Abstract
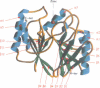
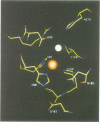
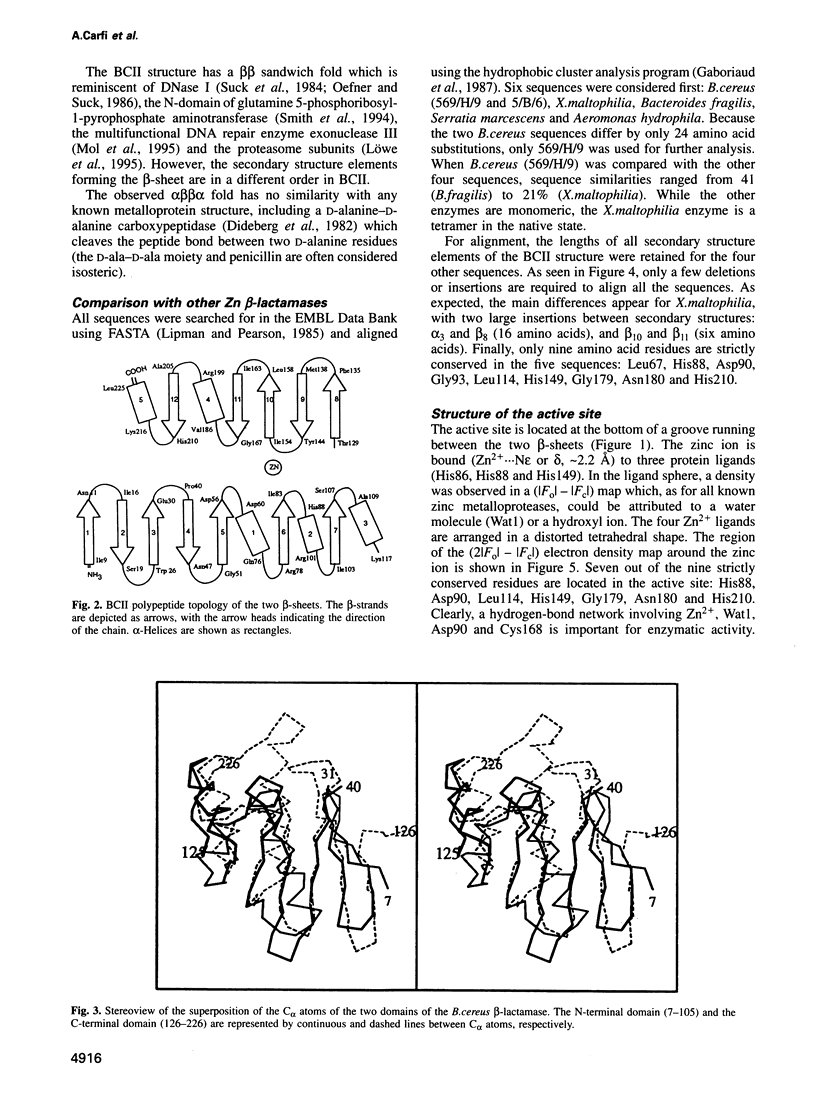
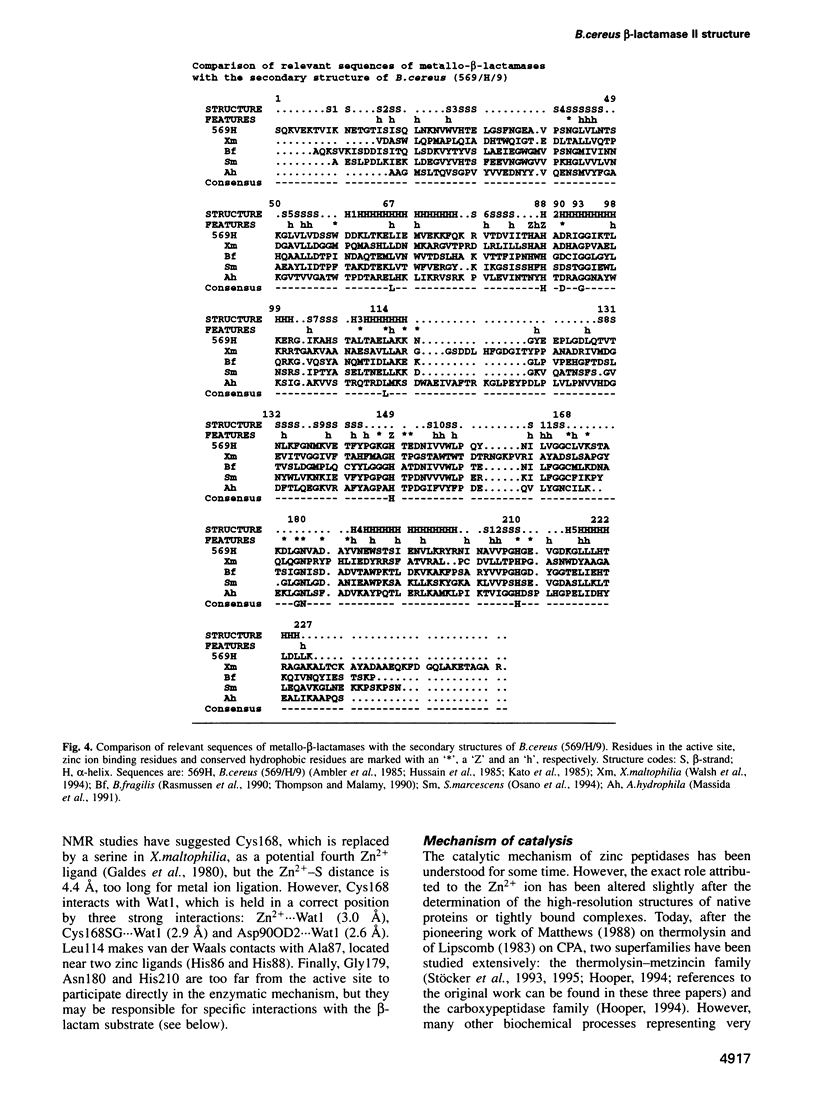
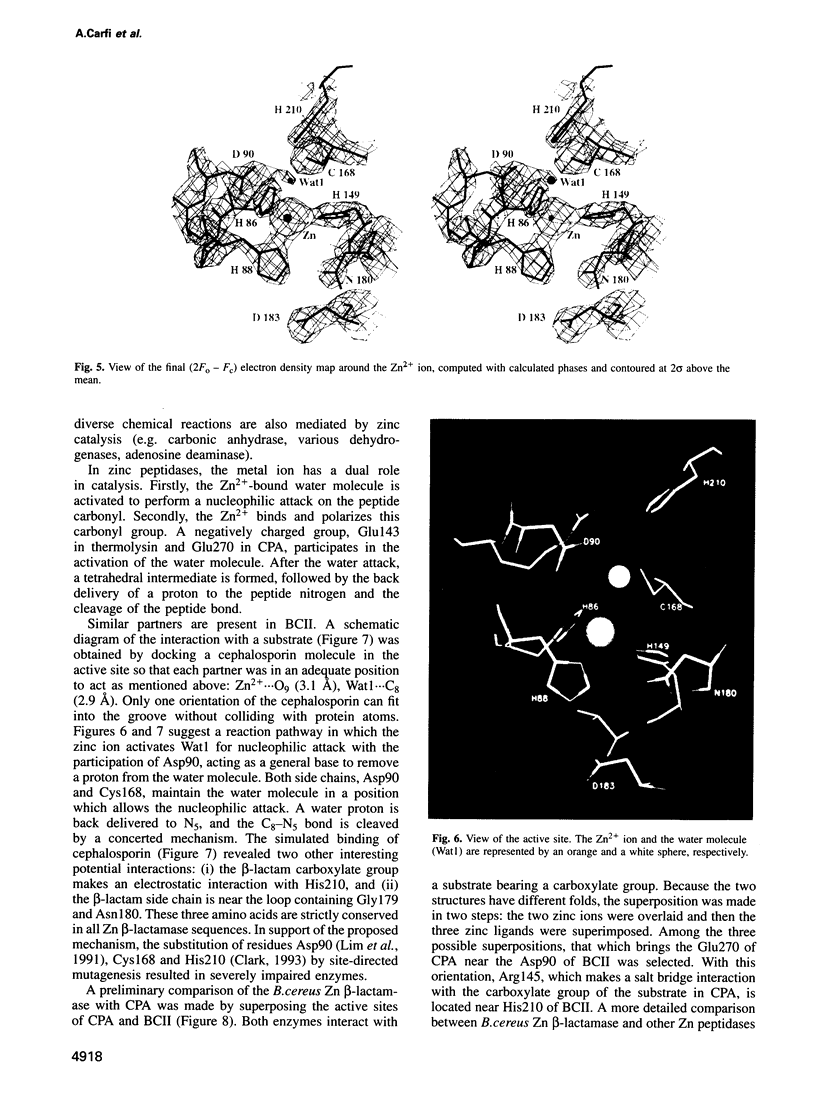
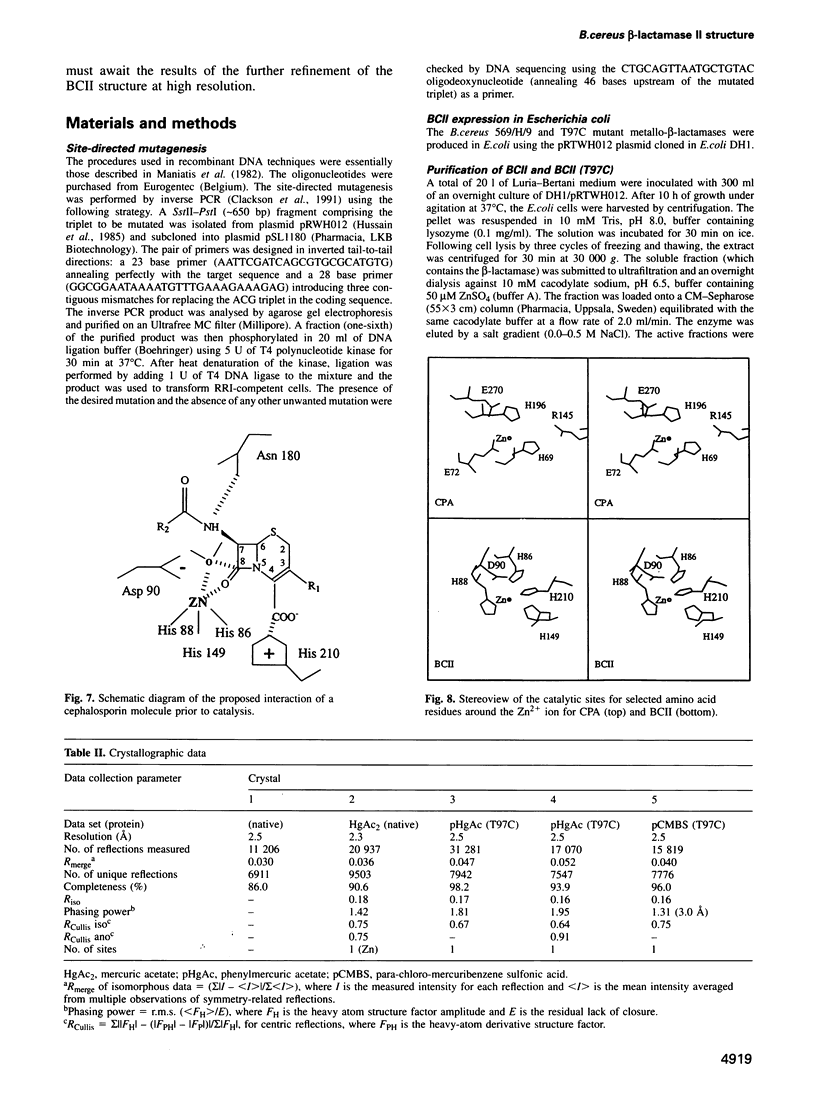
The 3-D structure of Bacillus cereus (569/H/9) beta-lactamase (EC 3.5.2.6), which catalyses the hydrolysis of nearly all beta-lactams, has been solved at 2.5 A resolution by the multiple isomorphous replacement method, with density modification and phase combination, from crystals of the native protein and of a specially designed mutant (T97C). The current model includes 212 of the 227 amino acid residues, the zinc ion and 10 water molecules. The protein is folded into a beta beta sandwich with helices on each external face. To our knowledge, this fold has never been observed. An approximate internal molecular symmetry is found, with a 2-fold axis passing roughly through the zinc ion and suggesting a possible gene duplication. The active site is located at one edge of the beta beta sandwich and near the N-terminal end of a helix. The zinc ion is coordinated by three histidine residues (86, 88 and 149) and a water molecule. A sequence comparison of the relevant metallo-beta-lactamases, based on this protein structure, highlights a few well-conserved amino acid residues. The structure shows that most of these residues are in the active site. Among these, aspartic acid 90 and histidine 210 participate in a proposed catalytic mechanism for beta-lactam hydrolysis.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P., Daniel M., Fleming J., Hermoso J. M., Pang C., Waley S. G. The amino acid sequence of the zinc-requiring beta-lactamase II from the bacterium Bacillus cereus 569. FEBS Lett. 1985 Sep 23;189(2):207–211. doi: 10.1016/0014-5793(85)81024-3. [DOI] [PubMed] [Google Scholar]
- Baldwin G. S., Galdes A., Hill H. A., Smith B. E., Waley S. G., Abraham E. P. Histidine residues of zinc ligands in beta-lactamase II. Biochem J. 1978 Nov 1;175(2):441–447. doi: 10.1042/bj1750441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin G. S., Waley S. G., Abraham E. P. Identification of histidine residues that act as zinc ligands in beta-lactamase II by differential tritium exchange. Biochem J. 1979 Jun 1;179(3):459–463. doi: 10.1042/bj1790459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell R., Emanuel E. L., Gagnon J., Waley S. G. The production and molecular properties of the zinc beta-lactamase of Pseudomonas maltophilia IID 1275. Biochem J. 1985 Aug 1;229(3):791–797. doi: 10.1042/bj2290791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell R., Schäffer A., Waley S. G., Auld D. S. Changes in the coordination geometry of the active-site metal during catalysis of benzylpenicillin hydrolysis by Bacillus cereus beta-lactamase II. Biochemistry. 1986 Nov 4;25(22):7208–7215. doi: 10.1021/bi00370a066. [DOI] [PubMed] [Google Scholar]
- Crompton I. E., Waley S. G. The determination of specificity constants in enzyme-catalysed reactions. Biochem J. 1986 Oct 1;239(1):221–224. doi: 10.1042/bj2390221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R. B., Abraham E. P. Metal cofactor requirements of beta-lactamase II. Biochem J. 1974 Oct;143(1):129–135. doi: 10.1042/bj1430129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dideberg O., Charlier P., Dive G., Joris B., Frère J. M., Ghuysen J. M. Structure of a Zn2+-containing D-alanyl-D-alanine-cleaving carboxypeptidase at 2.5 A resolution. Nature. 1982 Sep 30;299(5882):469–470. doi: 10.1038/299469a0. [DOI] [PubMed] [Google Scholar]
- Felici A., Amicosante G., Oratore A., Strom R., Ledent P., Joris B., Fanuel L., Frère J. M. An overview of the kinetic parameters of class B beta-lactamases. Biochem J. 1993 Apr 1;291(Pt 1):151–155. doi: 10.1042/bj2910151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriaud C., Bissery V., Benchetrit T., Mornon J. P. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987 Nov 16;224(1):149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- Galdes A., Hill H. A., Baldwin G. S., Waley S. G., Abraham E. P. The 1H nuclear-magnetic-resonance spectroscopy of cobalt(II)-beta-lactamase II. Biochem J. 1980 Jun 1;187(3):789–795. doi: 10.1042/bj1870789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper N. M. Families of zinc metalloproteases. FEBS Lett. 1994 Oct 31;354(1):1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- Hussain M., Carlino A., Madonna M. J., Lampen J. O. Cloning and sequencing of the metallothioprotein beta-lactamase II gene of Bacillus cereus 569/H in Escherichia coli. J Bacteriol. 1985 Oct;164(1):223–229. doi: 10.1128/jb.164.1.223-229.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato C., Kudo T., Watanabe K., Horikoshi K. Nucleotide sequence of the beta-lactamase gene of alkalophilic Bacillus sp. strain 170. J Gen Microbiol. 1985 Dec;131(12):3317–3324. doi: 10.1099/00221287-131-12-3317. [DOI] [PubMed] [Google Scholar]
- Lim H. M., Iyer R. K., Pène J. J. Site-directed mutagenesis of dicarboxylic acids near the active site of Bacillus cereus 5/B/6 beta-lactamase II. Biochem J. 1991 Jun 1;276(Pt 2):401–404. doi: 10.1042/bj2760401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Lipscomb W. N. Structure and catalysis of enzymes. Annu Rev Biochem. 1983;52:17–34. doi: 10.1146/annurev.bi.52.070183.000313. [DOI] [PubMed] [Google Scholar]
- Löwe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995 Apr 28;268(5210):533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- Massidda O., Rossolini G. M., Satta G. The Aeromonas hydrophila cphA gene: molecular heterogeneity among class B metallo-beta-lactamases. J Bacteriol. 1991 Aug;173(15):4611–4617. doi: 10.1128/jb.173.15.4611-4617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol C. D., Kuo C. F., Thayer M. M., Cunningham R. P., Tainer J. A. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995 Mar 23;374(6520):381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- Oefner C., Suck D. Crystallographic refinement and structure of DNase I at 2 A resolution. J Mol Biol. 1986 Dec 5;192(3):605–632. doi: 10.1016/0022-2836(86)90280-9. [DOI] [PubMed] [Google Scholar]
- Osano E., Arakawa Y., Wacharotayankun R., Ohta M., Horii T., Ito H., Yoshimura F., Kato N. Molecular characterization of an enterobacterial metallo beta-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob Agents Chemother. 1994 Jan;38(1):71–78. doi: 10.1128/aac.38.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne D. J. Metallo-beta-lactamases--a new therapeutic challenge. J Med Microbiol. 1993 Aug;39(2):93–99. doi: 10.1099/00222615-39-2-93. [DOI] [PubMed] [Google Scholar]
- RAMACHANDRAN G. N., RAMAKRISHNAN C., SASISEKHARAN V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963 Jul;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- Rasmussen B. A., Gluzman Y., Tally F. P. Cloning and sequencing of the class B beta-lactamase gene (ccrA) from Bacteroides fragilis TAL3636. Antimicrob Agents Chemother. 1990 Aug;34(8):1590–1592. doi: 10.1128/aac.34.8.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saino Y., Kobayashi F., Inoue M., Mitsuhashi S. Purification and properties of inducible penicillin beta-lactamase isolated from Pseudomonas maltophilia. Antimicrob Agents Chemother. 1982 Oct;22(4):564–570. doi: 10.1128/aac.22.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. L., Zaluzec E. J., Wery J. P., Niu L., Switzer R. L., Zalkin H., Satow Y. Structure of the allosteric regulatory enzyme of purine biosynthesis. Science. 1994 Jun 3;264(5164):1427–1433. doi: 10.1126/science.8197456. [DOI] [PubMed] [Google Scholar]
- Stöcker W., Gomis-Rüth F. X., Bode W., Zwilling R. Implications of the three-dimensional structure of astacin for the structure and function of the astacin family of zinc-endopeptidases. Eur J Biochem. 1993 May 15;214(1):215–231. doi: 10.1111/j.1432-1033.1993.tb17915.x. [DOI] [PubMed] [Google Scholar]
- Stöcker W., Grams F., Baumann U., Reinemer P., Gomis-Rüth F. X., McKay D. B., Bode W. The metzincins--topological and sequential relations between the astacins, adamalysins, serralysins, and matrixins (collagenases) define a superfamily of zinc-peptidases. Protein Sci. 1995 May;4(5):823–840. doi: 10.1002/pro.5560040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suck D., Oefner C., Kabsch W. Three-dimensional structure of bovine pancreatic DNase I at 2.5 A resolution. EMBO J. 1984 Oct;3(10):2423–2430. doi: 10.1002/j.1460-2075.1984.tb02149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton B. J., Artymiuk P. J., Cordero-Borboa A. E., Little C., Phillips D. C., Waley S. G. An X-ray-crystallographic study of beta-lactamase II from Bacillus cereus at 0.35 nm resolution. Biochem J. 1987 Nov 15;248(1):181–188. doi: 10.1042/bj2480181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. S., Malamy M. H. Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus beta-lactamase II. J Bacteriol. 1990 May;172(5):2584–2593. doi: 10.1128/jb.172.5.2584-2593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T. R., Hall L., Assinder S. J., Nichols W. W., Cartwright S. J., MacGowan A. P., Bennett P. M. Sequence analysis of the L1 metallo-beta-lactamase from Xanthomonas maltophilia. Biochim Biophys Acta. 1994 Jun 21;1218(2):199–201. doi: 10.1016/0167-4781(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Yang Y. J., Wu P. J., Livermore D. M. Biochemical characterization of a beta-lactamase that hydrolyzes penems and carbapenems from two Serratia marcescens isolates. Antimicrob Agents Chemother. 1990 May;34(5):755–758. doi: 10.1128/aac.34.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]