Abstract
Tumor recurrence after curative resection remains a major problem in patients with locally advanced colorectal cancer (CRC) treated with adjuvant chemotherapy. Genetic single nucleotide polymorphisms (SNPs) may serve as useful molecular markers to predict clinical outcomes in these patients and identify targets for future drug development. Recent in vitro and in vivo studies have demonstrated that the plastin genes, PLS3 and LCP1, are overexpressed in colon cancer cells and play an important role in tumor cell invasion, adhesion and migration. Hence, we hypothesized that functional genetic variations of plastin may direct effects on the progression and prognosis of locally advanced CRC. We tested whether functional tagging polymorphisms of PLS3 and LCP1 predict time to tumor recurrence (TTR) in 732 patients (training set: 234; validation set: 498) with stage II/III CRC. The PLS3 rs11342 and LCP1 rs4941543 polymorphisms were associated with a significantly increased risk for recurrence in the training set. PLS3 rs6643869 showed a consistent association with TTR in the training and validation set, when stratified by gender and tumor location. Female patients with the PLS3 rs6643869 AA genotype had the shortest median TTR compared to those with any G allele in the training set [1.7 vs. 9.4 years; HR2.84 (95% CI=1.32–6.1); p=0.005] and validation set [3.3 vs. 13.7 years; HR2.07 (95%CI=1.09–3.91); p=0.021]. Our findings suggest that several SNPs of the PLS3 and LCP1 genes could serve as gender and/or stage-specific molecular predictors of tumor recurrence in stage II/III CRC patients as well as potential therapeutic targets.
Keywords: PLS3, LCP1, colon cancer, tumor recurrence, gender
Introduction
Colorectal cancer (CRC) is the leading cause of death from gastrointestinal malignancy, with approximately 50,830 deaths expected in 2013 (1). Surgical resection affords the only chance for cure, and adjuvant chemotherapy is recommended for high-risk stage II and all stage III tumors as it reduces risk of recurrence and prolongs disease-free survival (2, 3). However, even with adjuvant chemotherapy, 20–30% of high-risk stage II and 50–65% of stage III patients relapse within 5 years. Disease recurrence and survival are related to tumor histology, grade and stage. Such conventional clinicopathological staging has served as the standard measure of prognosis for many years. We have yet to identify reliable molecular markers to predict tumor recurrence in patients with stage II–III disease who have received adjuvant chemotherapy.
While a number of genes have been implicated in colorectal cancer metastasis, the precise mechanisms directing the migration and invasion of tumor cells into specific organs remain elusive. Modulation of the actin cytoskeleton is critical for tumor cell migration and invasion (4), and changes in the actin cytoskeleton are accomplished by a variety of actin-binding proteins, such as plastins (4, 5). Moreover, increasing evidence suggests that overexpression of plastins in cultured epithelial cells can increase cell motility (6). Therefore, plastins may not only be a potential biomarker but also serve as a valuable target for inhibiting tumor cell invasion and metastasis.
The human plastin family is comprised of three protein isoforms: T-plastin (PLS3), L-plastin (LCP1) and I-plastin. I-plastin is specifically expressed in the small intestine and kidney. The cell-type-specific expression of the other isoforms (PLS3 and LCP1) is regulated under physiological conditions. However, ectopic expression of plastins in malignant cells has been observed in many studies (7, 8). PLS3 and LCP1, which have 80% amino acid homology, also share the unique property of cross-linking actin filaments into tight bundles. Although PLS3 and LCP1 are related in structure and function, they are encoded by two distinct genes located on chromosomes X and 13, respectively (9), and possess other unique features. Specifically, PLS3 is expressed in a wide variety of cells except for hematopoietic cells. Most neoplastic cells derived from solid tumors also express PLS3. Conversely, LCP1 is expressed in most human cancer and hematopoietic cell lines (7). PLS3 up-regulation by the lamina A filament protein has been correlated with increased invasiveness in CRC (10), and LCP1 overexpression plays a significant role in proliferation and invasion in colon cancer cell lines (11). Furthermore, LCP1 has been proposed to mediate cell adhesion and motility by binding actin (9, 12). However, little is known about the role of PLS3 and LCP1 in colorectal tumor metastasis.
Previously, data from our group and others have shown that certain genetic polymorphisms may predict clinical outcomes in colon cancer patients treated with adjuvant chemotherapy (13, 14). These common single nucleotide variations may alter gene transcription, translation or splicing, thereby causing different tumor recurrence and chemoresistance patterns. Since plastin genes play an important role in tumor cell invasion, adhesion and migration, functional variations in these genes may translate into direct effects on prognosis in patients with locally advanced colon cancer. To test this hypothesis, we performed a comprehensive study using both a training and validation set to evaluate associations between 6 functional PLS3 (4) and LCP1 (2) polymorphisms with clinical outcomes, including TTR, in colon cancer patients treated with adjuvant chemotherapy. Lastly, we explored whether PLS3 and LCP1 polymorphisms demonstrated gender or stage-specific differences and if such differences correlated with clinical outcomes.
Materials and Methods
Eligible Patients
A total of 234 patients with stage III and high-risk stage II colon cancer were included in this study as the training set (USC cohort) (Table 1). High risk stage II disease was defined by the presence of any of the following: T4 tumor, positive surgical margins, grade 3 or 4 histology in the absence of MSI-high status, perforation, obstruction, lymphovascular or perineural invasion, or less than 12 lymph nodes sampled at time of surgical resection (15). All patients were treated with 5-FU-based adjuvant chemotherapy at the Norris Comprehensive Cancer Center/University of Southern California (NCCC/USC) or the Los Angeles County/USC-Medical Center (LAC/USCMC) from 1987 to 2007. For the validation set (Table 1), a total of 498 patients with histologically confirmed stage II (both high risk and low risk tumors) and stage III colon cancer treated at the Division of Clinical Oncology, Department of Medicine, Medical University of Graz (MUG) from 2000 to 2009 were included. All patients were treated with a 5-fluorouracil (5-FU)-based regimen. All patients were included in the colon cancer surveillance program of NCCC/USC, LAC/USCMC, or MUG, providing the following information: history and physical examination and carcinoembryonic antigen (CEA) levels every 3 months for 3 years and every 6 months at years 4 and 5 after surgery; colonoscopy at year 1 and thereafter every 3–5 years; and computed tomography scans of chest and abdomen every 6 months for the first 3 years. Patient data were collected retrospectively through chart review. Whole blood was collected at the time of diagnosis and stored at −80 °C. The study was approved by the Institutional Review Boards of MUG and USC, and all study participants signed informed consent for the analysis of molecular correlates.
Table 1.
Colorectal cancer baseline patient characteristics
| Training set (USC cohort) | Validation set (Austrian cohort) | P value* | |||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Age, years | |||||
| <55 | 86 | 36.75 | 87 | 17.47 | |
| 55–64 | 82 | 35.04 | 138 | 27.71 | |
| ≥65 | 66 | 28.21 | 273 | 54.82 | <0.001 |
| Sex | |||||
| Female | 107 | 45.72 | 230 | 46.18 | |
| Male | 127 | 54.28 | 268 | 53.82 | 0.91 |
| T | |||||
| T1 | 2 | 0.85 | 10 | 2.01 | |
| T2 | 14 | 5.98 | 22 | 4.42 | |
| T3 | 187 | 79.91 | 358 | 71.89 | <0.001 |
| T4 | 27 | 11.54 | 108 | 21.69 | |
| Tx | 4 | 1.72 | 0 | ||
| N | |||||
| Negative | 105 | 44.87 | 192 | 38.55 | |
| N1 | 72 | 30.77 | 195 | 39.16 | |
| N2 | 57 | 24.36 | 111 | 22.29 | 0.085 |
| Grade | |||||
| Well | 11 | 2.18 | 25 | 5.02 | |
| Moderate | 151 | 64.53 | 326 | 65.46 | |
| Poor/undifferentiated | 54 | 23.08 | 145 | 29.12 | 0.51 |
| Missing | 18 | 10.21 | 2 | 0.40 | |
| Stage | |||||
| II | 105 | 44.87 | 192 | 38.55 | |
| III | 129 | 55.13 | 306 | 61.45 | 0.10 |
| N of resected lymph nodes | |||||
| ≤12 | 70 | 29.91 | 72 | 14.46 | |
| >12 | 145 | 61.97 | 426 | 85.54 | <0.001 |
| Missing | 19 | 8.12 | 0 | 0 | |
| Tumor side | |||||
| Left | 110 | 47.01 | 188 | 37.75 | |
| Right | 115 | 49.15 | 310 | 62.25 | 0.005 |
| Left and right | 4 | 1.71 | 0 | ||
| Missing | 5 | 2.13 | 0 | ||
| Adjuvant treatment | |||||
| 5-FU | 151 | 64.53 | 171 | 34.41 | |
| 5-FU/LV/Oxaliplatin | 60 | 25.64 | 326 | 65.59 | <0.001 |
| 5-FU/LV/Irinotecan | 23 | 9.83 | 0 | 0 | |
| Ethnicity | |||||
| Asian | 34 | 14.53 | 0 | 0 | |
| African American | 15 | 6.41 | 0 | 0 | |
| Caucasian | 123 | 52.56 | 498 | 100 | <0.001 |
| Hispanic | 62 | 26.5 | 0 | 0 | |
Based on χ2 test
Candidate Polymorphisms
Stringent pre-defined criteria were used to select candidate single nucleotide polymorphisms (SNPs) and included: (a) polymorphisms which could alter gene function in a biologically relevant manner (as predicted by the Functional-Single-Nucleotide-Polymorphism (F-SNP)database) (16, 17); (b) a minor allele frequency ≥10% in Caucasians (with relative allelic frequencies of polymorphisms in different ethnicities obtained from the genetics section in the Ensembl Genome Browser: http://uswest.ensembl.org/index.html). This study focused on testing the potentially functional genetic variants located at the 5′UTR, 3′UTR, or coding region, as well as tagging SNPs. (Supplementary Table 1).
Genotyping
Genomic DNA was extracted from peripheral blood using the QIAmp-kit (Qiagen) and according to the manufacturer’s instructions (www.qiagen.com). For the USC cohort, the candidate polymorphisms were tested using PCR-based direct DNA sequence analysis. Briefly, forward and reverse primers were used for PCR amplification. PCR fragments were sequenced on an ABI 3100A Capillary Genetic Analyzer (Applied Biosystems) and analyzed in either sense or antisense directions for the presence of the polymorphism. DNA sequence analyses were performed using the ABI Sequencing Scanner v1.0 (Applied Biosystems). The investigator analyzing the germline polymorphisms was blinded to the clinical dataset. For the MUG cohort, genotypes were centrally determined by a 5′-exonuclease assay (TaqMan) at the Medical University of Graz. Primer and probe sets were designed and manufactured using Applied Biosystems ‘Assay-by-Design’ custom service (Applera, Austria). General TaqMan reaction conditions were according to the manufacturer of the assays. End-point fluorescence was measured in a Lambda Fluoro 320 plus plate reader (MWG Biotech AG, Germany) using excitation/emission filters of 485/530 nm and 530/572 nm, respectively. In the plot, genotype groups were identified as separate and distinguishable clusters. To ensure methodological consistency and quality control, genotype determination was repeated in at least 96 randomly selected samples.
Statistical Analysis
The primary endpoint of this study was time to tumor recurrence (TTR), which was computed from the date of diagnosis of colon cancer to the date of the first documented tumor recurrence. If tumor recurrence was not observed, TTR was censored at the time of death or at the last follow-up. Deviation from Hardy-Weinberg equilibrium for allelic distributions of PLS3 and LCP1 SNPs were tested using a 1-degree-freedom χ2. The relationships between PLS3 and LCP1 SNPs and baseline patient and tumor characteristics were examined using χ2 or Fisher’s exact test whenever appropriate. The associations between PLS3 and LCP1 SNPs and TTR were assessed using Kaplan-Meier curves and log-rank tests in the univariate analysis, and Cox regression models adjusting for stage and type of adjuvant therapy, stratified by race in the multivariate analysis. As the PLS3 gene is located on X chromosome, the analyses for all PLS3 SNPs were conducted separately by gender. We tested the hypothesis that plastin polymorphisms predict time to recurrence in patients with stage II or III colon cancer in our USC institutional cohort first. These findings were then validated in an independent cohort. Concordance probability estimates (CPE) for the Cox proportional hazards model were calculated to evaluate the incremental contribution in discrimination provided by the PLS3 and LCP1 SNPs over the baseline prognostic factors in both cohorts (18).
SAS 9.3 (SAS Institute, Cary, NC, USA) and R package ‘CPE’ were used to perform all the analyses. All tests were 2-sided at a significance level of 0.05.
Results
The baseline characteristics of the training set and validation set patients included in this analysis are summarized in Table 1. All genotype frequencies for plastin polymorphisms analyzed were within Hardy-Weinberg equilibrium (HWE) (p>0.05).
Training Set
A total of 234 USC patients with stage II and III CRC were included in this analysis as the training set (14). All patients were diagnosed with stage III and high risk stage II CRC during the years of 1987 and 2007. The group consisted of 107 women (46%) and 127 men (54%), with a median age of 59 years (range 22–87 years). The median follow-up was 4.4 years (range 0.4–16.8 years). Of the 234 CRC patients, ninety (38.5%) patients showed tumor recurrence, with a stage III and high-risk stage II dependent 3-year recurrence probability of 0.45 ± 0.047 and 0.21 ± 0.043, respectively. The median TTR was 7.1 years (95% CI 4.9–16.8+ years), and forty-four (19%) of the 234 patients died. The median OS for this cohort has not yet been reached. Patients with stage III disease were more likely to present with recurrence, compared with patients with high risk stage II disease (log-rank p<0.001). We did not observe any significant associations between other demographic and clinicopathologic variables and TTR. In the USC cohort, plastin polymorphisms were not significantly associated with age, clinical (type of chemotherapy administered) or pathological characteristics (tumor stage, grade or lymph node status) (p>0.05).
Validation Set
In order to validate our results, we performed an analysis of the polymorphisms, PLS3 rs6643869 and LCP1 rs4941543, in an independent Austrian cohort of 498 patients with stage II and III CRC. Baseline patient characteristics, tumor biological factors and therapeutic modalities are shown in Table 1. The median age at time of diagnosis was 66 years (range 27–95 years). The median follow-up time was 64 months (range 1–186 months). The median OS of this cohort was 15.3 years (95%CI9.7–15.5+ years).
Single Nucleotide Polymorphism (SNP) Analysis
Training Set
Genotyping of PLS3 rs6643869 and LCP1 rs4941543 was successful in 180 (77%) of the 234 patients. In the remaining 54 (23%) cases, there was limited quantity and/or quality of extracted genomic DNA. As the PLS3 gene is located on the X chromosome, data were analyzed separately for men and women for the PLS3 polymorphism. In univariate analysis, PLS3 rs6643869 and LCP1 rs4941543 were significantly associated with time to tumor recurrence (TTR). Female patients with the PLS3 rs6643869 A/A genotype had a shorter median TTR of 1.7 years (95% CI 0.7–5.9+years) compared to those with the A/G or G/G genotypes, who had a median TTR of 9.4 years (95% CI 5.4–12.2+ years) (HR=2.84, 95% CI 1.32–6.10; p=0.005, log-rank test) (Figure 1, top panel; Table 2). We further analyzed female patients with PLS3 rs6643869 by tumor location. Both sides (right and left) have similar results (Supplementary Figure 1). All patients with the LCP1 rs4941543 C/C genotype had a longer median TTR of 10.7 years (95% CI 5.7–16.8+ years) compared to those with the C/T and T/T genotypes, who had a median TTR of 4.9 years (95% CI 2.0–9.0+ years) (HR=1.83, 95% CI 1.01–3.30; p=0.040, log-rank test) (Figure 1, bottom panel; Supplementary Table 2). In addition, male patients with the LCP1rs11342 T allele had a longer median TTR of 7.1 years (95% CI 4.9–11.4+ years), relative to men with the G/G genotype who had a median TTR2.3 years, (95% CI 1.5–6.8+ years) (HR=0.51, 95% CI 0.25–1.04; p=0.042) in univariate analysis (Supplementary Figure 2, Table 2).
Figure 1.
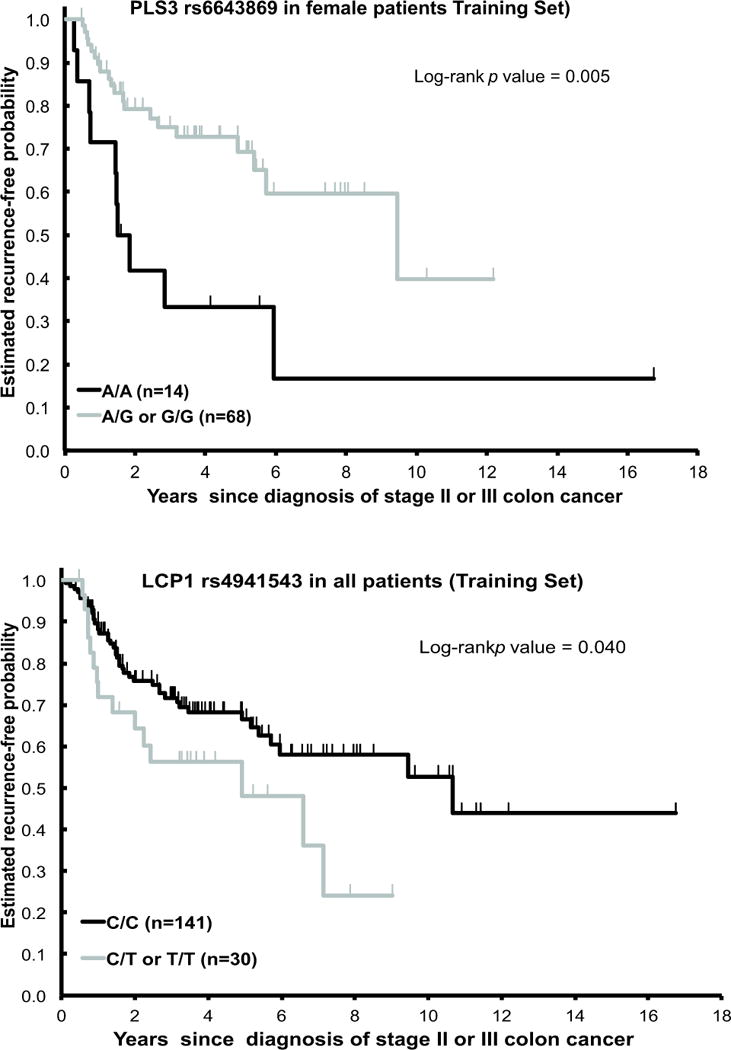
Time-to-tumor recurrence (TTR) by PLS3rs6643869 and LCP14941543 in USC cohort (Training set) stage II/III CRC patients treated with 5-fluorouracil (5-FU)-based chemotherapy.
Table 2.
PLS3 and LCP1 polymorphisms and outcome in patients with stage II or III colon cancer by sex (Training set)
| Males | Females | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | Median TTR, y (95% CI) | Prob ± SE* of 3-y recurrence | HR (95% CI) Univariate | HR (95% CI) Multivariate | N | Median TTR, y (95% CI) | Prob ± SE* of 3-y recurrence | HR (95% CI) Univariate | HR (95% CI) Multivariate | |
| PLS3 rs6643869 | ||||||||||
| G/G | 66 | 7.1 (3.5, 11.4+) | 0.33 ± 0.07 | 1 (Reference) | 1 (Reference) | 35 | 12.2+ (4.9, 12.2+) | 0.29 ± 0.08 | 1 (Reference) | 1 (Reference) |
| G/A | 27 | 5.2 (1.7, 10.7+) | 0.36 ± 0.10 | 1.38 (0.69, 2.75) | 1.48 (0.71, 3.08) | 33 | 9.4 (5.4, 9.4+) | 0.20 ± 0.08 | 0.89 (0.37, 2.14) | 0.75 (0.30, 1.92) |
| A/A | 0.35 | 0.30 | 14 | 1.7 (0.7, 5.9+) | 0.67 ± 0.13 | 2.69 (1.14, 6.35) | 1.96 (0.75, 5.13) | |||
| P† | 0.018 | 0.15 | ||||||||
| G/G or G/A | 68 | 9.4 (5.4, 12.2+) | 0.25 ± 0.06 | 1 (Reference) | 1 (Reference) | |||||
| A/A | 14 | 1.7 (0.7, 5.9+) | 0.67 ± 0.13 | 2.84 (1.32, 6.10) | 2.26 (0.97, 5.31) | |||||
| P† | 0.005 | 0.060 | ||||||||
| LCP rs11342 | ||||||||||
| G/G | 33 | 2.3 (1.5, 6.8+) | 0.52 ± 0.11 | 1 (Reference) | 1 (Reference) | 23 | 10.3+ | 0.23 ± 0.09 | 1 (Reference) | 1 (Reference) |
| G/T | 42 | 10.7 (5.2, 11.4+) | 0.17 ± 0.06 | 0.39 (0.17, 0.89) | 0.36 (0.14, 0.92) | 44 | 5.4 (1.8, 9.4+) | 0.40 ± 0.08 | 2.27 (0.86, 6.03) | 1.53 (0.50, 4.63) |
| T/T | 21 | 3.9 (1.4, 10.7+) | 0.41 ± 0.11 | 0.78 (0.34, 1.83) | 0.71 (0.29, 1.73) | 17 | 12.2+ (2.8, 12.2+) | 0.21 ± 0.11 | 1.00 (0.27, 3.71) | 0.68 (0.16, 2.85) |
| P† | 0.037 | 0.096 | 0.10 | 0.31 | ||||||
| G/T, T/T | 63 | 7.1 (4.9,11.4+) | 0.25 ± 0.06 | 0.51 (0.25,1.04) | 0.50 (0.23,1.10) | 61 | 5.7 (2.8,16.8+) | 0.35 ± 0.07 | 1.89 (0.72,4.93) | 1.29 (0.43,3.85) |
| P† | 0.042 | 0.085 | 0.19 | 0.65 | ||||||
| LCP rs4941543 | ||||||||||
| C/C | 75 | 10.7 (5.2, 11.4+) | 0.30 ± 0.06 | 1 (Reference) | 1 (Reference) | 66 | 9.4 (5.4, 16.8+) | 0.27 ± 0.06 | 1 (Reference) | 1 (Reference) |
| C/T‡ | 14 | 4.9 (1.0, 7.1+) | 0.35 ± 0.13 | 1.75 (0.77, 3.97) | 1.60 (0.64, 3.97) | 15 | 2.4 (0.7, 9.0+) | 0.52 ± 0.14 | 1.91 (0.81, 4.52) | 2.95 (1.10, 7.90) |
| T/T‡ | 1 | 0 | ||||||||
| P† | 0.17 | 0.32 | 0.13 | 0.031 | ||||||
Greenwood SE.
Estimates were not reached.
Based on log-rank test in the univariate analysis and based on Wald test within multivariate Cox proportional hazards model adjusting for stage and type of adjuvant therapy and stratified by race.
In the dominant model.
The multivariate model for the USC cohort was adjusted for stage and type of chemotherapy as covariates and stratified by race and gender. The A/A genotype (n=14) of PLS3 rs6643869 demonstrated a non-significant trend towards decreased TTR (HR=2.26, 95% CI 0.97–5.31; adjusted p=0.06) compared with patients with the A/G or G/G genotypes (n = 68) in female patients (Table 2). CPE was 0.728 (standard error (SE): 0.039) and 0.741 (SE: 0.037) for the multivariate model including baseline patient prognostic factors only (baseline model), and adding PLS3 rs6643869into the model among female patients, respectively. LCP1 rs4941543 was not associated with TTR in multivariate analysis (adjusted p=0.11) in any population. However, amongst female patients, those with the C/C genotype had a lower risk of recurrence than carriers of the T allele (HR=2.95, 95% CI1.10–7.90; adjusted p=0.031). CPE was 0.769 (SE: 0.034) for LCP1 rs4941543-added multivariate model and 0.744 (SE: 0.034) for the baseline model. There were no statistically significant differences between the PLS3 rs6643869, LCP1 rs4941543 and LCP1 rs11432 polymorphisms and TTR in male patients (Supplementary Figure 3; Table 2). These data suggest that the PLS3 rs6643869 and LCP1 rs4941543 polymorphisms may serve as gender-based prognostic factors. We did not observe statistically significant associations between the other tested genes with regards to time to disease relapse (Supplementary Table 3).
Validation Set
In the overall population analysis, the PLS3 rs6643869 and LCP1 rs4941543 polymorphisms were not associated with TTR in the validation set. However, when patients were stratified by gender, tumor location and stage, the PLS3 rs6643869 and LCP1 rs4941543 polymorphisms demonstrated the following significant relationships:
In right-sided CRC, female patients with any G allele of PLS3 rs6643869 had an increased TTR (13.7+ years) compared to the G/A and A/A genotypes (3.3 years) (Figure 2, top panel) in both univariate (HR=2.07, 95% CI1.09–3.91; p=0.021) and multivariate analyses (HR=2.41, 95% CI1.22–4.76; p=0.011) (Table 3, top right panel). CPE was 0.639 (SE: 0.037) and 0.672 (SE: 0.034) for the baseline multivariate model and PLS3 rs6643869-added model, respectively. In men, PLS3 rs6643869 was not significantly associated with TTR (Table 3, bottom left panel). These data suggest that the PLS3 rs6643869 polymorphism is a positive prognostic indicator whose influence depends upon both gender and tumor location.
Women with the LCP1 rs4941543 C/C genotype had a shorter TTR (10 years, 95% CI6.9–14.1+ years) compared to those with any T allele in univariate analysis (TTR 12.2+ years; HR=0.53, 95% CI 0.29–0.97; p=0.035) (Figure 2, bottom panel) and remained significantly associated in multivariate analysis (HR=0.53, 95% CI0.29–0.97; p=0.039) (Table 3, bottom panel). CPE was 0.650 (SE: 0.029) and 0.667 (SE: 0.028) for the baseline multivariate and LCP1 rs4941543-added model, respectively. These data are in contradiction to that found in our USC training cohort, which showed the C/C genotype to correlate with decreased risk of recurrence.
In stage III CRC, female patients with the LCP1 rs4941543 C/C genotype had a significantly decreased TTR (4.1 years) than carriers of the T allele in univariate analysis (10.6 years) (HR=0.47, 95% CI 0.23–0.96, p=0.031), with a similar but non-significant trend in multivariate analysis (HR=0.49, 95% CI 0.24–1.01; p=0.052) (Table 4). These data support the role of LCP1 rs4941543 as a prognosticator based on gender and tumor stage.
Figure 2.
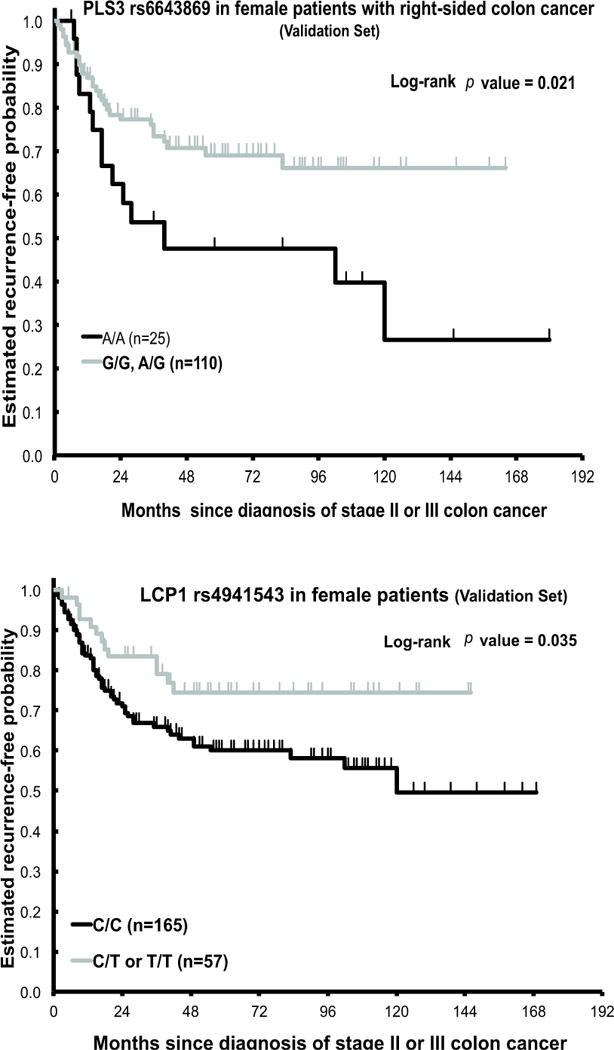
Time-to-tumor recurrence (TTR) by PLS3rs6643869 and LCP14941543 in Austrian cohort (Validation set) stage II/III CRC patients treated with 5-fluorouracil (5-FU)-basedchemotherapy.
Table 3.
Associations between Plastin polymorphisms and disease-free survival in Austrian female patients with stage II or III colon cancer by tumor site and gender (validation set)
| Left Colon | Right Colon | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | Median TTR, y (95% CI) | Probability ± SE* of 3-year recurrence | HR (95% CI) in univariate analysis | HR (95% CI) in multivariate analysis | N | Median TTR, y (95% CI) | Probability ± SE* of 3-year recurrence | HR (95% CI) in univariate analysis | HR (95% CI) in multivariate analysis | |
| PLS3 rs6643869 | ||||||||||
| G/G | 27 | 10.8+ (0.9, 10.8+) | 0.36 ± 0.10 | 1 (Reference) | 1 (Reference) | 52 | 13.7+ (4.6, 13.7+) | 0.25 ± 0.07 | 1 (Reference) | 1 (Reference) |
| G/A | 41 | 10.6+ (3.5, 10.6+) | 0.31 ± 0.08 | 0.84 (0.37, 1.90) | 0.67 (0.29, 1.58) | 58 | 13.2+ | 0.28 ± 0.06 | 0.93 (0.45, 1.90) | 0.99 (0.47, 2.06) |
| A/A | 12 | 11.6+ (1.7, 11.6+) | 0.11 ± 0.10 | 0.20 (0.03, 1.53) | 0.23 (0.03, 1.79) | 25 | 3.3 (1.4, 15.0+) | 0.46 ± 0.10 | 1.98 (0.94, 4.17) | 2.40 (1.09, 5.27) |
| P† | 0.23 | 0.31 | 0.068 | 0.040 | ||||||
| G | 68 | 10.8+ (3.7, 10.8+) | 0.33 ± 0.06 | 1 (Reference) | 1 (Reference) | 110 | 13.7+ | 0.26 ± 0.05 | 1 (Reference) | 1 (Reference) |
| A/A | 12 | 11.6+ (1.7, 11.6+) | 0.11 ± 0.10 | 0.22 (0.03, 1.61) | 0.29 (0.04, 2.15) | 25 | 3.3 (1.4, 15.0+) | 0.46 ± 0.10 | 2.07 (1.09, 3.91) | 2.41 (1.22, 4.76) |
| P† | 0.099 | 0.22 | 0.021 | 0.011 | ||||||
| LCP1 rs4941543 | ||||||||||
| C/C | 59 | 11.6+ (2.3, 11.6+) | 0.37 ± 0.07 | 1 (Reference) | 1 (Reference) | 106 | 10.0 (6.9, 14.1+) | 0.33 ± 0.05 | 1 (Reference) | 1 (Reference) |
| C/T‡ | 23 | 10.6+ | 0.17 ± 0.08 | 0.47 (0.18, 1.24) | 0.42 (0.16, 1.15) | 31 | 12.2+ | 0.23 ± 0.08 | 0.58 (0.27, 1.24) | 0.59 (0.27, 1.28) |
| T/T‡ | 1 | 2 | ||||||||
| P† | 0.11 | 0.092 | 0.15 | 0.18 | ||||||
| Male | Female | |||||||||
| N | Median TTR, y (95% CI) | Probability ± SE* of 3-year recurrence | HR (95% CI) in univariate analysis | HR (95% CI) in multivariate analysis | N | Median TTR, y (95% CI) | Probability ± SE* of 3-year recurrence | HR (95% CI) in univariate analysis | HR (95% CI) in multivariate analysis | |
| PLS3 rs6643869 | ||||||||||
| G/G | 139 | 14.4 (14.4, 14.5+) | 0.27 ± 0.04 | 1 (Reference) | 1 (Reference) | 79 | 13.7+ (6.9, 13.7+) | 0.29 ± 0.06 | 1 (Reference) | 1 (Reference) |
| G/A | 100 | 12.0+ (4.0, 12.0+) | 0.31 ± 0.05 | 1.36 (0.85, 2.19) | 1.42 (0.88, 2.30) | 99 | 13.2+ | 0.29 ± 0.05 | 0.91 (0.53, 1.55) | 0.88 (0.51, 1.52) |
| A/A | 37 | 10.0 (2.1, 15.0+) | 0.36 ± 0.08 | 1.24 (0.65, 2.37) | 1.43 (0.73, 2.80) | |||||
| P† | 0.19 | 0.15 | 0.61 | 0.33 | ||||||
| LCP1 rs4941543 | ||||||||||
| C/C | 196 | 14.4 (14.4, 15.0) | 0.28 ± 0.03 | 1 (Reference) | 1 (Reference) | 165 | 10.0 (6.9, 14.1+) | 0.34 ± 0.04 | 1 (Reference) | 1 (Reference) |
| C/T‡ | 58 | 8.6 (3.9, 11.4+) | 0.32 ± 0.07 | 1.16 (0.69, 1.95) | 1.26 (0.74, 2.13) | 54 | 12.2+ | 0.21 ± 0.06 | 0.53 (0.29, 0.97) | 0.53 (0.29, 0.97) |
| T/T‡ | 4 | 3 | ||||||||
| P† | 0.58 | 0.39 | 0.035 | 0.039 | ||||||
Greenwood SE.
Based on log-rank test in the univariate analysis and based on Wald test within multivariate Cox proportional hazards model adjusting for age, stage, and adjuvant therapy.
Estimates were not reached.
In the dominant model.
Table 4.
Associations between Plastin polymorphisms and disease-free survival in Austrian female patients with stage II or III colon cancer by stage (Validation set)
| Stage II | Stage III | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | Median TTR, y (95% CI) | Probability ± SE* of 3-year recurrence | HR (95% CI) in univariate analysis | HR (95% CI) in multivariate analysis | N | Median TTR, y (95% CI) | Probability ± SE* of 3-year recurrence | HR (95% CI) in univariate analysis | HR (95% CI) in multivariate analysis | |
| PLS3 rs6643869 | ||||||||||
| G/G | 28 | 12.2+ (4.6, 12.2+) | 0.18 ± 0.08 | 1 (Reference) | 1 (Reference) | 51 | 13.7+ (2.9, 13.7+) | 0.35 ± 0.07 | 1 (Reference) | 1 (Reference) |
| G/A | 37 | 13.2+ | 0.10 ± 0.06 | 0.42 (0.12, 1.49) | 0.40 (0.11, 1.41) | 62 | 10.6+ (2.3, 10.6+) | 0.39 ± 0.06 | 1.09 (0.60, 1.99) | 1.03 (0.56, 1.89) |
| A/A | 19 | 10.0 (3.3, 15.0+) | 0.17 ± 0.09 | 1.03 (0.32, 3.26) | 0.90 (0.28, 2.91) | 18 | 2.1 (1.1, 11.6+) | 0.58 ± 0.13 | 1.52 (0.68, 3.39) | 1.43 (0.63, 3.24) |
| P† | 0.29 | 0.31 | 0.57 | 0.66 | ||||||
| G | 65 | 13.2+ | 0.14 ± 0.05 | 1 (Reference) | 1 (Reference) | 113 | 13.7+ (3.5, 13.7+) | 0.37 ± 0.05 | 1 (Reference) | 1 (Reference) |
| A/A | 19 | 10.0 (3.3, 15.0+) | 0.17 ± 0.09 | 1.59 (0.57, 4.45) | 1.47 (0.52, 4.17) | 18 | 2.1 (1.1, 11.6+) | 0.58 ± 0.13 | 1.44 (0.70, 2.97) | 1.41 (0.68, 2.93) |
| P† | 0.36 | 0.47 | 0.31 | 0.36 | ||||||
| LCP1 rs4941543 | ||||||||||
| C/C | 64 | 14.1+ (8.5, 14.1+) | 0.15 ± 0.05 | 1 (Reference) | 1 (Reference) | 101 | 4.1 (1.8, 13.7+) | 0.46 ± 0.05 | 1 (Reference) | 1 (Reference) |
| C/T‡ | 23 | 12.2+ | 0.15 ± 0.08 | 0.75 (0.24, 2.31) | 0.64 (0.21, 1.98) | 31 | 10.6+ | 0.25 ± 0.08 | 0.47 (0.23, 0.96) | 0.49 (0.24, 1.01) |
| T/T‡ | 1 | 2 | ||||||||
| P† | 0.61 | 0.44 | 0.031 | 0.052 | ||||||
Greenwood SE.
Estimates were not reached.
Based on log-rank test in the univariate analysis and based on Wald test within multivariate Cox proportional hazards model adjusting for age, stage, and adjuvant therapy.
In the dominant model.
Discussion
In this study, we demonstrated that germline polymorphisms within the PLS3 and LCP1 genes are significantly associated with time to tumor recurrence in patients with locally advanced colon cancer treated with adjuvant chemotherapy. Moreover, when stratified by gender, the PLS3 rs6643869 and LCP1 rs4941543 polymorphisms correlate with TTR in women. The correlation between clinical outcomes and PLS3 rs6643869 was validated in an independent Austrian cohort. To our knowledge, this is the first report suggesting a relationship between PLS3 and LCP1 polymorphisms and clinical outcomes in stage II/III colon cancer patients that is dependent upon gender, tumor location, and stage. These findings suggest that plastins may serve as a useful prognostic biomarker and warrantfurther investigation.
Our study identified several SNPs in the PLS3 and LCP1 genes that may play a significant role in predicting outcomes of stage II/III colon cancer patients. Patients with the PLS3 rs6643869 polymorphism A/A genotype in the intron region of the PLS3 gene had a significantly higher risk of tumor recurrence compared with patients carrying at least one G allele in both the training and validation sets. Patients carrying the LCP1 rs4941543 polymorphism C/C genotype in the exon region of the LCP1 gene had a significantly lower risk of tumor recurrence compared with patients possessing either C/T or T/T in our training set. However, our validation set showed an opposite relationship between the C/C genotype and outcomes. This discrepancy could be attributed to two reasons. First, differences in ethnicity between the two populations may translate into contrasting pharmacogenomic interactions or activation of alternate pathways which affect tumor cell biology and behavior. Secondly, whereas the training set included only high risk stage II patients, the validation cohort included both high and low risk stage II patients (as we could not discern high or low risk status from the available clinical data). Low risk stage II patients may have a longer TTR and are more likely to live long enough to develop a recurrence.
The molecular mechanisms by which plastin polymorphisms affect tumor behavior and recurrence are under investigation. Modulations of transcription, interconnected stem cell pathways, and treatment-related factors have all been implicated. With regards to transcription, we know that the PLS3 rs6643869 polymorphism is located at intron 2 of PLS3 gene, and the SNP functional prediction tool (F-SNP) has shown that a single nucleotide change from A to G may alter transcription factor binding of this gene. For example, PLS3 rs6643869 containing the G allele but not the A allele binds two transcription factors, Tst-1 and HSF, which may affect subsequent transcriptional activity in a process known as intron-mediated enhancement. With regards to the LCP1 rs4941543 polymorphism located at exon 18, a missense mutation from C to T renders an amino acid change from lysine (K) to glutamine (E), which may lead to potential functional alterations. Moreover, our findings suggest that PLS3 rs6643869 and LCP1 rs4941543 are gender-specific markers, which may imply further hormonal interactions. The fact that PLS3 maps to X.q23 may partly account for its gender-specific effect, but additional studies are needed to better elucidate the mechanisms underlying these associations.
Once deranged transcription has occurred, plastins may then mediate colon cancer metastasis through two distinct mechanisms. The first involves the Epithelial-Mesenchymal Transition (EMT) which is critical for cancer development and metastasis (19, 20). A key effect or of this transition is E-cadherin, and E-cadherin expression levels have reliably demonstrated an inverse correlation with tumor invasiveness. It follows that loss of E-cadherin expression occurs at sites of EMT during tumor progression (21, 22). Therefore, one possibility is that overexpressed PLS3 and LCP1 in colon cancer cells lead to loss of E-cadherin (10, 11) and thereby promotes β-catenin release through the Wnt-signaling pathway (23, 24), facilitating passage through the EMT. Alternatively, plastin overexpression may enhance cell motility by modifying the actin cytoskeleton and, in doing so, increase metastatic potential.
Studies using CRC cell lines have shown a promising link between PLS3, the epithelial mesenchymal transition, and tumor metastasis. Willis and colleagues (10) showed that lamina A, a putative colon stem cell biomarker, can induce PLS3 upregulation, which in turn leads to E-cadherin downregulation and increased tumor invasiveness. Furthermore, a recent study led by Yokobori reported that PLS3 itself may be a novel circulating tumor cell (CTC) marker expressed during the epithelial mesenchymal transition (EMT), and the detection of PLS3-positive CTCs was found to be an independent prognostic factor in metastatic CRC patients (25). Our study showed that PLS3 plasma concentration was significantly higher in CRC patients compared to control patients with no evident disease (NED) (Supplementary Figure 4), which is consistent with previous studies, and suggests that PLS3 expression may serve as a surrogate for tumor invasiveness in stage II/III CRC.
Other studies have similarly demonstrated a connection between LCP1, E-cadherin mediated tumor spread, and poor prognosis in colon cancer. Elevated LCP1 levels in cancer cell lines and solid tumors have been correlated with increased migration, invasion and metastatic potential (12, 26, 27). In CRC, several independent studies have confirmed that L-plastin protein overexpression is also a marker for aggressive disease and poor prognosis (28, 29). Foran et al. (11) showed that CRC cell lines overexpressing LCP1 acquire invasiveness through E-cadherindownregulation, which induces dedifferentiation, invasion and metastasis. Furthermore, others (29, 30) have shown that LCP1 correlates with tumor protein expression and CRC progression using immunohistochemical analysis and ELISA, which is consistent with our results in the USC cohort. Our data showed that LCP1 plasma concentration was much higher in CRC patients than NED control patients. Moreover, LCP1 was higher in stage III than stage II patients, suggesting that LCP1 expression may be related to lymph node metastasis (Supplementary Figure 3).
In addition to affecting the intrinsic invasiveness of tumor cells, PLS3 has been implicated in modulating chemotherapeutic efficacy. Specifically, enhanced PLS3 expression has been associated with both chemotherapy and radiation resistance in cancer cells (31, 32). Hisano et al. reported that PLS3 downregulation led to increased sensitivity to cisplatin in platinum-resistant cell lines. In our future studies, we will focus on the correlation between plastin expression levels and the presence of LCP1or PLS3 polymorphisms. The PLS3rs6643869 and LCP1rs4941543 polymorphisms, in particular, may alter gene function/expression, thereby increasing PLS3 and LCP1 levels and promoting Wnt signaling through the E-cadherin/ß-catenin complex, ultimately resulting in CRC recurrence and progression.
In summary, this study provides the first evidence that germline genetic variants of the PLS3 and LCP1 genes predict tumor recurrence and time to disease relapse in a gender-and stage-specific manner. The PLS3 rs6643869 SNP retained a significant correlation with outcomes after adjusting for multiple variables and has now been confirmed in a validated cohort. Taken together, our data strengthen the role of plastin proteins in locally advanced CRC and may aid in the selection of patients who would benefit from plastin-targeted treatments in the future.
Supplementary Material
Acknowledgments
Financial disclosure: This study was funded by the NIH grant 5 P30CA14089-27S1 and Nancy Bernstein Research Fund to H.J. Lenz.
Footnotes
Disclosure of potential Conflicts of Interest: All authors have no potential conflicts of interest to disclose.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg RM, Tabah-Fisch I, Bleiberg H, de Gramont A, Tournigand C, Andre T, et al. Pooled analysis of safety and efficacy of oxaliplatin plus fluorouracil/leucovorin administered bimonthly in elderly patients with colorectal cancer. J Clin Oncol. 2006;24:4085–91. doi: 10.1200/JCO.2006.06.9039. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe T, Wu TT, Catalano PJ, Ueki T, Satriano R, Haller DG, et al. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med. 2001;344:1196–206. doi: 10.1056/NEJM200104193441603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 5.Samstag Y, Eibert SM, Klemke M, Wabnitz GH. Actin cytoskeletal dynamics in T lymphocyte activation and migration. J Leukoc Biol. 2003;73:30–48. doi: 10.1189/jlb.0602272. [DOI] [PubMed] [Google Scholar]
- 6.Giganti A, Plastino J, Janji B, Van Troys M, Lentz D, Ampe C, et al. Actin-filament cross-linking protein T-plastin increases Arp2/3-mediated actin-based movement. J Cell Sci. 2005;118:1255–65. doi: 10.1242/jcs.01698. [DOI] [PubMed] [Google Scholar]
- 7.Park T, Chen ZP, Leavitt J. Activation of the leukocyte plastin gene occurs in most human cancer cells. Cancer Res. 1994;54:1775–81. [PubMed] [Google Scholar]
- 8.Lin CS, Lau A, Yeh CC, Chang CH, Lue TF. Upregulation of L-plastin gene by testosterone in breast and prostate cancer cells: identification of three cooperative androgen receptor-binding sequences. DNA Cell Biol. 2000;19:1–7. doi: 10.1089/104454900314654. [DOI] [PubMed] [Google Scholar]
- 9.Lin CS, Chen ZP, Park T, Ghosh K, Leavitt J. Characterization of the human L-plastin gene promoter in normal and neoplastic cells. J Biol Chem. 1993;268:2793–801. [PubMed] [Google Scholar]
- 10.Willis ND, Cox TR, Rahman-Casans SF, Smits K, Przyborski SA, van den Brandt P, et al. Lamin A/C is a risk biomarker in colorectal cancer. PLoS One. 2008;3:e2988. doi: 10.1371/journal.pone.0002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foran E, McWilliam P, Kelleher D, Croke DT, Long A. The leukocyte protein L-plastin induces proliferation, invasion and loss of E-cadherin expression in colon cancer cells. Int J Cancer. 2006;118:2098–104. doi: 10.1002/ijc.21593. [DOI] [PubMed] [Google Scholar]
- 12.Klemke M, Rafael MT, Wabnitz GH, Weschenfelder T, Konstandin MH, Garbi N, et al. Phosphorylation of ectopically expressed L-plastin enhances invasiveness of human melanoma cells. Int J Cancer. 2007;120:2590–9. doi: 10.1002/ijc.22589. [DOI] [PubMed] [Google Scholar]
- 13.Ning Y, Manegold PC, Hong YK, Zhang W, Pohl A, Lurje G, et al. Interleukin-8 is associated with proliferation, migration, angiogenesis and chemosensitivity in vitro and in vivo in colon cancer cell line models. Int J Cancer. 2011;128:2038–49. doi: 10.1002/ijc.25562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerger A, Zhang W, Yang D, Bohanes P, Ning Y, Winder T, et al. Common cancer stem cell gene variants predict colon cancer recurrence. Clin Cancer Res. 2011;17:6934–43. doi: 10.1158/1078-0432.CCR-11-1180. [DOI] [PubMed] [Google Scholar]
- 15.Absenger G, Benhaim L, Szkandera J, Zhang W, Yang D, Labonte MJ, et al. The cyclin D1 (CCND1) rs9344 G>A polymorphism predicts clinical outcome in colon cancer patients treated with adjuvant 5-FU-based chemotherapy. Pharmacogenomics J. 2013 doi: 10.1038/tpj.2013.15. [DOI] [PubMed] [Google Scholar]
- 16.Lee PH, Shatkay H. F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2008;36:D820–4. doi: 10.1093/nar/gkm904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee PH, Shatkay H. An integrative scoring system for ranking SNPs by their potential deleterious effects. Bioinformatics. 2009;25:1048–55. doi: 10.1093/bioinformatics/btp103. [DOI] [PubMed] [Google Scholar]
- 18.Gonen M, Heller G. Concordance probability and discriminatory power in proportional hazards regression. Biometrika. 2005;92:965–70. [Google Scholar]
- 19.Cardiff RD. Epithelial to Mesenchymal Transition Tumors: Fallacious or Snail’s Pace? Clin Cancer Res. 2005;11:8534–7. doi: 10.1158/1078-0432.CCR-05-2250. [DOI] [PubMed] [Google Scholar]
- 20.Wu Y, Zhou BP. New insights of epithelial-mesenchymal transition in cancer metastasis. Acta Biochim Biophys Sin (Shanghai) 2008;40:643–50. doi: 10.1111/j.1745-7270.2008.00443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowin P, Rowlands TM, Hatsell SJ. Cadherins and catenins in breast cancer. Curr Opin Cell Biol. 2005;17:499–508. doi: 10.1016/j.ceb.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 22.Junghans D, Haas IG, Kemler R. Mammalian cadherins and protocadherins: about cell death, synapses and processing. Curr Opin Cell Biol. 2005;17:446–52. doi: 10.1016/j.ceb.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 24.Hulsken J, Birchmeier W, Behrens J. E-cadherin and APC compete for the interaction with beta-catenin and the cytoskeleton. J Cell Biol. 1994;127:2061–9. doi: 10.1083/jcb.127.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokobori T, Iinuma H, Shimamura T, et al. Plastin3 Is a Novel Marker for Circulating Tumor Cells Undergoing the Epithelial–Mesenchymal Transition and Is Associated with Colorectal Cancer Prognosis. Cancer Res. 2013;73(7):2059–69. doi: 10.1158/0008-5472.CAN-12-0326. [DOI] [PubMed] [Google Scholar]
- 26.Lapillonne A, Coue O, Friederich E, Nicolas A, Del Maestro L, Louvard D, et al. Expression patterns of L-plastin isoform in normal and carcinomatous breast tissues. Anticancer Res. 2000;20:3177–82. [PubMed] [Google Scholar]
- 27.Li MX, Xiao ZQ, Liu YF, Chen YH, Li C, Zhang PF, et al. Quantitative proteomic analysis of differential proteins in the stroma of nasopharyngeal carcinoma and normal nasopharyngeal epithelial tissue. J Cell Biochem. 2009;106:570–9. doi: 10.1002/jcb.22028. [DOI] [PubMed] [Google Scholar]
- 28.Samstag Y, Klemke M. Ectopic expression of L-plastin in human tumor cells: diagnostic and therapeutic implications. Adv Enzyme Regul. 2007;47:118–26. doi: 10.1016/j.advenzreg.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Zhao R. Expression and clinical significance of L-plastin in colorectal carcinoma. J Gastrointest Surg. 2011;15:1982–8. doi: 10.1007/s11605-011-1678-4. [DOI] [PubMed] [Google Scholar]
- 30.Hao Y, Yu Y, Wang L, Yan M, Ji J, Qu Y, et al. IPO-38 is identified as a novel serum biomarker of gastric cancer based on clinical proteomics technology. J Proteome Res. 2008;7:3668–77. doi: 10.1021/pr700638k. [DOI] [PubMed] [Google Scholar]
- 31.Hisano T, Ono M, Nakayama M, Naito S, Kuwano M, Wada M. Increased expression of T-plastin gene in cisplatin-resistant human cancer cells: identification by mRNA differential display. FEBS Lett. 1996;397:101–7. doi: 10.1016/s0014-5793(96)01150-7. [DOI] [PubMed] [Google Scholar]
- 32.Higuchi Y, Kita K, Nakanishi H, Wang XL, Sugaya S, Tanzawa H, et al. Search for genes involved in UV-resistance in human cells by mRNA differential display: increased transcriptional expression of nucleophosmin and T-plastin genes in association with the resistance. Biochem Biophys Res Commun. 1998;248:597–602. doi: 10.1006/bbrc.1998.8978. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


