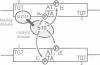
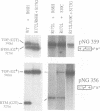
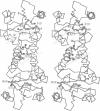
Abstract
The resolvase protein of the gamma delta transposon is a site-specific recombinase that acts by a concerted break-and-join mechanism. To analyse the role of individual resolvase subunits in DNA strand cleavage, we have directed the binding of catalytic mutants to specific recombination crossover sites or half-sites. Our results demonstrate that the resolvase subunit bound at the half-site proximal to each scissile phosphodiester bond provides the Ser10 nucleophile and Arg8, Arg68 and Arg71 residues essential for cleavage and covalent attachment to the DNA. Several other residues near the presumptive active site are also shown to act in cis. Double-strand cleavage at one crossover site can proceed independently of cleavage at the other site, although interactions between the resolvase dimers bound at the two crossover sites remain essential. An appropriately oriented heterodimer of active and inactive protomers can in most cases mediate either a 'top' or 'bottom' single-strand cleavage, suggesting that there is no obligatory order of strand cleavages. Top-strand cleavage is associated with the topoisomerase I activity of resolvase, suggesting that a functional asymmetry may be imposed on the crossover site by the structure of the active synapse.
Full text
PDF
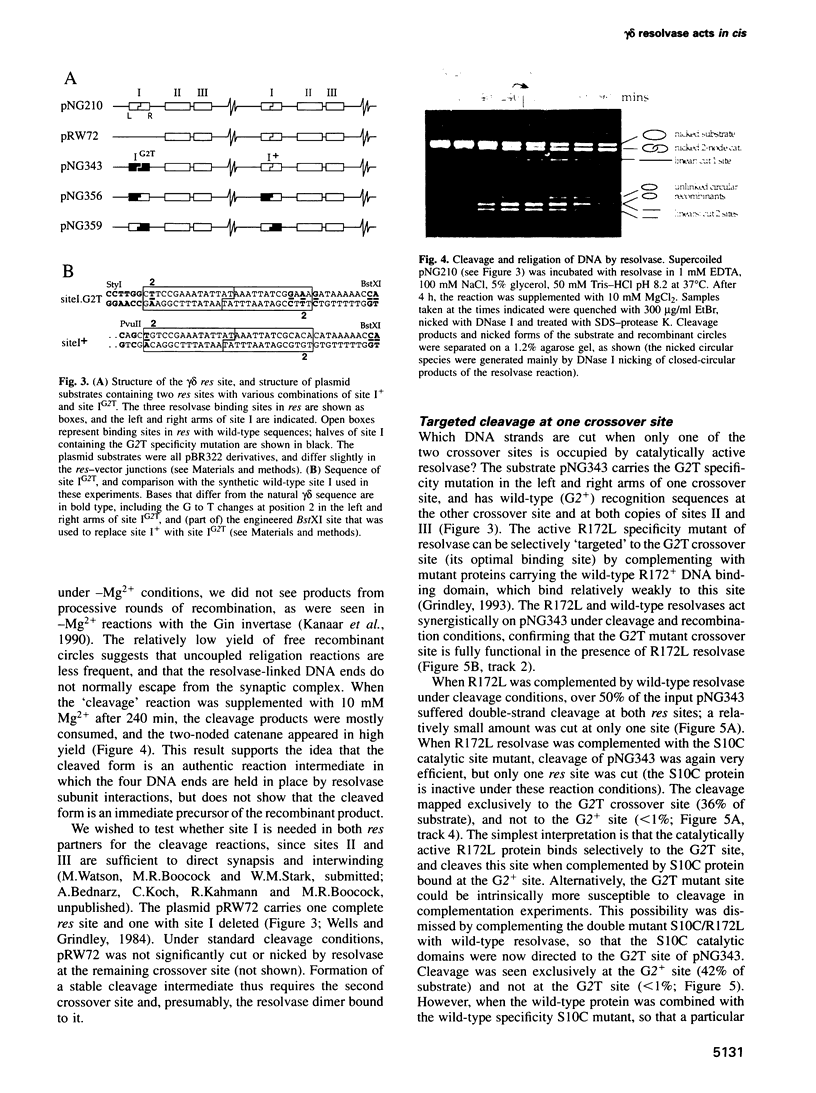
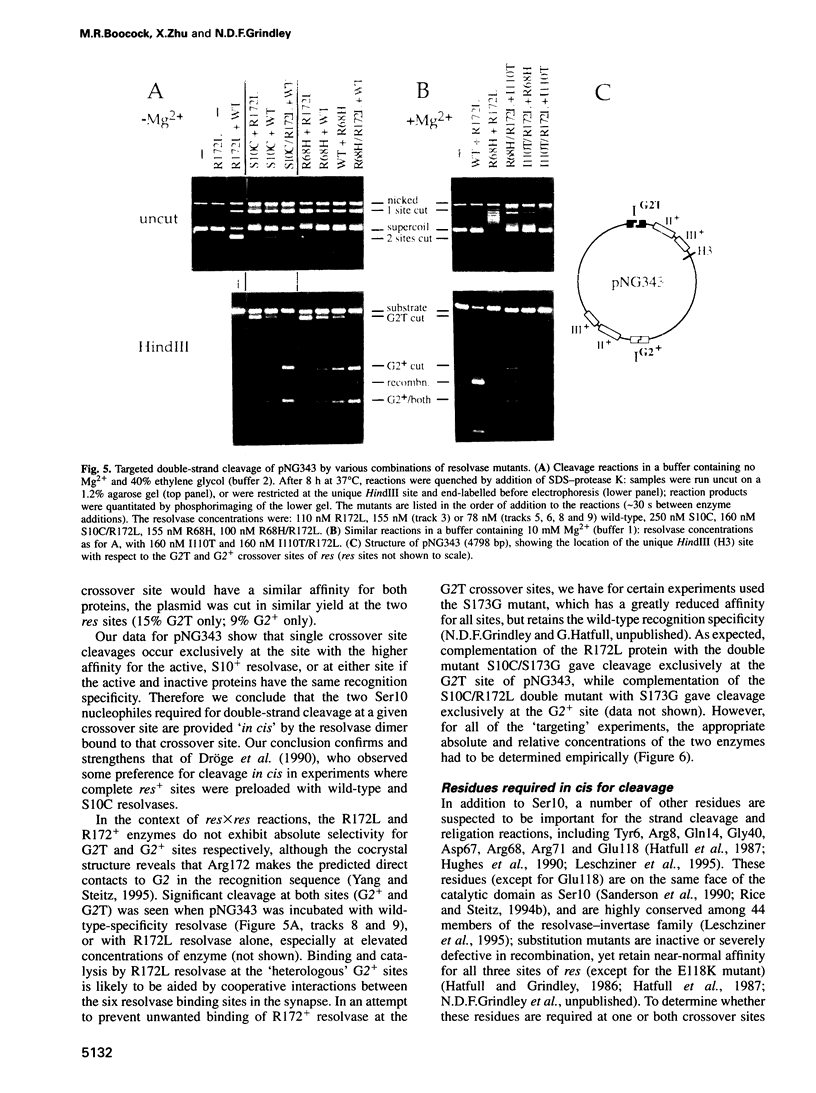








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdel-Meguid S. S., Grindley N. D., Templeton N. S., Steitz T. A. Cleavage of the site-specific recombination protein gamma delta resolvase: the smaller of two fragments binds DNA specifically. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2001–2005. doi: 10.1073/pnas.81.7.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciszewska L. K., Sherratt D. J. Xer site-specific recombination in vitro. EMBO J. 1995 May 1;14(9):2112–2120. doi: 10.1002/j.1460-2075.1995.tb07203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. A., Mizuuchi M., Savilahti H., Mizuuchi K. Division of labor among monomers within the Mu transposase tetramer. Cell. 1993 Aug 27;74(4):723–733. doi: 10.1016/0092-8674(93)90519-v. [DOI] [PubMed] [Google Scholar]
- Bednarz A. L., Boocock M. R., Sherratt D. J. Determinants of correct res site alignment in site-specific recombination by Tn3 resolvase. Genes Dev. 1990 Dec;4(12B):2366–2375. doi: 10.1101/gad.4.12b.2366. [DOI] [PubMed] [Google Scholar]
- Benjamin H. W., Cozzarelli N. R. Geometric arrangements of Tn3 resolvase sites. J Biol Chem. 1990 Apr 15;265(11):6441–6447. [PubMed] [Google Scholar]
- Blake D. G., Boocock M. R., Sherratt D. J., Stark W. M. Cooperative binding of Tn3 resolvase monomers to a functionally asymmetric binding site. Curr Biol. 1995 Sep 1;5(9):1036–1046. doi: 10.1016/s0960-9822(95)00208-9. [DOI] [PubMed] [Google Scholar]
- Blakely G., May G., McCulloch R., Arciszewska L. K., Burke M., Lovett S. T., Sherratt D. J. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993 Oct 22;75(2):351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- Castell S. E., Halford S. E. DNA supercoiling determines the activation energy barrier for site specific recombination by Tn21 resolvase. Nucleic Acids Res. 1989 Sep 12;17(17):7045–7058. doi: 10.1093/nar/17.17.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. W., Lee J., Jayaram M. DNA cleavage in trans by the active site tyrosine during Flp recombination: switching protein partners before exchanging strands. Cell. 1992 May 15;69(4):647–658. doi: 10.1016/0092-8674(92)90228-5. [DOI] [PubMed] [Google Scholar]
- Derbyshire V., Freemont P. S., Sanderson M. R., Beese L., Friedman J. M., Joyce C. M., Steitz T. A. Genetic and crystallographic studies of the 3',5'-exonucleolytic site of DNA polymerase I. Science. 1988 Apr 8;240(4849):199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- Dröge P., Hatfull G. F., Grindley N. D., Cozzarelli N. R. The two functional domains of gamma delta resolvase act on the same recombination site: implications for the mechanism of strand exchange. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5336–5340. doi: 10.1073/pnas.87.14.5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falvey E., Grindley N. D. Contacts between gamma delta resolvase and the gamma delta res site. EMBO J. 1987 Mar;6(3):815–821. doi: 10.1002/j.1460-2075.1987.tb04824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falvey E., Hatfull G. F., Grindley N. D. Uncoupling of the recombination and topoisomerase activities of the gamma delta resolvase by a mutation at the crossover point. Nature. 1988 Apr 28;332(6167):861–863. doi: 10.1038/332861a0. [DOI] [PubMed] [Google Scholar]
- Grindley N. D. Analysis of a nucleoprotein complex: the synaptosome of gamma delta resolvase. Science. 1993 Oct 29;262(5134):738–740. doi: 10.1126/science.8235593. [DOI] [PubMed] [Google Scholar]
- Han Y. W., Gumport R. I., Gardner J. F. Complementation of bacteriophage lambda integrase mutants: evidence for an intersubunit active site. EMBO J. 1993 Dec;12(12):4577–4584. doi: 10.1002/j.1460-2075.1993.tb06146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull G. F., Grindley N. D. Analysis of gamma delta resolvase mutants in vitro: evidence for an interaction between serine-10 of resolvase and site I of res. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5429–5433. doi: 10.1073/pnas.83.15.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull G. F., Noble S. M., Grindley N. D. The gamma delta resolvase induces an unusual DNA structure at the recombinational crossover point. Cell. 1987 Apr 10;49(1):103–110. doi: 10.1016/0092-8674(87)90760-4. [DOI] [PubMed] [Google Scholar]
- Hatfull G. F., Sanderson M. R., Freemont P. S., Raccuia P. R., Grindley N. D., Steitz T. A. Preparation of heavy-atom derivatives using site-directed mutagenesis. Introduction of cysteine residues into gamma delta resolvase. J Mol Biol. 1989 Aug 20;208(4):661–667. doi: 10.1016/0022-2836(89)90156-3. [DOI] [PubMed] [Google Scholar]
- Hughes R. E., Hatfull G. F., Rice P., Steitz T. A., Grindley N. D. Cooperativity mutants of the gamma delta resolvase identify an essential interdimer interaction. Cell. 1990 Dec 21;63(6):1331–1338. doi: 10.1016/0092-8674(90)90428-h. [DOI] [PubMed] [Google Scholar]
- Hughes R. E., Rice P. A., Steitz T. A., Grindley N. D. Protein-protein interactions directing resolvase site-specific recombination: a structure-function analysis. EMBO J. 1993 Apr;12(4):1447–1458. doi: 10.1002/j.1460-2075.1993.tb05788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Bruist M. F. Intermediates in Hin-mediated DNA inversion: a role for Fis and the recombinational enhancer in the strand exchange reaction. EMBO J. 1989 May;8(5):1581–1590. doi: 10.1002/j.1460-2075.1989.tb03542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R., Klippel A., Shekhtman E., Dungan J. M., Kahmann R., Cozzarelli N. R. Processive recombination by the phage Mu Gin system: implications for the mechanisms of DNA strand exchange, DNA site alignment, and enhancer action. Cell. 1990 Jul 27;62(2):353–366. doi: 10.1016/0092-8674(90)90372-l. [DOI] [PubMed] [Google Scholar]
- Klippel A., Cloppenborg K., Kahmann R. Isolation and characterization of unusual gin mutants. EMBO J. 1988 Dec 1;7(12):3983–3989. doi: 10.1002/j.1460-2075.1988.tb03286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klippel A., Kanaar R., Kahmann R., Cozzarelli N. R. Analysis of strand exchange and DNA binding of enhancer-independent Gin recombinase mutants. EMBO J. 1993 Mar;12(3):1047–1057. doi: 10.1002/j.1460-2075.1993.tb05746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klippel A., Mertens G., Patschinsky T., Kahmann R. The DNA invertase Gin of phage Mu: formation of a covalent complex with DNA via a phosphoserine at amino acid position 9. EMBO J. 1988 Apr;7(4):1229–1237. doi: 10.1002/j.1460-2075.1988.tb02935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnow M. A., Cozzarelli N. R. Site-specific relaxation and recombination by the Tn3 resolvase: recognition of the DNA path between oriented res sites. Cell. 1983 Apr;32(4):1313–1324. doi: 10.1016/0092-8674(83)90312-4. [DOI] [PubMed] [Google Scholar]
- Landy A. Mechanistic and structural complexity in the site-specific recombination pathways of Int and FLP. Curr Opin Genet Dev. 1993 Oct;3(5):699–707. doi: 10.1016/s0959-437x(05)80086-3. [DOI] [PubMed] [Google Scholar]
- Lavoie B. D., Chan B. S., Allison R. G., Chaconas G. Structural aspects of a higher order nucleoprotein complex: induction of an altered DNA structure at the Mu-host junction of the Mu type 1 transpososome. EMBO J. 1991 Oct;10(10):3051–3059. doi: 10.1002/j.1460-2075.1991.tb07856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Whang I., Lee J., Jayaram M. Directed protein replacement in recombination full sites reveals trans-horizontal DNA cleavage by Flp recombinase. EMBO J. 1994 Nov 15;13(22):5346–5354. doi: 10.1002/j.1460-2075.1994.tb06869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H. M. Analysis of subunit interaction by introducing disulfide bonds at the dimerization domain of Hin recombinase. J Biol Chem. 1994 Dec 9;269(49):31134–31142. [PubMed] [Google Scholar]
- Liu T., Liu D., DeRose E. F., Mullen G. P. Studies of the dimerization and domain structure of gamma delta resolvase. J Biol Chem. 1993 Aug 5;268(22):16309–16315. [PubMed] [Google Scholar]
- Mazzarelli J. M., Ermácora M. R., Fox R. O., Grindley N. D. Mapping interactions between the catalytic domain of resolvase and its DNA substrate using cysteine-coupled EDTA-iron. Biochemistry. 1993 Mar 30;32(12):2979–2986. doi: 10.1021/bi00063a008. [DOI] [PubMed] [Google Scholar]
- Newman B. J., Grindley N. D. Mutants of the gamma delta resolvase: a genetic analysis of the recombination function. Cell. 1984 Sep;38(2):463–469. doi: 10.1016/0092-8674(84)90501-4. [DOI] [PubMed] [Google Scholar]
- Nunes-Düby S. E., Tirumalai R. S., Dorgai L., Yagil E., Weisberg R. A., Landy A. Lambda integrase cleaves DNA in cis. EMBO J. 1994 Sep 15;13(18):4421–4430. doi: 10.1002/j.1460-2075.1994.tb06762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. N., Halford S. E. Dynamics of long-range interactions on DNA: the speed of synapsis during site-specific recombination by resolvase. Cell. 1991 Aug 23;66(4):781–791. doi: 10.1016/0092-8674(91)90121-e. [DOI] [PubMed] [Google Scholar]
- Reed R. R., Grindley N. D. Transposon-mediated site-specific recombination in vitro: DNA cleavage and protein-DNA linkage at the recombination site. Cell. 1981 Sep;25(3):721–728. doi: 10.1016/0092-8674(81)90179-3. [DOI] [PubMed] [Google Scholar]
- Reed R. R., Moser C. D. Resolvase-mediated recombination intermediates contain a serine residue covalently linked to DNA. Cold Spring Harb Symp Quant Biol. 1984;49:245–249. doi: 10.1101/sqb.1984.049.01.028. [DOI] [PubMed] [Google Scholar]
- Reed R. R. Transposon-mediated site-specific recombination: a defined in vitro system. Cell. 1981 Sep;25(3):713–719. doi: 10.1016/0092-8674(81)90178-1. [DOI] [PubMed] [Google Scholar]
- Rimphanitchayakit V., Grindley N. D. Saturation mutagenesis of the DNA site bound by the small carboxy-terminal domain of gamma delta resolvase. EMBO J. 1990 Mar;9(3):719–725. doi: 10.1002/j.1460-2075.1990.tb08165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson M. R., Freemont P. S., Rice P. A., Goldman A., Hatfull G. F., Grindley N. D., Steitz T. A. The crystal structure of the catalytic domain of the site-specific recombination enzyme gamma delta resolvase at 2.7 A resolution. Cell. 1990 Dec 21;63(6):1323–1329. doi: 10.1016/0092-8674(90)90427-g. [DOI] [PubMed] [Google Scholar]
- Soultanas P., Oram M., Halford S. E. Site-specific recombination at res sites containing DNA-binding sequences for both Tn21 and Tn3 resolvases. J Mol Biol. 1995 Jan 20;245(3):208–218. doi: 10.1006/jmbi.1994.0017. [DOI] [PubMed] [Google Scholar]
- Stark W. M., Boocock M. R., Sherratt D. J. Catalysis by site-specific recombinases. Trends Genet. 1992 Dec;8(12):432–439. [PubMed] [Google Scholar]
- Stark W. M., Boocock M. R. The linkage change of a knotting reaction catalysed by Tn3 resolvase. J Mol Biol. 1994 May 27;239(1):25–36. doi: 10.1006/jmbi.1994.1348. [DOI] [PubMed] [Google Scholar]
- Stark W. M., Sherratt D. J., Boocock M. R. Site-specific recombination by Tn3 resolvase: topological changes in the forward and reverse reactions. Cell. 1989 Aug 25;58(4):779–790. doi: 10.1016/0092-8674(89)90111-6. [DOI] [PubMed] [Google Scholar]
- Wasserman S. A., Dungan J. M., Cozzarelli N. R. Discovery of a predicted DNA knot substantiates a model for site-specific recombination. Science. 1985 Jul 12;229(4709):171–174. doi: 10.1126/science.2990045. [DOI] [PubMed] [Google Scholar]
- Wells R. G., Grindley N. D. Analysis of the gamma delta res site. Sites required for site-specific recombination and gene expression. J Mol Biol. 1984 Nov 15;179(4):667–687. doi: 10.1016/0022-2836(84)90161-x. [DOI] [PubMed] [Google Scholar]
- Yang W., Steitz T. A. Crystal structure of the site-specific recombinase gamma delta resolvase complexed with a 34 bp cleavage site. Cell. 1995 Jul 28;82(2):193–207. doi: 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]