Abstract
The sequencing of marine metagenomic fosmids led to the discovery of several new complete phage genomes. Among the 21 major sequence groups, 10 totally novel groups of marine phages could be identified. Some of these represent the first phages infecting large marine prokaryotic phyla, such as the Verrucomicrobia and the recently described Ca. Actinomarinales. Coming from a single deep photic zone sample the diversity of phages found is astonishing, and the comparison with a metavirome from the same location indicates that only 2% of the real diversity was recovered. In addition to this large macro-diversity, rich micro-diversity was also found, affecting host-recognition modules, mirroring the variation of cell surface components in their host marine microbes.
Keywords: metagenomics, metavirome, marine phages, deep chlorophyll maximum, constant-diversity, red-queen, phage evolution, pan-selectome, pan-genome
Introduction
The problem of retrieving microbes in pure culture has hampered enormously the development of microbiology. For many years now, it has been clear that the microbes stored in culture collections only represent a tiny slice of the real diversity of microbes in nature.1 Furthermore, the recent development of comparative genomics has revealed that the gene diversity displayed by an isolate is a very poor representation of the enormous genetic diversity contained in prokaryotic2 and apparently, although at much smaller scale, also in some protist3 species, the so-called pan-genome. It seems clear now that a good representation of the genomic diversity of microbes cannot be acquired within a sensible time frame by the classical approach of culturing and sequencing individual isolates. A new avenue was found by pioneers such as Edward DeLong and J Craig Venter that applied genomic shot gun sequencing to microbial communities providing information about their collective genomes, the metagenome.4,5 Later the developments in high throughput, new generation sequencing technologies have opened the field of metagenomics to most microbiologists decreasing the cost and technical difficulties.6,7 A major step forward to understand microbes in nature is in the making.
However, a fundamental gap was hitherto left largely unfilled. If the cultivation of cellular microbes is demanding, what of their accompanying non-cellular viral predators? In aquatic environments there are, as a rule of thumb 10 phages per cell.8,9 Even assuming that the typical phage genome is ca. 100 times smaller than the one of the host the amount of genetic diversity that they carry is sizeable.10 In addition, phages are an essential component of microbial communities that contribute to the evolution and diversity of the prey species.11 However, the development of a parallel description of the community virome (metavirome) has been hampered by the relatively small amount of DNA available in the viral fraction. The use of genomic amplification based approaches (mostly multiple displacement amplification or MDA) bias heavily most available metaviromes.12 In addition, the annotation of phage genes, always challenging, becomes extremely difficult using the small reads generated by the presently available high throughput sequencing techniques.13
Serendipitously, we have found an alternative to dig into the natural diversity of phages. Very early in the development of metagenomics the presence of typical viral genes in cellular metagenomes was described.14,15 Gradually it became evident that this was just the result of the retrieval of cells undergoing the lytic cycle16 in the cellular fraction. Many phages replicate forming long genome concatamers17 and when the DNA from the cells in which they are replicating is collected, large amounts of the phage genome becomes available, actually amplified in a natural way. We described some years ago a method to sequence fosmid clones by NGS.18 Since the insert in a fosmid clone is similar to the size of many phage genomes (30–40 Kb), fosmids provide a convenient way to generate natural contigs derived from these phage genomes amplified in the metagenomic DNA. We did a pilot study in which a set of 130 randomly selected clones were sequenced and 16 long contigs that clearly belonged to bacteriophages could be assembled.16 Further analyses suggested that these phage-derived contigs actually belonged to five different sets of highly related siphoviruses and they likely infected picocyanobacteria, particularly Synechococcus. Importantly, three of these represented complete genomes.
Given the success in this relatively straightforward approach to obtaining phage genomes, we sequenced a much larger set of fosmids (ca. 6000), which provided more than a thousand contigs, of which 208 were complete phage genomes from a single sample taken from the deep (50 m) chlorophyll maximum in the Western Mediterranean.19 This is the region of maximum cell density in the water column where infection rate is probably high. Actually, ca. 10–15% of fosmid sequences retrieved from the cellular fraction DNA corresponded to phage genomes.19
The Local Diversity
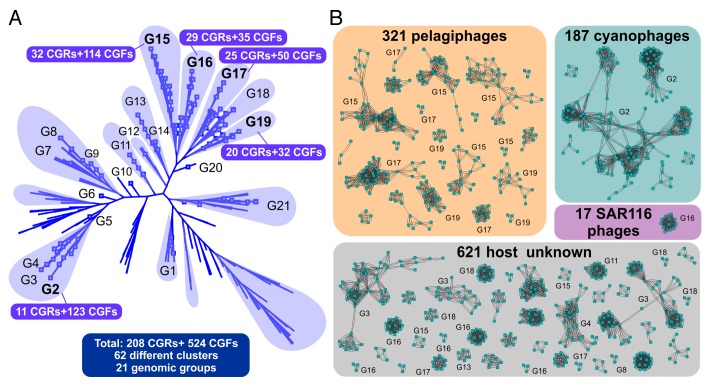
The 1148 phage contigs were organized into increasingly larger groups using three different stages of clustering.19 In the first stage, contigs with > 95% nucleotide identity and > 95% coverage were clustered to identify nearly identical sequences (the largest number of identical contigs was 22 and the mean 5). In the second, the coverage criterion was reduced (from > 95% to > 20%) to gather more contigs for each previous cluster. These two stages enabled creating groups of contigs that had very high nucleotide identity to each other and also likely originating from the same lineage of phage genome. Even though the majority of the contigs (913) could be clustered with a high nucleotide identity (95%) in 117 different clusters, 235 contigs were singletons and remained unclustered. The overlaps within these contigs that probably derive from phage replicating concatamers also enabled us to assess genome completeness, and 208 complete genomes could be identified. The third clustering stage was based on protein sequence similarities in order to identify much more distant relationships within the 208 complete phage genomes to reference phage genomes, what allowed us to define 10 novel yet undescribed groups of phages within this collection (Fig. 1).
Figure 1. (A) All-vs-all genomic comparison of several reference tailed bacteriophage genomes (non-labeled branches) and the 208 complete phage genomes retrieved in Mizuno et al., 2013 (represented as squares at the end of the branches). Clustering is based on a sequence similarity derived metric. The new genomic groups are labeled as G1 to G21. CGR: Complete Genomic Representative, sequences that had been assumed to represent complete phage genomes (Mizuno et al., 2013). CGF: Complete Genomic Fragment, sequences that from the comparison with the CGRs are known to be incomplete phage genomes. (B) Network view of similarity relationships within the novel phage groups. Each phage contig (complete genome or incomplete) is represented by a node and edges between any two nodes indicate a high sequence identity (> 95%) and coverage of at least 20% of the shorter contig. The groups are boxed by the predicted host. The numbers of some of the genomic groups (G1-G21) to which the cluster of contigs belong are also indicated. Only clusters with more than two members are shown, 235 singletons and 47 doublets were also found but are not depicted in the figure.
The largest clusters could be associated with cyanophages (largest cluster with 102 contigs) and pelagiphages (largest cluster with 40 contigs) (also shown in Fig. 1). Such contigs are the most likely to belong to active phages that were replicating by concatamer formation when the sample was taken (though they may not be exactly the most abundant in the sample). Apart from phages infecting picocyanobacteria and the SAR11 group microbes, we also identified contigs that likely infect marine Verrucomicrobia20 (2 contigs, 1 complete genome) and low GC Actinobacteria21 (a single contig, incomplete genome), that represent the first phages from the marine habitat for these two groups.
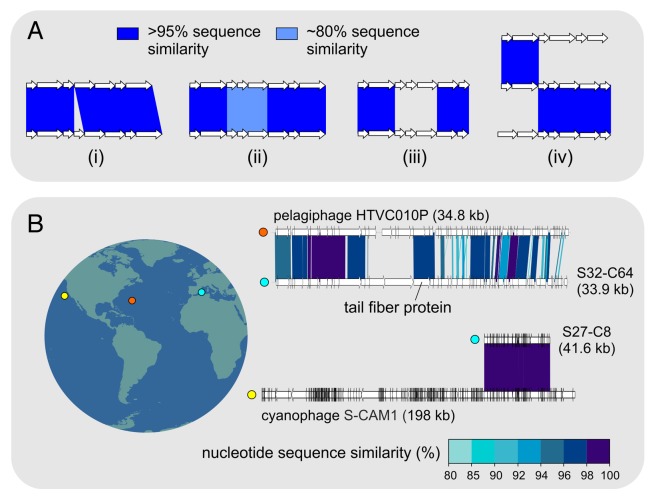
The availability of large numbers of highly related contigs in these clusters allowed examining genuinely concurrent phage populations, something that with cultured isolates has only been possible to a limited extent.22,23 Broadly, aside from the identical fragments derived from a single concatamer, three different patterns in phage genome diversity were discernible (Fig. 2). The first was the recovery of phage genome fragments that were nearly identical to each other along their entire lengths except for small single nucleotide polymorphisms and indels. Such highly related versions may represent the lowest levels in phage genome population diversity. These are possibly the closest approximations of a natural phage population to a clonally derived genome in pure culture. Next, we found genomes sharing long, nearly identical regions, but with significant sequence divergence in smaller regions, that although often preserving synteny had much lower or no similarity (Fig. 2). Typically these regions contained host-recognition genes (e.g., tail fiber) but also others (e.g., terminases and capsids). Though this might suggest that such divergence is the result of gradually evolving host-specificity, other explanations are possible (see below). These “genomic islands” actually have a clear parallel in prokaryotic genomes, although in them gene content seems to be more variable i.e., synteny is less preserved. An apparent feature that emerged from these comparisons, apart from the patterns discussed above, was the mosaic nature of the genomic architecture in these concurrent populations (Fig. 2). Given the extremely high nucleotide identity (> 95%) in large parts of these contigs, the presence of such a hybrid architecture can only be explained by recombination during mixed infections of the common host.
Figure 2. (A) Patterns in Local Diversity: Schematic representation of four types of patterns seen in concurrent phage genomes (i) syntenic, nearly identical contigs with small insertions/deletions (ii) syntenic, nearly identical contigs with smaller, more divergent regions and (iii) flexible genomic islands in highly related contigs, (iv) typical mosaic architecture displayed by multiple, related contigs. (B) Global distribution of nearly identical phage fragments: On the left a map of the world is shown where three locations are marked in different colors, from left to right, South California Coast, (Pacific Ocean, yellow), Bermuda (North Atlantic, orange) and the Mediterranean (blue). On the right, genome fragments originating from different sources are compared (nucleotide sequence identity is shown). The filled circles next to the contigs indicate the isolation location (as shown in the map). The names of both genomic fragments originated from the Mediterranean are abbreviated, e.g., S32-C64 for uvMED-CGR-C28-MedDCM-OCT-S32-C64. Length of the fragments and are also indicated.
In any case, it seems clear that even with the more than a thousand large genomic fragments described we were far from covering even a small fraction of the diversity of phages present in this single Mediterranean sample. Comparing the assembled phage contigs with a metavirome from the same location we have calculated that barely 2% of the diversity found in the metavirome (15 Gb) was represented in the combined 26 Mb of sequences assembled in the fosmids.
The Same Phages Everywhere
A controversial issue in microbiology is the existence of a biogeography of microbes. Recent evidence from metagenomics and genomics of marine bacteria indicates a weak geographic association at the strain level, although most species are global along the broad band of oligotrophic temperate and tropical ocean around the Earth.24,25 Regarding phages there seems to be evidence of widespread distribution, but based only on short overlaps.13 We have retrieved examples19 that indicate a global distribution of phages throughout these ocean water masses (altogether close to half the surface of the Earth). For example the recovery of a cyanophage genome fragment more than 40 Kb and 97% similarity (nucleotides) to an isolate obtained off the coast of Southern California (Fig. 2). Along the same lines, the finding of nearly identical versions of the hyper-diverse (see below) tail fiber protein (Fig. 2) in a pelagiphage from Bermuda26 and our Mediterranean sample all point toward very similar populations found at vastly separated ocean sites. Of course the global ocean is a continuum in which eventually all the water gets mixed, but it is an extremely slow process that takes in the range of thousands of years.27 Airborne transportation in extreme meteorological events might provide faster connection among marine microbial communities that travel in the aerosols formed for example in large storms, but once they arrive to the distant places they would face a huge dilution compared with the local strains. It is possible also that these populations are conserved under the similar conditions of the deep photic zone worldwide because they are selected for28 (see below).
Is the Red Queen Running or is it a Zoetrope?
The presence of simultaneous, closely related lineages displaying enormous microdiversity, either in prokaryotic or phage genomes gives rise to two different interpretations. One in which populations are viewed as fast-evolving assemblages where both, microbes and their phages, are in a constant arms-race (the Red-Queen hypothesis29), and another that asserts that microbial or phage lineages are stable, though very numerous conveying, like in a zoetrope, an illusion of a fast-paced evolutionary scenario. (A zoetrope is a 19th century device that produces an effect of apparent motion by rapidly displaying static pictures in succession).
Actually the question is much more relevant than just what is the time frame for change in the prokaryotic world. Some of us have proposed a Constant-Diversity (C-D) model30 in which we posit that there is a high diversity of concurrent clonal lineages within a single bacterial or archaeal population (within the same species). This diversity optimizes resource utilization by increasing the range of ecological niches that can be exploited. For example, in heterotrophic bacteria different concurrent clones carry different sets of membrane transporters, increasing the range of organic compounds that the population can utilize efficiently. The result is a better exploitation of resources by a smaller number of different species. However, since prokaryotes reproduce clonally, that is, producing exact copies of themselves, there is the risk that one clone could be more successful (for example by acquiring an unusually increased share of the resources, or because of a transient expansion of its niche). Such clonal sweeps are a known phenomenon in laboratory cultures in which a clone normally predominates in the long-term.31 Here is when phages come to the rescue. Since phage infection rate is density dependent, the inordinate increase of a clone would immediately make it more efficiently predated upon by the populations of specific phages adapted to its receptors, following a classical Lotka-Volterra predator/prey equilibrium. This idea has repercussions at multiple levels, from biotechnology to food microbiology or human health (through the microbiome).
This model can be taken beyond the purely mechanistic level since it implies that the bacterial population actually includes its associated phages as a sort of tax collectors that equate and preserve the efficiency of the system making this consortium of cell clones, plus the phages preying on them, a sort of meta-clonal organism that can be selected upon or extinguished i.e., a unit of selection or selectome.28 Our results support this view, as the local diversity found at a single marine water column sample is astonishing. Even within relatively similar genomes abundant microdiversity could be established and still some hyper-diversified regions are found identical in genomes isolated from distant locations including the Atlantic and Pacific Oceans.
However, some results appear puzzling from a C-D perspective. For example, most phage genomes recruit unevenly from a metavirome from the same location,32 as is the case of prokaryotic genomes, and as the C-D model would predict.30,33 On the other hand, some groups (specifically putative pelagiphages) recruited much more evenly and had no equivalent of metagenomic islands. It is possible that these are generalistic phages that do not have hyper-diversified recognition modules.34 The under-recruiting genes were often involved in host recognition, for example the tail fiber genes, as predicted by C-D. But other under-recruiting genes, such as some terminases, appeared totally disconnected from host recognition.
Maybe we are underestimating the complexity of phage behavior. For example, different strategies of phage survival have been proposed in which some are slow growers with small burst size while others produce large burst sizes.35 Some are specialized in infecting a very narrow range of host while others have a wider range with the risk of failed infection or inferior productive yield.34,36 It is certainly possible that some genes that are diverse within the population are involved in the different intracellular environments of the host strains at multiple levels. Different terminases might help packaging different types of concatamers or with different types of capsids. The in-depth study of such hyper-diverse genes might help in understanding phage-host interactions at these different levels.
A Million Phage Genomes
With the advent of next-generation sequencing, we have already witnessed an exponential increase in the number of microbial genomes that are being sequenced. However, phage genomics has remained largely untouched by this sequencing revolution. Given the advances in sequencing that are now unfolding, it is not unreasonable to finally tap the largest genetic reservoir on the planet. The methods described here, in combination with new methods of single-molecule sequencing actually provide finally an uncomplicated way to interrogate complete communities and their associated phages. It is only a matter of time before we will be on the threshold of being able to track evolutionary processes in real-time in natural habitats.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by projects MAGYK (BIO2008-02444), MICROGEN (Programa CONSOLIDER-INGENIO 2010 CDS2009-00006), CGL2009-12651-C02-01 from the Spanish Ministerio de Ciencia e Innovación, DIMEGEN (PROMETEO/2010/089) and ACOMP/2009/155 from the Generalitat Valenciana and MaCuMBA Project 311975 of the European Commission FP7. FEDER funds supported this project. RG was supported by a Juan de la Cierva scholarship from the Spanish Ministerio de Ciencia e Innovación.
Citation: Rodriguez-Valera F, Mizuno CM, Ghai R. Tales from a thousand and one phages. Bacteriophage 2014; 4:e28265; 10.4161/bact.28265
Footnotes
Previously published online: www.landesbioscience.com/journals/bacteriophage/article/28265
References
- 1.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–7. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 2.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A. 2005;102:13950–5. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Read BA, Kegel J, Klute MJ, Kuo A, Lefebvre SC, Maumus F, Mayer C, Miller J, Monier A, Salamov A, et al. Emiliania huxleyi Annotation Consortium Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature. 2013;499:209–13. doi: 10.1038/nature12221. [DOI] [PubMed] [Google Scholar]
- 4.DeLong EF. Preface. Microbial metagenomics, metatranscriptomics, and metaproteomics. Methods Enzymol. 2013;531:xxi. doi: 10.1016/B978-0-12-407863-5.09983-4. [DOI] [PubMed] [Google Scholar]
- 5.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 6.Ghai R, Martin-Cuadrado AB, Molto AG, Heredia IG, Cabrera R, Martin J, Verdú M, Deschamps P, Moreira D, López-García P, et al. Metagenome of the Mediterranean deep chlorophyll maximum studied by direct and fosmid library 454 pyrosequencing. ISME J. 2010;4:1154–66. doi: 10.1038/ismej.2010.44. [DOI] [PubMed] [Google Scholar]
- 7.Ghai R, Rodriguez-Valera F, McMahon KD, Toyama D, Rinke R, Cristina Souza de Oliveira T, Wagner Garcia J, Pellon de Miranda F, Henrique-Silva F. Metagenomics of the water column in the pristine upper course of the Amazon river. PLoS One. 2011;6:e23785. doi: 10.1371/journal.pone.0023785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Proctor LM, Okubo A, Fuhrman JA. Calibrating estimates of phage-induced mortality in marine bacteria: Ultrastructural studies of marine bacteriophage development from one-step growth experiments. Microb Ecol. 1993;25:161–82. doi: 10.1007/BF00177193. [DOI] [PubMed] [Google Scholar]
- 9.Suttle CA. Marine viruses--major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–12. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 10.Rohwer F, Edwards R. The Phage Proteomic Tree: a genome-based taxonomy for phage. J Bacteriol. 2002;184:4529–35. doi: 10.1128/JB.184.16.4529-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paul JH, Sullivan MB. Marine phage genomics: what have we learned? Curr Opin Biotechnol. 2005;16:299–307. doi: 10.1016/j.copbio.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Casas V, Rohwer F. Phage metagenomics. Methods Enzymol. 2007;421:259–68. doi: 10.1016/S0076-6879(06)21020-6. [DOI] [PubMed] [Google Scholar]
- 13.Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, Martinez A, Sullivan MB, Edwards R, Brito BR, et al. Community genomics among stratified microbial assemblages in the ocean’s interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 15.Martín-Cuadrado AB, López-García P, Alba JC, Moreira D, Monticelli L, Strittmatter A, Gottschalk G, Rodríguez-Valera F. Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. PLoS One. 2007;2:e914. doi: 10.1371/journal.pone.0000914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizuno CM, Rodriguez-Valera F, Garcia-Heredia I, Martin-Cuadrado AB, Ghai R. Reconstruction of novel cyanobacterial siphovirus genomes from Mediterranean metagenomic fosmids. Appl Environ Microbiol. 2013;79:688–95. doi: 10.1128/AEM.02742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maniloff J, Ackermann HW. Taxonomy of bacterial viruses: establishment of tailed virus genera and the order Caudovirales. Arch Virol. 1998;143:2051–63. doi: 10.1007/s007050050442. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Heredia I, Martin-Cuadrado AB, Mojica FJ, Santos F, Mira A, Antón J, Rodriguez-Valera F. Reconstructing viral genomes from the environment using fosmid clones: the case of haloviruses. PLoS One. 2012;7:e33802. doi: 10.1371/journal.pone.0033802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizuno CM, Rodriguez-Valera F, Kimes NE, Ghai R. Expanding the marine virosphere using metagenomics. PLoS Genet. 2013;9:e1003987. doi: 10.1371/journal.pgen.1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freitas S, Hatosy S, Fuhrman JA, Huse SM, Welch DBM, Sogin ML, Martiny AC. Global distribution and diversity of marine Verrucomicrobia. ISME J. 2012;6:1499–505. doi: 10.1038/ismej.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghai R, Mizuno CM, Picazo A, Camacho A, Rodriguez-Valera F. Metagenomics uncovers a new group of low GC and ultra-small marine Actinobacteria. Sci Rep. 2013;3:2471. doi: 10.1038/srep02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angly F, Youle M, Nosrat B, Srinagesh S, Rodriguez-Brito B, McNairnie P, Deyanat-Yazdi G, Breitbart M, Rohwer F. Genomic analysis of multiple Roseophage SIO1 strains. Environ Microbiol. 2009;11:2863–73. doi: 10.1111/j.1462-2920.2009.02021.x. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Heredia I, Rodriguez-Valera F, Martin-Cuadrado AB. Novel group of podovirus infecting the marine bacterium Alteromonas macleodii. Bacteriophage. 2013;3:e24766. doi: 10.4161/bact.24766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, González JM, Luo H, Wright JJ, Landry ZC, Hanson NW, et al. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci U S A. 2013;110:11463–8. doi: 10.1073/pnas.1304246110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.López-Pérez M, Gonzaga A, Rodriguez-Valera F. Genomic diversity of “deep ecotype” Alteromonas macleodii isolates: evidence for Pan-Mediterranean clonal frames. Genome Biol Evol. 2013;5:1220–32. doi: 10.1093/gbe/evt089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y, Temperton B, Thrash JC, Schwalbach MS, Vergin KL, Landry ZC, Ellisman M, Deerinck T, Sullivan MB, Giovannoni SJ. Abundant SAR11 viruses in the ocean. Nature. 2013;494:357–60. doi: 10.1038/nature11921. [DOI] [PubMed] [Google Scholar]
- 27.Bollmann M, Bosch T, Colijn F, Ebinghaus R, Froese R, Guessow K, Khalilian S, Krastel S, Koertzinger A, Lagenbuch M. World ocean review 2010: living with the oceans. 2010 [Google Scholar]
- 28.Rodriguez-Valera F, Ussery DW. Is the pan-genome also a pan-selectome? F1000Res. 2012;1:16. doi: 10.12688/f1000research.1-16.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stern A, Sorek R. The phage-host arms race: shaping the evolution of microbes. Bioessays. 2011;33:43–51. doi: 10.1002/bies.201000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasić L, Thingstad TF, Rohwer F, Mira A. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–36. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 31.Atwood KC, Schneider LK, Ryan FJ. Periodic selection in Escherichia coli. Proc Natl Acad Sci U S A. 1951;37:146–55. doi: 10.1073/pnas.37.3.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizuno CM, Ghai R, Rodriguez-Valera F. Evidence for Metaviromic Islands in Marine phages. Frontiers in Microbiology 2014; 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasić L, Rodriguez-Mueller B, Martin-Cuadrado AB, Mira A, Rohwer F, Rodriguez-Valera F. Metagenomic islands of hyperhalophiles: the case of Salinibacter ruber. BMC Genomics. 2009;10:570. doi: 10.1186/1471-2164-10-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flores CO, Meyer JR, Valverde S, Farr L, Weitz JS. Statistical structure of host-phage interactions. Proc Natl Acad Sci U S A. 2011;108:E288–97. doi: 10.1073/pnas.1101595108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koskella B, Meaden S. Understanding bacteriophage specificity in natural microbial communities. Viruses. 2013;5:806–23. doi: 10.3390/v5030806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flores CO, Valverde S, Weitz JS. Multi-scale structure and geographic drivers of cross-infection within marine bacteria and phages. ISME J. 2013;7:520–32. doi: 10.1038/ismej.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]