Summary
The tetrapyrroles heme, bacteriochlorophyll and cobalamin (B12) exhibit a complex interrelationship regarding their synthesis. In this study, we demonstrate that AerR functions as an antirepressor of the tetrapyrrole regulator CrtJ. We show that purified AerR contains B12 that is bound to a conserved histidine (His145) in AerR. The interaction of AerR to CrtJ was further demonstrated in vitro by pull down experiments using AerR as bait and quantified using microscale thermophoresis. DNase I DNA footprint assays show that AerR containing B12 inhibits CrtJ binding to the bchC promoter. We further show that bchC expression is greatly repressed in a B12 auxotroph of Rhodobacter capsulatus and that B12 regulation of gene expression is mediated by AerR’s ability to function as an antirepressor of CrtJ. This study thus provides a mechanism for how the essential tetrapyrrole, cobalamin controls the synthesis of bacteriochlorophyll, an essential component of the photosystem.
Keywords: cobalamin, tetrapyrrole gene regulation, Rhodobacter capsulatus, CrtJ/PpsR
Introduction
Cobalamin (vitamin B12) is one of the most complex nonpolymeric biomolecules synthesized by cells. B12 is important cofactor of many enzymes, such as methyltransferases, reductases, and isomerases, which catalyze a range of biological processes (Gruber et al., 2011). There are two forms of B12 that are commonly used in vivo, adenosylcobalamin (AdoB12) and methylcobalamin (MeB12) that typically have 5’-deoxyadenosyl or a methyl group coordinated as an upper axial ligand to a corrin bound cobalt, respectively. The upper carbon-cobalt bond is relatively weak, generating a reactive leaving group with a homolytic bond disassociating energy of ~30 kcal mol−1 (Halpern, 1985, Waddington and Finke, 1993). Both AdoB12 and MeB12 undergo spontaneous homolytic cleavage of the carbon-cobalt bond resulting in the generation of hydroxocobalamin (OHB12) when photoexcited at room temperature in aqueous solutions (Hogenkamp, 1966, Weissbach et al., 1960).
Only certain members of bacteria and archaea synthesize B12 de novo (Perlman, 1959). The metabolically diverse photosynthetic α-proteobacterium Rhodobacter capsulatus is capable of synthesizing B12 under both aerobic and anaerobic conditions (McGoldrick et al., 2002). In this species heme, bacteriochlorophyll (Bchl) and B12 biosynthesis share common early intermediates from δ-aminolevulinic acid (ALA) to uroporphyrinogen III at which point the B12 pathway branches off from the heme/Bchl pathways. The heme and Bchl pathways further diverge at protoporphyrin IX, where iron is chelated to form heme or magnesium is chelated to form the first dedicated intermediate in the Bchl pathway.
Among photosynthetic organisms such as R. capsulatus, the levels of heme and B12 stay relatively stable while the amount of Bchl is altered dramatically in response to different environmental conditions. Under aerobic conditions only trace amounts of Bchl are synthesized as expression of genes coding for enzymes for the Bchl branch are repressed by the aerobic repressor CrtJ (Ponnampalam and Bauer, 1997, Ponnampalam et al., 1995). Under anaerobic conditions the amount of Bchl is significantly higher as bch genes are no longer repressed by CrtJ, and are additionally anaerobically activated by the RegB/RegA two component system (Bird et al., 1999, Du et al., 1998, Willett et al., 2007).
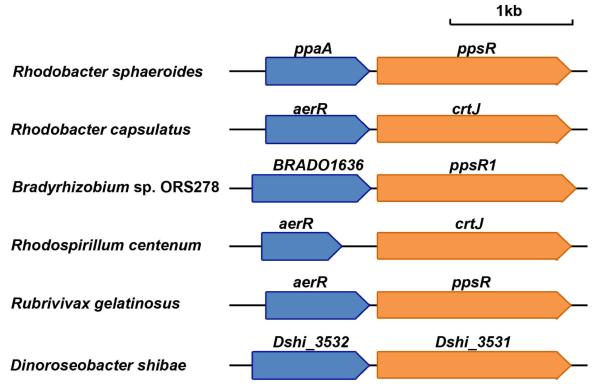
CrtJ is a major regulator of genes coding for enzymes involved in the biosynthesis of heme, Bchl and carotenoids, as well as structural proteins of light harvesting-II complex (Penfold and Pemberton, 1991, 1994, Ponnampalam et al., 1995). Previous studies have demonstrated that CrtJ binds promoters containing two conserved palindromes (TGTN12ACA) (Ponnampalam and Bauer, 1997, Ponnampalam et al., 1995), and that redox regulated binding is dependent on oxidation of a conserved cysteine (C420) located in the helix-turn-helix DNA binding motif (Cheng et al., 2012, Masuda et al., 2002). In many proteobacteria, crtJ homologs are immediately preceded by a gene (termed aerR in R. capsulatus) that encodes a regulator of photosynthesis gene expression (Fig. 1). AerR does not have any identifiable DNA binding domains so it has been assumed that it may control gene expression by affecting the DNA binding activity of another protein with CrtJ being the most likely candidate(Gomelsky et al., 2003, Masuda et al., 2008). Interestingly, AerR has significant sequence similarity to B12 binding domains suggesting that it may be capable of interacting with B12 (Gomelsky et al., 2003, Ludwig and Matthews, 1997). However, the tetrapyrrole specificity of AerR is somewhat controversial as there is a report that a maltose binding domain fused to a truncated variant of an homolog from R. sphaeroides preferentially binds heme over B12 (Moskvin et al., 2010).
Fig. 1. Diagram showing the conserved chromosomal positioning of aerR homologs upstream of crtJ homologs in various species.
Blue arrows represent homologs of aerR; orange arrows represent homologs of crtJ.
In addition to the role of B12 as an enzyme cofactor, this complex tetrapyrrole is also sometimes involved in regulating gene expression. For example, B12 is a ligand for RNA-based riboswitches that typically regulate synthesis of enzymes involved in B12 biosynthesis (Mandal and Breaker, 2004, Nou and Kadner, 2000). In Myxococcus xanthus CarH functions as a B12-dependent repressor of carotenogenic genes (Ortiz-Guerrero et al., 2011). AdoB12 containing CarH binds an operator as a tetramer to repress crt gene expression in the dark. Photolysis of AdoB12 to OHB12 disassembles the tetramer and causes dissociation of CarH from the operator (Ortiz-Guerrero et al., 2011). The involvement of cobalamin in regulating photosystem genes from R. capsulatus was initially reported by Pollich et al (Pollich et al., 1993, Pollich and Klug, 1995). These studies demonstrated that light harvesting and reaction center photosystem gene expression were significantly impaired in a B12 auxotroph. Normal photosystem gene expression could be recovered by exogenous addition of B12, however the mechanism whereby B12 regulates expression of photosystem genes was not established.
In this study, we demonstrate that the aerR open reading frame located immediately upstream of crtJ codes for a B12 binding antirepressor of CrtJ. This study provides the first example where B12 regulates gene expression by controlling the interaction of an anti-repressor with a transcription repressor. It also provides a molecular mechanism for the control of photosystem gene expression based on the availability of B12.
Results
AerR is required for photosystem synthesis
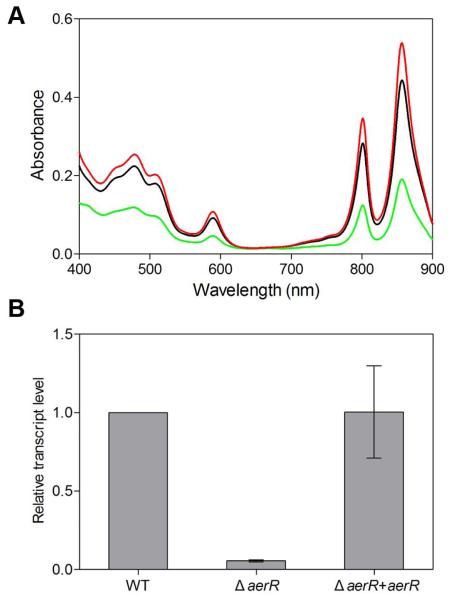
A chromosomal deletion of the aerR gene was generated by removing its entire coding sequence with the exception of retention of the start codon (the correct start codon was determined as described in Supporting Information) that remained in frame with the stop codon. The ΔaerR strain exhibits a much lighter pigmentation in comparison to wild-type R. capsulatus and also grows significantly slower under photosynthetic conditions. Spectral analyses of the ΔaerR cell lysate revealed that photosystem synthesis is reduced ~2.5 fold in this strain when grown under anaerobic photosynthetic conditions (Fig. 2A). QRT-PCR analysis of bchC expression, which is a CrtJ regulated gene that codes for an enzyme in the bacteriochlorophyll biosynthetic pathway, is also consistent with spectral analyses (Fig. 2B). Specifically, we observed that the ΔaerR strain exhibits a ~18 fold reduction in anaerobic bchC expression over that observed with wild-type cells (Fig 2B).
Fig. 2. AerR and B12 activate photosystem synthesis.
A. Spectral scans of crude cell extracts from anaerobically grown wild-type parent strain (SB1003) (black), the aerR deletion strain (ΔaerR) (green) and the aerR deletion strain complemented with a plasmid encoded copy of aerR (red).
B. Quantitative real-time QRT-PCR showing the relative expression level of bchC in thethe aerR deletion strain (ΔaerR), and ΔaerR complementation strain (ΔaerR+aerR) as compared to the wild-type SB1003 strain (defined as a transcript level of 1) under photosynthetic growth condition. Error bars represent standard error of the mean (n = 3).
aerR is located immediately upstream of crtJ in R. capsulatus as well as in nearly all sequenced species of purple photosynthetic bacteria that contain CrtJ (Fig. 1). To address whether removal of aerR could cause a polar effect on downstream crtJ expression, a FLAG-epitope-tagged chromosomal version of crtJ was introduced in a ΔaerR strain as described previously (Dong et al., 2002). ΔaerR/Flag-crtJ and SB1003/Flag-crtJ were grown to 100 Klett units under anaerobic photosynthetic conditions and cell extracts were subjected to Western blot analysis with monoclonal antibody to the FLAG epitope tag in CrtJ. Levels of CrtJ protein in the ΔaerR and wild-type parental strains are comparable indicating that deletion of aerR did not affect the cellular level of CrtJ (Fig. S1). Finally, we also complemented the ΔaerR strain by introducing a plasmid-born aerR gene in trans. As shown in Fig. 2A, the complemented strain exhibits nearly normal amount of photosystem synthesis. QRT-PCR results also indicate that the addition of AerR in trans also restored expression of bchC to normal levels (Fig. 2B). We conclude, therefore, that the reduction of photopigment synthesis observed by the ΔaerR strain is caused by the absence of AerR and not due to a polar effect on the downstream expression of the crtJ gene.
AerR is a B12 binding protein
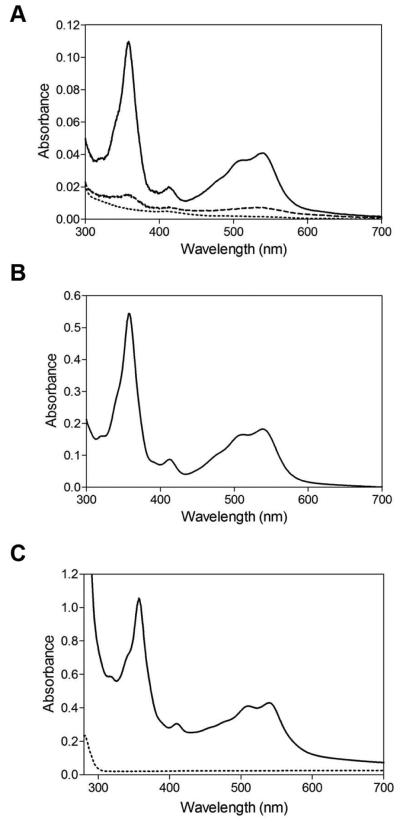
Analysis of the primary amino acid sequence of AerR shows the presence of a putative cobalamin binding site Asp/Glu-X-His-X-X-Gly-(41)-Ser(Thr)-X-Leu-(26-28)-Gly-Gly (Fig. S2A) (Ludwig and Matthews, 1997). To test whether AerR is capable of binding B12, various forms of B12 were added to purified AerR and after 5 min incubation, unbound B12 was then removed by passing the mixture through a desalting column. Spectral analysis of the eluted protein (Fig. 3A) shows that purified AerR exhibits no observable interaction with AdoB12 or with MeB12. Interestingly, exposure of the AerR and AdoB12 mixture to white light, which promotes upper ligand photolysis to generate OHB12, leads to binding of B12 (Fig. 3A). This is confirmed by the direct addition of OHB12, to purified apo-AerR which subsequently binds stoichiometric amounts of OHB12 even under dark conditions (Fig. 3B). Finally, tight binding of OHB12 to AerR can also be observed by the addition of 10 μM AdoB12 to the cell lysate of an E.coli strain that overexpressed AerR followed by exposure to white light. Spectral analysis of AerR isolated from the light excited lysate exhibits an absorption spectrum that reflects excellent binding of light-generated OHB12 (Fig. 3C, solid line).
Fig. 3. AerR binding to OHB12.
A. Light converts AdoB12 to OHB12 and enables AerR association. Dash line is a spectral scan of AerR incubated with AdoB12 in dark for 5 min followed by removal of unbound AdoB12; Dotted line is a spectral scan of AerR incubated with MeB12 in dark for 5 min followed by removal of unbound MeB12; Solid line is a spectral scan of AerR incubated with AdoB12 under illumination (~1000 μmol·m−2·sec−1 of white light) for 5 min followed by removal of unbound AdoB12.
B. Spectral scan of AerR incubated with OHB12 in the dark for 5 min followed by removal of unbound OHB12.
C. Spectral scan of WT AerR and AerR H145A mutant protein purified from E.coli with AdoB12 added to the growth medium and lysis buffer with the lysate exposed to ~1000 μmol·m−2·sec−1 of white light for 5 min prior to protein purification. Solid line is WT AerR; dotted line is AerR H145A.
Proteins used for spectral analysis were SUMO tagged.
In well-studied B12-dependent enzymes such as methionine synthase and methylmalonyl-coenzyme A mutase, a conserved histidine in the B12 binding domain displaces the lower dimethylbenzimidazole ligand to form a histidine lower axial ligand with the cobalt atom of B12 (Drennan et al., 1994). An alignment of the B12 binding domain from methionine synthase to four AerR homologs shows the presence of a conserved histidine (His145) in AerR that aligns with the lower axial cobalt linkage histidine of methionine synthase (Fig S2). To test if His145 is involved in binding of OHB12, we constructed an alanine substitution at this position and assayed whether this mutant protein could bind B12 as discussed above. Spectral analysis shows that purified AerR H145A protein indeed does not bind any detectable amounts of B12 (Fig. 3C, dotted line).
We further characterized B12 binding to AerR by performing liquid chromatography electrospray ionization mass spectrometry (LC-ESI-MS) on the AerR:B12 complex. ESI-MS has been widely used to study protein-ligand complexes (Loo, 1997) with an example of methionine synthase-B12 complex successfully detected by ESI-MS (Drummond et al., 1993). The deconvoluted mass spectrum generated by MassLynx software of purified AerR:B12 complex treated with 6 M urea gives two distinct species (Fig. 4A) with masses of 26892 ± 2 Da and 28221 ± 2 Da. The mass of the smaller species matches very well with the expected 26893.2 Da mass of apo-AerR as calculated by its protein sequence plus a sodium atom. The mass difference between the two species is 1329 ± 1 Da, which is consistent with the mass of the B12 prosthetic group (1329.4 Da). When 0.04 M HCl was added to the purified AerR:B12 complex, the LC-ESI-MS spectrum still showed the same two species (Fig. 4B). The AerR:B12 bound form at mass 28221 Da is still present, though less intense following the HCl treatment than in the urea treated sample. This indicates that B12 remains attached to a portion of the protein even under strong acidic conditions and suggests that the interaction between B12 and AerR is stronger than the expected noncovalent axial interaction between cobalt and H145.
Fig. 4.
Mass spectrometric analyses of AerR and AerR mutant proteins. The deconvoluted spectrum of full length protein LC-ESI-MS spectrum of (A) 6 M urea treated WT AerR protein; (B) 0.04M HCl treated WT AerR protein; (C) 6 M urea treated AerR H10A protein; (D) 0.04 M HCl treated AerR H10A protein. (E) ~ (G) are LC-ESI-MS/MS analyses of a B12 containing peptide (residues 6~16: VELEHGSTGCL) obtained by pepsin digestion. (E) Extracted ion chromatogram (EIC) of the triply-charged peptide containing B12 at m/z 824.7 (* indicates the peak of interest); the corresponding UV-Vis spectrum of this peptide is shown in the inset. (F) High-resolution mass spectrum of this B12 containing peptide ion at m/z 824.7 (2 ppm mass error). (G) Collision induced dissociation (CID) spectrum of this peptide ion at m/z 824.7. (E) and (G) were obtained using the Bruker HCT ion trap MS instrument whereas (F) was obtained using the Thermo LTQ-Orbitrap MS instrument. (H) Structure and CID fragmentation of the pepsin-generated WT AerR peptide containing B12 attached to His10. The experimental data for this peptide are shown in Fig. 4E-G.
To address which amino acid residue in AerR is forming an upper ligand with B12, we utilized liquid chromatography electrospray tandem ionization mass spectrometric analysis (LC-ESI-MS/MS). We first digested purified AerR:B12 complex with pepsin in the presence of 0.04 M HCl overnight at 37°C and then analyzed the resulting peptides with LC-ESI-MS/MS. We did not find any peptides containing H145 that also had an attached B12 but did observe a peptide corresponding to residues 6-16 (VELEHGSTGCL) of AerR that exhibited a UV-Vis spectrum consistent with an attached B12 (Fig. 4E inset). The CID spectrum of this peptide yielded fragment ions (Fig. 4G) that are consistent with a B12 containing peptide and with the fragment ion assignments in Fig. 4H. These results suggest that an amino acid residue in this segment of AerR (residues 6-16) forms the tight upper axial cobalt ligand with B12 (Fig. 4E~G). Since there are two amino acid residues in this peptide that could potentially form an upper axial ligand to cobalt (His10 and Cys15), we mutated each of these two residues to alanine either individually or in combination and then analyzed these mutants for peptides containing B12 using LC-ESI-MS/MS. The C15A mutant protein still yielded a B12-containing peptide as shown in Fig. S3 while the H10A mutant and the H10A-C15A double mutant proteins did not yield any B12 containing peptides. These results suggest that His10 is the upper axial cobalt-coordinating residue present above the corrin plane and that it forms a stronger bond to cobalt than does the proposed lower axial His145 bond.
We also used LC-ESI-MS analysis to confirm that the proposed lower linkage between His145 and cobalt is weaker than the proposed upper linkage to His10. For this analysis we observed that LC-ESI-MS analysis of full length AerR H10A:B12 gave rise to two species after treatment with 6 M urea: a higher mass (28154 ± 2 Da) that is consistent with that of AerR H10A containing an attached B12 plus a sodium atom (28156.6 Da) and a lower mass (26826 ± 2 Da) that is of AerR H10A without bound B12 plus a sodium atom (26827.2 Da) (Fig. 4C). However treatment of AerR H10A:B12 protein with 0.04 M HCl, which would protonate the ε-nitrogen of His145 and result in it losing its coordination to cobalt, gave rise only to apo-H10A without bound B12 (26826 ± 2 Da) (Fig. 4D). This is distinctly different from what was observed with wild-type AerR:B12 that has both proposed upper and lower cobalt coordinating His145 and His10 and is still capable of retaining bound B12 even under acidic conditions (Fig. 4B). These results confirm that both His10 and His145 are coordinating to cobalt and that the proposed lower axial bond between cobalt and His145 is weaker than that of the proposed upper axial bond between cobalt and His10.
AerR is an antirepressor of the tetrapyrrole regulator CrtJ
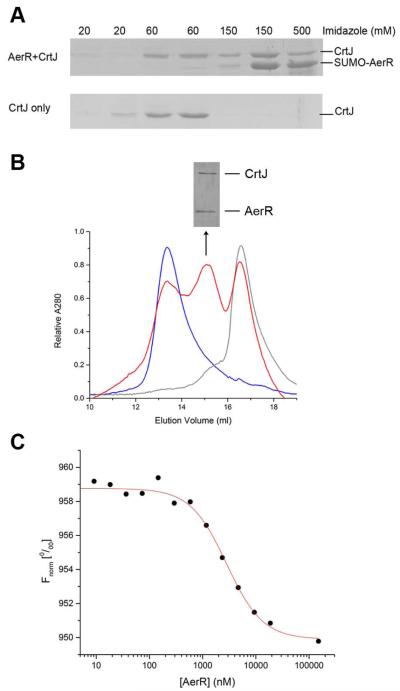
We undertook several independent studies to ascertain whether AerR can interact with CrtJ. In one assay, we performed in vitro pull down experiments using resin bound SUMO-AerR containing B12 as “bait” for the binding of tag-less purified CrtJ. SUMO-AerR:B12 was loaded on the Ni2+ column followed by extensive washing to remove unbound SUMO-AerR:B12. Purified tag-less CrtJ was then loaded onto the column followed by extensive washing and then SUMO-AerR:B12 was eluted with a gradient of imidazole. Fig. 5A showed that CrtJ co-eluted with SUMO-AerR at an imidazole concentration of 150 mM. As a control, we observed that addition of just tag-less CrtJ to the Ni2+ column resulted in CrtJ eluting at a lower imidazole concentration of 60 mM.
Fig. 5. CrtJ and AerR:B12 form a complex in vitro.
A. CrtJ-AerR pull down experiments. Top panel is SUMO-AerR:B12 used as “bait” to pull down tag-less CrtJ. Bottom panel is tag-less CrtJ used as a control without SUMO-AerR:B12. The imidazole concentration of buffer used in each step was indicated above the picture.
B. Absorbance traces of CrtJ (blue line), AerR:B12 (grey line) and mixture of CrtJ and AerR:B12 (red line), respectively, that were subjected to Superdex 200 chromatography. SDS-PAGE analysis of the middle red peak at volume 15 is shown above the Superdex 200 chromatography absorbance trace.
C. Representative CrtJ-AerR:B12 binding isotherm generated by microscale thermophoresis assay. Continuous line through binding isotherm represents a non-linear regression analysis fitted to a Hill equation (Equation 1).
We also performed Superdex 200 size exclusion chromatography to determine whether AerR and CrtJ form a stable complex that co-elutes. The elution profile of CrtJ alone showed that it chromatographs as a single peak with an apparent molecular weight (MW) of 127.3 ± 4.0 kDa which is close to the size of a CrtJ dimer that has a calculated MW of 101.0 kDa (Fig. 5B blue line). The elution profile of AerR:B12 alone also shows a single major peak with a calculated MW of 27.3 ± 4.0 kDa which is close to the size of an AerR monomer containing B12 that has a calculated MW of 28.2 kDa (Fig. 5B grey line). After incubating CrtJ with AerR:B12 (~20 μM, respectively) at 22 °C for 1 hour, the protein mixture was subjected to chromatography with a resulting elution profile containing three peaks (Fig. 5B red line). One peak corresponds to the same size of a CrtJ dimer, a second peak corresponds to the AerR monomer with a third unique peak present with a calculated MW of 73.0 ± 4.3kDa. This size corresponds well to the calculated MW of 78.7 kDa for a 1:1 CrtJ-AerR:B12 complex. SDS-PAGE analysis of the peak corresponding to the AerR-CrtJ complex fraction is shown above the absorbance trace. The CrtJ and AerR protein bands have similar intensity further indicating that CrtJ binds to AerR:B12 with a 1:1 stoichiometry.
We also measured the binding affinity for the interaction of CrtJ and AerR:B12, by performing microscale thermophoresis analysis (Monolith NT.115, Nanotemper Technologies GmbH). Microscale thermophoresis measures a change in a protein hydration shell caused by a change in protein conformation or by the formation of a stable interaction between two proteins (Wienken et al., 2010). The binding affinity as measured using replicates of this procedure was calculated to be EC50 = 3.3 ± 0.7 μM (Fig. 5C).
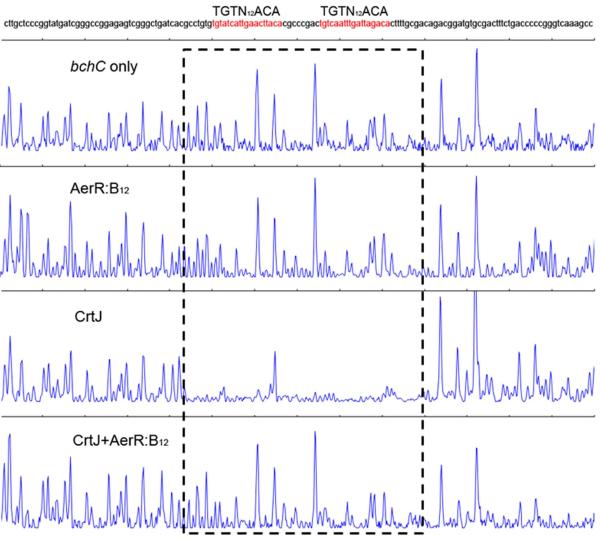
The effect of AerR:B12 binding to CrtJ on the ability of CrtJ to bind operator DNA was addressed by undertaking DNase I DNA footprint assay of CrtJ binding to a pair of CrtJ recognition sequences present in the bchC promoter (Fig. 6). Footprint analysis of CrtJ alone shows excellent protection of the tandem CrtJ binding sites (TGTN12ACA) that overlaps the -35 and -10 promoter recognition sequences. However, when CrtJ was pre-incubated with AerR:B12 at 22°C for one hour and then used for footprint analysis, these two protection sites disappeared. As a control, AerR:B12 alone does not show any protection on the bchC promoter. These results indicate that AerR containing bound B12 effectively inhibits the ability of CrtJ to bind to its target sequence.
Fig. 6. AerR:B12 inhibits CrtJ binding to the bchC promoter.
DNase I footprint analysis of CrtJ binding to the bchC promoter region in the absence of AerR:B12 (third panel), in the presence of AerR:B12 (fourth panel). The first panel is the DNase I digestion pattern of the bchC promoter without adding any protein and the second panel is AerR:B12 alone with the bchC promoter, which used as a control. CrtJ binding sites are indicated with dashed boxes.
AerR senses B12 in vivo
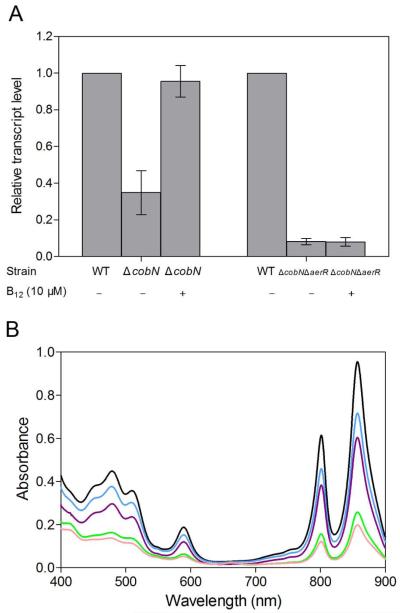
We addressed whether the presence of cobalamin has an effect on in vivo photosystem gene expression by constructing a ΔcobN B12 auxotroph strain that contains a kanamycin resistance gene cassette inserted into the cobN gene that encodes the N subunit of cobalt chelatase. Cells were grown under photosynthetic conditions in the presence of 10 μM exogenous B12 or under B12 limiting condition. Expression of the bacteriochlorophyll biosynthesis bchC gene was then assayed using QRT-PCR as bchC is known to be repressed by CrtJ (Ponnampalam et al., 1995, Ponnampalam et al., 1998). The QRT-PCR results shown in Fig. 7A demonstrate that bchC gene expression is lowered (~3 fold) when B12 is limiting and that this effect is reversed by the addition of 10 μM exogenous B12. Similar studies were carried out in a ΔcobNΔaerR double deletion strain. In this strain bchC expression remains low compared to WT strain (~13 fold lower) with or without exogenous B12 in the growth medium. This indicates that the ΔcobNΔaerR strain can no longer control bchC gene expression in vivo in response to B12 availability.
Fig. 7. AerR senses B12 in vivo.
A . QRT-PCR showing the relative expression level of bchC in the cobN disruption strain (ΔcobN) and the cobN aerR double deletion strain (ΔcobNΔaerR) under photosynthetic condition in the presence of exogenous 10 μM B12 or under B12 limited conditions, as compared with wild-type SB1003 strain (defined as a transcript level of 1).
B. Spectral scans of crude cell extracts from anaerobically grown wild-type parent strain (SB1003) (black), the aerR deletion strain (ΔaerR) (green), the aerR H10A mutant strain (blue), the aerR H145A mutant strain (purple) and the aerR H10A H145A double mutant strain (pink).
To test the effect of disrupting the putative upper and lower cobalt binding ligands on AerR’s activity in vivo we recombined aerR-H10A, aerR-H145A and aerR H10A H145A mutation into the cognate position in the chromosome and assayed for photosystem synthesis. As shown by the spectral analysis in Fig. 7B, the aerR H10A (blue spectrum) and H145A strain (purple spectrum) exhibits photosystem production that is intermediate from that of the AerR deletion strain (green spectrum) and the wild-type parent strain (black spectrum). Interestingly, photosystem production of a strain that contains a H10A H145A double mutation (pink spectrum) is quite similar to the aerR deletion strain (green spectrum), indicating that both of the two His residues play a role in the control of AerR’s function.
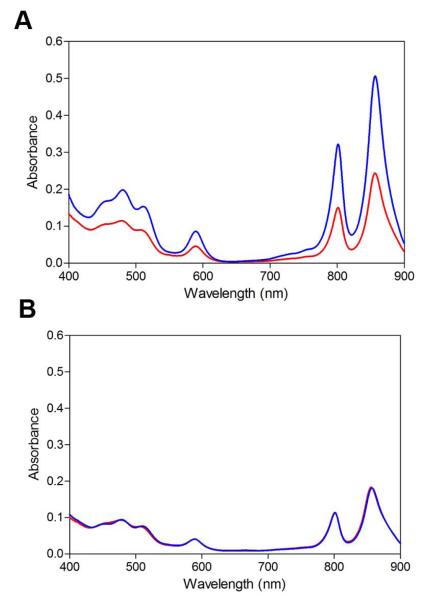
Finally, we tested if there is an effect of disrupting the AerR lower ligand to OHB12 on the light sensing capabilities of these cells. As shown in Fig. 8A, growth of wild-type R. capsulatus under high light intensity (~1000 μmol·m-2·sec-1) leads to an ~50% reduction in pigment synthesis relative to the amount of pigments synthesized when growth under low light (~5 μmol·m−2·sec−1) conditions. In contrast, growth of H145A mutant cells under similar high and low light conditions (Fig. 8B) results in cells that contain the same intermediate amounts of pigment production at levels that are similar to that observed in wild-type cells under high light growth conditions. The implications of this phenotype are discussed below.
Fig. 8. Spectral scans of crude cell extracts from photosynthetically grown cells under different light intensities.
A. Wild-type strain SB1003 grown under high light (red line) with ~1000 μmol·m−2·sec−1 of white light and low light (blue line) with ~5 μmol·m-2·sec-1 of white light.
B. aerR H145A mutant strain under high (red line) and low (blue line) light intensity.
Discussion
The involvement of B12 in the control of photosystem synthesis has been a long observed feature of photosynthetic microorganisms. Nearly 5 decades ago, Pfennig and Lippert reported the isolation of several strains of purple bacteria that required B12 for growth (Pfennig and Lippert, 1966). These strains exhibited severely reduced photosystem synthesis under B12 limiting conditions. Decades later this observation was followed by studies by Pollich and Klug which demonstrated that photosystem gene expression is six- to seven-fold lower in a R. capsulatus B12 auxotroph than in the wild-type strain (Pollich et al., 1993, Pollich and Klug, 1995). A molecular mechanism for B12 dependence of photosystem gene expression has, however, remained unclear.
A major finding of our study is that AerR functions as a B12-sensing antirepressor of CrtJ. The ability of AerR to bind B12, and subsequently interact with CrtJ, is clearly shown by pull down experiments using AerR:B12 as “bait”, by coelution of AerR:B12 with CrtJ during size exclusion chromatography, and finally by the determination of the AerR binding affinity to CrtJ (Fig. 5). DNase I DNA footprint assays also demonstrate that AerR:B12 inhibits the binding of CrtJ to the bchC promoter which clearly shows that AerR is functioning as an antirepressor of CrtJ (Fig. 6). The in vitro B12 binding and antirepressor activity of AerR is also supported by in vivo assays which show that a ΔcobN B12 auxotroph has three-fold reduction in bchC expression under B12 limiting conditions with resumption of normal bchC expression occurring after addition of 10 μM B12 only when there is functional AerR (Fig. 7A). These results suggest that AerR indeed controls the DNA binding activity of CrtJ in response to B12 in vivo, which provides the first molecular understanding as to why B12 is required for optimal photosystem gene expression in this species. Furthermore, all sequenced strains of purple photosynthetic bacteria that contain a crtJ homolog also contain a homolog of aerR (Fig. 1). This indicates that B12 control of photosystem gene expression via AerR is most likely a conserved feature of this group of organisms.
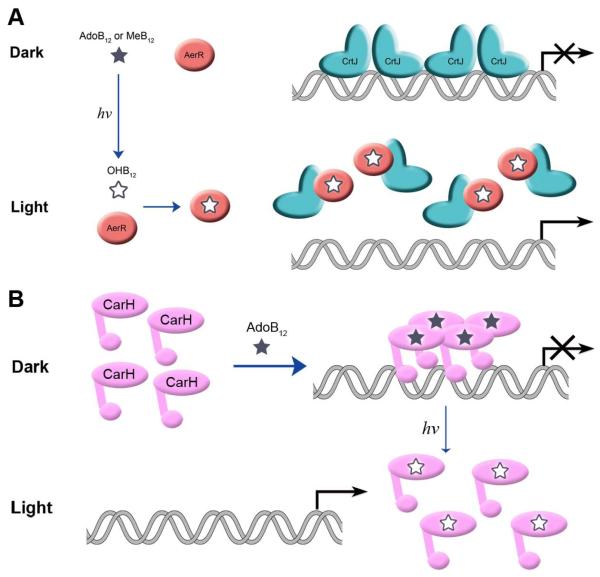
Until recently, B12 was considered to be a simple metabolite used as a cofactor for enzyme reactions. However, B12 is also known to be involved in controlling expression of genes coding for enzymes involved in B12 biosynthesis by functioning as a metabolite that is sensed by RNA-based riboswitches that are present upstream of a number of cob genes (Nahvi et al., 2002, Rodionov et al., 2003). Most recently it has been observed that M. xanthus also contains a DNA binding transcription factor called CarH that utilizes AdoB12 as a cofactor to regulate carotenoid gene expression. CarH containing bound AdoB12 forms a stable tetramer that represses carotenoid gene expression in the dark. Exposure of M. xanthus to light promotes photolysis of AdoB12 to OHB12 which causes disassembly of the CarH tetramer and subsequent dissociation of CarH from the carotenoid promoters (Ortiz-Guerrero et al., 2011). This light mediated photolysis of the AdoB12::CarH tetramer to the OHB12::CarH monomer subsequently allows M. xanthus to synthesize carotenoids in the light (Fig. 9B). In the case of R. capsulatus, AerR also appears to utilize B12 as a light sensor at some level as it only binds OHB12 that would presumably be elevated under light excitation growth conditions (Fig. 9A). Interestingly the H145A mutant does seem to lack the ability to regulate pigment production in response to alterations in light intensity (Fig. 8). However, this mutant only synthesizes intermediate amount of pigments which is not dim light pigment levels that one would expect if it were only defective in sensing OHB12 production (Fig. 8). Clearly additional studies will have to be undertaken to determine what role, if any, that wild-type AerR has on adjusting the amount of pigment production in response to alterations in light intensity and whether such a response is due to light driven changes in OHB12 production.
Fig. 9. Models for the light-responsive B12-based regulatory switches in R. capsulatus and M. xanthus.
A. Light converts AdoB12 to OHB12 which enables OHB12 binding to AerR and thus binding to CrtJ and derepresses photosynthesis gene expression in R. capsulatus.
B. Light mediated photolysis of the AdoB12::CarH tetramer to the OHB12::CarH monomer allows M. xanthus to synthesize carotenoids in the light (Ortiz-Guerrero et al., 2011).
CrtJ is also known to contain a redox regulated cysteine in the helix-turn-helix DNA binding domain that when oxidized promotes an increase in DNA binding activity. Under anaerobic growth conditions this Cys is present in a reduced thiol form that reduces CrtJ DNA binding affinity by ~20-fold (Cheng et al., 2012). Mutagenesis of the redox active Cys to Ala leads to constitutive derepression while mutating the redox active Cys to Ser leads to constitutive repression (Cheng et al., 2012). The phenotype of the aerR deletion strain is somewhat surprising in that it has severely reduced bchC expression even in the absence of oxygen. This suggests that CrtJ is capable of repressing target gene expression under reducing conditions in strains where aerR is deleted. It has also been shown that the CrtJ homolog from R. sphaeroides, PpsR, binds heme and regulates expression of heme biosynthesis genes in response to heme availability (Yin et al., 2012). Preliminary studies with R. capsulatus CrtJ also indicates that it is also capable of binding heme in a manner similar to that described for PpsR indicating that heme is a likely input that controls CrtJ’s activity as is described for PpsR (Z. Cheng and C. E. Bauer, unpublished observation). Thus, the regulation of pigment production by CrtJ is highly complex involving an interplay of numerous inputs; redox, potentially heme, and now B12 via AerR. Furthermore, two of these inputs, heme and B12 availability involve end products of a biosynthetic pathway that is regulated by CrtJ. Determining the mechanism(s) whereby CrtJ responds to these very different inputs remains a distinct challenge going forward.
The similar phenotypes of the aerR H145A and H10A mutant strains is also surprising as gene expression and photosystem synthesis of these two mutant are intermediate between that of wild-type cells and that of the aerR deletion strain (Fig. 7B). The phenotype of the H145A point mutation is also not as severe as observed with the B12 auxotroph suggesting that even though this point mutation is not capable of loading B12 in vitro, it is not a null mutation in vivo. Either this mutation is still capable of binding some form of B12 in vivo or it suggests that apo-AerR may also have a role in controlling gene expression. Unfortunately, apo-AerR is very unstable in vitro, which has hampered our ability to purify enough protein without B12 to undertake biochemical analysis of its function, if any, in affecting the DNA binding activity of CrtJ.
Sequence alignment of AerR homologs from numerous purple bacteria shows that there is significant homology with the B12 binding domain from E. coli methionine synthase as well as to the presumptive B12 binding region of CarH from M. xanthus (Fig. S2A). In free B12, the 5,6-dimethylbenzimidazole base forms an axial ligand to cobalt at the lower side of the corrin macrocycle. However when bound to methionine synthase, the 5,6-dimethylbenzimidazole unbinds and swings away to allow histidine to form a lower axial ligand to cobalt. This swapping of the lower ligand is called a base-off His-on conformation (Drennan et al., 1994). All characterized B12-dependent methyltransferases have the conserved B12 binding sequence Asp/Glu-X-His-X-X-Gly-(41)-Ser(Thr)-X-Leu-(26-28)-Gly-Gly which is also present in AerR (Fig. S2A) (Ludwig and Matthews, 1997). This suggests that B12 bound to AerR also has the dimethylbenzimidazole base-off His-on conformation and that His145 may be forming a lower axial ligand with bound cobalt. This lower ligand is also supported by modeling an AerR structure based on the methyltransferases structure that shows the proper positioning of His145 (Fig. S2B), and by our mutational studies which show that a His145 to Ala mutation is incapable of binding B12 in vitro.
In methyltransferases the upper cobalt ligand is a methyl group that functions as a leaving group for methylation of substrates (Gruber et al., 2011). AerR does not bind MeB12 and instead binds OHB12 that has a bound water as an upper ligand. Mass spectrometric (MS) analysis shows that AerR tightly binds OHB12 presumably by displacement of the upper OH-cobalt linkage present in OHB12 with His10. This upper axial ligand is extremely tight and indeed remains bound during MS analyses even under conditions where the protein is denatured and digested by proteases. Mammalian B12 transporter transcobalamin is also an example of a B12 binding protein that uses a histidine to form an upper ligand to corrin bound cobalt (Wuerges et al., 2006). The binding of the OHB12 to transporter transcobalamin also includes the displacement of water with a histidine residue (Fedosov et al., 2000). However, this B12 transporter differs in that it is highly selective for the base-on form of B12 which presumably facilitates its release upon transport. The overall structure of transporter transcobalamin, and the orientation of the bound B12 is also quite different from other B12 binding enzymes that have the classic B12 binding sequence as observed with AerR, CarH and methyltransferases (Gruber et al., 2011). AerR from R. capsulatus is neither an enzyme nor a B12 transporter so it is interesting that it combines the ligand binding features of these two different B12 binding proteins.
AerR and its homologs from other species also have considerable homology to the SCHIC domain of a well-characterized antirepressor from Rhodobacter sphaeroides called AppA (Gomelsky et al., 2003, Moskvin et al., 2007). Along the same lines as AerR, AppA is known to inhibit the DNA binding activity of PpsR, the CrtJ homolog in R. sphaeroides (Masuda and Bauer, 2002, Winkler et al., 2013). Interestingly, the AppA SCHIC domain preferentially binds heme instead of B12 as occurs with AerR (Han et al., 2007, Moskvin et al., 2007, Yin et al., 2013). The recent crystal structure of the AppA SCHIC domain, however, highlights several important differences between the putative heme binding pocket in the SCHIC domain and the B12 binding pocket present in the methionine synthase structure (Drennan et al., 1994, Winkler et al., 2013, Yin et al., 2013). Specifically, the His that forms a lower axial ligand to heme in AppA is offset two residues in AppA relative to the location of the lower ligand His present in methyltransfeerase and for AerR (Yin et al., 2013). In addition, methionine synthase has a space in the core of the MetH:B12 Rossmann fold that anchors the bulky lower nucleotide tail that attached to B12 (Drennan et al., 1994). In the SCHIC domain, the access to this docking space is blocked by Glu289 (Yin et al., 2013). In contrast to the heme binding SCHIC domain, AerR and its homologs have retained extensive homology to region of MetH that allows insertion of the lower nucleotide tail of B12 into the Rossmann fold. Presumably, this difference is promoting tetrapyrrole specificity differences between AerR and AppA.
Given that AerR is a B12 binding protein, it is also surprising that a previous study on an AerR homolog from R. sphaeroides called PpaA was shown to bind heme in vitro (Moskvin et al., 2010). We have repeatedly attempted to observe heme binding by apo-AerR without success, which is contrasted by the ease at which AerR is capable of binding OHB12. A major difference between our studies and that of Moskvin et al is that we have analyzed the tetrapyrrole binding activity of full length tag-less AerR while the Gomelsky study utilized a truncated variant of PpaA that only contained homology to the SCHIC domain of AppA. The analyzed truncated variant of PpaA was further fused to a maltose binding domain. Ongoing analysis of tetrapyrrole binding by PpaA and other AerR homologs by our laboratory indicates that tetrapyrrole specificity of this class of B12 binding proteins involves residues outside of the SCHIC region of homology. This region was missing in the prior study with PpaA (Vermeulen and Bauer, unpublished).
In conclusion, this study, in conjunction with the recent CarH study from M. xanthus, indicates that B12 has an important role in controlling gene expression in several species by modulating the DNA binding activity of transcription factors. This occurs either directly as is the case of CarH or indirectly with an antirepressor as is the case of AerR and CrtJ (Fig. 9). Proteins that contain B12 binding domain similar to AerR or CarH are present in a number of sequenced bacterial genomes (Ortiz-Guerrero et al., 2011), so it is quite possible that this newly discovered form of regulating gene expression may not be an uncommon mechanism.
Experimental procedures
Strains, media and growth conditions
In vivo studies were undertaken with R. capsulatus SB1003. E. coli strains DH5α and DH10β were used for cloning. E. coli TEC5 (Taylor et al., 1983) was used in conjugation to introduce various plasmids to R. capsulatus. E. coli S17-1λpir (Miller and Mekalanos, 1988) was used to facilitate the transfer of suicide plasmids into R. capsulatus. AerR overexpression was carried out in E. coli BL21 (DE3). Various R. capsulatus strains were grown in PY salts liquid medium as described previously (Cheng et al., 2012).
Construction of aerR mutations
An aerR in-frame deletion was constructed by amplifying ~500 base pairs upstream of aerR and ~500 downstream of aerR with PCR, using the primers shown in Table S1 (aerRupSacI-F and aerRupEcoRV-R; aerRdownEcoRV-F and aerRdownXbaI-R). The upstream ~500bp fragment was cloned into SacI and EcoRV sites while the downstream ~500bp fragment was cloned into EcoRV and XbaI sites of pBlueskript SK+ (Stratagene). This generated a ~1kb fragment that contains the upstream and downstream sequences of aerR with the only remaining aerR coding sequence ATGGATATCTGA (underlined sequence introduced an EcoRV restriction site). The SacI-XbaI fragment was subsequently subcloned into corresponding sites in suicide plasmid pZJD29a (Masuda and Bauer, 2004), generating pZJD29a-ΔaerR, and then conjugated into R. capsulatus SB1003. pZJD29a-ΔaerR genome recombinants were selected for the presence of gentamycin resistance and sucrose sensitivity. Cells carrying the in-frame deletion of aerR gene were further verified by colony-PCR. The strain carrying aerR in-frame deletion was designated as ΔaerR.
A chromosomally located aerR H145A point mutation was generated by constructing a ~1.6kb fragment covering the aerR gene and both the upstream and downstream ~500bp region by PCR amplification with primers aerRupSacI-F and aerRdownXbaI-R (Table S1). The PCR product was digested with SacI and XbaI and inserted into pBlueskript SK+ (Stratagene). This plasmid was then used as a template for generating a point mutation via PCR mutagenesis as using a Stratagene QuikChange kit with Turbo Pfu DNA polymerase. A pair of primers aerRH145A-f and aerRH145A-r (Table S1) was subsequently used to introduce the point mutation. The mutation was confirmed by sequencing before subcloning into pZJD29a. pZJD29a-aerRH145A was transformed into S17-1 λpir and subsequently conjugated into the ΔaerR strain. Procedures to select for first and second recombination events were the same as used in generating the ΔaerR strain. The presence of the mutated version of aerR gene in the genome was confirmed by PCR amplification of the aerR region followed by sequence analysis of the PCR segment. aerR H10A point mutant and aerR H10A H145A double mutant strain were generated using the same procedure as used for aerR H145A mutant strain (primers used are shown in Table S1).
AerR overexpression and purification
A 750bp fragment of aerR gene was PCR amplified using primers aerRSUMOBbsI-f and aerRSUMOBamHI-r (Table S1). The PCR product was cloned into pCR4Blunt-TOPO plasmid as described using an Invitrogen Zero Blunt TOPO PCR Cloning Kit. After the cloned fragment was checked by sequencing analysis it was then digested with restriction enzymes BbsI and BamHI followed by sub-cloning into a SUMOpro vector plasmid (Lifesensors Inc.). The resulting ORF encodes AerR with an N-terminal His6-tag and a SUMO-tag, which enhances solubility of AerR. The resulting plasmid pSUMO-AerR was then transformed into BL21 (DE3) for protein overexpression.
To overexpress SUMO tagged AerR, E. coli strain BL21 (DE3)/pSUMO-AerR was grown in LB medium at 37°C in the presence of 25 μg ml−1 kanamycin to an OD600 of 0.7~0.8. Cultures were cooled to 16°C and then 50 μM of isopropyl-β-D-thiogalactopyranoside (IPTG) and 1 μM AdoB12 were added with AerR expression continued at 16 °C for 16~20 hours. Cells were harvested by centrifugation and resuspended in a buffer containing 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 5 mM imidazole, 10% glycerol and 10 μM AdoB12. Cells were then lysed by passing the cell suspension through a chilled French Press cell three times at 18,000 psi. The lysate was then clarified by centrifugation at 30,000 xg for 30 min at 4 °C with the resulting supernatant illuminated with white light at intensity of ~1000 μmol·m−2·sec−1 for 5 min. The supernatant was then passed through a 45 μM membrane filter (Millipore) and loaded onto a 1 ml HisTrap column (GE) using an ÄKTA chromatography system (GE), washed with 20-column volume of wash buffer containing 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 20 mM imidazole, and 10% glycerol. SUMO-AerR was then eluted with a gradient of 20 mM imidazole to 500 mM imidazole in the loading buffer over 15 column volumes total. Eluted protein was then digested with SUMO protease Ulp1 in presence of 1 mM DTT at 22 °C for 2 hours, followed by a desalting column (Bio-Red, Econo-Pac® 10DG Column) against wash buffer. SUMO tag was then removed from the protein solution by passing through 1 ml Ni Sepharose high performance resin (GE Healthcare) in an Econo-Pac chromatography column (Bio-Rad) and wash buffer was used to wash the residual protein on the resin. Tag-less AerR was further purified by Superose 12 (S-12) a 16 × 700 mm size exclusion chromatography in 20 mM Tris-HCl (pH 8.0) and 200 mM NaCl at a flow rate of 1 ml min−1. Peak fractions were analyzed by SDS-PAGE with AerR containing fractions collected and pooled. AerR stayed as a monomer in solution, as calculated from its size exclusion chromatography profile.
pSUMO-AerR plasmid was used as a template to generate aerR point mutations (primers used are shown in Table S1) and mutant proteins were purified using the same procedure as that for WT AerR.
Complementation analysis
A 1.2 kb fragment containing the aerR gene as well as 600bp upstream was PCR amplified with primers aerRupSacI-F and aerRSUMOBamHI-r (Table S1). This fragment was cloned into pCR4Blunt-TOPO plasmid (Invitrogen) and then cleaved with HindIII and EcoRV restriction sites located on pCR4Blunt-TOPO plasmid. The HindIII-EcoRV fragment was then inserted into pBBR-MCS2 that was pre-digested with the same enzymes. Plasmid pBBR-MCS2-aerR was then transformed into S17-1 λpir and then conjugated into the ΔaerR strain.
Spectral analysis
R. capsulatus parent strain SB1003 and the mutant strains ΔaerR and aerRH145A were grown photosynthetically either under low light intensity (~5 μmol·m−2·sec−1), normal light intensity (~30 μmol·m−2·sec−1), or under high light intensity (~1000 μmol·m−2·sec−1) to a density of 75 klett units in PY salts medium. 10 ml of cultures were chilled on ice and then harvested by centrifugation at 4,400 xg for 10 min at 4 °C. Cell pellets were resuspended in 1 ml of buffer containing 20 mM Tris-HCl pH 8.0, NaCl 150 mM and lysed by sonication 10 sec x 3. The cell lysate was clarified by centrifugation at 15,600 xg for 10 min. The protein concentration of each supernatant was determined using the Bradford assay (Advanced Protein Assay Reagent, Cytoskeleton, Inc.) with BSA as a standard. Each sample was measured three times and the average concentration was obtained. Absorption spectrum was scanned from 400 nm to 900 nm on a DU640 Beckman spectrophotometer with each supernatant adjusted by its individual protein concentration.
For analysis of B12 binding, purified WT and H145 AerR protein were scanned on a DU640 Beckman spectrophotometer from 300 nm to 700 nm. The AdoB12, MeB12 and OHB12 binding experiments were performed with SUMO tagged AerR as tag-less AerR is unstable without bound B12. Apo SUMO tagged protein was purified using the same procedure as used for SUMO-AerR:B12, except that no AdoB12 was added to the LB medium and lysis buffer and the supernatant was not illuminated. The absence of B12 in the purified SUMO-AerR was confirmed by spectral analysis. SUMO-AerR was incubated with different B12 in the buffer containing 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 20 mM imidazole, and 10% glycerol in the dark or with white light for 5 min (light intensity ~1000 μmol·m-2·sec−1) before passing through a 13 × 26 mm PD MidiTrap G-25 desalting chromatography column (GE). Eluted samples were scanned for spectral absorption.
Mass spectrometry
Purified wild-type and mutant AerR proteins containing bound B12 (~20 μM) were treated with either 6 M urea or 0.04 M HCl for 15 min and then used for liquid chromatography electrospray ionization mass spectrometry (LC-ESI-MS) analysis, using a Waters/Micromass LCT Classic time of flight (Waters Corp., Milford, MA) mass spectrometer with a Waters CapLC inlet. 5 μl of sample was loaded onto a 0.5×50 mm Thermo BioBasic C8 reverse-phase column (300 Å pore size , 5 μm particles size, Thermo Scientific, West Palm Beach, FL) in Solvent A (5% acetonitrile, 95% water, 0.1% formic acid) and analyzed in positive ion mode. The elution was carried out in a 20 min linear gradient from 10% solvent B to 90% Solvent B (95% acetonitrile, 5% water, 0.1% formic acid) at 15 uL min−1. UV absorption at 216 nm and mass spectra from m/z 600-2300 were collected and analyzed using MassLynx software.
Purified wild-type and mutant AerR proteins (~20 μM) were digested with either 20-fold (weight ratio) pepsin (Promega) or chymotrypsin (Roche Applied Science) overnight at 37 °C for pepsin or 22 °C for chymotrypsin. The resulting samples were then analyzed by LC-ESI-MS/MS analysis using the LTQ Orbitrap instrument. LC-ESI-MS/MS analysis was performed using an Eksigent nano LC system (AB SCIEX, Farmingham, MA) interfaced to an externally calibrated LTQ Orbitrap hybrid mass spectrometer equipped with a nanospray ion source (Thermo Scientific, San Jose, CA). a 4-μl aliquot of the peptide digest dissolved in 20 μl of mobile phase A was washed with mobile phase A for 5 min at a flow rate of 5 μl min−1 using an in-house packed reversed-phase C18 (Micrhom Magic C18, 200 Å pore size, 5 μm particle size, Bruker-Michrom, Fremont, CA) trap column made from a 15 mm × 100 μm Integrafrit column (New Objective, Woburn, MA). The desalted peptides were then separated using an in-house packed reversed-phase C18 (Magic C18, 100 Å, 5 μm particle size, Bruker-Michrom) column (150 mm × 75 μm, 15 μm tip) (New Objective, Woburn, MA) and analyzed in positive ion mode. Mobile phase A consisted of 0.1 % formic acid in acetonitrile:water (3%:97%, volume:volume) and mobile phase B consisted of 0.1 % formic acid in acetonitrile:water (97%:3%, volume:volume). The flow rate was 300 nl min−1 with the following gradient: 3% B from 0-1 min, ramp to 10% B from 1-9 min, ramp to 30% B from 9-28 min, ramp to 40% B from 28-31 min, ramp to 80% B from 31-35 min, hold at 90% B from 36-43 min, ramp to 3% B from 43-44 min, then re-equilibrate the column at 3% B for 14 min (58 min total run time). The mass spectrometer was operated in an automated data dependent mode alternating between an MS scan in the Orbitrap and five collision-induced dissociation (CID) scans in the linear ion trap. The Orbitrap monitored precursor ions from m/z 300 to m/z 2000 at 15000 resolving power. The five most intense precursor ions in each scan were sequentially isolated in the linear trap with a 2 m/z isolation width and were collisionally activated at 35% normalized collision energy. The cycle was continuously repeated throughout the entire separation with the dynamic exclusion set to 45 sec with a repeat count of 2. The Orbitrap provided high mass accuracy (< 5 ppm) for the precursor ions while the CID scan in the linear ion trap provided nominal mass information (± 0.25 m/z) for the tandem mass spectra. B12 containing peptides were found by searching the tandem mass spectra for B12 diagnostic fragment ions as shown in Fig. 4E~G and Fig S3. The presence of B12 was further confirmed by coincidence of the B12 UV absorption peak at 360 nm with the parent m/z values and diagnostic fragment ions of peptides containing B12 using liquid chromatography-UV/Vis spectroscopy-tandem mass spectrometry (LC-UV/Vis-MS/MS). The chromatographic separation was performed with an Agilent 1200 capillary liquid pump (Agilent Technologies, Santa Clara, CA) equipped with a micro well-plate autosampler and a diode array UV-VIS absorbance detector that had a 500 nl flow cell. The column was 0.3 mm i.d., 150 mm long and packed with 5 μm Zorbax SBC18-300 particles (Agilent). 8 μl of sample was injected with the peptides separated at 4 μl min−1 using the same gradient conditions as detailed above. The UV-VIS detector recorded absorption spectra from 250-750 nm at 2.5 Hz with individual chromatograms at 216 and 360 nm recorded. Tandem mass spectra of the peptides were recorded in manual MS2 mode with the instrument set to obtain CID spectra for specific m/z values corresponding to the B12 containing peptides previously observed using the Orbitrap. A Bruker HCT-Ultra PTM ion trap mass spectrometer (Bruker Daltonics, Billerica, MA) after the diode array detector used 1.0 V activation energy on the endcaps for fragmentation and recorded data from m/z 400-1800 in UltraScan mode (50 second delay measured between UV and MS).
RNA isolation, quantitative real-time PCR
R. capsulatus wild-type and mutant strains were grown as described above for spectral analysis. 1 ml cell cultures were harvested with total RNA of each sample extracted using the ISOLATE RNA Kit (Bioline). RNA samples were then treated with TURBO DNase (Ambion) with final OD260 to OD280 ratio of RNA samples (approximately 2.0) measured using NanoDrop spectrophotometer (Thermo Scientific). Quantitative real-time (QRT-PCR) analysis of gene expression utilized SensiFASTTM SYBR Hi-ROX One-Step Kit (Bioline) with 8 ng of RNA sample used in a 20 ul reaction. cDNA synthesis was at 45°C for 10 min followed by polymerase activation at 95°C for 2 min. A two-step PCR reaction was used, consisting of product melting at 95°C for 5 sec, followed by primer annealing and DNA polymerase elongation at 60°C for 20 sec. As an internal control, the house-keeping gene rpoZ that encodes DNA-directed RNA polymerase omega subunit was used. All primers used in QRT-PCR were as previously described (Cheng et al., 2012).
CrtJ-AerR interaction studies
In vitro pull down experiments were performed with SUMO-AerR that was purified as describe above using tag-less CrtJ as purified previously (Cheng et al., 2012). 1 ml Ni Sepharose high performance resin (GE Healthcare) was loaded onto an Econo-Pac chromatography column (Bio-Rad) and equilibrated with wash buffer (20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 20 mM imidazole, and 10% glycerol). SUMO-AerR protein was loaded onto the Ni Sepharose column as “bait” then the column was washed with 20 ml of wash buffer at at 22 °C. The purified tag-less CrtJ was then loaded onto the column and subjected to a gradient imidazole step elution using a buffer comprised of 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 10% glycerol with increasing imidazole concentrations. Tag-less CrtJ was used as a control without the loading of SUMO-AerR. Protein composition in each fraction was visualized by SDS-PAGE.
An AerR-CrtJ interaction was also probed using co-elution size exclusion chromatography on a column that was calibrated using commercial gel filtration standards (Bio-Rad). Purified CrtJ (~20 μM) and/or AerR (~20 μM) were incubated (molar ratio 1:1) at 22 °C for 1 hr before loading on a prepacked Superdex 200 column (10 × 300 mm) that was equilibrated. The column was equilibrated and eluted with 20 mM Tris-HCl (pH 8.0), 150 mM NaCl at 0.5 ml min−1.
Microscale thermophoresis assays
20 μM purified CrtJ was labeled using Monolith NT™ Protein Labeling Kit RED-NHS (Nano Temper Technologies Inc.). 500 nM labeled CrtJ was incubated with different concentration of AerR in a buffer containing 20 Tris-HCl (pH 8.0), 200 mM NaCl for 1 hr at 22°C. The reactions were loaded on Monolith NT.115™ Standard Treated Capillaries (Nano Temper Technologies Inc.) and analyzed with a Monolith NT.115 Blue Red microscale thermophoresis (MST) instrument. Thermophoresis with no jump was used to plot the binding isotherm. The resulting data were fitted to a Hill equation (Equation 1) with OriginPro 8.6.
| Equation 1 |
Parameters of the fit included normalized fluorescence intensities of free and bound CrtJ protein (FC and FCB). Hill coefficient (n) and the apparent dissociation constants (Kapp) were both obtained. Kapp= (EC50)n; A: AerR concentration. EC50 corresponds to the AerR concentration when 50% of CrtJ are bound to AerR.
DNase I DNA footprint assay
The bchC promoter region (325 bp) was amplified using a 5-HEX labeled forward primer and a 6-FAM labeled reverse primer (Table S1). After DNA agarose gel electrophoresis the PCR product was purified using the QIAquick Gel extraction kit (Qiangen) and used as DNA probe. The concentration of the DNA probe was determined spectrophotometrically, using the appropriate extinction coefficient. 10 nM DNA probe was incubated with different concentrations of AerR or CrtJ for 30 min at 22 °C in a buffer containing 25 mM Tris-HCl (pH 8.0), 200 mM NaCl, 2mM MgCl2 and 500 μg ml−1 heparin (250-fold weight excess over probe). For reactions incubated with CrtJ and AerR co-complex, CrtJ and AerR were mixed and incubated for 1 hr at 22 °C before adding to the probe. The final concentrations for CrtJ and AerR were at 100 nM and 1 μM, respectively. The reaction was set up in 20 μl volume to which 3 μl of 125 U ml−1 DNase I (New England BioLabs) was added and incubated for 15 min at 22 °C to digest DNA. 23 μl of 0.5 M EDTA (pH 8.0) was added to stop the reaction. DNA digestion product was recovered using Min Elute PCR purification kit (Qiangen), with sample eluted with 15 μl EB buffer from the kit plus 1 μl of 500 LIZTM size standards (Applied Biosystems). The eluted samples were separated and detected with a 3730 DNA analyzer (Applied Biosystems) and the results analyzed using Peak Scanner v1.0 (Applied Biosystems).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by an NIH grant R37 GM040941 awarded to CEB. The METACyt Biochemical Analysis Center was supported by a grant from the Eli Lilly Endowment.
References
- Bird TH, Du S, Bauer CE. Autophosphorylation, phosphotransfer, and DNA-binding properties of the RegB/RegA two-component regulatory system in Rhodobacter capsulatus. J Biol Chem. 1999;274:16343–16348. doi: 10.1074/jbc.274.23.16343. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Wu J, Setterdahl A, Reddie K, Carroll K, Hammad LA, et al. Activity of the tetrapyrrole regulator CrtJ is controlled by oxidation of a redox active cysteine located in the DNA binding domain. Mol Microbiol. 2012;85:734–746. doi: 10.1111/j.1365-2958.2012.08135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Elsen S, Swem LR, Bauer CE. AerR, a second aerobic repressor of photosynthesis gene expression in Rhodobacter capsulatus. J Bacteriol. 2002;184:2805–2814. doi: 10.1128/JB.184.10.2805-2814.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drennan CL, Huang S, Drummond JT, Matthews RG, Lidwig ML. How a protein binds B12: A 3.0 Å X-ray structure of B12-binding domains of methionine synthase. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- Drummond JT, Loo RR, Matthews RG. Electrospray mass spectrometric analysis of the domains of a large enzyme: observation of the occupied cobalamin-binding domain and redefinition of the carboxyl terminus of methionine synthase. Biochemistry. 1993;32:9282–9289. doi: 10.1021/bi00087a004. [DOI] [PubMed] [Google Scholar]
- Du S, Bird TH, Bauer CE. DNA binding characteristics of RegA. A constitutively active anaerobic activator of photosynthesis gene expression in Rhodobacter capsulatus. >J Biol Chem. 1998;273:18509–18513. doi: 10.1074/jbc.273.29.18509. [DOI] [PubMed] [Google Scholar]
- Fedosov SN, Fedosova NU, Nexo E, Petersen TE. Conformational changes of transcobalamin induced by aquocobalamin binding. Mechanism of substitution of the cobalt-coordinated group in the bound ligand. J Biol Chem. 2000;275:11791–11798. doi: 10.1074/jbc.275.16.11791. [DOI] [PubMed] [Google Scholar]
- Gomelsky L, Sram J, Moskvin OV, Horne IM, Dodd HN, Pemberton JM, et al. Identification and in vivo characterization of PpaA, a regulator of photosystem formation in Rhodobacter sphaeroides. Microbiology. 2003;149:377–388. doi: 10.1099/mic.0.25972-0. [DOI] [PubMed] [Google Scholar]
- Gruber K, Puffer B. and Krautler, B. Vitamin B12-derivatives-enzyme cofactors and ligands of proteins and nucleic acids. Chem Soc Rev. 2011;40:4346–4363. doi: 10.1039/c1cs15118e. [DOI] [PubMed] [Google Scholar]
- Halpern J. Mechanisms of coenzyme B12-dependent rearrangements. Science. 1985;227:869–875. doi: 10.1126/science.2857503. [DOI] [PubMed] [Google Scholar]
- Han YC, Meyer MHF, Keusgen M, Klug G. A haem cofactor is required for redox and light signalling by the AppA protein of Rhodobacter sphaeroides. Mol Microbiol. 2007;64:1090–1104. doi: 10.1111/j.1365-2958.2007.05724.x. [DOI] [PubMed] [Google Scholar]
- Hogenkamp HP. The photolysis of methylcobalamin. Biochemistry. 1966;5:417–422. doi: 10.1021/bi00866a005. [DOI] [PubMed] [Google Scholar]
- Loo JA. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Ludwig ML, Matthews RG. Structure-based perspectives on B12-dependent enzymes. Annu Rev Biochem. 1997;66:269–313. doi: 10.1146/annurev.biochem.66.1.269. [DOI] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- Masuda S, Bauer CE. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell. 2002;110:613–623. doi: 10.1016/s0092-8674(02)00876-0. [DOI] [PubMed] [Google Scholar]
- Masuda S, Bauer CE. Null mutation of HvrA compensates for loss of an essential relA/spoT-like gene in Rhodobacter capsulatus. J Bacteriol. 2004;186:235–239. doi: 10.1128/JB.186.1.235-239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Berleman J, Hasselbring BM, Bauer CE. Regulation of aerobic photosystem synthesis in the purple bacterium Rhodospirillum centenum by CrtJ and AerR. Photochem Photobiol Sci. 2008;7:1267–1272. doi: 10.1039/b802365b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Dong C, Swem D, Setterdahl AT, Knaff DB, Bauer CE. Repression of photosynthesis gene expression by formation of a disulfide bond in CrtJ. Proc Natl Acad Sci U S A. 2002;99:7078–7083. doi: 10.1073/pnas.102013099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoldrick H, Deery E, Warren M, Heathcote P. Cobalamin (vitamin B12) biosynthesis in Rhodobacter capsulatus. Biochem Soc Trans. 2002;30:646–648. doi: 10.1042/bst0300646. [DOI] [PubMed] [Google Scholar]
- Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer-membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskvin OV, Gilles-Gonzalez MA, Gomelsky M. The PpaA/AerR regulators of photosynthesis gene expression from anoxygenic phototrophic proteobacteria contain heme-binding SCHIC domains. J Bacteriol. 2010;192:5253–5256. doi: 10.1128/JB.00736-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskvin OV, Kaplan S, Gilles-Gonzalez MA, Gomelsky M. Novel heme-based oxygen sensor with a revealing evolutionary history. Journal of Biological Chemistry. 2007;282:28740–28748. doi: 10.1074/jbc.M703261200. [DOI] [PubMed] [Google Scholar]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- Nou X, Kadner RJ. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci U S A. 2000;97:7190–7195. doi: 10.1073/pnas.130013897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Guerrero JM, Polanco MC, Murillo FJ, Padmanabhan S, Elias-Arnanz M. Light-dependent gene regulation by a coenzyme B12-based photoreceptor. Proc Natl Acad Sci U S A. 2011;108:7565–7570. doi: 10.1073/pnas.1018972108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfold RJ, Pemberton JM. A gene from the photosynthetic gene cluster of Rhodobacter sphaeroides induces trans suppression of bacteriochlorophyll and carotenoid levels in R. sphaeroides and R. capsulatus. Curr Microbiol. 1991;23:259–263. [Google Scholar]
- Penfold RJ, Pemberton JM. Sequencing, chromosomal inactivation, and functional expression in Escherichia coli of ppsR, a gene which represses carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J Bacteriol. 1994;176:2869–2876. doi: 10.1128/jb.176.10.2869-2876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D. Microbial synthesis of cobamides. Adv Appl Microbiol. 1959;1:87–122. doi: 10.1016/s0065-2164(08)70476-3. [DOI] [PubMed] [Google Scholar]
- Pfennig N, Lippert KD. Über das Vitamin B12-Bedürfnis phototropher Schwefelbakterien. Arch Mikrobiol. 1966;55:245–256. [Google Scholar]
- Pollich M, Jock S, Klug G. Identification of a gene required for the oxygen-regulated formation of the photosynthetic apparatus of Rhodobacter capsulatus. Mol Microbiol. 1993;10:749–757. doi: 10.1111/j.1365-2958.1993.tb00945.x. [DOI] [PubMed] [Google Scholar]
- Pollich M, Klug G. Identification and sequence analysis of genes involved in late steps in cobalamin (vitamin B12) synthesis in Rhodobacter capsulatus. J Bacteriol. 1995;177:4481–4487. doi: 10.1128/jb.177.15.4481-4487.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnampalam SN, Bauer CE. DNA binding characteristics of CrtJ. A redox-responding repressor of bacteriochlorophyll, carotenoid, and light harvesting-II gene expression in Rhodobacter capsulatus. J Biol Chem. 1997;272:18391–18396. doi: 10.1074/jbc.272.29.18391. [DOI] [PubMed] [Google Scholar]
- Ponnampalam SN, Buggy JJ, Bauer CE. Characterization of an aerobic repressor that coordinately regulates bacteriochlorophyll, carotenoid, and light harvesting-II expression in Rhodobacter capsulatus. J Bacteriol. 1995;177:2990–2997. doi: 10.1128/jb.177.11.2990-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnampalam SN, Elsen S, Bauer CE. Aerobic repression of the Rhodobacter capsulatus bchC promoter involves cooperative interactions between CrtJ bound to neighboring palindromes. J Biol Chem. 1998;273:30757–30761. doi: 10.1074/jbc.273.46.30757. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem. 2003;278:41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- Taylor DP, Cohen SN, Clark WG, Marrs BL. Alignment of genetic and restriction maps of the photosynthesis region of the Rhodopseudomonas capsulata chromosome by a conjugation-mediated marker rescue technique. J Bacteriol. 1983;154:580–590. doi: 10.1128/jb.154.2.580-590.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington MD, Finke RG. Neopentylcobalamin (NeopentylB12) cobalt-carbon bond thermolysis products, kinetics, activation parameters, and bond dissociation energy: a chemical-model exhibiting 106 of the 1012 enzymatic activation of coenzyme B12's cobalt-carbon bond. J Am Chem Soc. 1993;115:4629–4640. [Google Scholar]
- Weissbach H, Ladd JN, Volcani BE, Smyth RD, Barker HA. Structure of the adenylcobamide coenzyme: degradation by cyanide, acid, and light. J Biol Chem. 1960;235:1462–1473. [PubMed] [Google Scholar]
- Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1 doi: 10.1038/ncomms1093. DOI: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- Willett J, Smart JL, Bauer CE. RegA control of bacteriochlorophyll and carotenoid synthesis in Rhodobacter capsulatus. J Bacteriol. 2007;189:7765–7773. doi: 10.1128/JB.00853-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler A, Heintz U, Lindner R, Reinstein J, Shoeman RL, Schlichting I. A ternary AppA-PpsR-DNA complex mediates light regulation of photosynthesis-related gene expression. Nat Struct Mol Biol. 2013;20:859–867. doi: 10.1038/nsmb.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuerges J, Garau G, Geremia S, Fedosov SN, Petersen TE, Randaccio L. Structural basis for mammalian vitamin B12 transport by transcobalamin. Proc Natl Acad Sci U S A. 2006;103:4386–4391. doi: 10.1073/pnas.0509099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Dragnea V, Bauer CE. PpsR, a regulator of heme and bacteriochlorophyll biosynthesis, is a heme-sensing protein. J Biol Chem. 2012;287:13850–13858. doi: 10.1074/jbc.M112.346494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Dragnea V, Feldman G, Hammad LA, Karty JA, Dann CE, 3rd, Bauer CE. Redox and Light Control the Heme-Sensing Activity of AppA. MBio. 2013;4:e00563–13. doi: 10.1128/mBio.00563-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.