Abstract
The modification-dependent restriction endonuclease AspBHI recognizes 5-methylcytosine (5mC) in the double-strand DNA sequence context of (C/T)(C/G)(5mC)N(C/G) (N = any nucleotide) and cleaves the two strands a fixed distance (N12/N16) 3′ to the modified cytosine. We determined the crystal structure of the homo-tetrameric AspBHI. Each subunit of the protein comprises two domains: an N-terminal DNA-recognition domain and a C-terminal DNA cleavage domain. The N-terminal domain is structurally similar to the eukaryotic SET and RING-associated (SRA) domain, which is known to bind to a hemi-methylated CpG dinucleotide. The C-terminal domain is structurally similar to classic Type II restriction enzymes and contains the endonuclease catalytic-site motif of DX20EAK. To understand how specific amino acids affect AspBHI recognition preference, we generated a homology model of the AspBHI-DNA complex, and probed the importance of individual amino acids by mutagenesis. Ser41 and Arg42 are predicted to be located in the DNA minor groove 5′ to the modified cytosine. Substitution of Ser41 with alanine (S41A) and cysteine (S41C) resulted in mutants with altered cleavage activity. All 19 Arg42 variants resulted in loss of endonuclease activity.
Mammalian DNA cytosine methylation is an important epigenetic modification1. It remains unclear how cytosine methylation within particular sequences is initiated, maintained and particularly, recognized. Epigenetic DNA modification is dynamic, and differences are found in the epigenomes of cells during normal development2, aging and mental health, and during pathologic processes such as cancer, among many others3. To learn more about the role of epigenetic modification in development and disease, and to understand the mechanisms that control its locations and levels in the human genome, the genomic locations of modified cytosines must be mapped with accuracy, to single-base resolution. Newly identified ‘modification-dependent’ restriction endonucleases are proving useful for this purpose4,5 and for understanding how specific recognition of modified cytosine occurs.
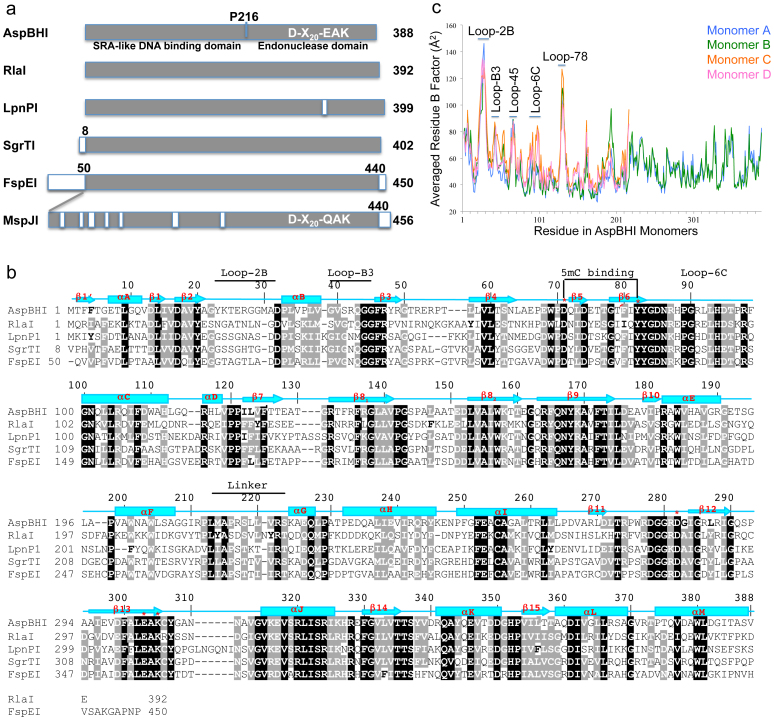
AspBHI from Azoarcus sp. BH72 belongs to a family of modification-dependent restriction endonucleases that recognize 5-methylcytosine (5mC) in the context of specific DNA sequences and cleave N12/N16 3′ downstream of the modified cytosine4,6. These proteins vary in length from 388 amino acids (AspBHI) to 456 (MspJI), and include a conserved core region of ~390 amino acids (Fig. 1a). FspEI has an additional amino-terminal 50 amino acids not present in other family members, whereas MspJI has insertions in multiple locations7. Besides MspJI, the other family members share sequence conservation throughout the entire region, with invariant (~26%) or conservatively substituted positions (~30%) scattered throughout the conserved core (Fig. 1b). Only one insertion of six residues was found in the conserved core of LpnPI (residues 316–321).
Figure 1. AspBHI is a member of MspJI family.
(a) Schematic representation of AspBHI and members of MspJI family. The conserved region is shown in dark grey and insertions are shown in open boxes. (b) Sequence alignment of AspBHI and members of MspJI family. The AspBHI residue numbering is shown above the sequence alignment. The pairwise comparison of AspBHI and MspJI was shown previously7. Amino acids highlighted are either invariant (white against black) among the five proteins or similar (white against grey) as defined by the following groupings: V, L, I, and M; F, Y, and W; K and R, E and D; Q and N; E and Q; D and N; S and T; and A, G, and P. Helices are labeled αA-αM; strands are labeled β1–β15 (strand β8 is subdivided into β81 and β82 owing to a discontinuity in this strand). (c) Distribution of averaged crystallographic thermal B factor per residue.
Previously we reported the tetrameric structure of MspJI which recognizes (5mC)NN(G/A)7. Here we report the structure of AspBHI which recognizes (C/T)(C/G)(5mC)N(C/G)4 and we confirm that it also forms a tetramer. To understand how specific amino acids of AspBHI determine its substrate recognition preference, we generated a homology model of the AspBHI-DNA complex, and probed the importance of a number of individual amino acids by mutagenesis.
Results
Tetrameric form of AspBHI
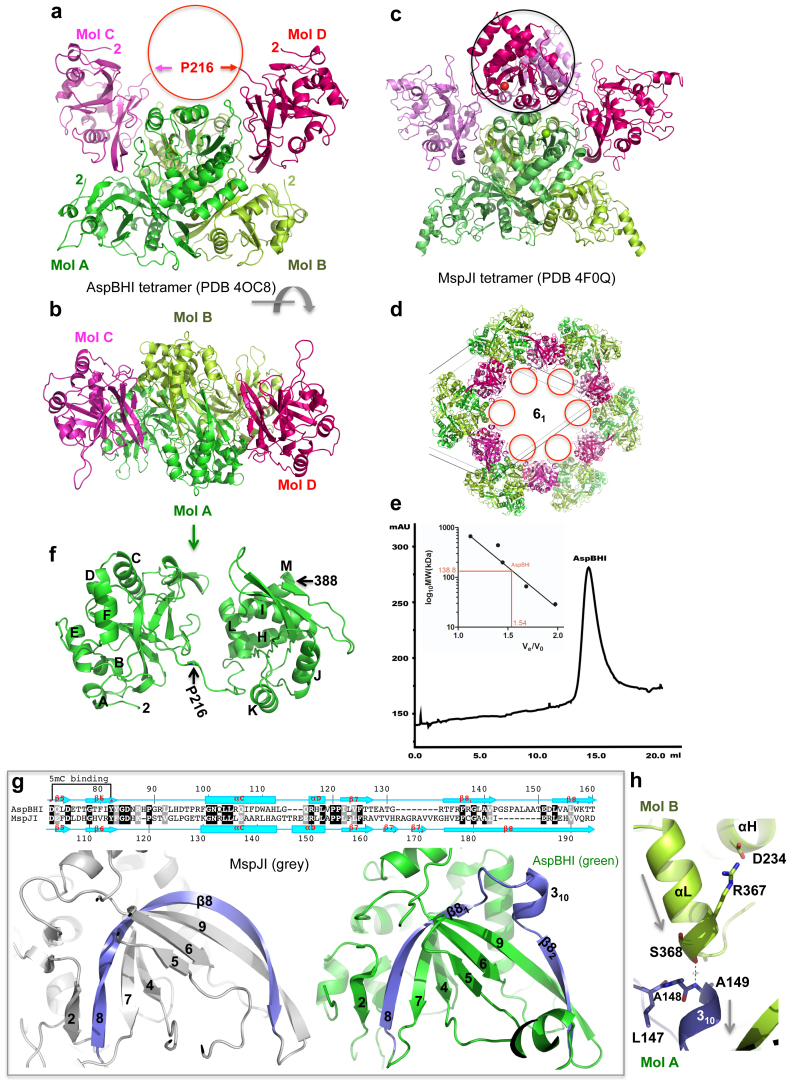
We determined the structure of AspBHI at the resolution of 2.8 Å (Table 1). Like MspJI7, AspBHI is assembled into a tetramer, formed by molecules A, B, C, and D (Fig. 2a–b). Molecules A and B form a closed dimer with high quality electron densities observed for all 388 residues. Interestingly, molecules C and D have an intact N-terminal (DNA-recognition) domain up to Pro216, but the entire C-terminal (DNA-cleavage) domain could not be traced due to discontinuous residual densities. We inferred the general location of the C-terminal domains of molecules C and D by comparison with those of MspJI (Fig. 2c), and found them to be in a void along the crystallographic 6-fold axis with a diameter of 100 Å (Fig. 2d). Absence of crystal packing forces may allow the C-terminal domains of molecules C and D to be mobile and thus unobservable. Analytical gel-filtration measurement confirmed that AspBHI exists as a tetramer in solution (Fig. 2e). An “invisible” domain in a protein crystal structure is not a common occurrence, but several examples have been observed8,9,10. In these structures, as in ours, a large space is found where a domain connected to another by a linker can move as a rigid body owing to the absence of any intra-molecular or inter-molecular crystal-packing interactions.
Table 1. Summary of Diffraction and refinement statistics of AspBHI crystals.
| Data collection | Native | Hg-soaked (L228M) | Se-substituted (L228M) |
|---|---|---|---|
| Space group | P62 | P62 | P62 |
| Cell dimensions | α = β = 90°, γ = 120° | α = β = 90°, γ = 120° | α = β = 90°, γ = 120° |
| a (Å) = | 195.09 | 193.55 | 194.48 |
| b (Å) = | 195.09 | 193.55 | 194.48 |
| c (Å) = | 81.45 | 80.96 | 82.03 |
| Beamline (SERCAT) | APS 22-BM | APS 24-ID-E | APS 24-ID-E |
| Wavelength (Å) | 1.06296 | 0.97919 | 0.97919 |
| Resolution (Å)* | 30.74–2.89(2.99–2.89) | 33.33–2.79(2.89–2.79) | 34.95–3.22(3.34–3.22) |
| aRmerge* | 0.116 (0.820) | 0.136 (0.663) | 0.124(0.590) |
| b<I/σI>* | 19.7 (3.0) | 13.9 (3.1) | 16.9(3.8) |
| Completeness (%)* | 100.0 (100.0) | 100.0 (100.0) | 100.0(100.0) |
| Redundancy* | 10.9 (10.4) | 7.0 (6.8) | 10.0(8.9) |
| Observed reflections | 437,647 | 304,887 | 288,668 |
| Unique reflections* | 40,065 (3973) | 43,261(4292)(40,583 I+ and I− pairs) | 28,794 (2873)(27,087 I+ and I− pairs) |
| Mean FOM (SAD) after refinement: | 0.28 | 0.26 | |
| FOM (MIRAS): | 0.36 | ||
| Density Modification (MIRAS), R-factor: | 0.2693 | ||
| Refinement | |||
| Resolution (Å) | 2.89 | ||
| No. reflections | 39,996 | ||
| cRwork/dRfree | 0.196/0.236 | ||
| No. Atoms | |||
| Protein | 6046 (A and B) and 3377 (C and D) | ||
| Phosphate Ion | 40 | ||
| Water | 53 | ||
| B Factors (Å2) | |||
| Protein | 51.7 (A and B) and 57.1 (C and D without the C-termianl disordered domains) | ||
| Phosphate Ion | 75.0 | ||
| Water | 45.1 | ||
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.004 | ||
| Bond angles (°) | 0.87 | ||
*Values in parenthesis correspond to highest resolution shell.
aRmerge = |I − <I>|/I, where I is the observed intensity and <I> is the averaged intensity from multiple observations.
b<I/σI> = averaged ratio of the intensity (I) to the error of the intensity (σI).
cRwork = |Fobs − Fcal|/|Fobs|, where Fobs and Fcal are the observed and calculated structure factors, respectively.
dRfree was calculated using a randomly chosen subset (5%) of the reflections not used in refinement.
Figure 2. Structure of AspBHI.
(a) Four AspBHI monomers, A, B, C and D, form a tetramer. Molecules C and D have mobile C-terminal domains (indicated by a circle). (b) AspBHI tetramer, rotated ~90° from the view of panel (a). (c) For comparison, MspJI has an intact tetramer showing in a similar orientation of panel (a). (d) The disordered C-terminal domains of molecules C and D of AspBHI tetramer were located in the void space along the crystallographic 6-fold axis with a diameter of 100 Å. (e) Elution profile of AspBHI on Superdex 200™10/300 GL (GE Healthcare). The column buffer was 20 mM Tris-HCl (pH 7.5), 300 mM NaCl and 1 mM DTT, and 150 ng of AspBHI was loaded onto the column. The inset shows the standardization of the size exclusion column using a Gel Filtration Markers Kit for Protein Molecular Weights (SIGMA-ALDRICH, Cat. No. MWGF1000) at the time AspBHI was profiled using the same buffer. (f) Monomeric AspBHI contains two domains connected by a linker. (g) AspBHI has a discontinuity in strand β8 owing to the insertion of a 310 helix (right panel), whereas MspJI has a corresponding 20-residue-long curved strand β8 (left panel). Pairwise sequence alignment is shown above the panels. (h) The 310 helix of molecule A is involved in the dimer interface with the C-terminal helix αL of molecule B. The amino end of the 310 helix (Ala149 of molecule A) interacts with the carboxyl end of helix αL (Ser368 of molecule B). Arrows indicate helical dipoles.
Monomeric AspBHI structure
Focusing on molecules A and B, the monomeric AspBHI contains two domains, connected by a 10-residue linker (residues 212 to 221) including residue Pro216 (Fig. 2f). Among the family members, AspBHI is the smallest in length (388 residues), while MspJI is the largest (456 residues) (Fig. 1a). Superimposing the AspBHI and MspJI structures revealed that MspJI has seven insertions of five to eight residues in the N-terminal DNA binding domain, mostly in the loops, and a 15-residue extension at the C-terminus (Fig. 1a)7. One interesting difference lies in the 20-residue-long curved strand β8 in MspJI, where AspBHI has an 8-residue insertion that breaks the strand into two parts (Fig. 2g). The insertion includes a 310 helix that protrudes into the C-terminal helix bundle of molecule B (Fig. 2h). The main chain carbonyl oxygen of Ser368 of molecule B forms a hydrogen bond with the main chain amide nitrogen of Ala149 of molecule A, connecting helix αL of molecule B with the 310 helix of molecule A (Fig. 2h).
A model of the N-terminal SRA-like DNA-binding domain in complex with DNA
Like MspJI7, the N-terminal domain of AspBHI is structurally similar to the eukaryotic SET and RING-associated (SRA) domain of UHRF1 (Fig. 3a–b), which binds to hemi-methylated 5mCpG dinucleotide sequences11,12. The C-terminal domain of AspBHI is structurally similar to several prokaryotic Type II endonucleases (Fig. 3c–d). We created a model of the AspBHI N-terminal SRA-like domain bound to DNA, using the coordinates of the mouse SRA–DNA complex13. After superimposing the protein components, the bound DNA was positioned over the mostly basic surface of AspBHI except for an apparent acidic pocket. An equivalent pocket is present in the SRA–DNA complex where it forms the binding site for the methylated cytosine, which is flipped out from the DNA helix (Fig. 3b). The flipped 5mC models accurately into the AspBHI pocket, in a position to interact with Asp71 via two hydrogen bonds and Tyr82 via planar stacking contact. Asp71 is part of the loop between strand β4 and β5 and the last residue prior to strand β5. Tyr82 is part of the strand β6, which is anti-parallel to strand β5 and is positioned alongside Asp71. These two amino acids are conserved among the AspBHI family enzymes (Fig. 1a) and also among known SRA domains13, where Asp474 and Tyr483 of mouse UHRF1 interact with the flipped 5mC in the same way. The methyl group of 5mC interacts with the Cα and Cβ atoms of Ser486 in UHRF1 (Fig. 3b)13, and likely does the same with Asp85 of AspBHI, the side chain of which points away from the binding pocket (Fig. 3b). Mutating Asp71, Tyr82 or Asp85 to alanine abolished AspBHI activity (Fig. 4, lanes 9–11), indicating that these residues are essential for binding the flipped 5mC nucleotide, for subsequent endonuclease catalysis, or for both.
Figure 3. A model of AspBHI in complex with DNA.
(a) Superimposition of the AspBHI N-terminal domain (in green) with the SRA domain of mouse UHRF1 (in yellow; PDB 3FDE). (b) The flipped 5mC nucleotide can be docked into the binding pocket of AspBHI. (c) Superimposition of the AspBHI C-terminal endonuclease domain (in green) and the HindIII–DNA complex (conserved secondary elements in yellow and additional in grey) (PDB 2E52). (d) The scissile phosphate group (shown as an orange ball) is near the proposed catalytic residues (Glu303 and Lys305 in AspBHI). The side chain of conserved Asp282 in AspBHI, pointing away from the active site, might undergo conformational change upon DNA binding. (e) A model of the AspBHI N-terminal domain docked with a DNA (taken from PDB 3FDE) containing a flipped 5mC (which is faded in the background). The opposite guanine is labeled. The Loop-B3 occupies the DNA minor groove 5′ to the 5mC, while the Loop-2B occupies the minor groove 3′ to the 5mC.
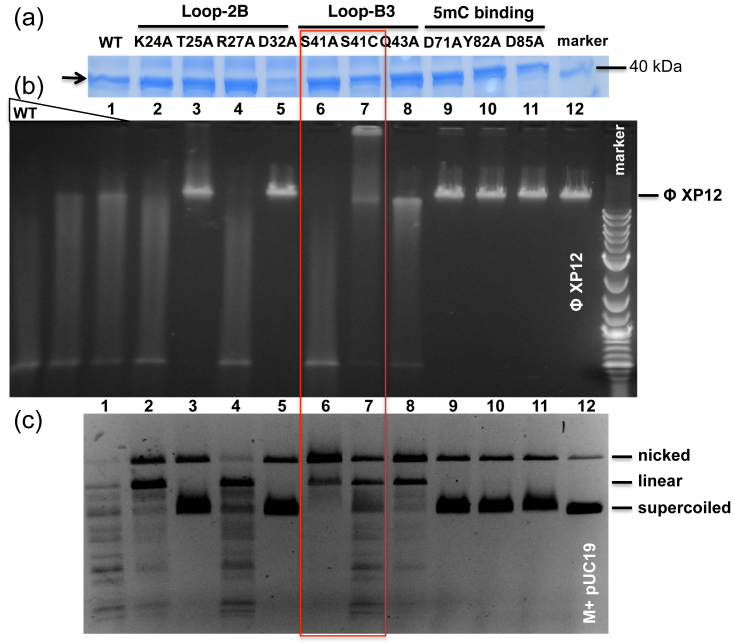
Figure 4. AspBHI variants and activity assays on modified plasmid and phage DNA substrates.
(a) SDS-PAGE analysis of partially purified His-tagged AspBHI WT and its variants after nickel-chelated affinity chromatography. Arrow indicates the AspBHI protein band. (b) Endonuclease activity assay on phage XP12 DNA containing 5mC. Three concentration of WT AspBHI (~0.57 pmoles, with 2-fold serial dilution) were used in the digestion. Mutant enzyme concentrations were estimated at 0.29 to 0.57 pmoles. The smearing may result from partial digestions of the phage DNA. We note that S41C protein tends to precipitate in conditions with <0.2 M NaCl. (c) Endonuclease activity assay on Dcm+ and M.HpaII modified pUC19 DNA.
In order to hydrogen bond with the ring atom N3 and the exocyclic amino group N4 (NH2) of the flipped 5mC (Fig. 3b), the side chain carboxylate group of Asp71 must be in the protonated state, even though the pKa of this group in solution (3.9) is well below the pH (7.9) at which the enzyme is active. The same must be true for Asp474 of UHRF1, and also for the conserved binding pocket glutamate of motif V (‘ENV’) of the 5mC-methyltransferases14,15,16,17 which likewise hydrogen bonds with the flipped substrate cytosine preparatory to methyl transfer.
Our model of the AspBHI N-terminal domain bound to DNA, derived from the UHRF1 SRA-DNA complex, suggests that three loops (Loops 2B, B3 and 6C) might intrude into the DNA minor or major grooves (Fig. 3a and 3e) and provide the interactions needed for AspBHI to recognize its DNA substrate sequence. Loop-2B (residues 23–31 between strand β2 and helix αB) could make base-specific contacts in the minor groove on the 3′ side of the flipped 5mC, where N(C/G) is recognized, and Loop-B3 (residues 39–43 between helix αB and strand β3) could make base-specific contacts in the minor groove on the 5′ side where (T/C)(C/G) is recognized. Loop-2B is unique to AspBHI in sequence among the family members (Fig. 1b) as well as in length compared with UHRF1. The corresponding loop in UHRF1 is a one-residue sharp turn13. Alanine mutations of potential contact residues within Loop-2B were constructed and tested. K24A and R27A cleaved phage DNA similarly to WT AspBHI (Fig. 4, lanes 2 and 4), but plasmid digestion was somewhat reduced, especially for K24A. T25A and D32A [Asp32 is an invariant residue within the family, Fig. 1b] abolished cleavage activity altogether (Fig. 4, lanes 3 and 5).
Loop-B3 contains Ser41 and Arg42 that are unique to AspBHI (Fig. 1b). The corresponding loop in UHRF1 also approaches the DNA from the minor groove and contains Val451, which occupies the space left behind by the flipped 5mC, and His450, which interacts with the 5′ base pair13. To examine the effects of Loop-B3 mutations, we changed Ser41 and Arg42 to all 19 other amino acids (the results are discussed below). The third loop, Loop-6C is between strand β6 and helix αC (residues 84–99). The corresponding loop in UHRF1 contains Arg496, which hydrogen bonds from the major groove with the intra-helical orphaned guanine (Fig. 3a)13. Loop-6C is six-residue shorter than its UHRF1 counterpart, and it adopts a different conformation due perhaps to the absence of DNA (Fig. 3a), making it too short to reach the DNA major groove in the current model. Nevertheless, Loop-6C is a prime candidate for making base specific interaction in the major groove if the substrate DNA and/or protein undergo structural rearrangement during binding.
S41A and S41C variants have altered cleavage activities
Substitutions of Ser41 by other amino acids drastically reduced enzyme activity (data not shown) except for the alanine (S41A) and cysteine (S41C) replacements. These two variants showed somewhat different cleavage properties towards modified plasmid or phage DNA compared to the WT enzyme (Fig. 4, lanes 6–7): S41A cleaved phage XP12 DNA similarly to WT enzyme (Fig. 4b, lane 6), but barely cleaved pUC19 DNA, except for converting supercoiled DNA to nicked intermediate (only one strand cut) and linear form (one double-strand cut) (Fig. 4c, lane 6). S41C demonstrated the opposite effect: it cleaved phage XP12 DNA much less efficiently than pUC19. The phage DNA appears to be trapped by the S41C protein precipitation (Fig. 4b, lane 7, the band near the top loading well), although it is not clear whether the bound DNA had been cleaved.
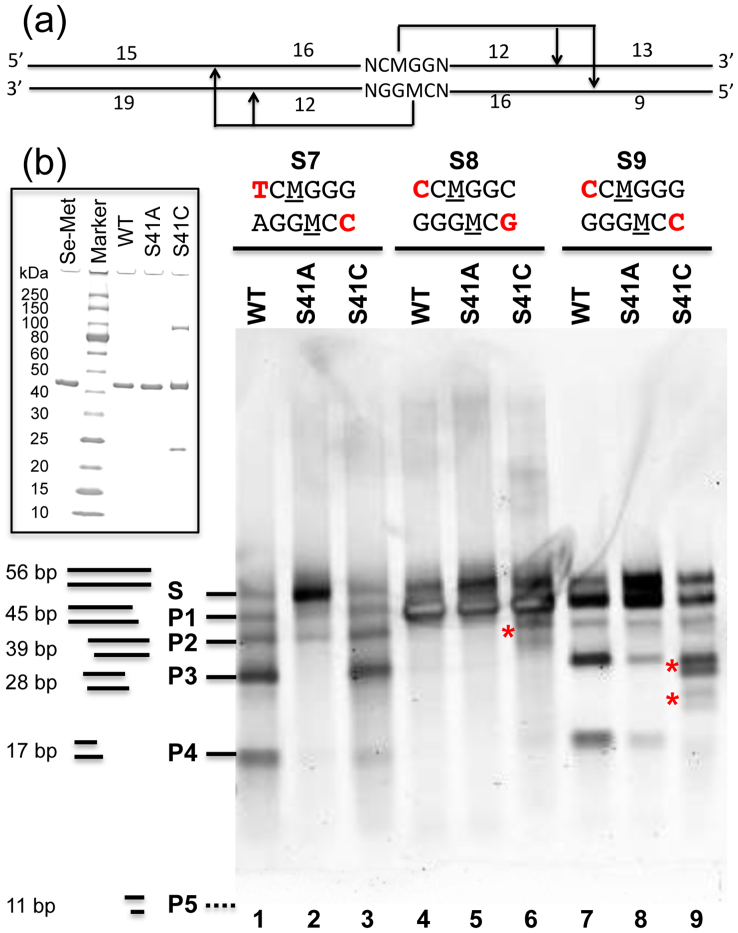
To investigate the specificity of the S41A and S41C variants, we used three 56-bp synthetic duplexes containing the symmetric sequence 5′-NC(5mC)GGN-3′ (Fig. 5a), methylated on both strands. If the enzyme recognizes the top strand methylated site, cleavage on the 3′ side N12/N16 away will result in two products of 43-bp and 9-bp, both with a 4-bp overhang. We termed these products as P1 and P5 with averaged lengths of 45-bp and 11-bp (Fig. 5b). [The product P5 was not observed probably because it was too small to be stained or the small duplex (9 bp + 4 nt overhang) dissociated at 37°C after cleavage and the two short single-stranded oligonucleotides ran out of the gel.] If the enzyme recognizes the bottom strand methylated site, cleavage will result in two products of 39-bp (P2) and 17-bp (P4). And if the enzyme recognizes both top and bottom strand methylated sites, cleavage on both sides will result in three products of averaged lengths of 28-bp (P3), 17-bp (P4), and 11-bp (P5). The cleavage products were resolved using 20% native PAGE (Fig. 5b). The results indicate that AspBHI is capable of cleaving the substrates having a 5′ pyrimidine base (T or C) (lanes 1 and 7) but not a guanine (or adenine4): lane 4 of Fig. 5b only shows top strand (with a 5′ C) recognition products, P1 and P5 (not visible), but not the bottom strand (with a G) recognition products P2 and P4.
Figure 5. S41A and S41C activity assays on methylated oligonucleotide substrates.
(a) Schematic diagram of the fully methylated oligonucleotide substrates (M = 5mC) used for analyzing possible cleavage products (P1–P5 shown in panel b). (b) Duplex oligonucleotides (20 ng) were incubated at 37°C for 2 hours with 0.5 μg (0.29 pmoles) of WT, S41A, or S41C. Products were resolved on a 20% TBE native PAGE gel and visualized with Sybr Gold staining. Inserted is a 10–20% gradient SDS-PAGE showing the proteins used for crystallization (Se-Met) and for activity (WT, S41A and S41C). NEB protein ladder was used as molecular weight markers.
S41A variant showed lower activity in cleaving all three substrates as a significant amount of full-length duplex oligonucleotides remained (Fig. 5b, lanes 2, 5 and 8). However, it appeared to prefer the S9 substrate, with the two 5′ most positions being a C on both strands, compared with substrate S7 that has 5′ T or 5′ C on each strand (comparing lanes 2 and 8). This is in contrast to the WT enzyme that cleaved substrate S7 better (comparing lanes 1 and 7), suggesting a potential change of substrate specificity. On the other hand, an approximately equal amount of P1 and P2 products were generated by S41A on S7 substrate (lane 2), suggesting S7 might be a poor substrate for S41A, regardless of a 5′ T or 5′ C. The S41C variant had a digestion pattern similar to that of the WT enzyme. However, in addition to the predominant cleavage position at N12/N16 from the modified cytosine, S41C appears to have additional cleavage positions (as marked with asterisk in lanes 6 and 9) – an observation previously observed as wobble cleavage4.
Arg42 is essential for activity
A total of 19 variants R42X (natural amino acids other than arginine) were constructed by site-directed mutagenesis. All 19 variants were purified through nickel-chelated and heparin affinity chromatography. All were inactive in cleaving modified plasmid DNA, including the conservative Arg42-to-lysine substitution (data not shown). Arg42 might interact with the target 5mC:G base pair (the only unambiguous base pair within the recognition sequence) during the initial protein-DNA encounter or stabilize the flipped 5mC via interaction with the orphaned guanine for enhanced recognition and tightening of the protein-DNA complex and thereby promoting cleavage. The precise way in which Arg42 and Ser41 mediate specific DNA recognition awaits the solution of a protein-DNA complex structure.
Discussion
The wide diversity of restriction enzymes18, from the smallest dimeric PvuII19, to tetrameric Type IIF enzymes20, and the polymerized SgrAI21, make them versatile tools for laboratory experimentation, and fascinating subjects for studies of molecular architecture22. Here we show structurally that the modification-dependent restriction enzyme AspBHI comprises two domains, one typically eukaryotic and the other typically prokaryotic. The N-terminal part of AspBHI (residues 1–211) resembles an SRA-like 5-methylcytosine binding domain in structure and function. It recognizes 5mC within the specific DNA sequence context. The C-terminal part of AspBHI (residues 222–388) resembles a classic Type II restriction endonuclease of the PD-(D/E)XK superfamily23,24,25. It is attached to the N-terminal domain by a 10-residue loop, and cleaves duplex DNA outside of the recognition sequence on one side, N12/N16 3′ downstream of the 5mC, somewhat like a Type IIs restriction enzyme.
FokI, the best-known Type IIs enzyme, has a similar domain organization comprising an N-terminal recognition domain and a C-terminal catalytic domain. It also recognizes an asymmetric sequence and cleaves downstream N9/N13, but there the similarities stop. FokI is monomeric in solution and double-strand (ds) cleavage occurs by transient dimerization between the catalytic domains of neighboring molecules at least one of which is bound to a recognition site26,27. AspBHI (and MspJI7), in contrast, assembles into a tetramer, even in the absence of DNA, with two centers for ds DNA cleavage (i.e. two catalytic-domain mediated dimers) and four 5mC-recognition domains. A complex model based on structural and biochemical evidence has been proposed for MspJI7 - and likely also applies to AspBHI - in which three monomers of the tetramer are involved, respectively, in binding modified cytosine, making the first proximal N12 cleavage in the same strand, and then making the second distal N16 cleavage in the opposite strand. In contrast to AspBHI, the N6-methyladenine dependent restriction enzyme DpnI, comprises an N-terminal combined recognition and catalytic domain and a C-terminal non-catalytic DNA-binding domain28 (opposite of the domain arrangement of AspBHI and MspJI), and is monomeric.
The variety of restriction enzymes also makes them fascinating subjects for studying protein-DNA interactions among enzymes with a common basic function – highly specific DNA recognition and cleavage. Surprisingly, even for very well characterized restriction enzymes such as EcoRV29,30,31,32,33,34,35,36,37,38, the mechanistic features that determine specificity and selectivity are difficult to model on the basis of the available structural information39. Other than requiring a 5mC:G base pair, AspBHI is promiscuous in the bases it recognizes on either side of the modified cytosine: 5′-(C/T)(C/G)(5mC)N(C/G)-3′. For example, the 5′ most base can be a thymine or cytosine but not a guanine (or adenine) (Fig. 5b). We attempted to relax specificity further on the 5′ side of the 5mC by targeted mutagenesis of Ser41 and Arg42, but we were unsuccessful. Arg42, which is not conserved among family members (Fig. 1b), was found nevertheless to be essential for enzyme activity, and all Arg42 mutants were inactive. Ser41 mutants were likewise inactive except S141A and C. Interestingly, S41A, which loses the ability to make hydrogen bonds, showed somewhat different cleavage properties towards modified oligonucleotides with variation at the outermost 5′ (C/T) position. Although considerable progress has been made regarding the mechanisms of action of restriction enzymes, many challenges remain, the most ambitious perhaps being the engineering of enzyme variants with new specificities.
Methods
All enzymes, plasmids and bacterial strains, if not otherwise specified, were obtained from New England Biolabs (NEB). Escherichia coli codon optimized AspBHI with an N-terminal 6xHis tag was cloned into a pUC19 derivative pZZ1 (Z. Zhu, NEB) between NdeI and BamHI sites4. Site-directed mutagenesis was carried out by inverse PCR using Vent® DNA polymerase and mutagenic primers designed with NEB in-house software. The entire alleles in AspBHI variants were sequenced to confirm the desired mutation.
Protein expression and purification
Wild type (WT) and mutant AspBHI with N-terminal 6xHis tags were expressed in a Dcm-deficient E. coli strain T7 Express (C2566). Cells were grown at 30°C in 10 mL (small scale) or 0.5 to 1 L (medium scale) in LB + Amp to OD600 0.3–0.6 and induced with a final concentration of 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). Induced cultures were grown overnight at 25°C, harvested and then kept at −20°C. His-tagged proteins (small scale) were partially purified using Qiagen Ni-NTA spin kit as recommended by the supplier and used in the experiments shown in Fig. 4. For medium-scale production cells were lysed using sonication in 20 mM Tris-HCl, pH 7.5, 400 mM NaCl, 20 mM imidazole. Clarified cell extract was loaded over a gravity column using a Ni-NTA resin (Qiagen). Protein was eluted with 500 mM imidazole. Pooled fractions were then diluted by 10 fold in 20 mM Tris-HCl, pH 7.5, 20 mM NaCl and loaded over a 5 mL Hi-Trap Heparin column using an AKTA FPLC machine (GE Healthcare). The proteins were eluted at ~250–290 mM NaCl with a linear gradient of 20 mM to 1 M NaCl. Fractions containing AspBHI were identified on 10–20% gradient Tris-Glycine gels (Novex/Life Technologies) with the protein appearing as the major band (purity approximately 95%; Fig. 5b insert). Proteins were diluted to a working stock of 0.5–1 mg ml−1 and used in the experiments shown in Fig. 5b.
Crystallography
For crystallization of AspBHI, 12 L of IPTG-induced E. coli cultures were harvested and the non-tagged enzyme was purified to homogeneity by chromatography through Heparin DM, Bio-Gel HTP hydroxyapatite, Mono Q, and Heparin TSK columns. Alternatively, further purification was performed via tandem HiTrap Q/SP (GE Healthcare) and a sizing column Superdex 200 (GE Healthcare). The position of the protein peak in the Superdex 200 column suggests the protein to be a tetramer (Fig. 2e).
Final concentrations of the protein are between 6–20 mg ml−1 in 20 mM Tris-HC1 (pH 8.0), 150 mM NaCl, 10% glycerol, 1 mM ethylenediaminetetraacetic acid (EDTA), and 1 mM dithiothreitol (DTT). Crystallizations were carried out by the hanging-drop vapor-diffusion method at 16°C using equal amounts of protein and well solutions. Conditions giving large and well-diffracting AspBHI crystals were (i) 12% polyethylene glycol 3350 with 0.5 M K2HPO4/Na2HPO4 (pH 7.4) and (ii) 6–15% polyethylene glycol MME 5000, 5% Tacsimate (Hampton Research), and 100 mM HEPES (pH 6.2–7.4). The AspBHI crystal structure was solved by multi-wavelength anomalous diffraction phasing methods40 using three datasets: a native AspBHI dataset, a Se anomalous dataset from a selenium-methionine (SeMet) labeled Leu228-to-Met (L228M) mutant crystal, and a Hg anomalous dataset from L228M mutant crystal soaked with ~5 mM K2HgI4 overnight (Table 1).
AspBHI contains two methionines at residues 30 and 214 in addition to the N-terminal methionine. To increase the phasing potential of SeMet labeled crystals, we mutated Leu228-to-Met because other family members (RlaI and LpnP1) have a methionine at the corresponding position (Fig. 1b) and the mutant protein was utilized for phasing purposes. A total of ten Se atoms were found in the asymmetric unit of the selenium-methionine labeled crystal, three each for molecules A and B and two each for molecules C and D (L228M located in disordered C-terminal domains of molecules C and D were not detected). In the Hg derivative, a total of four Hg2+ atoms were found in the asymmetric unit, two of which reacted to Cys255 and Cys306 of molecules A or B. All the data sets were processed using the program HKL200041, which calculated values of Rmerge and <I/σI> (Table 1). Phasing, map production, and model refinement were conducted using the PHENIX software suite42. The AutoSol Wizard43 of PHENIX used RESOLVE44 to carry out density modification and applied non-crystallographic symmetry (NCS) calculated from positions of heavy-atom sites45, resulting in the multi isomorphous replacement with anomalous scattering (MIRAS) electron density map with superior quality compared to either single anomalous diffraction (SAD) map. Maps and model were visualized with COOT46 as well as manual model manipulation during refinement rounds without the disordered C-terminal domains of molecules C and D. Individual thermal B-factors were refined only at the end stages of refinement, with the averaged root-mean-square deviation of 3.7 Å2 for main chain atoms and 5.1 Å2 for side chain atoms and did not vary significantly for any ordered domain of the modeled monomers. Distribution of averaged crystallographic thermal B-factor pre residue for the four monomers is shown in Figure 1C, with the highest B-factors occur in the loops.
DNA cleavage assays using methylated plasmids and phage DNA
Dcm+ pUC19 (100 μg) was incubated with various methyltransferases (M.AluI, M.SssI, M.HaeIII, M.HpaII, M.HhaI, or M.MspI) overnight at 37°C in the presence of 32 mM AdoMet (160 mM AdoMet for M.SssI) in a total reaction volume of 500 μL. Reactions were treated with 5 μL Proteinase K (10 mg ml−1) for 1 h at 37°C. Plasmids were then purified by spin column (Qiagen) and the DNA concentration was measured using the Nanodrop.
For plasmid digestions, 100 to 300 ng of DNA was digested with 1–5 μg of AspBHI (1 mg ml−1) in NEB buffer 4 in the presence of 15 μM of a self-annealed stem-loop activator (5′ CTCCMAGGATCTTTTTTGATCMTGGGAG-3′ where M = 5mC)4. Adding an activator with the recognition sequence in trans can accelerate the slow reactions by the AspBHI family members4. Titrations of AspBHI were done using dilution buffer (diluent B, NEB). Enzyme titration was carried out to make sure that the AspBHI concentration used in digestion was not inhibitory. Digestions were carried out for 2 h at 37°C and then treated with 2 μL proteinase K for 15 min. Digestion products were resolved and visualized after running on a 1% agarose gel (Figure 4).
Phage XP12 DNA (bacterial host Xanthomonas oryzae) was a gift from Dr. Peter Weigele (NEB). XP12 phage particles were purified from lysate by CsCl gradient centrifugation and its DNA was further purified by phenol-CHCl3 extraction and ethanol precipitation. The phage DNA contains 5-methylcytosine, which serves as a substrate for modification-dependent restriction enzymes47. The endonuclease digestion was terminated by addition of a loading dye with ethylenediaminetetraacetic acid (EDTA), sodium dodecyl sulfate (SDS), and glycerol. We used both XP12 phage DNA (which is methylated at every cytosine) and 5mC-modified pUC19 (which is methylated at the specific sites) to corroborate the mutant activity. In general, most of the mutant activity is consistent on both substrates except for S41C as shown in Figure 4.
Digestion of fully methylated oligonucleotides
Three sets of 56-base pair (bp) oligonucleotides containing NCMGGN (M = 5mC, N = A, T, C or G) was used for digestion as described4:
5′-CGGCGTTTCCGGGTTCCATAGGCTCCGCNCMGGNCTCTGATGACCAGGGCATCACA-3′
3′-GCCGCAAAGGCCCAAGGTATCCGAGGCGNGGMCNGAGACTACTGGTCCCGTAGTGT-5′
Duplex oligonucleotide substrates (20 ng) were incubated with 0.5 μg of AspBHI (WT, S41A, or S41C) in NEB buffer 4 with a final volume of 10 μL at 37°C for 2 h and then treated with 0.5 μL proteinase K for 15 min. Digestion products were resolved on a 20% native TBE PAGE gel (Life Technologies), stained with Sybr Gold (Life Technologies) and visualized using a Typhoon 9400 imager (GE) (Fig. 5b).
Author Contributions
J.R.H. performed crystallographic work, R.L.N. and A.L. performed site-directed mutagenesis, R.L.N., A.L. and S.Y.X. purified mutants and performed activity assays, M.Y.M. constructed and assessed all 19 mutants of R42X, A.F. performed over-expression of non-tagged AspBHI used for crystallography, D.C.K. contributed enzyme reagents, R.M.G. and X.Z. performed purification and crystallization trials, G.G.W. performed structural analysis, suggested structure-based mutagenesis and assisted in preparing the manuscript, X.C., Y.Z. and S.Y.X. organized and designed the scope of the study, and all were involved in analyzing data and preparing the manuscript.
Additional information
Accession codes The X-ray structure (coordinates and structure factor files) of AspBHI has been submitted to the Protein Data Bank as entry 4OC8.
Acknowledgments
We thank Siu-Hong Chan for help with protein purification, Thomas Buswell for assistance in site-directed mutagenesis, Derrick Xu for the original AspBHI plasmid, Bill Jack for reading the manuscript. New England Biolabs supported the mutagenesis work and the National Institutes of Health (Grant GM049245-20) supported the crystallographic work. X.C. is a Georgia Research Alliance Eminent Scholar. The Department of Biochemistry at the Emory University School of Medicine supported the use of the Southeast Regional Collaborative Access Team synchrotron beamlines at the Advanced Photon Source of Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U.S. Department of Energy under Contract W-31-109-Eng-38.
Footnotes
Restriction enzymes, and modification-dependent restriction enzymes, mentioned in this article are products of New England Biolabs.
References
- Kohli R. M. & Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R. et al. Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H. & Laird P. W. Interplay between the cancer genome and epigenome. Cell 153, 38–55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Karni D. et al. The MspJI family of modification-dependent restriction endonucleases for epigenetic studies. Proc. Natl. Acad. Sci. U S A 108, 11040–11045 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z. et al. High-resolution enzymatic mapping of genomic 5-hydroxymethylcytosine in mouse embryonic stem cells. Cell reports 3, 567–576 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y. et al. A unique family of Mrr-like modification-dependent restriction endonucleases. Nucleic Acids Res. 38, 5527–5534 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J. R. et al. Structure and cleavage activity of the tetrameric MspJI DNA modification-dependent restriction endonuclease. Nucleic Acids Res. 40, 9763–9773 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo E. J. et al. Structural mechanism for inactivation and activation of CAD/DFF40 in the apoptotic pathway. Mol. Cell 14, 531–539 (2004). [DOI] [PubMed] [Google Scholar]
- Turkenburg J. P. et al. Structure of a pullulanase from Bacillus acidopullulyticus. Proteins 76, 516–519 (2009). [DOI] [PubMed] [Google Scholar]
- Nakatani Y., Cutfield S. M., Cowieson N. P. & Cutfield J. F. Structure and activity of exo-1,3/1,4-beta-glucanase from marine bacterium Pseudoalteromonas sp. BB1 showing a novel C-terminal domain. Febs J. 279, 464–478 (2012). [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Horton J. R., Zhang X. & Cheng X. UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications. Epigenetics 4, 8–14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J. & Koseki H. Recruitment of Dnmt1: roles of the SRA protein Np95 (Uhrf1) and other factors. Prog. Mol. Biol.Transl. Sci. 101, 289–310 (2011). [DOI] [PubMed] [Google Scholar]
- Hashimoto H. et al. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455, 826–829 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. C. & Santi D. V. Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem. 262, 4778–4786 (1987). [PubMed] [Google Scholar]
- Baker D. J., Kan J. L. & Smith S. S. Recognition of structural perturbations in DNA by human DNA(cytosine-5)methyltransferase. Gene 74, 207–210 (1988). [DOI] [PubMed] [Google Scholar]
- Liu L. & Santi D. V. Mutation of asparagine 229 to aspartate in thymidylate synthase converts the enzyme to a deoxycytidylate methylase. Biochemistry 31, 5100–5104 (1992). [DOI] [PubMed] [Google Scholar]
- O'Gara M., Klimasauskas S., Roberts R. J. & Cheng X. Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaI methyltransferase-DNA-AdoHcy complexes. J. Mol. Biol. 261, 634–645 (1996). [DOI] [PubMed] [Google Scholar]
- Pingoud A., Fuxreiter M., Pingoud V. & Wende W. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 62, 685–707 (2005). [DOI] [PubMed] [Google Scholar]
- Cheng X., Balendiran K., Schildkraut I. & Anderson J. E. Structure of PvuII endonuclease with cognate DNA. Embo J. 13, 3927–3935 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siksnys V., Grazulis S. & Huber R. Structure and function of the tetrameric restriction enzymes. Nucleic Acids Mol. Biol. 14, 237–259 (2004). [Google Scholar]
- Lyumkis D. et al. Allosteric regulation of DNA cleavage and sequence-specificity through run-on oligomerization. Structure 21, 1848–1858 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden D. T. The architecture of restriction enzymes. Structure 21, 1720–1721 (2013). [DOI] [PubMed] [Google Scholar]
- Niv M. Y. et al. Topology of Type II REases revisited; structural classes and the common conserved core. Nucleic Acids Res. 35, 2227–2237 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski J. & Bujnicki J. M. Structural and evolutionary classification of Type II restriction enzymes based on theoretical and experimental analyses. Nucleic Acids Res. 36, 3552–3569 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steczkiewicz K., Muszewska A., Knizewski L., Rychlewski L. & Ginalski K. Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res. 40, 7016–7045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanamee E. S., Santagata S. & Aggarwal A. K. FokI requires two specific DNA sites for cleavage. J. Mol. Biol. 309, 69–78 (2001). [DOI] [PubMed] [Google Scholar]
- Wah D. A., Bitinaite J., Schildkraut I. & Aggarwal A. K. Structure of FokI has implications for DNA cleavage. Proc. Natl. Acad. Sci. U S A 95, 10564–10569 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwek W., Czapinska H., Bochtler M., Bujnicki J. & Skowronek K. Crystal structure and mechanism of action of the N6-methyladenine-dependent type IIM restriction endonuclease R.DpnI. Nucleic Acids Res. 40, 7563–7572 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielking V. et al. Site-directed mutagenesis studies with EcoRV restriction endonuclease to identify regions involved in recognition and catalysis. Biochemistry 30, 6416–6422 (1991). [DOI] [PubMed] [Google Scholar]
- Wenz C. et al. Protein engineering of the restriction endonuclease EcoRV: replacement of an amino acid residue in the DNA binding site leads to an altered selectivity towards unmodified and modified substrates. Biochim. Biophys. Acta 1219, 73–80 (1994). [DOI] [PubMed] [Google Scholar]
- Wenz C., Jeltsch A. & Pingoud A. Probing the indirect readout of the restriction enzyme EcoRV. Mutational analysis of contacts to the DNA backbone. J. Biol. Chem. 271, 5565–5573 (1996). [DOI] [PubMed] [Google Scholar]
- Stahl F., Wende W., Jeltsch A. & Pingoud A. Introduction of asymmetry in the naturally symmetric restriction endonuclease EcoRV to investigate intersubunit communication in the homodimeric protein. Proc. Natl. Acad. Sci. U S A 93, 6175–6180 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanio T. et al. EcoRV-T94V: a mutant restriction endonuclease with an altered substrate specificity towards modified oligodeoxynucleotides. Protein engineering 9, 1005–1010 (1996). [DOI] [PubMed] [Google Scholar]
- Wenz C., Hahn M. & Pingoud A. Engineering of variants of the restriction endonuclease EcoRV that depend in their cleavage activity on the flexibility of sequences flanking the recognition site. Biochemistry 37, 2234–2242 (1998). [DOI] [PubMed] [Google Scholar]
- Stahl F., Wende W., Wenz C., Jeltsch A. & Pingoud A. Intra- vs intersubunit communication in the homodimeric restriction enzyme EcoRV: Thr 37 and Lys 38 involved in indirect readout are only important for the catalytic activity of their own subunit. Biochemistry 37, 5682–5688 (1998). [DOI] [PubMed] [Google Scholar]
- Stahl F., Wende W., Jeltsch A. & Pingoud A. The mechanism of DNA cleavage by the type II restriction enzyme EcoRV: Asp36 is not directly involved in DNA cleavage but serves to couple indirect readout to catalysis. Biol. Chem. 379, 467–473 (1998). [DOI] [PubMed] [Google Scholar]
- Lanio T., Jeltsch A. & Pingoud A. Towards the design of rare cutting restriction endonucleases: using directed evolution to generate variants of EcoRV differing in their substrate specificity by two orders of magnitude. J. Mol. Biol. 283, 59–69 (1998). [DOI] [PubMed] [Google Scholar]
- Schottler S., Wenz C., Lanio T., Jeltsch A. & Pingoud A. Protein engineering of the restriction endonuclease EcoRV--structure-guided design of enzyme variants that recognize the base pairs flanking the recognition site. Euro. J. Bioc./FEBS 258, 184–191 (1998). [DOI] [PubMed] [Google Scholar]
- Lanio T., Jeltsch A. & Pingoud A. On the possibilities and limitations of rational protein design to expand the specificity of restriction enzymes: a case study employing EcoRV as the target. Protein engineering 13, 275–281 (2000). [DOI] [PubMed] [Google Scholar]
- Hendrickson W. A., Horton J. R. & LeMaster D. M. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. Embo J. 9, 1665–1672 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Borek D., Majewski W. & Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr. A 59, 228–234 (2003). [DOI] [PubMed] [Google Scholar]
- Adams P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T. C. et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T. C. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374, 22–37 (2003). [DOI] [PubMed] [Google Scholar]
- Terwilliger T. C. Rapid automatic NCS identification using heavy-atom substructures. Acta Crystallogr. D Biol. Crystallogr. 58, 2213–2215 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P. & Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- Adouard V. et al. The accessibility of 5-methylcytosine to specific antibodies in double-stranded DNA of Xanthomonas phage XP12. Euro. J. Bioc./FEBS 152, 115–121 (1985). [DOI] [PubMed] [Google Scholar]