or added to a lithium heparin tube for clinical chemistry analysis using the comprehensive diagnostic panel analyzed on a Vetscan (Abaxis, Union City, CA). Evaluated hematology parameters included the following parameters: white blood cells (WBC), total number and percentage of neutrophils (NE), lymphocytes (LY), monocytes (MO), eosinophils (EO), and basophils (BA), red blood cells (RBC), hemoglobin (HGB), hematocrit (HCT), mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), red blood cell distribution width (RDW), platelets (PLT), and mean platelet volume (MPV). The clinical chemistry parameters evaluated by the Vetscan included the following: albumin (ALB), alkaline phosphatase (ALP), alanine aminotransferase (ALT), amylase (AMY), blood urea nitrogen (BUN), calcium (Ca++), creatinine (CRE), globulin (GLOB), glucose (GLU), potassium (K+), sodium (Na+), phosphorus (PHOS), total bilirubin (Tbil), and total protein (TP).
Cytokine quantification
Whole blood was collected by cardiac puncture at necropsy from mice infected with B. pseudomallei K96243 or HPUB10303a at day 1, 3, 7, 10, and 14 post-infection. Blood was added to a collection tube without anti-coagulant, allowed to clot at room temperature for 10–30 minutes and then centrifuged for 10–15 minutes. Separated serum was frozen at −80°C followed by γ-irradiation using a JL Shepherd Model 109–68 Cobalt-60 Research Irradiator (JL Shepherd & Associates, San Fernando, CA 91340). Samples were irradiated on dry ice until 5 MRAD of exposure was reached and sterility was verified by plating 10% of the serum volume on Ashdown agar. To assess the chemokine/cytokine levels in the collected serum, a murine bioplex ELISA kit (BioRad Bio-Plex Pro™ Mouse Cytokine 23-plex Assay) was used according to the manufacturer's recommendations using serum samples diluted 1:3 with diluent. Target molecules included IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-17A, eotaxin, G-CSF, GM-CSF, IFN-γ KC, MCP-1 (MCAF), MIP-1α, MIP-1β, RANTES, and TNF-α. A quantitative assessment of each molecule was performed using a standard curve generated via standards provided with the kit. Serum samples from control (non-infected) mice were analyzed in parallel and used to establish baseline ranges across treatment days. Data values were calculated in reference to the standard curve by linear regression as recommended by the manufacturer and as we have previously described52. Sample results that were out of range (OOR) high (>OOR) were assigned to the highest extrapolated value to provide a conservative estimate that allowed statistical analysis.
Statistical analysis
Survival curves were calculated by Kaplan Meier survival analysis with log-rank tests between groups. Student's t-test or one-way analysis of variance was performed using a 95% confidence interval. All analyses were done using GraphPad Prism 4.0 (GraphPad Software Inc., San Diego, CA). A P-value of 0.05 or less was considered significant.
Author Contributions
Author contributions: S.M., J.W.P., T.B., J.W.L., J.J.E. and A.G.T. designed research; S.M., L.A.Y., C.A.B., S.V., E.S., J.J.E. and A.G.T. performed research; L.A.Y., C.A.B., S.V., E.S. and J.J.E. contributed new reagents/analytic tools; S.M., L.A.Y., E.S., J.W.P., T.B., J.W.L., J.J.E. and A.G.T. analyzed data; and S.M., L.A.Y., E.S., T.B., J.J.E. and A.G.T. wrote the paper.
Supplementary Material
Supplementary Information
Acknowledgments
We want to thank R. Katie Johnston, Barbara Judy and Tiffany Mott for technical assistance; Drs. Lisa Morici, Vanessa Sperandio, Mary Burtnick and Paul Brett for insightful constructive comments; and BARDA officials for criticisms. This Project has been funded with federal funds from Biomedical Advanced Research and Development Authority, Department of Health and Human Services under contract: HHSO100201100015: Task Order No. HHSO10033002T.
References
- Currie B. J., Fisher D. A., Anstey N. M. & Jacups S. P. Melioidosis: acute and chronic disease, relapse and re-activation. Trans R Soc Trop Med Hyg 94, 301–304 (2000). [DOI] [PubMed] [Google Scholar]
- Currie B. J., Dance D. A. & Cheng A. C. The global distribution of Burkholderia pseupseudomallei and melioidosis: an update. Trans R Soc Trop Hyg 102 Suppl 1, S1–4 (2008). [DOI] [PubMed] [Google Scholar]
- Wiersinga W. J., Currie B. J. & Peacock S. J. Melioidosis. N Engl J Med 367, 1035–1044 (2012). [DOI] [PubMed] [Google Scholar]
- Dance D. A. Melioidosis. Curr Opin Infect Dis 15, 127–132 (2002). [DOI] [PubMed] [Google Scholar]
- Limmathurotsakul D. et al. Systematic review and consensus guidelines for environmental sampling of Burkholderia pseudomallei. PLoS Neg Trop Dis 7, e2105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie B. J. et al. Endemic melioidosis in tropical northern Australia: a 10-year prospective study and review of the literature. Clin Infect Dis 31, 981–986 (2000). [DOI] [PubMed] [Google Scholar]
- Cheng A. C. et al. Clinical definitions of melioidosis. Am J Trop Med Hyg 88, 411–413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngauy V., Lemeshev Y., Sadkowski L. & Crawford G. Cutaneous melioidosis in a man who was taken as a prisoner of war by the Japanese during World War II. J Clin Microbiol 43, 970–972 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A. C. & Currie B. J. Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev 18, 383–416 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie B. J. Melioidosis: an important cause of pneumonia in residents of and travellers returned from endemic regions. Eur Respir J 22, 542–550 (2003). [DOI] [PubMed] [Google Scholar]
- Waag D. & DeShazer D. in Biological Weapons of Defense: Infectious Diseases and Counterbioterrorism. (ed Lebeda FJ, Lindler LE, Korch GW) 209–237, (Humana Press, 2005).
- Carr-Gregory B. & Waag D. in Medical Aspects of Biological Warfare. (ed Dembek Z. F.) 121–146, (Borden Institute, 2007).
- Lehavi O., Aizenstien O., Katz L. H. & Hourvitz A. [Glanders–a potential disease for biological warfare in humans and animals]. Harefuah 141 Spec No, 88–91, 119. (2002). [PubMed] [Google Scholar]
- Wheelis M. in Biological and Toxin Weapons Research, Development, and Use from the Middle Ages to 1945 (ed Moon JEvC Geissler E.) 35–72 (Oxford University Press, 1999).
- Thibault F. M., Hernandez E., Vidal D. R., Girardet M. & Cavallo J. D. Antibiotic susceptibility of 65 isolates of Burkholderia pseudomallei and Burkholderia mallei to 35 antimicrobial agents. J Antimicro Chemother 54, 1134–1138 (2004). [DOI] [PubMed] [Google Scholar]
- Viktorov D. V. et al. High-level resistance to fluoroquinolones and cephalosporins in Burkholderia pseudomallei and closely related species. Trans R Soc Trop Med Hyg 102 Suppl 1, S103–110 (2008). [DOI] [PubMed] [Google Scholar]
- Van Zandt K. E., Tuanyok A., Keim P. S., Warren R. L. & Gelhaus H. C. An objective approach for Burkholderia pseudomallei strain selection as challenge material for medical countermeasures efficacy testing. Front Cell Infect Microbiol 2, 120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz D. L., Vogel P., Brown D. R. & Waag D. M. The hamster model of intraperitoneal Burkholderia mallei (glanders). Vet Pathol 36, 276–291 (1999). [DOI] [PubMed] [Google Scholar]
- Warawa J. & Woods D. E. Type III secretion system cluster 3 is required for maximal virulence of Burkholderia pseudomallei in a hamster infection model. Fems Microbiol Lett 242, 101–108 (2005). [DOI] [PubMed] [Google Scholar]
- Brett P. J., Deshazer D. & Woods D. E. Characterization of Burkholderia pseudomallei and Burkholderia pseudomallei-like strains. Epidemiol Infect 118, 137–148 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leakey A. K., Ulett G. C. & Hirst R. G. BALB/c and C57Bl/6 mice infected with virulent Burkholderia pseudomallei provide contrasting animal models for the acute and chronic forms of human melioidosis. Microb Pathog 24, 269–275 (1998). [DOI] [PubMed] [Google Scholar]
- Bondi S. K. & Goldberg J. B. Strategies toward vaccines against Burkholderia mallei and Burkholderia pseudomallei. Exp Rev Vaccines 7, 1357–1365 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warawa J. M. Evaluation of surrogate animal models of melioidosis. Frontiers in microbiology 1, 141 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titball R. W. et al. Burkholderia pseudomallei: animal models of infection. Trans R Soc Trop Med Hyg 102 Suppl 1S111–116 (2008). [DOI] [PubMed] [Google Scholar]
- Judy B. M., Whitlock G. C., Torres A. G. & Estes D. M. Comparison of the in vitro and in vivo susceptibilities of Burkholderia mallei to Ceftazidime and Levofloxacin. BMC Microbiol 9, 88 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock G. C. et al. Burkholderia mallei cellular interactions in a respiratory cell model. J Med Microbiol 58, 554–562 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey S. et al. In vivo Bioluminescence Imaging of Burkholderia mallei Respiratory Infection and Treatment in the Mouse Model. Fron Microbiol 2, 174 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyear A. et al. Protection from pneumonic infection with Burkholderia species by inhalational immunotherapy. Infection and Immunity 77, 1579–1588 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever M. S., Nelson M., Stagg A. J., Beedham R. J. & Simpson A. J. Experimental acute respiratory Burkholderia pseudomallei infection in BALB/c mice. Int J Exp Pathol 90, 16–25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaus H. C. et al. Efficacy of post exposure administration of doxycycline in a murine model of inhalational melioidosis. Sci Rep 3, 1146 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh J. A., Fritz D. L., Waag D. M., Hartings J. M. & Andrews G. P. Biodefense-Driven Murine Model of Pneumonic Melioidosis. Infection and Immunity 71, 584–587 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes K. B. et al. Trimethoprim/sulfamethoxazole (co-trimoxazole) prophylaxis is effective against acute murine inhalational melioidosis and glanders. Int J Antimicrob Agents 41, 552–557 (2013). [DOI] [PubMed] [Google Scholar]
- Tan G. Y. et al. Burkholderia pseudomallei aerosol infection results in differential inflammatory responses in BALB/c and C57Bl/6 mice. J Med Microbiol 57, 508–515 (2008). [DOI] [PubMed] [Google Scholar]
- Reed LJ M. H. A simple method for estimating fifty percent end points. Am J Hyg, 27, 493–497 (1938). [Google Scholar]
- Lafontaine E. R. et al. Use of a Safe, Reproducible, and Rapid Aerosol Delivery Method to Study Infection by Burkholderia pseudomallei and Burkholderia mallei in Mice. PloS One 8, e76804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilhante R. S. et al. Clinical-epidemiological features of 13 cases of melioidosis in Brazil. J Clin Microbiol 50, 3349–3352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis T. J., Rolim D. B. & Sousa Ade Q. Melioidosis in the Americas. Am J Trop Med Hyg 75, 947–954 (2006). [PubMed] [Google Scholar]
- Rolim D. B. et al. Melioidosis, northeastern Brazil. Emerg Infect Dis 11, 1458–1460 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenson B., Fuxench Z., Morales J. A., Suarez-Villamil R. A. & Souchet L. M. Severe community-acquired pneumonia and sepsis caused by Burkholderia pseudomallei associated with flooding in Puerto Rico. Bol Asoc Med P R 95, 17–20 (2003). [PubMed] [Google Scholar]
- Dorman S. E., Gill V. J., Gallin J. I. & Holland S. M. Burkholderia pseudomallei infection in a Puerto Rican patient with chronic granulomatous disease: case report and review of occurrences in the Americas. Clin Infect Dis 26, 889–894 (1998). [DOI] [PubMed] [Google Scholar]
- Josephson J. Melioidosis: an emerging tropical health problem. Environ Sci Technol 35, 454A–457A (2001). [DOI] [PubMed] [Google Scholar]
- West T. E., Myers N. D., Liggitt H. D. & Skerrett S. J. Murine pulmonary infection and inflammation induced by inhalation of Burkholderia pseudomallei. Int J Exp Pathol 93, 421–428 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. J. et al. Particle-size dependent effects in the Balb/c murine model of inhalational melioidosis. Fron Cell Infect Micrbobiol 2, 101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieves W. et al. A naturally derived outer-membrane vesicle vaccine protects against lethal pulmonary Burkholderia pseudomallei infection. Vaccine 29, 8381–8389 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler P. et al. Early granuloma formation after aerosol Mycobacterium tuberculosis infection is regulated by neutrophils via CXCR3-signaling chemokines. Eur J Immunol 33, 2676–2686 (2003). [DOI] [PubMed] [Google Scholar]
- Pankla R. et al. Genomic transcriptional profiling identifies a candidate blood biomarker signature for the diagnosis of septicemic melioidosis. Genome Biol 10, R127 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowska E. & Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol 13, 159–175 (2013). [DOI] [PubMed] [Google Scholar]
- Serbina N. V., Jia T., Hohl T. M. & Pamere E. G. Monocyte-mediated defense against microbial pathogens. Annu Rev Immunol 26, 421–452 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila I. S. et al. Efficient cytokine-induced IL-13 production by mast cells requires both IL-33 and IL-3. J Allergy Clin Immun 132, 704–712 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asquith K. L., Ramshaw H., Lopez A. & Foster P. S. The IL-3/IL-5/GM-CSF Common beta Receptor Plays a Pivotal Role in Regulating Th2 Immunity and Allergic Airway Inflammation. Faseb J 22, 1199–1206 (2008). [DOI] [PubMed] [Google Scholar]
- Fogli L. K. et al. T cell-derived IL-17 mediates epithelial changes in the airway and drives pulmonary neutrophilia (vol 191, pg 3100, 2013). J Immunol 191, 5318–5318 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judy B. M. et al. Prophylactic application of CpG oligonucleotides augments the early host response and confers protection in acute melioidosis. PloS One 7, e34176 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information