Abstract
DNA and porphyrin based therapeutics are important for anti-cancer treatment. The present studies demonstrate single-stranded DNA (ssDNA) assembles with meso-tetra-4-pyridyl porphine (MTP) forming porphyrin:DNA nano-complexes (PDN) that are stable in aqueous solution under physiologically relevant conditions and undergo dissociation with DNA release in hydrophobic environments, including cell membranes. PDN formation is DNA-dependent with the ratio of porphyrin:DNA being approximately two DNA nucleobases per porphyrin. PDN produce reactive oxygen species (ROS) in a light-dependent manner under conditions that favor nano-complex dissociation in the presence of hydrophobic solvents. PDN induce light-dependent cytotoxicity in vitro and anti-tumor activity towards bladder cancer xenografts in vivo. Light-dependent, PDN-mediated cell death results from ROS-mediated localized membrane damage due to lipid peroxidation with mass spectrometry indicating the generation of the lipid peroxidation products 9- and 13-hydroxy octadecanoic acid. Our results demonstrate that PDN have properties useful for therapeutic applications, including cancer treatment.
Keywords: Multi-modality nanoparticle, Cancer therapy, Photodynamic therapy, Porphyrin, DNA assembly
Background
Porphyrins are a group of compounds containing the porphin ring structure that is important in biology and medicine and that has recently been studied for their propensity to form nano-materials,1,2 including nanotubes3 through non-covalent chemistry. The principal driving force for porphyrin self-assembly is hydrophobic interactions4 resulting in vertical stacking of porphyrins in aqueous solutions in a pH-dependent manner.5,6 Previous studies demonstrating nanoparticle assembly of porphyrins were conducted in pure water and self-assembly was optimal at acidic pH, limiting potential biological applications.5,6 Porphyrin assembly has been shown to be modulated by biological molecules, such as poly-glutamic acid,7 however these complexes also have limited stability under physiologically relevant conditions. The preparation of porphyrin-containing nanostructures that are stable under biologically relevant conditions, but that dissociate in a predicable manner could be useful for drug delivery, photodynamic therapy (PDT),8–12 and other biologically relevant processes.13
The present studies focus on developing porphyrin:DNA nanoparticles (PDN) that are stable under physiological conditions of pH, salt, and temperature and that have potential use for PDT as well as in nucleic acid delivery. In principle, hydrophobic stacking and hydrogen bonding interactions between appropriately modified porphyrins and DNA (or RNA) nucleobases could provide an additional interaction complementary to porphyrin aggregation potentially resulting in production of porphyrin:DNA nanoparticles (PDN) that have both the light-mediated cell killing properties of porphyrin photosensitizers (PS) and the capacity to deliver a nucleic acid payload with therapeutic utility. Our studies utilize meso-tetra-4-pyridyl porphin (MTP) interacting with the single-stranded DNA (ssDNA) (GT)20. In the absence of DNA, MTP forms amorphous aggregates in aqueous solution under physiological conditions, although acidification of the solution (< pH 3.0) dissociates the aggregates and re-generates non-complexed porphyrin. The pyridyl groups of MTP have the potential to base stack and/or form hydrogen bonds with nucleobases of DNA at physiological pH and the phosphodiester backbone of DNA may confer aqueous solubility to the complex. We demonstrate that unlike previous porphyrin:DNA complexes,14–18 the PDN developed with this approach form discrete porphyrin:DNA nanoparticles that are of the appropriate size for drug-delivery applications via the enhanced permeability and retention (EPR) effect19,20 and that readily dissociate upon cell internalization to deliver a therapeutic payload.
The PDN prepared by self-assembly of MTP and ssDNA have the potential to be highly effective agents for cancer treatment. We demonstrate that PDN are capable of exerting light-dependent cytotoxicity towards bladder cancer cells both in tissue culture models and in vivo. The light-dependent cytotoxicity of PDN occurs with generation of the lipid oxidation products 9- and 13-hydroxy octadecanoic acid (HODE) and with damage to the integrity of the plasma membrane consistent with generation of singlet oxygen via a type II photochemical reaction.21 PDN initially associate with the plasma membrane and are internalized into endocytic vesicles where free DNA and porphyrin may release, recovering the functionality of the constituent materials while leaving no extraneous carrier that may impart systemic toxicities. Importantly, localized PDN-treatment combined with light-activation significantly reduced tumor volumes in vivo.
Methods
Preparation of porphyrin:DNA nanoparticles
Meso-tetra-4-pyridyl porphine (MTP; ~0.25 mg – Frontier Scientific) was suspended in 5 mL 20 mM Phosphate Buffer (pH 7.4) containing 5 nmol (0.025 mg/mL) d(GT)20 and sonicated in a bath sonicator for 1.5 h with temperature maintained at 5–7 °C. A second addition of MTP (~0.25 mg) was followed by an additional 1 h sonication, followed by addition of 5 nmol DNA and further sonication for 1 h. The mixture was then centrifuged and filtered with membrane filter tube (Amicon Ultra, MWCO 100 KDa) to remove any free DNA. The resulting brownish-yellow aqueous suspension was used for subsequent studies. The porphyrin:DNA ratio for PDN (~19:1) was calculated by acid dissolution of the nanoparticle followed by spectrophotometric determination.
Biophysical characterization of PDN
UV-Vis spectra were collected under ambient conditions using a DU800 UV-Vis spectrophotometer (Beckman Coulter). Fluorescence spectra were acquired using a Perkin-Elmer-F1000 fluorometer with excitation at 420 nm and emission scanned over the range 550–900 nm. Transmission electron microscopy (TEM) images of PDN were acquired using a FEI Thcnai-Spirit TEM. TEM images were analyzed and the distribution of PDN length and diameter was determined using Matlab software. Dynamic light scattering (DLS) was performed under ambient conditions using a Malvern Zetasizer nano series ZEN-1600 in particle size and measurement mode. Each sample was read for 60 s using a 442.0 Kcps count rate. Data were analyzed using Malvern software.
Hydrophobicity-dependent DNA release and endosomal uptake of PDN
PDN were prepared as a suspension in 20 mM phosphate buffer (250 μg/mL) which was then diluted to 15 μg/mL in 20 mM phosphate buffer mixture containing either 0%, 20% or 40% acetonitrile. The desired pH was obtained by adding concentrated HCl or NaOH. The pH of all mixtures was further confirmed using a pH meter after addition of all components. After addition of all components, mixtures were incubated at room temperature overnight followed by filtration through membrane filter tubes (Amicon Ultra; 100KDa MWCO). 25 μL of each filtrate including a 2.5 μM d(GT)20 standard was then analyzed by polyacrylamide gel electrophoresis (15% native gel in TBE buffer, pH 7.4, 50 mA for 90 min). The gel was stained using Syber Gold solution and scanned using a Typhoon FLA 9500. The image of the gel was analyzed with Image Quant 5.2 software. The pH- and hydrophobicity-dependent DNA dissociation was also quantified by investigating absorbance of the filtrates at 260 nm. The concentration of DNA filtrates was quantified using the absorbance of a solution of 1.275 μM d(GT)20 as standard that represents the total DNA content of PDN (calculated from PDN concentration with ratiometric quantification).
Endocytosis-mediated cellular uptake of PDN was evaluated by co-localization with FITC-labeled dextran. Human bladder cancer cells (ATCC 5637) were cultured in sterile chamber slides and treated with complete media containing PDN (2 μg/mL) and FITC-labeled dextran (1 mg/mL), then incubated overnight at 37 °C, and, next morning, washed with sterile PBS and imaged using a Zeiss Axiovert LSM-510 microscope. PDN were excited using the 633 nm laser and emission was collected with the 650 nm long-pass filter set. FITC-labeled dextran was excited using the 488 nm argon laser and emission collected with 505–530 nm bandpass filter set. DIC images of cells were collected in a separate channel for overlay. The co-localization analysis was done using LSM 510 software (Carl Zeiss).
Tissue culture cytotoxicity and apoptosis assays
Human bladder cancer cells (ATCC 5637) were cultured in complete media (RPMI + 10% FBS). The cells were then treated with complete media containing nanoparticle at final concentrations of 0, 1, 2, 5 μg/mL followed by incubation overnight then washed with PBS. One of two plates was then exposed to 420 nm blue light (Trophy Skin Blue MD) at a power density of 2.3 μW/cm2 for 10 min followed by incubation for 24 h. Cell viability was assessed using the CellTiter 96 Proliferation Assay reagent (Promega) following the manufacturer’s instructions. Apoptosis was assessed using the Caspase Glo 3/7 assay (Promega) with similar procedures except PDN concentration was evaluated at 0 or 2 μg/mL. Luminescence was measured using GENios (TECAN) microplate reader. Each set of data (net absorbance) was expressed as a percentage, considering the no treatment group as 100%.
Light-dependent membrane damage by PDN
Localized membrane damage of PDN upon blue light irradiation was evaluated using confocal microscopy to detect calcein-AM retention, a cytoplasm-localizing fluorescent dye. Cells were cultured in each of two sterile chamber slides and treated with complete media containing PDN (0 or 2 μg/mL), incubated overnight at 37 °C, washed with PBS followed by addition of Calcein-AM (Invitrogen) at final concentration 2.5 μM, and incubated for 15 min at 37 °C. One of the two chamber slides was then exposed to 420 nm blue light for 10 min. Confocal microscopy was performed as mentioned in the FITC-dextran experiment.
Light-dependent ROS generation and membrane lipid peroxidation by PDN
PDN suspension was prepared in 20 mM phosphate buffer (pH 7.4) as described above to a final concentration of 250 μg/mL. Acetonitrile buffer mixture was prepared by adding appropriate amount of 100 mM sodium phosphate and acetonitrile with water. PDN suspension was added to the mixture (5 μg/mL) followed by addition of the dye C11 Bodipy to final concentration 20 μM in a 96 well black flat bottom plate (Costar). Mixtures were prepared in triplicates in two plates. One of the plates was then exposed to the blue light for 2 min immediately followed by scanning with Tecan-Safire-II microplate reader in fluorescence detection mode with 480 nm excitation and 500–800 nm emission. Slit opening was set to 10 nm, and reading was average for 5 actuations.
Membrane lipid peroxidation upon blue light irradiation of cancer cells treated with PDN was evaluated using confocal microscopy detecting a membrane localizable fluorescent dye (C11-Bodipy). Microscopy was performed as described for confocal microscopy experiments including an additional channel for detecting non-oxidized dye, excited with a 543 nm He-Ne laser and emission was collected using 565–615 bandpass filter.
Detection of light-dependent membrane damage by PDN
PDN-mediated membrane damage upon blue light irradiation was detected by Transmission Election Microscopy (TEM). Cell culture and PDN treatment, including blue light irradiation, were performed in 6-well clear bottom plates (Costar) following the same procedure as for the in vitro cytotoxicity assay with the PDN concentration 0 or 2 μg/mL. Cells were fixed with 2.5% glutaraldehyde and treated with 2% osmium tetroxide, then dehydrated gradually followed by embedding in the resin and sectioning for TEM investigation.
Antitumor activity of PDN
All animal experiments were performed in accordance with protocols approved by the Wake Forest School of Medicine Animal Care and Use Committee. Six week old female nude mice were ordered from NCI. Tumor xenografts were generated by subcutaneous injection of 1.5 × 106 human bladder cancer cells suspended in 200 μL of 1:1 PBS/Matrigel (BD bioscience) in both flanks of 10 female nude mice. Mice were used for experimental procedures 3 weeks following inoculation with tumor cells, after tumor size had reached approximately 75 mm3. Each of 10 mice was injected with 100 μL of 250 μg/mL PDN suspension in one flank and 100 μL saline in the other flank. Approximately 12 h later, tumors on both flanks of 6 mice were irradiated with a blue beam of 420 nm for 3 min. The source laser for these studies was a Mira 900 (Coherent Inc., Santa Clara, CA), a mode-locked femtosecond Ti:Sapphire laser. The pulses were approximately 90 femtoseconds at a wavelength of 840 nm with an average power of 600 mW. The beam was transmitted directly into the tumor using a multimode optical fiber (SFS105/125Y, Thorlabs Inc., Newton, NJ). The output power at the final end of the fiber was 100 mW. The remaining four mice were treated identically but were not exposed to the laser. The laser irradiation was performed every 5th day for a total of five doses. Tumor sizes were measured using calipers and tumor volumes were calculated using the formula xy2 π/6 (where x and y are the long and short diameters of the tumor, respectively). The tumors were analyzed as four independent groups: i) no treatment; ii) light-only; iii) PDN-only; iv) PDN + light. Relative tumor volumes (V/Vo) were graphed vs. time (where V is the present tumor volume and Vo was the tumor volume when treatment started). Repeated measures mixed models were fit to compare tumor volumes between groups. In these models, animals were treated as random effects and group (four-levels) and time (days) were treated as fixed-effects. The group-by-time interaction was examined to determine whether the rate of change in tumor volume differed over time among the four groups. All statistical analyses were performed using SAS 9.1.
Histopathological analysis of tumor tissue
At the conclusion of the study animals were sacrificed and tumors were excised and placed in 4% paraformaldehyde solution overnight at ambient temperature. The following day, tumors were embedded in molten paraffin and thin sections were prepared from different layers of tumors and placed on glass slides followed by H&E staining.
Results
Biophysical properties of porphyrin:DNA nanoparticles
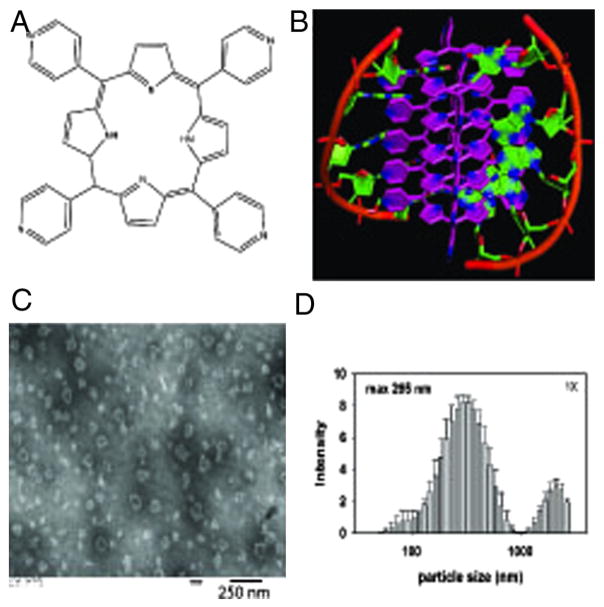
Meso-tetra-4-pyridyl porphine (MTP – Figure 1, A) is soluble in aqueous solution only at acidic pH (< 5.5) and forms aggregates in solution near physiological pH. Sonication of MTP in the presence of single-stranded DNA at pH 7.4, however, permitted recovery of discrete, nano-sized particles composed of both porphyrin and DNA (porphyrin:DNA nanoparticles – PDN; Figure 1, B). Production of nanoparticles (Figure S1, Supporting Information) was DNA-dependent and the ratio of porphyrin to DNA (d(GT)20) was determined to be ~19:1 (mol/mol) or about two DNA nucleobases per MTP based upon the UV absorbances calculated from the acid-dissolved nanoparticles (Table S1 and Figure S2, Supporting Information). The size and shape of the PDN were investigated using transmission electron microscopy (TEM – Figure 1, C) and the hydrodynamic properties of PDN were investigated using dynamic light scattering (DLS – Figure 1, D). TEM images revealed irregular shaped particles with average length and diameter ~60 nm (Figure S3, Supporting Information) while DLS revealed PDN in aqueous solution had an average hydrodynamic radius of ~295 nm (Figure 1, D).
Figure 1.
Discrete porphyrin:DNA nanoparticles (PDN) form upon sonication of MTP porphyrin with ssDNA. (A) Structure of the MTP porphyrin used for these studies. (B) Molecular model of PDN nano-complex showing both porphyrin-porphyrin and porphyrin:DNA interactions. (C) TEM image of PDN nanoparticles. (D) DLS analysis of PDN hydrodynamic radius distribution.
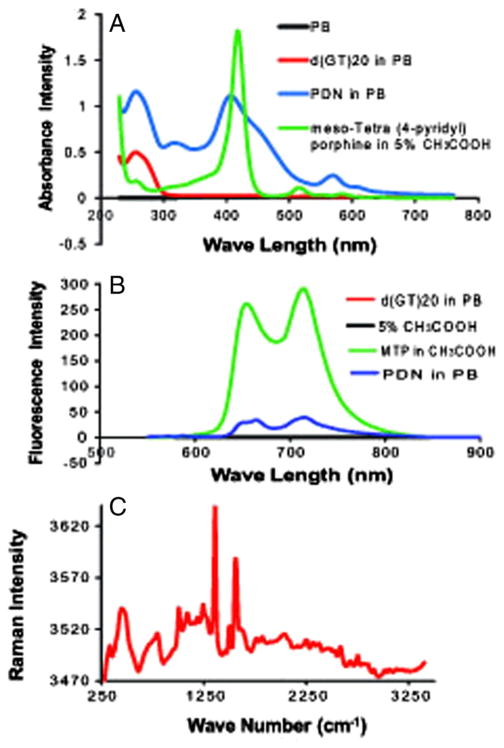
PDN were characterized by UV-Vis, fluorescence, and Raman spectroscopies to determine to what extent the context of the PDN complex altered the electronic properties of the constituent porphyrin and DNA. UV-Vis spectra for PDN revealed the absorbance at 260 nm from the DNA was little changed relative to free DNA while the Soret (S0->S2) band22 was slightly red-shifted and broadened for PDN relative to MTP (Figure 2, A). The Q band (S0->S1)23 at 535 nm was also enhanced and sharpened for the PDN relative to the free porphyrin. Fluorescence spectra for PDN with excitation at 420 nm showed that the emission maxima at 660 and 725 nm for the MTP monomer were substantially quenched for PDN with emission maxima reduced ~25-fold (Figure 2, B). Raman spectra (457.9 absorption; Figure 2, C) revealed two sharp peaks at 1360 and 1555 cm−1 for PDN similar to resonances assigned previously to (ν(Xβ − Xβ) + (Xβ − H)) and (ν(Xα − N) + δ(Xβ − H)) for tetrasulfonated tetraphenyl porphyrin (TSSP) upon electrode binding,24 consistent with surface-enhancement of Raman spectra for MTP in the context of PDN. Together, the spectroscopic properties reveal an altered electronic structure for MTP in the context of PDN that is consistent with π – π stacking of porphyrins with DNA nucleobases contributing significantly to PDN assembly.
Figure 2.
Spectroscopic characterization of PDN reveals the physical properties of the porphyrin are altered in the PDN complex. (A) Absorbance spectra for PDN (blue), MTP monomer (green), DNA (red), and phosphate buffer (black). (B) Fluorescence spectra for MTP monomer (green), PDN (blue), DNA (black), and 5% acetic acid (magenta). (C) Raman spectrum for PDN.
Hydrophobicity-dependent DNA release and endosomal uptake of PDN
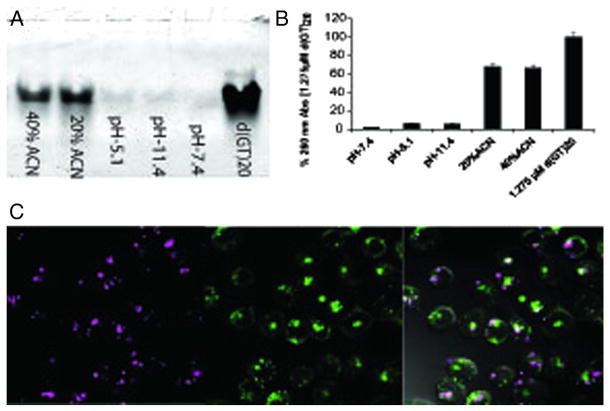
The stability of PDN as a function of pH and solvent hydrophobicity was investigated to gain insight into the nature of forces promoting complex stability. PDN stability at physiologically-relevant pH is mainly dependent on hydrophobic interactions (Figure 3, A and B and S4, S5, Supporting Information). Acidification of PDN to pH 5.1 may also enhance its dissociation process, as indicated by small amount of DNA release, due to pyridyl nitrogen protonation. Approximately 70% of total DNA was released upon PDN dissociation in 20% and 40% acetonitrile solution; while pH 5.1 and 11.4 induced ~7% of total DNA release in the 100% aqueous environment (Figure 3, A and B) and no DNA release was observed in pH 7.4 at 100% aqueous environment. However in vitro acidification of PDN to pH 3.0 or below leads to complete PDN dissociation immediately due to full protonation of the porphyrin molecules including central pyrrol rings.25,26 While the hydrophobicity-dependence of PDN dissociation is consistent with disruption of hydrophobic base stacking between DNA and MTP, the DNA release from PDN was favored by increasing hydrophobicity of the solvent system by acetonitrile addition (Figure 3, A and B and S4, Supporting Information). Thus, the data are consistent with π – π stacking between DNA nucleobases and pyridyl side chains of MTP significantly contributing to PDN stability. The hydrophobicity-dependence of PDN dissociation in vitro caused us to investigate to what extent PDN dissociate in hydrophobic environments in vivo. Significantly, hydrophobic environment of plasma membrane potentially promotes PDN disassembly by dynamic hydrophobic interactions (Figure S5, Supporting Information).27,28
Figure 3.
PDN-disassembly as a function of pH and solvent hydrophobicity. (A) 15% native polyacrylamide gel electrophoresis of DNA filtrates released upon incubation at pH 7.4, 11.4, 5.1 and at 20%, 40% acetonitrile containing mixture. A solution of 2.5 μM d(GT)20 was included as standard. (B) Representative graphs of 260 nm UV absorbance of DNA filtrates as mentioned above. A solution of 1.275 μM d(GT)20 was included as representative of total DNA content of the PDN suspension calculated from ratiometric quantification. (C) Confocal microscopy images evaluating endosomal uptake of PDN. Cells were co-treated with PDN (magenta; left) and FITC labeled dextran (green; middle). Endosomal PDN uptake was demonstrated by colocalization both fluorescence in overlay with DIC image of the cell (right).
Endosomal uptake of PDN was demonstrated by co-localization of PDN with endosome-localizing FITC-dextran complex.29–31 Co-localization data showed approximately 45% of the PDN gets internalized by endocytosis (overlap coefficient ~0.7)32,33 (Figure 3, C). While about half of the PDN undergoes endocytosis, a significant fraction remains associated with the cell membrane as demonstrated by the co-localization of PDN with membrane localizing dye Bodipy in a circular peripheral pattern (Figure S6, supporting Information). Endosomes are acidic in nature and may promote PDN dissociation upon cellular internalization in addition to interactions with the hydrophobic lipid membrane. Increased acidity, associated with the tumor microenvironment raises the possibility that PDN may dissociate in tumor tissue. We performed confocal microscopy experiments with fluorescently-(6-FAM-) labeled DNA and evaluated DNA release upon PDN dissociation (Figure S5, Supporting Information). Confocal microscopy analysis demonstrated DNA dissociation with a co-localization coefficient 0.083 indicating less than 10% of total DNA co-localizes with porphyrin.
Light-dependent cytotoxicity of PDN
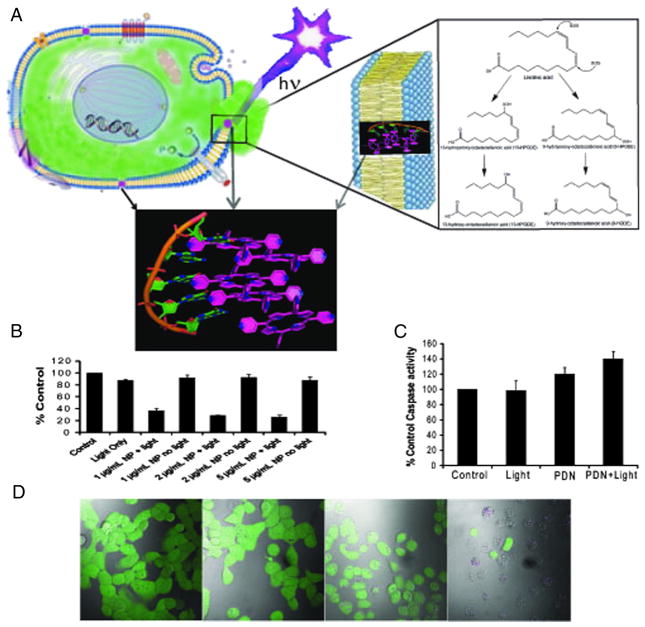
The cytotoxicity of PDN towards human bladder cancer cells was evaluated to determine to what extent these nano-sized complexes are cytotoxic towards malignant cells in a light-dependent manner (Figure 4 and Figures S8–S10, Supporting Information). ROS resulting from PDN dissociation upon interacting with the plasma membrane could, upon light stimulation, result in lipid peroxidation producing subsequent loss of membrane integrity and cell death (Figure 4, A). Cell viability assays demonstrated PDN are cytotoxic to bladder cancer cells in a light-dependent manner (Figure 4, B). Interestingly, the light-dependent cytotoxicity of PDN towards bladder cancer cells is relatively independent of PDN concentration consistent with a threshold level of membrane damage inducing cell death. The light-dependent cytotoxicity of PDN was accompanied by only a slight increase in apoptosis (Figure 4, C), with little DNA damage (Figure S14, Supporting Information) and no effect of z-VAD rescue consistent with cell death being predominantly non-apoptotic.
Figure 4.
PDN are cytotoxic to bladder cancer cells via localized damage to the plasma membrane as a consequence of light-activated oxidation of membrane lipids. (A) Depiction of a cancer cell with PDN localized to the plasma membrane and with light (hν) irradiation; inset displays an enlargement of the lipid bilayer of the plasma membrane with a PDN complex internalized in the membrane and shows further structures for the 9-HODE and 13-HODE lipid peroxidation products detected by mass spectrometry following PDN/light treatment. The green efflux reflects loss of calcein-AM dye resulting from membrane damage. (B) Cytotoxicity assay results showing PDN/light reduces bladder cancer viability. (C) Apoptosis assay results showing PDN-mediated cell death is predominantly non-apoptotic. (D) Confocal microscopy images displaying overlay of calcein-AM (green) fluorescence on DIC images for bladder cancer cells following treatment with (left – right) no-treatment, light-only, PDN-only, PDN/light. Treatment with PDN/light resulted in efflux of calcein-AM.
To gain further insight into the nature of cell death, bladder cancer cells were pre-treated with the cytoplasm-localizing dye calcein-AM and the effects of PDN and light on calcein retention were investigated. Treatment with PDN and light resulted in dye efflux for nearly all cells (Figure 4, D) while treatment with PDN-only or light-only did not stimulate dye efflux (Figure 4, D). The results indicate cell death is accompanied by a loss of membrane integrity. Interestingly, the free radical scavenger N-acetyl cysteine (NAC) could not rescue cells from the light-mediated cytotoxicity of PDN (Figure S10, Supporting Information). The results are consistent with PDN exerting localized cell damage that is not affected by the REDOX state of the cell.
In vitro heating and ROS production of PDN
Porphyrins are widely used for PDT34 in which absorbed light is used for production of cytotoxic ROS. The UV-Vis spectra for PDN indicated UV/Vis absorption was attenuated by ~2 fold relative to the same concentration of free MTP (Figure 2, A) while fluorescence for the PDN complex is quenched by ~25 fold (Figure 2, B) consistent with absorbed light being dissipated as heat and/or used for ROS production. The time- and concentration-dependence for heating of aqueous solutions of PDN was evaluated to determine whether temperature increases of a magnitude required for cell-killing could be induced (Figure S7, Supporting Information). PDN are not efficient transducers of heat although measurable temperature increases were detected upon photo-stimulation.
Cell death is mediated by oxidation of membrane lipids causing localized damage
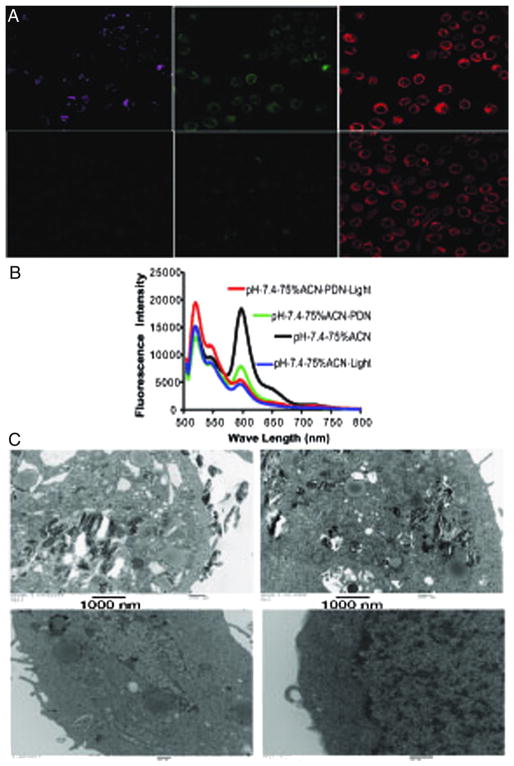
The light-dependent cytotoxic mechanism of PDN was further investigated using confocal microscopy, TEM, and mass spectrometry to determine if physical damage to the plasma membrane and endosomal compartments were important for PDN-mediated cell death (Figure 5 and Figure S11–13, Supporting Information). The nature of the observed membrane damage was investigated using the fluorescent membrane-localizing dye Bodipy35,36 to visualize the occurrence of oxidized lipids in the plasma membrane of bladder cancer cells treated with PDN and light (Figure 5, A). The dye undergoes a change in absorbance from red to green upon oxidation. The results demonstrate that treatment of bladder cancer cells with PDN and light, but not light-only, resulted in increased levels of oxidized lipids as indicated by the elevated level of green fluorescence (Figure 5, A). The ROS production by PDN upon light stimulation was further confirmed in vitro. Experimental results evidenced for PDN mediated C11-Bodipy oxidation occurred selectively in a light-dependent manner in solvent systems that favored PDN dissociation such as the presence of acetonitrile (Figure 5, B). TEM images revealed PDN were associated with the plasma membrane and internalized, likely in endosomes, and that substantial membrane damage and vacuole formation was observed proximal to nanoparticles in a light-dependent manner (Figure 5, C and S11, Supporting Information). Localized membrane damage, caused by PDN/light mediated oxidation, leads to necrotic cell death.37,38 The presence of oxidized lipids in the plasma membrane was investigated further using mass spectrometry. Mass spectrometry confirmed that elevated levels of 9-HODE and 13-HODE, products resulting from lipid peroxidation,39 occurred selectively in cells treated with PDN and light (Figure S12, S13). Together, these results indicate cell death following treatment with PDN and light occurred from localized membrane damage leading to a loss of plasma membrane integrity.
Figure 5.
PDN/Light treatment results in oxidation of plasma membrane lipids and localized membrane damage. (A) Confocal microscopy images displaying free porphyrin (left – magenta), oxidized membrane lipids detected with Bodipy (center – green), and non-oxidized membrane lipids detected with Bodipy (right-red). The top panels are from bladder cancer cells treated with PDN/light while the bottom panels are from light-only cells. (B) Representative fluorescence spectrum of dye c11 bodipy in presence of PDN + light (red); PDN (green); light only (blue) and control (black) with excitation 480 nm in pH 7.4 in 75% ACN + 25% aqueous solution mixture. (C) TEM images of bladder cancer cells treated with PDN/light (top-left), PDN-only (top-right), light-only (bottom-left), and no-treatment (bottom-right).
PDN display light-dependent antitumor activity in vivo
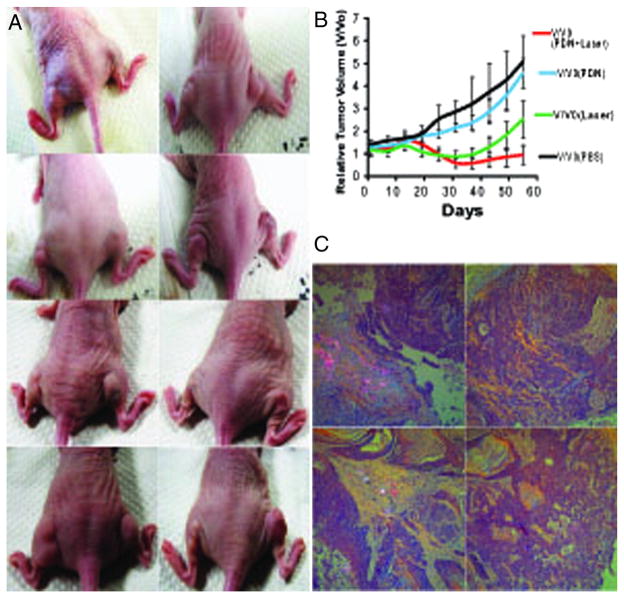
The light-dependent cytotoxicity of PDN towards bladder cancer cells in tissue culture invites the question of whether PDN can be used for light-dependent reduction or eradication of tumors in vivo. To address this question, we formed xenograft tumors bilaterally on the flanks of nude mice (Figure 6, A and Figure S15, Supporting Information). Initial tumor volumes were ~75 mm3 and there was no difference among the treatment groups at baseline. Tumors were treated by direct injection of either PDN or a saline solution. The tumor groups were further divided into light-treated and mock-treated groups to create four groups: i) no treatment; ii) light-only; iii) PDN-only; iv) PDN + light. Light treatment was achieved by inserting a fiber-optic cable into the tumor followed by treatment with 420 nm laser light for 3 min. Tumors that were not light-treated underwent a similar number of identical procedures introducing the fiber-optic cable without light exposure. The time-dependence of tumor growth and regression is shown in Figure 6, B. Beginning around day 20 tumors treated with either light or PDN + light displayed tumor regression while the untreated tumors began to display more rapid growth. Tumors treated with PDN and light displayed significant tumor regression that was superior to no treatment or treatment with PDN-only beginning on day 20 and achieving statistical significance at day 30 and persisting until the conclusion of the study (P < 0.05). Tumor regression following treatment with PDN and light was significantly greater than for light-only beginning on day 40 and achieving statistical significance at day 49 and persisting until completion of the study (P < 0.05). The results demonstrate that PDN display light-dependent anti-tumor activity in vivo.
Figure 6.
PDN/light treatment displays significant anti-tumor activity in vivo. (A) Representative tumors from the four treatment groups at day 0 (top row – prior to any treatment), day 1 (2nd row – after initial treatments), day 25 (3rd row – at the conclusion of light treatment), and day 55 (bottom row – end of study). The left mouse, left flank (L, L) received no treatment; the left mouse, right flank (L-R) received PDN-only. The right mouse, left flank (R, L) received PDN/light and the right mouse, right flank (R, R) received light-only. (B) Plot of relative volumes for bladder cancer xenografts over time following treatment with PDN/light (red), light-only (green), PDN-only (blue), no treatment (black). (C) Histopathological analysis of tumor tissue. Polarization detection as part of histological analysis for PDN in tissues excised from animals at the conclusion of the study receiving the following treatments: (top-left) – PDN/light; (top-right) – light-only; (bottom-left) – PDN-only; (bottom-right) – no treatment.
Histopathological analysis of tumor tissue following treatment with PDN
At the conclusion of the in vivo study, tumor tissue was removed from sacrificed mice and sectioned for morphologic evaluation. In all study groups, sections showed subcutaneous tumoral deposits comprised of urothelial carcinoma with high-grade cytology and partial keratinization. Tumor tissue from mice in the control group that received neither nanoparticle nor light showed only subcutaneous tumoral deposits. Polarization revealed no polarizable nanoparticles (Figure 6, C and Figure S16, Supporting Information) and there was minimal fibrosis, chronic inflammation, or reparative changes. Tumor tissue from mice treated with light-only revealed similar features as those mice in the control group. For mice in both the PDN-only and the PDN/light groups, polarization revealed the presence of nano-particles confirming the persistence of PDN at the site of injection for several weeks allowing multiple exposures from single administered dose (Figure 6, C and Figure S16, Supporting Information). For the PDN-only group, tumor was absent in areas where nanoparticle was present demonstrating PDN having a native mild tumor abortive effect even in the absence of light. For the PDN/light group, tumor was also absent in the areas where nanoparticle was present with marked fibrosis, chronic inflammation, and reparative changes. This supports a potent tumor abortive effect likely enhanced by light excitation; a result which already accentuates the native anti-tumor activity of PDN. Some areas of tumor not displaying polarizable nanoparticles, however, showed tumor re-growth consistent with incomplete penetration of nanoparticles. Another advantage appears that the range of killing activity is confined to zones around PDN deposition, allowing for precise margins with this therapeutic approach.
Discussion
The present studies demonstrate that discrete nano-sized particles can be obtained from non-covalent assembly of MTP and DNA under controlled conditions. Discrete nanoparticles do not form from MTP in the absence of DNA indicating DNA: porphyrin interactions are critical for PDN assembly. The ratio of porphyrin to DNA in PDN (~19:1) (mol/mol) is consistent with two nucleobases from each DNA 40-mer interacting with each porphyrin. Thus, approximately half the pyridyl side chains in each MTP (Figure 1, A) may be engaged in interactions with DNA nucleobases (Figure 1, B). The spectroscopic results are consistent with π – π stacking between pyridyl side chains and DNA nucleobases as being a principal driving force for nanoparticle assembly.40,41 PDN display quenching of porphyrin fluorescence and increased intensity and sharpness for Raman spectra similar to that observed upon surface-enhancement for free porphyrins. Dissociation of PDN was favored in the hydrophobic solvent system, consistent with PDN stability result from π – π stacking between PDN and DNA with aqueous solubility conferred by the phosphodiester backbone of DNA.42,43 The nature of PDN disassembly in the hydrophobic environment, releasing the DNA component, could be significant for DNA-based therapeutics delivery upon PDN dissociation in the hydrophobic milieu of cell membrane.
We have demonstrated that PDN cause cell death predominantly via localized membrane damage resulting in necrosis. While conventional photosensitizers can induce membrane damage upon light stimulation, a variety of other cellular organelles, including the mitochondria,44 are also potential sites for PDT-induced damage. Our studies demonstrate PDN localize to the plasma membrane (Figure S6, Supporting Information) and also undergo internalization via endocytosis (Figure 3, C). Upon interaction with cellular components the fluorescence of FAM-labeled DNA dissociates from porphyrin fluorescence (Figure S5, Supporting Information) consistent with PDN disassembly in the hydrophobic environment of cell membranes or vesicles. Photo-mediated damage by porphyrin results in loss of plasma membrane integrity and vacuole formation (Figure 4, D and C and S11, Supporting Information). The plasma membrane damage is caused by lipid oxidation (Figure 5, A and C and S11, Supporting Information) and specifically generation of the oxidized fatty acids 9-HODE and 13-HODE (Figure S12, S13, Supporting Information).
Importantly, PDN/light treatment of bladder cancer xenografts resulted in significant reduction in tumor volumes in vivo (Figure 6, A and B). Histopathological analysis of tumor tissue revealed, PDN/light displayed strong tumorpathic properties and elicited inflammatory and reparative changes associated with an immune response (Figure 6, C and S16, Supporting Information). No tumor tissue was identified in regions where PDN were localized indicating PDN/light treatment exerts powerful anti-tumor effects with very tight margins. Although mild anti-tumor activity was observed in case of PDN-only treatment probably due to basal level ROS production and immune response,45,46 this mild anti-tumor effect is not likely to be sufficient for the treatment of aggressively growing tumors in advanced cases.47,48 These results indicate PDN/light may be highly effective for cancer treatment in humans. Localized retention of the PDN provides the opportunity for multiple light exposures as required to eliminate remaining or re-growing malignant tissue. However, the concern of uniform delivery of PDN needs to be addressed to achieve a significant overall anti-tumor effect.
The present studies demonstrate the potential application of PDN for treatment of solid tumors via intra-tumoral injection.49–51 However, in future studies, cancer cell specific DNA or RNA aptamers52,53 could be conjugated to the PDN for molecular targeting upon systemic injection. PDN could be developed as multi-modality nanoparticles54 by replacing the presently used d(GT)20 with cytotoxic DNA like FdUMP[10]55 thus DNA-release may contribute to the overall therapeutic response. Presently performed experiments indicated PDN readily dissociate upon cellular internalization as a consequence of the hydrophobic environment in the cell membrane releasing DNA payload. However detailed, time-dependent DNA dissociation kinetics should be examined in future studies. Dissociated therapeutic nucleic acid, if used, may exert additional anti-tumor activity providing a therapeutic advantage not realizable using conventional photosensitizers or nanoparticles that do not include a DNA component. Importantly, PDN are biocompatible and elicited no apparent morbidity upon in vivo administration.56,57
Supplementary Material
Acknowledgments
Funding Resources: This work was supported by DOD PCRP 093606 (WHG) and NIH-NCI P30CA012197 (WHG).
The authors acknowledge Dr. Baidyanath Saha for helping with TEM image analysis; Chris Stuart for helping with in vivo experiments; Michael Samuel for helping with mass-spectrometry experiment; Dr. David A. Horita for helping with nano-particle model building; and WFU Department of Physics for use of laser facilities.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.nano.2013.07.019.
References
- 1.Gong X, Milic T, Xu C, Batteas JD, Drain CM. Preparation and characterization of porphyrin nanoparticles. J Am Chem Soc. 2002;124:14290–1. doi: 10.1021/ja027405z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang XL, Li XD, Yue XL, Dai ZF. Conjugation of porphyrin to nanohybrid cerasomes for photodynamic diagnosis and therapy of cancer. Angew Chem Int Edit. 2011;50:11622–7. doi: 10.1002/anie.201103557. [DOI] [PubMed] [Google Scholar]
- 3.Wang ZC, Medforth CJ, Shelnutt JA. Porphyrin nanotubes by ionic self-assembly. J Am Chem Soc. 2004;126:15954–5. doi: 10.1021/ja045068j. [DOI] [PubMed] [Google Scholar]
- 4.Drain CM, Smeureanu G, Patel S, Gong XC, Garno J, Arijeloye J. Porphyrin nanoparticles as supramolecular systems. New J Chem. 2006;30:1834–43. [Google Scholar]
- 5.De Napoli M, Nardis S, Paolesse R, Vicente MG, Lauceri R, Purrello R. Hierarchical porphyrin self-assembly in aqueous solution. J Am Chem Soc. 2004;126:5934–5. doi: 10.1021/ja0494757. [DOI] [PubMed] [Google Scholar]
- 6.Lauceri R, De Napoli M, Mammana A, Nardis S, Romeo A, Purrello R. Hierarchical self-assembly of water-soluble porphyrins. Synthetic Met. 2004;147:49–55. [Google Scholar]
- 7.Bellacchio E, Lauceri R, Gurrieri S, Scolaro LM, Romeo A, Purrello R. Template-imprinted chiral porphyrin aggregates. J Am Chem Soc. 1998;120:12353–4. [Google Scholar]
- 8.Anand S, Ortel BJ, Pereira SP, Hasan T, Maytin EV. Biomodulatory approaches to photodynamic therapy for solid tumors. Cancer Lett. 2012;326:8–16. doi: 10.1016/j.canlet.2012.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davila ML. Photodynamic therapy. Gastrointest Endosc Clin N Am. 2011;21:67–79. doi: 10.1016/j.giec.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Ethirajan M, Chen Y, Joshi P, Pandey RK. The role of porphyrin chemistry in tumor imaging and photodynamic therapy. Chem Soc Rev. 2011;40:340–62. doi: 10.1039/b915149b. [DOI] [PubMed] [Google Scholar]
- 11.Josefsen LB, Boyle RW. Unique diagnostic and therapeutic roles of porphyrins and phthalocyanines in photodynamic therapy, imaging and theranostics. Theranostics. 2012;2:916–66. doi: 10.7150/thno.4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Connor AE, Gallagher WM, Byrne AT. Porphyrin and nonporphyrin photosensitizers in oncology: preclinical and clinical advances in photodynamic therapy. Photochem Photobiol. 2009;85:1053–74. doi: 10.1111/j.1751-1097.2009.00585.x. [DOI] [PubMed] [Google Scholar]
- 13.Walton SP, Wu M, Gredell JA, Chan C. Designing highly active siRNAs for therapeutic applications. FEBS J. 2010;277:4806–13. doi: 10.1111/j.1742-4658.2010.07903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ananyan G, Avetisyan A, Aloyan L, Dalyan Y. The stability of DNA-porphyrin complexes in the presence of Mn(II) ions. Biophys Chem. 2011;156:96–101. doi: 10.1016/j.bpc.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Bennett M, Krah A, Wien F, Garman E, Mckenna R, Sanderson M, et al. A DNA-porphyrin minor-groove complex at atomic resolution: The structural consequences of porphyrin ruffling. P Natl Acad Sci USA. 2000;97:9476–81. doi: 10.1073/pnas.160271897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D’Urso A, Mammana A, Balaz M, Holmes AE, Berova N, Lauceri R, et al. Interactions of a tetraanionic porphyrin with DNA: from a Z-DNA sensor to a versatile supramolecular device. J Am Chem Soc. 2009;131:2046–7. doi: 10.1021/ja808099u. [DOI] [PubMed] [Google Scholar]
- 17.Geacintov NE, Ibanez V, Rougee M, Bensasson RV. Orientation and linear dichroism characteristics of porphyrin DNA complexes. Biochemistry. 1987;26:3087–92. doi: 10.1021/bi00385a021. [DOI] [PubMed] [Google Scholar]
- 18.Lipscomb LA, Zhou FX, Presnell SR, Woo RJ, Peek ME, Plaskon RR, et al. Structure of a DNA—Porphyrin complex. Biochemistry. 1996;35:2818–23. doi: 10.1021/bi952443z. [DOI] [PubMed] [Google Scholar]
- 19.Jain RK, Stylianopoulos T. Delivering nanomedicine to solid tumors. Nat Rev Clin Oncol. 2012;7:653–64. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda H. Macromolecular therapeutics in cancer treatment: The EPR effect and beyond. J Control Release. 2012;164:138–44. doi: 10.1016/j.jconrel.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 21.Plaetzer K, Krammer B, Berlanda J, Berr F, Kiesslich T. Photophysics and photochemistry of photodynamic therapy: fundamental aspects. Lasers Med Sci. 2009;24:259–68. doi: 10.1007/s10103-008-0539-1. [DOI] [PubMed] [Google Scholar]
- 22.Nemykin VN, Hadt RG. Interpretation of the UV-Vis spectra of the meso(ferrocenyl)-containing porphyrins using a TDDFT approach: is Gouterman’s classic four-orbital model still in play? J Phys Chem A. 2012;114:12062–6. doi: 10.1021/jp1083828. [DOI] [PubMed] [Google Scholar]
- 23.Lan MZH, Yuan H, Jiang C, Zuo S, Jiang Y. Absorption and EPR spectra of some porphyrins and metalloporphyrins. Dyes Pigments. 2007;74:357–62. [Google Scholar]
- 24.Cotton T, Schultz SG, van Duyne RP. Surface-enhanced resonance Raman scattering from water-soluble adsorbed on a silver electrode. J Am Chem Soc. 1982;104:6528–32. [Google Scholar]
- 25.Cunderlikova B, Kaalhus O, Cunderlik R, Mateasik A, Moan J, Kongshaug M. pH-dependent modification of lipophilicity of porphyrin-type photosensitizers. Photochem Photobiol. 2004;79:242–7. doi: 10.1562/0031-8655(2004)79<242:pmolop>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 26.Stepanek P, Andrushchenko V, Ruud K, Bour P. Porphyrin protonation studied by magnetic circular dichroism. J Phys Chem A. 2012;116:778–83. doi: 10.1021/jp2105192. [DOI] [PubMed] [Google Scholar]
- 27.Chandler D. Interfaces and the driving force of hydrophobic assembly. Nature. 2005;437:640–7. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 28.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–6. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 29.Bayer N, Schober D, Prchla E, Murphy RF, Blaas D, Fuchs R. Effect of bafilomycin A1 and nocodazole on endocytic transport in HeLa cells: implications for viral uncoating and infection. J Virol. 1998;72:9645–55. doi: 10.1128/jvi.72.12.9645-9655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lencer WI, Weyer P, Verkman AS, Ausiello DA, Brown D. FITC-dextran as a probe for endosome function and localization in kidney. Am J Physiol. 1990;258:C309–17. doi: 10.1152/ajpcell.1990.258.2.C309. [DOI] [PubMed] [Google Scholar]
- 31.Shurety W, Stewart NL, Stow JL. Fluid-phase markers in the basolateral endocytic pathway accumulate in response to the actin assembly-promoting drug Jasplakinolide. Mol Biol Cell. 1998;9:957–75. doi: 10.1091/mbc.9.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manders FJVEMM, Aten JA. Measurement of co-localization of objects in dual-colour confocal images. J Microsc. 1993;169:375–82. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 33.Zinchuk V, Zinchuk O, Okada T. Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: pushing pixels to explore biological phenomena. Acta Histochem Cytochem. 2007;40:101–11. doi: 10.1267/ahc.07002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allison RR, Sibata CH. Oncologic photodynamic therapy photosensitizers: a clinical review. Photodiagnosis Photodyn Ther. 2010;7:61–75. doi: 10.1016/j.pdpdt.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Drummen GP, van Liebergen LC, Op den Kamp JA, Post JA. C11-BODIPY(581/591), an oxidation-sensitive fluorescent lipid peroxidation probe: (micro)spectroscopic characterization and validation of methodology. Free Radic Biol Med. 2002;33:473–90. doi: 10.1016/s0891-5849(02)00848-1. [DOI] [PubMed] [Google Scholar]
- 36.Carlsen CU, Kurtmann L, Bruggemann DA, Hoff S, Risbo J, Skibsted LH. Investigation of oxidation in freeze-dried membranes using the fluorescent probe C11-BODIPY(581/591) Cryobiology. 2009;58:262–7. doi: 10.1016/j.cryobiol.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 37.Ahn WS, Bae SM, Huh SW, Lee JM, Namkoong SE, Han SJ, et al. Necrosis-like death with plasma membrane damage against cervical cancer cells by photodynamic therapy. Int J Gynecol Cancer. 2004;14:475–82. doi: 10.1111/j.1048-891x.2004.14308.x. [DOI] [PubMed] [Google Scholar]
- 38.Peeters S, Kitz M, Preisser S, Wetterwald A, Rothen-Rutishauser B, Thalmann GN, et al. Mechanisms of nanoparticle-mediated photomechanical cell damage. Biomed Opt Express. 2012;3:435–46. doi: 10.1364/BOE.3.000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Catala A. Lipid peroxidation of membrane phospholipids generates hydroxy-alkenals and oxidized phospholipids active in physiological and/or pathological conditions. Chem Phys Lipids. 2009;157:1–11. doi: 10.1016/j.chemphyslip.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 40.Acharya A, Ramanujam B, Mitra A, Rao CP. Nanofibers formed through pi…pi stacking of the complexes of glucosyl-C2-salicyl-imine and phenylalanine: characterization by microscopy, modeling by molecular mechanics, and interaction by alpha-helical and beta-sheet proteins. ACS Nano. 2010;4:4061–73. doi: 10.1021/nn1006286. [DOI] [PubMed] [Google Scholar]
- 41.Koren AB, Curtis MD, Francis AH, Kampf JW. Intermolecular interactions in pi-stacked conjugated molecules. Synthesis, structure, and spectral characterization of alkyl bithiazole oligomers. J Am Chem Soc. 2003;125:5040–50. doi: 10.1021/ja029216m. [DOI] [PubMed] [Google Scholar]
- 42.Kato Y, Inoue A, Niidome Y, Nakashima N. Thermodynamics on soluble carbon nanotubes: how do DNA molecules replace surfactants on carbon nanotubes? Sci Rep. 2012;2:733. doi: 10.1038/srep00733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Z, Kang H, Clarke ML, Lacerda SH, Zhao M, Fagan JA, et al. Water-soluble DNA-wrapped single-walled carbon-nanotube/quantum-dot complexes. Small. 2009;5:2149–55. doi: 10.1002/smll.200801932. [DOI] [PubMed] [Google Scholar]
- 44.Rapozzi V, Miculan M, Xodo LE. Evidence that photoactivated pheophorbide a causes in human cancer cells a photodynamic effect involving lipid peroxidation. Cancer Biol Ther. 2009;8:1318–27. doi: 10.4161/cbt.8.14.8628. [DOI] [PubMed] [Google Scholar]
- 45.Lavi R, Shainberg A, Friedmann H, Shneyvays V, Rickover O, Eichler M, et al. Low energy visible light induces reactive oxygen species generation and stimulates an increase of intracellular calcium concentration in cardiac cells. J Biol Chem. 2003;278:40917–22. doi: 10.1074/jbc.M303034200. [DOI] [PubMed] [Google Scholar]
- 46.Lavi R, Shainberg A, Shneyvays V, Hochauser E, Isaac A, Zinman T, et al. Detailed analysis of reactive oxygen species induced by visible light in various cell types. Lasers Surg Med. 2010;42:473–80. doi: 10.1002/lsm.20919. [DOI] [PubMed] [Google Scholar]
- 47.Gaffney DK, Winter K, Dicker AP, Miller B, Eifel PJ, Ryu J, et al. Efficacy and patterns of failure for locally advanced cancer of the cervix treated with celebrex (celecoxib) and chemoradiotherapy in RTOG 0128. Int J Radiat Oncol Biol Phys. 2007;69:111–7. doi: 10.1016/j.ijrobp.2007.02.050. [DOI] [PubMed] [Google Scholar]
- 48.Lin CS, Jen YM, Cheng MF, Lin YS, Su WF, Hwang JM, et al. Squamous cell carcinoma of the buccal mucosa: an aggressive cancer requiring multimodality treatment. Head Neck. 2006;28:150–7. doi: 10.1002/hed.20303. [DOI] [PubMed] [Google Scholar]
- 49.Ghosh S, Dutta S, Gomes E, Carroll D, D’Agostino R, Jr, Olson J, et al. Increased heating efficiency and selective thermal ablation of malignant tissue with DNA-encased multiwalled carbon nanotubes. ACS Nano. 2009;3:2667–73. doi: 10.1021/nn900368b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shikanov A, Shikanov S, Vaisman B, Golenser J, Domb AJ. Paclitaxel tumor biodistribution and efficacy after intratumoral injection of a biodegradable extended release implant. Int J Pharm. 2008;358:114–20. doi: 10.1016/j.ijpharm.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 51.Shikanov A, Shikanov S, Vaisman B, Golenser J, Domb AJ. Cisplatin tumor biodistribution and efficacy after intratumoral injection of a biodegradable extended release implant. Chemother Res Pract. 2011;2011:175054. doi: 10.1155/2011/175054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hrkach J, Von Hoff D, Mukkaram Ali M, Andrianova E, Auer J, Campbell T, et al. Preclinical development and clinical translation of a PSMA-targeted docetaxel nanoparticle with a differentiated pharmacological profile. Sci Transl Med. 2012;4:128ra39. doi: 10.1126/scitranslmed.3003651. [DOI] [PubMed] [Google Scholar]
- 53.Kasten BB, Liu T, Nedrow-Byers JR, Benny PD, Berkman CE. Targeting prostate cancer cells with PSMA inhibitor-guided gold nanoparticles. Bioorg Med Chem Lett. 2013;23:565–8. doi: 10.1016/j.bmcl.2012.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fabbro C, Ali-Boucetta H, Da Ros T, Kostarelos K, Bianco A, Prato M. Targeting carbon nanotubes against cancer. Chem Commun (Camb) 2012;48:3911–26. doi: 10.1039/c2cc17995d. [DOI] [PubMed] [Google Scholar]
- 55.Liao ZY, Sordet O, Zhang HL, Kohlhagen G, Antony S, Gmeiner WH, et al. A novel polypyrimidine antitumor agent FdUMP[10] induces thymineless death with topoisomerase I-DNA complexes. Cancer Res. 2005;65:4844–51. doi: 10.1158/0008-5472.CAN-04-1302. [DOI] [PubMed] [Google Scholar]
- 56.Fiorella AM, Callari L. Luigi Monsù Scolaro, Ludovico Valli and Salvatore Sortino Biocompatible nanoparticles of amphiphilic cyclodextrins entangling porphyrins as suitable vessels for light-induced energy and electron transfer. J Mater Chem. 2008;18:802–5. [Google Scholar]
- 57.Meenakshisundaram G, Eteshola E, Pandian RP, Bratasz A, Selvendiran K, Lee SC, et al. Oxygen sensitivity and biocompatibility of an implantable paramagnetic probe for repeated measurements of tissue oxygenation. Biomed Microdevices. 2009;11:817–26. doi: 10.1007/s10544-009-9298-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.