Abstract
Our understanding of congenital heart defects has been recently advanced by whole exome sequencing projects, which have identified de novo mutations in many genes encoding epigenetic regulators. Notably, multiple subunits of SWI/SNF chromatin-remodeling complexes have been identified as strong candidates underlying these defects because they physically and functionally interact with cardiogenic transcription factors critical to cardiac development, such as TBX5, GATA-4, and NKX2-5. While these studies indicate a critical role of SWI/SNF complexes in cardiac development and congenital heart disease, many exciting new discoveries have identified their critical role in the adult heart in both physiological and pathological conditions involving multiple cell types in the heart, including cardiomyocytes, vascular endothelial cells, pericytes, and neural crest cells. This review summarizes the role of SWI/SNF chromatinremodeling complexes in cardiac development, congenital heart disease, cardiac hypertrophy, and vascular endothelial cell survival. Although the clinical relevance of SWI/SNF mutations has traditionally been focused primarily on their role in tumor suppression, these recent studies illustrate their critical role in the heart whereby they regulate cell proliferation, differentiation, and apoptosis of cardiac derived cell lines.
Keywords: SWI/SNF, BRG1, BRM, BAF250a, cardiovascular, epigenetics
Introduction
Our traditional focus on the regulation of cardiac disease and congenital causes of cardiovascular disease involves a focus on the cell signaling pathways and downstream transcription factors involved. However, the emerging field of epigenetics is offering a new way to look how development, congenital cardiac disease, and the diseased heart is regulated through chromatin remodeling under different physiological and pathological conditions. Chromatin modeling and histone modifications play an essential role in cardiac development and disease, including pathologic cardiac hypertrophy. These chromatin remodeling complexes, including the SWI/SNF (switching defective/sucrose non-fermenting) complex, are comprised of 9-12 subunits, including ATPase subunits encoded by either brm (brahma) or brg1 (brahma-related gene 1) [1]. In this review, we discuss our current understanding of SWI/SNF complexes, their regulation of in congenital cardiac defects, cardiac development, and cardiac disease states. We then discuss new studies implicating, for the first time, their role in the maintenance of the healthy adult heart. The use of the new classes of drugs that regulate SWI/SNF associated histone acetylation, including histone deacetylase (HDAC) inhibitors, will be considered for their possible unintended affects in the cardiovascular system.
De Novo Mutations in Epigenetic Regulators Cause Congenital Heart Defects
Developmental cardiac defects represent the most common serious birth defects, affecting ~2% of newborns with abnormalities that can range from mild, where the effects might not be observed until adulthood, to severe, with immediate morbidity or mortality [2]. Congenital heart defects affect 1.35 million patients each year, and they are also identified in 10% of stillbirths [3], where they are presumed to be a common cause of fetal demise. The importance of genetics in congenital heart disease is supported by a growing list of genes that are mutated [4]. Genes encoding cardiogenic transcripton factors, such as tbx1, tbx5, tbx20, gata4, and gata6have received particular attention for good reason considering their important role in the transcriptional regulation of downstream target genes [4]. However, transcription factors generally function in combination with histone-modifying enzymes and chromatin-remodeling complexes to epigenetically regulate gene expression. For this reason, it stands to reason that many yet to be identified mutations occur in epigenetic regulators [5].
Indeed, the advent of high-throughput sequencing platforms has led to whole-exome sequencing projects that have recently identified mutations in many genes encoding histone-modifying enzymes and components of chromatin-remodeling complexes [6–13]. An important feature of these sequencing projects is that parent-offspring trios were sequenced in which the proband was a child with severe congenital cardiac defects who had no first-degree relatives with structural heart disease. Importantly, the mutations in these epigenetic regulatory genes occurred de novowhich implies that they have dosage-sensitive cardiac functions and explains why these genes were not identified in previous pedigree analyses. Unlike cardiogenic transcription factors, these epigenetic regulators are broadly expressed, interact with many diverse transcription factors to regulate hundreds to thousands of genes, and have pleiotropic functions both within and outside of the heart. As a result, a sub-set of heterozygous loss-of-function mutations result in severe cardiac defects that preclude the viability and fertility necessary for a pedigree to be established. Consequently, one might expect exome sequencing projects to continue identifying novel genes that cause congenital heart defects.
Mutations in SWI/SNF Subunits Result in Congential Syndromes that Include Cardiac Defects
Several independent exome sequencing projects have recently identified de novo mutations in 4 different SWI/SNF subunits in three congenital syndromes that include cardiac defects: Coffin-Siris syndrome (CSS), Nicolaides-Baraitser syndrome (NCBRS), and ARID1B-related intellectual disability (ID) syndrome [9–13]. Patients with CSS, NCBRS, and ID syndromes display a wide variety of symptoms including severe intellectual deficits and cardiac defects such as atrial/ventricular septal defects, patent ductus arteriosus (PDA), mitral and pulmonary atresia, mitral and tricuspid regurgitation, aortic stenosis, coarctation of the aorta, and single right ventricle [14].
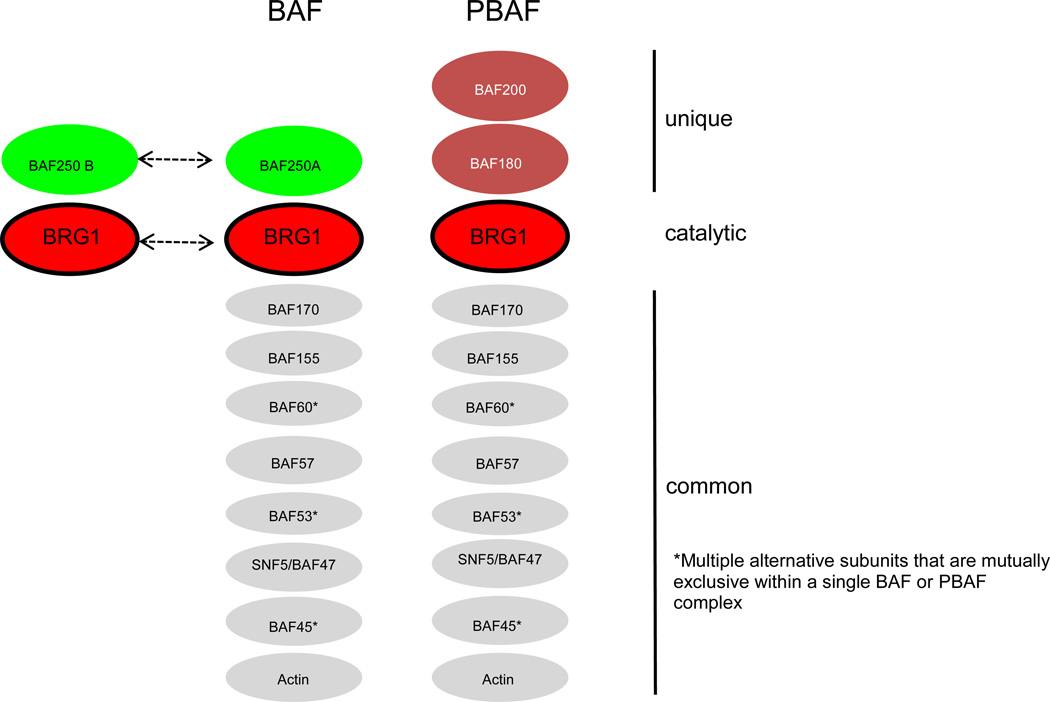
SWI/SNF chromatin-remodeling complexes consist of 9–12 subunits and are recruited by sequence-specific transcription factors to the promoters of numerous target genes, where they slide or evict nucleosomes near the transcripton start site (TSS) to regulate RNA Polymerase II occupancy and transcriptional initiation (Figures 1 and 2). Depending on whether a transcriptional activator or repressor recruits SWI/SNF, transcription can be upregulated or downregulated. Each SWI/SNF complex utilizes either BRG1 (also known as SMARCA4) or BRM (also known as SMARCA2) as alternative catalytic subunits with DNA-dependent ATPase activity [15]. The energy of ATP hydrolysis is harnessed to disrupt histone-DNA contacts and move nucleosomes away from the TSS or toward the TSS. BRG1 and BRM represent 2 of the 4 SWI/SNF subunits that are known to be mutated in CSS and NCBRS. The non-catalytic subunits of SWI/SNF are often referred to as BAFs (BRG1 or BRM associated factors with a number referring to the molecular mass of the protein). Each SWI/SNF complex contains a single ARID (AT-rich interacting domain)-containing subunit. SWI/SNF complexes are subdivided into BAF and PBAF complexes based on their catalytic and ARID subunits (Figure 1). BAF complexes are catalyzed by either BRG1 or BRM and incorporate either BAF250a or BAF250b (also known as ARID1a and ARID1b, respectively), whereas PBAF complexes are exclusively catalyzed by BRG1 and incorporate BAF200 (also known as ARID2). The ARID subunits are arguably the next best understood subunits within SWI/SNF complexes. Each ARID subunit can bind to DNA in a non-specific manner and is believed to influence SWI/SNF recruitment by physically associating with different transcription factors [16, 17]. BAF250a and BAF250b are the other 2 subunits mutated in CSS and NCBRS, and BAF250b is also mutated in ID syndrome. The clinical importance of the catalytic and ARID-containing subunits is underscored by the observation that BRG1, BRM, BAF250a, and BAF250b are important tumor-suppressor genes that are recurrently mutated or silenced in a variety of human primary tumors [11, 18].
Figure 1. Mammalian SWI/SNF chromatin-remodeling complexes are subdivided into BAF and PBAF complexes.
BAF complexes are defined by either BAF250A or BAF250B as the ARID-containing subunit (light green ovals), which are mutually exclusive (as denoted by dashed arrow with dual arrowheads), while PBAF complexes utilize BAF200 (blue oval) as the ARID-containing subunit. PBAF complexes also incorporate BAF180 (blue oval), which has 6 bromodomains that can bind to acetylated histones. BAF complexes are catalyzed by either BRG1 or BRM (red ovals), but no single complex contains both as denoted by dashed arrow with dual arrowheads. In contrast, PBAF is catalyzed exclusively by BRG1 (red oval). A number of other common subunits (BAFs for BRG1 or BRM associated factors) are also present (gray ovals from largest to smallest with numbers referring to protein size in KDa), and some of these subunits are represented by multiple isoforms (as indicated by asterisks) that are encoded by separate genes although only 1 isoform is present in a single complex similar to BAF250A-BAF250B and BRG1-BRM.
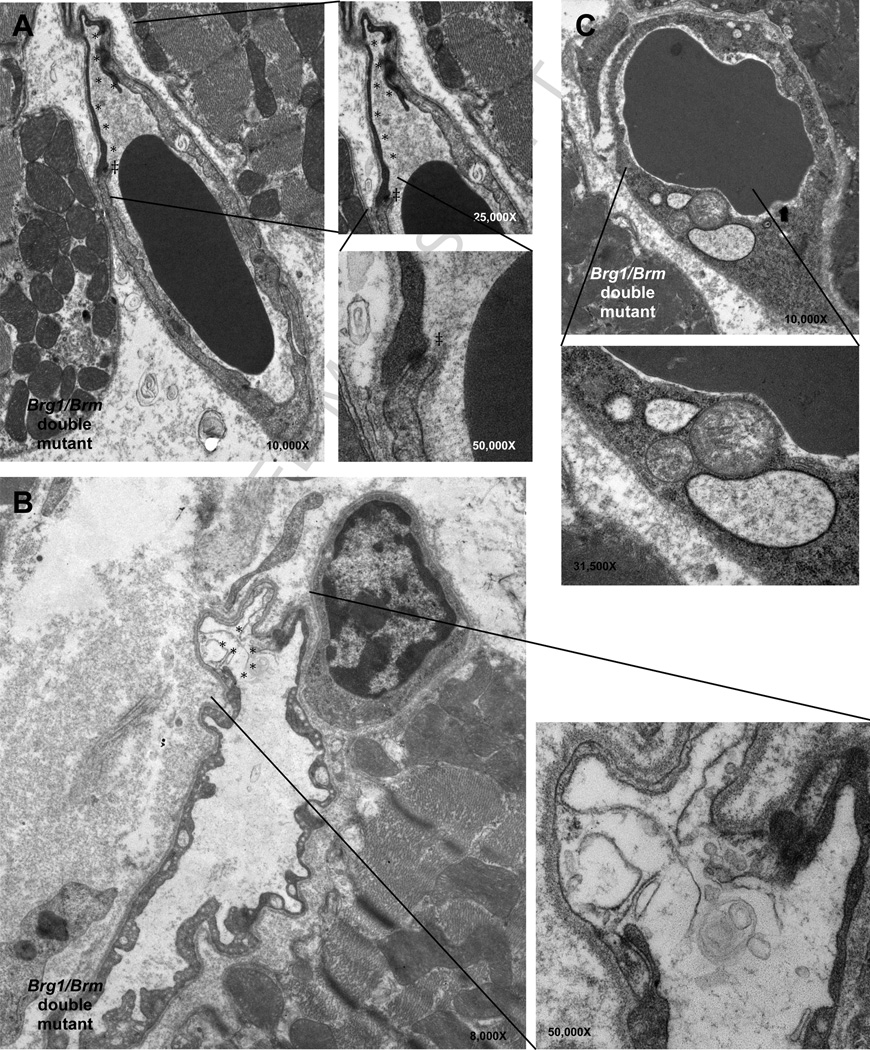
Figure 2. Functional compensation of BRG1 and BRM in adult vascular endothelial cells of the heart.
Transmission electron micrographs reveal 2 ultrastructural defects of cardiac endothelial cells in many of the capillary vessel walls 25 days after temporal induction of the Brg1 mutation. A and B, Dying endothelial cells are present in the capillaries of treated BRG1/BRM double mutant mice (asterisks). Throughout the cardiac capillary bed, gap junctions between endothelial cells were intact between healthy cells as well as between affected cells. Note that even between viable and injured cells the gap junctions are intact (double-daggers). C, Vascular endothelial cells from treated BRG1/BRM double-mutant mice have swollen mitochondria (white daggers) and rough endoplasmic reticulum (black daggers), which are characteristic changes found in cell death. A and C correspond to the epicardium; B corresponds to the endocardium. Reprinted from: Willis, et al., 2012 [41], with permission.
Congenital cardiac defects can arise from abnormal development of cardiomyocytes as well as vascular endothelial cells, pericytes (i.e., a distinct population of vascular smooth muscle cells), and neural crest-derived structures such as semilunar valves and the septum. It is noteworthy that SWI/SNF complexes play an important role in each of these lineages based on gene-targeting experiments in mice. For example, constitutive null mutations of Baf180 and Baf60c result in mid-gestation lethality due to cardiac defects. Constitutive null mutations of Brg1, Baf250a/Arid1a, Baf155and Baf47/Snf5 result in even earlier lethality at the peri-implantation stage. Although this early embryonic lethality occurs before the onset of cardiac development, Cre/loxP conditional mutations have established a role in cardiovascular development. The sections that follow will summarize this work for each cell lineage and emphasize mechanistic insights that are clinically relevant.
BRG1 and cardiomyocyte development
Brg1 genetically interacts with tbx5 [19], and the BAF60c subunit facilitates a physical interaction between brg1 and the cardiogenic transcription factors TBX4, GATA4, and NKX2-5 [20]. Furthermore, increased expression of BAF60c plus TBX5 and GATA4 can program non-cardiac mesoderm to become beating cardiomyocytes [21]. Based on these findings, it is not surprising that BRG1 and SWI/SNF complexes play an important role in cardiomyocyte development. Experimentally, a conditional mutation of Brg1 in myocardial progenitors within the secondary heart field resulted in hypoplastic right ventricles and outflow tracts, which culminated in embryonic lethality at E10.5. A second Brg1 conditional mutation in the myocardium resulted in a thin myocardium, lack of a septum, and embryonic lethality at E11.5 [22]. This phenotype may be explained, in part, by BRG1 activating Bmp10 mRNA levels to promote myocardial proliferation. A third conditional mutation of Brg1which was restricted to the developing endocardium, also resulted in embryonic lethality at E10.5-11.5 [23]. These endocardium mutants exhibited hypotrabeculation and a lack of cardiac jelly. This work revealed a role for BRG1 in the transcriptional repression of Adamts1which encodes an extracellular protease, to promote myocardial trabeculation and prevent the premature degradation of cardiac jelly.
BAF- and PBAF-specific subunits in cardiomyocyte development
Because BRG1 catalyzes both BAF and PBAF complexes, the aforementioned Brg1 conditional mutations cannot provide insight into the relative importance of these two categories of SWI/SNF complexes. To evaluate BAF versus PBAF functions, it is necessary to analyze mutations in subunits that are specifically incorporated into BAF or PBAF complexes but not both. As mentioned above, BAF250a is exclusively incorporated into BAF complexes. A conditional mutation of Baf250a in myocardial progenitors within the secondary heart field caused a phenotype similar to Brg1 mutants with delayed development of the right ventricle, decreased trabeculation, defective formation of the ventricular septum and outflow tract, and embryonic lethality around E13 [24].
By mutating Baf250a in ES cells, it was shown that BAF250a is crucial for the differentiation of cardiac progenitor cells into mature cardiomyocytes [24]. Gene expression was altered with decreased Bmp10, ErbB2and ErbB4 mRNA levels, which, in combination, may have contributed to the decreased trabeculation and myocardial development [24–28]. Nkx2.5 and Mef2c mRNA levels were also decreased, and this suggests that SWI/SNF might be part of an autoregulatory loop to maintain the expression of cardiogenic transcription factors. ChIP assays demonstrated that BRG1 occupancy at the Nkx2.5, Mef2c and Bmp10 promoters was diminished in Baf250a mutant cells, which suggests that BRG1 is recruited to these sites in a BAF250a-dependent manner. Consistent with the idea that SWI/SNF complexes alter chromatin structure, DNase I hypersensitivity assays showed that decreased expression of target genes in Baf250a mutant cells correlated with a more “closed” chromatin configuration (i.e., indicative of higher nucleosome density) of the corresponding promoter regions. [24].
A point mutation in the ARID domain of BAF250a has also been reported that behaves as a hypomorph with similar cardiac defects including a lack of ventricular trabeculation and ventricular septal defects with mid-gestation lethality at E10.5-12.5 [16]. This work also suggests that BAF250a is important for SWI/SNF recruitment to target genes including thrombospondin 1 (Thbs1). This is an interesting target gene because the thrombospondin family has been implicated as cardioprotective factors in numerous incidences of cardiovascular pathology [29, 30].
In contrast to BAF250a, BAF180 as well as BAF200 are unique to PBAF complexes. BAF180 does not have an ARID domain but instead has 6 bromodomains that bind to acetylated histones. This is believed to influence the recruitment of PBAF complexes (which stands for polybromo BAF complexes because of BAF180) such that it has different targets than BAF complexes. As a result, PBAF complexes can potentiate the transcriptional activation mediated by certain nuclear receptors such as RXRα, VDR and PPARγ. Constitutive null mutants of Baf180 have a variety of cardiac defects including hypoplastic ventricles and ventricular-septal defects with embryonic lethality occurring at E12.5-E15.5 [31, 32]. Consistent with PBAF cooperating with RXRα, a number of retinoic acid target genes such as RXRβ2 and CRABPII were altered in Baf180 mutants. Therefore, both BAF and PBAF complexes are required for cardiomyocyte development although additional work will be necessary to understand how they are different functionally.
BAF60c and cardiomyocyte development
Although BAF60c is one of three alternative BAF60 subunits, it is expressed in a tissue-specific manner with high levels in the developing heart [20]. As a result, it is the predominant BAF60 subunit in both BAF and PBAF complexes. An RNAi-mediated approach was used to perturb BAF60c in all embryonic tissues, and this resulted in a variety of cardiac defects including abnormal looping, hypoplastic ventricles, hypotrabeculation, and a shortened outflow tract with lethality at E10-11 [20]. BAF60c activates Notch and Nodal signaling pathways, which is believed to be important for the looping phenotype in Baf60c knockdown embryos. BAF60c is also noteworthy because it interacts with Tbx5, Nkx2.5, and GATA-4, and increased expression of BAF60c, TBX5, and GATA4 in non-cardiac mesoderm can drive their differentiation into beating cardiomyocytes[21].
Considering that BRG1, BAF250a, BAF180, and BAF60c are subunits of SWI/SNF complexes, it is reassuring that there is so much overlap among the mutant phenotypes. It is also reassuring that the defects that have been observed, such as defective outflow tract remodeling, is similar to what is observed in human congenital cardiac defects. Cre/loxP conditional mutations have also been used to investigate the role of BRG1 in the development of vascular endothelial cells, pericytes, and neural crest derived cardiac structures that when perturbed contribute to human congenital cardiac defects. The role of BRG1 in each of these cell types is discussed below.
Vascular endothelial cells
A Tie2-Cre conditional mutation of Brg1 resulted in embryonic lethality at E11.5. Although a defect in erythropoiesis is believed to be the cause of death, there were widespread defects in VEC development within the yolk sac [33]. The mutant vessels were thin, disconnected, and did not sprout/regress properly. Vasculogenesis was relatively normal in the embryo proper, including the developing heart, but the early embryonic lethality may have precluded these defects from being observed at a slightly later stage. Characterization of the mutant phenotype revealed dual roles for BRG1 in Wnt signaling [34, 35], which is consistent with several other reports implicating SWI/SNF in Wnt signaling, and that BRG1 also promotes COUP-TFII expression and venous specification during development [36]. Yolk sac angiogenesis defects have also been observed in Baf155 constitutive mutants that develop beyond the peri-implantation stage because of partial rescue from a Baf155 transgene [37].
In addition to having cardiomyocyte defects, Baf180 constitutive mutants have defective coronary vessel development around the ventricle and within the myocardium [31, 32]. These vessels are derived from epicardial cells, and the coronary vessel defects are proposed to be due to a problem with the epithelial-to-mesenchymal transition and the migration of epicardial cells.
Pericytes
A conditional mutation of Brg1 in smooth muscle resulted in a subset of the mutants dying during the early postnatal stage due to cardiopulmonary defects [38]. The mutants exhibited patent ductus arteriosus (PDA), ventricular septal derfects, and had dilated cardiac chambers consistent with left-right shunt and volume overload congestive heart failure. This phenotype is consistent with in vitro experiments demonstrating that BRG1 and BRM are required for maintaining the expression of several smooth muscle specific protein-coding genes and microRNAs [39, 40]. This transcriptional regulatory mechanism was mediated, in part, by enabling myocardin to increase serum response factor (SRF) at the target genes.
A critical component of the vascular remodeling that occurs in the pathophysiology of vascular diseases include the phenotypic transition of vascular smooth muscle cells (VSMCs) to proliferate, migrate, and invade [41–43]. The gasotransmitter H2S exerts a wide range of control on the proliferation and survival of VSMCs through the MAPK pathway [44, 45]. Recent studies have identified that BRG1 is crucial for H2S-induced inhibition of vascular smooth muscle cell proliferation using overexpression and knockdown of BRG1 [46]. Interestingly, H2S in return inhibits BRG1 transcription and expression by preventing its recruitment to the promoters of BRG1-regulated genes, including Pcna, Ntf3, and Pdgfα [46]. By altering the chromatic remodeling in vascular smooth muscle cells, H2S inhibits BRG1 regulation, altering the phenotype that would be expected in disease.
Neural crest
Neural crest cells originate from the dorsal neural tube and migrate to many sites in the developing embryo where they differentiate into diverse cell types including cardiovascular structures. For example, neural crest cells contribute to the cardiac outflow tract, vascular smooth muscle cells, and the muscular septum of the aorta and pulmonary trunk. A conditional mutation of Brg1 in neural crest cells resulted in embryonic lethality at E11.5-12.5, and these mutants had abnormal vascular development and shortened outflow tracts [47]. This phenotype was associated with aberrant apoptosis and cell proliferation, and BRG1 suppressed apoptosis signal regulating kinase 1 (Ask1) and the cycle-dependent kinase inhibitor p21. BRG1 also promoted myosin heavy chain 11 (Myh11) to differentiate neural crest cells into vascular smooth muscle cells. Finally, the Brg1 neural crest phenotype resembled the congential heart defects observed in human CHARGE syndrome. CHARGE syndrome is due to mutations in the CHD7 chromatin remodeler [7], and BRG1 and CHD7 both bind to the PlexinA2 promoter and co-regulate its expression, which encodes a semaphoring to guide neural crest cells to the outflow tract [47].
BRG1 and pathological cardiac hypertrophy
BRG1 expression is increased in cardiomyocytes in response to cardiac stress [22], and this finding parallels another study that reported increased expression of BRG1, BAF180, and BAF60c during the cardiac hypertrophy [48]. However, all of the mouse models described above conferred embryonic lethality, which precluded a functional assessment of SWI/SNF in this process. Therefore, to circumvent embryonic lethality, Brg1 was inducibly mutated in the myocardium of adult mice, and this revealed a crucial role in cardiac hypertrophy relevant to human heart disease [22]. The temporally-induced Brg1 mutants were viable and appeared normal under basal conditions. However, when transverse aortic constriction (TAC banding) was use to pressure overload the heart and induce cardiac hypertrophy, the mutants exhibited minor hypertrophy compared to controls.
The normal response to hypertrophy involves extensive transcriptional and metabolic reprogramming to a fetal-like state. Transcriptionally, the cardiogenic transcription factors, which are normally expressed only during development, are re-expressed during hypertrophy, and there is also a switch in myosin heavy chain (MHC) isoforms from α-MHC to β-MHC. Metabolically, cardiomyocytes modulate their primary use of fatty acids as their preferred energy source and revert to using glucose to generate ATP. Considering that there is not a cardiac stem population capable of significant regeneration, these transcriptional and metabolic changes may represent an attempt by cardiomyocytes to switch from a quiescent, adult-type state to a proliferative, fetal-like state for the purpose of regeneration. Not only was BRG1 shown to be required for hypertrophy, but BRG1 has been found to occupy the MHC promoters to regulate their expression [22]. BRG1 formed a complex with PARP1 and several histone deacetylases (HDACs) to induce the α- to β-MHC switch in hearts from TAC-banded mice. These findings are clinically relevant to humans because certain patients with hypertrophic cardiomyopathy had BRG1 levels that were correlated with disease severity and changes in myosin heavy chain (MHC) isoforms [22].
Functional compensation of homologous SWI/SNF subunits in post-natal homeostasis
While BAF and PBAF complexes are defined by their catalytic and ARID-domain containing subunits (Figure 1), the BAF complexes can be further subdivided. These complexes are catalyzed by either BRG1 or BRM and contain either BAF250a or BAF250b (Figure 1). Although the BRG1-BRM and BAF250a-BAF250b homolog pairs are highly conserved (75% and 50% amino-acid identity, respectively), they are functionally distinct. BRG1 and BAF250a are essential for embryonic development [49, 50], whereas BRM and BAF250b are dispensable since homozygotes are viable and fertile [32, 51].
The difference between BRG1 and BRM may be due to divergent N-termini, which influences the transcription factors they can physically interact with and the target genes they are recruited to [52]. Another possibility is that BRG1 is known to be expressed at higher levels than BRM in the early embryo so BRG1 may compensate for the loss of BRM but not vice versa [50, 53]. However, the gap between BRG1 and BRM expression levels does not apply to many differentiated tissues, and BRM may therefore be important for post-natal cardiovascular function and homeostasis. This is, in fact, the case for vascular endothelial cells. Brg1 is required for the development of this lineage in embryos, whereas Brm is not. Furthermore the Brg1 mutant phenotype is not exacerbated by BRM deficiency[33, 35]. Yet an inducible mutation of Brg1 in vascular endothelial cells did not confer an obvious phenotype in adult mice. This lack of a mutant phenotype was due to Brm redundancy because when the same Brg1 mutation was induced on a Brm-deficient background, the double mutants exhibited a vascular endothelial cell defect most evident in the heart (Figure 2), resulting in death within 1 month [54]. Mechanistically, the absence of BRG1 and BRM in VECS resulted vascular leakage, cardiac hemorrhage, and secondary cardiomyocyte cell death due to ischemia and ventricular dissections were identified as the underlying pathogenesis (Figure 2) [54].
Conclusions
As our understanding of new ways to regulate transcription factors and gene expression expands, so does our understanding of cardiac development, congenital heart disease, cardiac disease and the maintenance of the healthy heart. In this review, we summarized the emerging field of epigenetic regulation of transcription factors by chromatin remodeling and histone modification by the ATP-dependent SWI/SNF (switching defective/sucrose non-fermenting) complex, comprised of ATPase subunits encoded by either BRG1 (brahma-related gene 1) or BRM (brahma). While these subunits have been implicated in cardiac development by regulating transcription factors, mutations in these subunits can result in human congenital heart disease because of defects in many parallel transcription factors including TBX5 and GATA4 (see Table 1). A growing appreciation of BRG1 and BRM in the development of heart failure and the maintenance of pericytes and vascular endothelial cells in the healthy heart illustrate the important of SWI/SNF in maintaining an intact cardiovascular system for survival. While we are beginning to understand the specific transcription factors and genes regulated by different combinations of SWI/SNF complexes in vivoour understanding of the ATPase subunits BRG1 and BRM by miRNA and post-translational modification by ubiquitin and SUMO is in its infancy [55, 56]. Since human SWI/SNF complexes interacts with HDAC proteins with histone deacetylase activities (HDAC) [57], care should be taken with the increased used of HDAC inhibitors as cancer therapies for their unintended effects on cardiac function maintenance, in light of recent studies implicating the importance of BRG1 and BRM in the adult heart experimentally [58].
Table 1.
SWI/SNF subunits with cardiovascular phenotypes based on mouse knockout studies
| Subunit | Cardiomyocytes | Vascular Endothelial Cells |
Pericytes | Neural Crest |
|---|---|---|---|---|
| BRG1 | Hypoplastic right ventricle, thin myocardium Shortened outflow tract Lack of septum Hypotrabeculation Lack of cardiac jelly |
Yolk sac defects Coronary vessel death in adults if also BRM deficient |
Patent ductus arteriosus Ventricular -septal defects Dilated cardiac chambers |
Shortened outflow tracts Vascular defects |
| BAF250A | Developmental delay of right ventricle Defective formation of outflow tract Defective formation of ventricular septum Hypotrabeculation Cardiac progenitor cell differentiation in vitro |
N.D. | N.D. | N.D. |
| BAF180 | Hypoplastic ventricles Defective formation of ventricular septum |
Coronary vessel defects | N.D. | N.D. |
| BAF60C | Hypoplastic ventricles Shortened outflow tract Hypotrabeculation Abnormal looping |
N.D. | N.D. | N.D. |
| BAF155 | Yolk sac defects | N.D. | N.D. |
N.D. Does not necessarily infer there is no function for that cell type but can instead reflect either a lack of the appropriate conditional tissue-specific conditional mutation or insufficient phenotypic analysis.
Acknowledgements
The authors thank Sarah Edwards for her editorial assistance with this manuscript.
Non-Standard Abbreviations
- BAF
BRG1/BRM-associated factor
- BRG1
brahmarelated gene 1
- BRM
brahma
- SMC
smooth muscle cell
- SWI/SNF
mating type, switching/sucrose non-fermenting
- VEC
vascular endothelial cell
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None.
References
- 1.Han P, Hang CT, Yang J, Chang CP. Chromatin remodeling in cardiovascular development and physiology. Circulation research. 2011;108:378. doi: 10.1161/CIRCRESAHA.110.224287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annual review of physiology. 2012;74:41. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman JI. Incidence of congenital heart disease: II. Prenatal incidence. Pediatric cardiology. 1995;16:155. doi: 10.1007/BF00794186. [DOI] [PubMed] [Google Scholar]
- 4.Fahed AC, Gelb BD, Seidman JG, Seidman CE. Genetics of congenital heart disease: the glass half empty. Circulation research. 2013;112:707. doi: 10.1161/CIRCRESAHA.112.300853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Y, Kim J, Yuan X, Braun T. Epigenetic modifications of stem cells: a paradigm for the control of cardiac progenitor cells. Circulation research. 2011;109:1067. doi: 10.1161/CIRCRESAHA.111.243709. [DOI] [PubMed] [Google Scholar]
- 6.Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nature genetics. 2004;36:955. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 8.Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nature genetics. 2010;42:790. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosho T, Okamoto N, Ohashi H, Tsurusaki Y, Imai Y, Hibi-Ko Y, et al. Clinical correlations of mutations affecting six components of the SWI/SNF complex: detailed description of 21 patients and a review of the literature. American journal of medical genetics Part A. 2013;161:1221. doi: 10.1002/ajmg.a.35933. [DOI] [PubMed] [Google Scholar]
- 10.Tsurusaki Y, Okamoto N, Ohashi H, Kosho T, Imai Y, Hibi-Ko Y, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nature genetics. 2012;44:376. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- 11.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nature reviews Genetics. 2013;14:347. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santen GW, Aten E, Sun Y, Almomani R, Gilissen C, Nielsen M, et al. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nature genetics. 2012;44:379. doi: 10.1038/ng.2217. [DOI] [PubMed] [Google Scholar]
- 13.Van Houdt JK, Nowakowska BA, Sousa SB, van Schaik BD, Seuntjens E, Avonce N, et al. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nature genetics. 2012;44:445. doi: 10.1038/ng.1105. [DOI] [PubMed] [Google Scholar]
- 14.Kosho T, Kuniba H, Tanikawa Y, Hashimoto Y, Sakurai H. Natural history and parental experience of children with trisomy 18 based on a questionnaire given to a Japanese trisomy 18 parental support group. American journal of medical genetics Part A. 2013;161A:1531. doi: 10.1002/ajmg.a.35990. [DOI] [PubMed] [Google Scholar]
- 15.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandler RL, Brennan J, Schisler JC, Serber D, Patterson C, Magnuson T. ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Molecular and cellular biology. 2013;33:265. doi: 10.1128/MCB.01008-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagl NG, Jr, Wang X, Patsialou A, Van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. The EMBO journal. 2007;26:752. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nature reviews Cancer. 2011;11:481. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 19.Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguin P, Holloway AK, et al. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nature communications. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nature genetics. 2013;45:592. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI, et al. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Developmental cell. 2008;14:298. doi: 10.1016/j.devcel.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei I, Gao X, Sham MH, Wang Z. SWI/SNF protein component BAF250a regulates cardiac progenitor cell differentiation by modulating chromatin accessibility during second heart field development. The Journal of biological chemistry. 2012;287:24255. doi: 10.1074/jbc.M112.365080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, Shi S, Acosta L, Li W, Lu J, Bao S, et al. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development. 2004;131:2219. doi: 10.1242/dev.01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee KF, Simon H, Chen H, Bates B, Hung MC, Hauser C. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature. 1995;378:394. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- 27.Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, Klein R, et al. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature. 1995;378:390. doi: 10.1038/378390a0. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Bressan M, Hassel D, Huisken J, Staudt D, Kikuchi K, et al. A dual role for ErbB2 signaling in cardiac trabeculation. Development. 2010;137:3867. doi: 10.1242/dev.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stenina-Adognravi O. Thrombospondins: old players, new games. Current opinion in lipidology. 2013 doi: 10.1097/MOL.0b013e3283642912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynch JM, Maillet M, Vanhoutte D, Schloemer A, Sargent MA, Blair NS, et al. A thrombospondin-dependent pathway for a protective ER stress response. Cell. 2012;149:1257. doi: 10.1016/j.cell.2012.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang X, Gao X, Diaz-Trelles R, Ruiz-Lozano P, Wang Z. Coronary development is regulated by ATP-dependent SWI/SNF chromatin remodeling component BAF180. Developmental biology. 2008;319:258. doi: 10.1016/j.ydbio.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Zhai W, Richardson JA, Olson EN, Meneses JJ, Firpo MT, et al. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes & development. 2004;18:3106. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffin CT, Brennan J, Magnuson T. The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development. 2008;135:493. doi: 10.1242/dev.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curtis CD, Griffin CT. The chromatin-remodeling enzymes BRG1 and CHD4 antagonistically regulate vascular Wnt signaling. Molecular and cellular biology. 2012;32:1312. doi: 10.1128/MCB.06222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffin CT, Curtis CD, Davis RB, Muthukumar V, Magnuson T. The chromatinremodeling enzyme BRG1 modulates vascular Wnt signaling at two levels. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2282. doi: 10.1073/pnas.1013751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis RB, Curtis CD, Griffin CT. BRG1 promotes COUP-TFII expression and venous specification during embryonic vascular development. Development. 2013;140:1272. doi: 10.1242/dev.087379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han D, Jeon S, Sohn DH, Lee C, Ahn S, Kim WK, et al. SRG3, a core component of mouse SWI/SNF complex, is essential for extra-embryonic vascular development. Developmental biology. 2008;315:136. doi: 10.1016/j.ydbio.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 38.Zhang M, Chen M, Kim JR, Zhou J, Jones RE, Tune JD, et al. SWI/SNF complexes containing Brahma or Brahma-related gene 1 play distinct roles in smooth muscle development. Molecular and cellular biology. 2011;31:2618. doi: 10.1128/MCB.01338-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou J, Zhang M, Fang H, El-Mounayri O, Rodenberg JM, Imbalzano AN, et al. The SWI/SNF chromatin remodeling complex regulates myocardin-induced smooth muscle-specific gene expression. Arteriosclerosis, thrombosis, and vascular biology. 2009;29:921. doi: 10.1161/ATVBAHA.109.187229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen M, Herring BP. Regulation of microRNAs by Brahma-related gene 1 (Brg1) in smooth muscle cells. The Journal of biological chemistry. 2013;288:6397. doi: 10.1074/jbc.M112.409474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerthoffer WT. Mechanisms of vascular smooth muscle cell migration. Circulation research. 2007;100:607. doi: 10.1161/01.RES.0000258492.96097.47. [DOI] [PubMed] [Google Scholar]
- 42.Lacolley P, Regnault V, Nicoletti A, Li Z, Michel JB. The vascular smooth muscle cell in arterial pathology: a cell that can take on multiple roles. Cardiovascular research. 2012;95:194. doi: 10.1093/cvr/cvs135. [DOI] [PubMed] [Google Scholar]
- 43.Korshunov VA, Schwartz SM, Berk BC. Vascular remodeling: hemodynamic and biochemical mechanisms underlying Glagov's phenomenon. Arteriosclerosis, thrombosis, and vascular biology. 2007;27:1722. doi: 10.1161/ATVBAHA.106.129254. [DOI] [PubMed] [Google Scholar]
- 44.Yang G, Cao K, Wu L, Wang R. Cystathionine gamma-lyase overexpression inhibits cell proliferation via a H2S-dependent modulation of ERK1/2 phosphorylation and p21Cip/WAK-1. The Journal of biological chemistry. 2004;279:49199. doi: 10.1074/jbc.M408997200. [DOI] [PubMed] [Google Scholar]
- 45.Du J, Hui Y, Cheung Y, Bin G, Jiang H, Chen X, et al. The possible role of hydrogen sulfide as a smooth muscle cell proliferation inhibitor in rat cultured cells. Heart and vessels. 2004;19:75. doi: 10.1007/s00380-003-0743-7. [DOI] [PubMed] [Google Scholar]
- 46.Li L, Liu D, Bu D, Chen S, Wu J, Tang C, et al. Brg1-dependent epigenetic control of vascular smooth muscle cell proliferation by hydrogen sulfide. Biochimica et biophysica acta. 2013;1833:1347. doi: 10.1016/j.bbamcr.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 47.Li W, Xiong Y, Shang C, Twu KY, Hang CT, Yang J, et al. Brg1 governs distinct pathways to direct multiple aspects of mammalian neural crest cell development. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:1738. doi: 10.1073/pnas.1218072110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mehotra A, Joe B, de la Serna IL. Enhanced SWI/SNF enzyme recruitment and chromatin remodeling on fetal cardiac gene promoters is associated with cardiac hypertrophy in a genetic rat model of hypertension. Journal of cellular physiology. 2013 doi: 10.1002/jcp.24404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6656. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Molecular cell. 2000;6:1287. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 51.Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) The EMBO journal. 1998;17:6979. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Molecular cell. 2003;11:377. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 53.Smith-Roe SL, Bultman SJ. Combined gene dosage requirement for SWI/SNF catalytic subunits during early mammalian development. Mammalian genome : official journal of the International Mammalian Genome Society. 2013;24:21. doi: 10.1007/s00335-012-9433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willis MS, Homeister JW, Rosson GB, Annayev Y, Holley D, Holly SP, et al. Functional redundancy of SWI/SNF catalytic subunits in maintaining vascular endothelial cells in the adult heart. Circulation research. 2012;111:e111. doi: 10.1161/CIRCRESAHA.112.265587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Y, Price BD. Chromatin dynamics and the repair of DNA double strand breaks. Cell cycle. 2011;10:261. doi: 10.4161/cc.10.2.14543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khanna-Gupta A. Sumoylation and the function of CCAAT enhancer binding protein alpha (C/EBP alpha) Blood cells, molecules & diseases. 2008;41:77. doi: 10.1016/j.bcmd.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Current opinion in genetics & development. 2003;13:136. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 58.Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annual review of pharmacology and toxicology. 2009;49:243. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]