Abstract
In clinical settings, biopsies are routinely used to determine cancer type and grade based on tumor cell morphology, as determined via histochemical or immunohistochemical staining. Unfortunately, in a significant number of cases, traditional biopsy results are either inconclusive or do not provide full subtype differentiation, possibly leading to inefficient or ineffective treatment. Glycomic profiling of the cell membrane offers an alternate route towards cancer diagnosis. In this study, isomer-sensitive nano-LC/MS was used to directly obtain detailed profiles of the different N-glycan structures present on cancer cell membranes. Membrane N-glycans were extracted from cells representing various subtypes of breast, lung, cervical, ovarian, and lymphatic cancer. Chip-based porous graphitized carbon nano-LC/MS was used to separate, identify, and quantify the native N-glycans. Structure-sensitive N-glycan profiling identified hundreds of glycan peaks per cell line, including multiple isomers for most compositions. Hierarchical clusterings based on Pearson correlation coefficients were used to quickly compare and separate each cell line according to originating organ and disease subtype. Based simply on the relative abundances of broad glycan classes (e.g. high mannose, complex/hybrid fucosylated, complex/hybrid sialylated, etc.) most cell lines were readily differentiated. More closely-related cell lines were differentiated based on several-fold differences in the abundances of individual glycans. Based on characteristic N-glycan profiles, primary cancer origins and molecular subtypes could be distinguished. These results demonstrate that stark differences in cancer cell membrane glycosylation can be exploited to create an MS-based biopsy, with potential applications towards cancer diagnosis and direction of treatment.
Keywords: mass spectrometry, LC/MS, N-glycans, cell membrane, cancer, molecular subtype
INTRODUCTION
The mammalian cell membrane is coated with a dense thicket of glycans — complex sugar chains that are post-translationally attached to most membrane proteins. These glycans serve as a primary point of contact between the cell and the rest of the world, mediating cell interactions with small molecules, microorganisms, and even other cells.1 As a result, changes in glycosylation are often indicative of irregularities in the cellular machinery, and have previously been correlated with a number of human diseases — most famously, cancer. Several decades of research have now firmly established aberrant glycosylation as one of the hallmarks of cancer.2, 3 Glycans have been shown to mediate many cancer-related processes,4–8 and cell membrane glycans, in particular, play primary roles in tumorigenesis,9–11 metastatic colonization,12, 13 and chemotherapeutic drug resistance.14, 15
Numerous studies on the human serum glycome (i.e. from secreted glycoproteins) have already identified potential biomarkers for multiple cancer types.16–19 However, the cell membrane glycome remains a largely untapped source of potential biomarkers, due in large part to the difficulty of isolating cell membrane glycoproteins from those found in the cytoplasm, nucleoplasm, or cellular organelles. Previous studies on cell membrane glycosylation have been limited in scope, relying largely on affinity interactions (either lectin- or antibody-based) to detect specific glycosylation motifs or epitopes.12, 20–22 Unfortunately, because these affinity interactions typically target only terminal monosaccharides, the analytical data gleaned from affinity-based studies is often inconclusive or subject to extensive interpretation. Additionally, due to the extraordinary structural heterogeneity of glycans, many glycan structures simply cannot be detected (or distinguished) by currently-available affinity-based methods.
Recently-developed LC/MS-based methods of glycomic profiling, in contrast, offer direct and comprehensive access to cell membrane glycosylation. An et al recently developed and validated a method to isolate and enrich cell membrane N-glycans, enabling quantitative, reproducible, and (most importantly) LC/MS-compatible analysis.23 LC/MS compatibility is crucial to glycomic analyses, particularly when complex, structurally heterogenous analytes (such as cell membrane glycans) are involved. Over 200 glycosyltransferases and glycosidases work in concert to decorate proteins with glycosylation, and even slight perturbations in this delicate balance can result in widespread changes.24 Glycomic profiling of these changes can be an extremely sensitive indicator of disease progression; however, detailed analytical methods are necessary in order to accurately capture the full structural complexity of the glycome. LC/MS fits this requirement exceedingly well, providing two-dimensional separation of complex glycan mixtures according to accurate mass, size, and three-dimensional conformation.
Nano-LC/MS methods for separation and profiling of N-glycans have recently been developed and optimized by Hua et al.16 These methods utilize chip-based porous graphitized carbon (PGC) nano-LC to separate complex glycan mixtures in a structure-sensitive manner prior to high resolution, accurate mass TOF-MS analysis. In tests on serum N-glycans, the optimized PGC nano-LC/TOF-MS platform provided detailed and comprehensive glycan identification, miniscule sample consumption, and dramatically increased sensitivity over previous MALDI-MS or high-flow LC/MS methods.25
The present study combines cell membrane N-glycan enrichment procedures with detailed nano-LC/MS glycan profiling to provide the first comprehensive survey of cancer cell N-glycosylation. Cell membrane N-glycan profiles were obtained for fifteen different cell lines isolated from ovarian, breast, lung, cervical, and lymphatic cancer patients (as well as one non-cancerous fibrocystic breast cell line). Cell lines were grouped and compared according to biologically- or clinically-relevant characteristics (e.g. cancerous vs non-cancerous cell lines, lung carcinoma vs cervical carcinoma cell lines, etc). Significant changes in membrane glycan profiles were confirmed by statistical tests such as Pearson correlations and hierarchical clustering. Major differences were uncovered amongst the membrane glycan profiles of the analyzed cell lines, in terms of both overall glycan biosynthetic class and individual glycan signals. LC/MS-based membrane glycan profiling was shown to be a powerful platform for differentiation and identification of different cancer cell types.
METHODS
Cell culture
Cell lines (Hs-578T, ZR-75-1, MCF-7, MCF-7/HER2, MCF10A, Raji, Ramos, NCI-H929, BCBL-1, ES-2, NCI-H358, A549, C-33A, CaSki, and HeLa) were obtained from ATCC. Cell cultures were performed using the ATCC-recommended growth media, subcultivation ratios, and medium renewal interval (available online at www.atcc.org).
During cell harvest, in order to preserve membrane glycoprotein integrity, cells were scraped rather than enzymatically lifted. For each cell sample, approximately 10 million cells were counted and collected. Duplicate samples were collected from the lymphoma (Raji; Ramos; NCI-H929; BCBL-1), cervical carcinoma (33A; CaSki; HeLa), and lung carcinoma (NCI-H358; A549) cell cultures; however, due to cell growth limitations, only single samples were collected from the breast (Hs-578T; ZR-75-1; MCF-7; MCF-7/HER2; MCF10A) and ovarian (ES-2) cell cultures.
Plasma membrane extraction
Isolation of the plasma membrane was performed according to optimized procedures published by An et al.23 Briefly, cells were suspended a homogenization buffer consisting of 0.25 M sucrose; 20 mM HEPES-KOH, pH 7.4; and 1:100 protease inhibitor mixture (EMD Millipore) prior to sonication. Cell lysates were ultracentrifuged at 1,000 g and 200,000 g to remove the nuclear and cytosolic fractions, respectively. Supernatants, consisting of enriched plasma membranes, were collected for glycan extraction.
N-glycan release and enrichment
Enzymatic release and solid-phase extraction of N-glycans were performed according to optimized procedures published by Kronewitter et al.26 Briefly, membrane glycoproteins were denatured by rapid thermal cycling (25–100 °C) in an aqueous solution of 100 mM ammonium bicarbonate and 5 mM dithiothreitol. Next, 2.0 μL (or 1000 U) of peptide N-glycosidase F (New England Biolabs) were added and the mixture was incubated in a microwave reactor (CEM Corporation) for 10 minutes at 20 watts. Following the addition of 800 μL of cold ethanol, the mixture was chilled at −80 °C for 1 hour, then centrifuged in order to precipitate out the deglycosylated proteins. The glycan-rich supernatant fraction was collected and dried in vacuo.
Graphitized carbon solid-phase extraction was performed using an automated liquid handler (Gilson). Graphitized carbon cartridges (150 mg, 4.0 mL, Grace Davison) were washed with 80% acetonitrile / 0.10% trifluoroacetic acid (v/v) in water, then conditioned with pure water. Aqueous N-glycan solutions were loaded onto the cartridges and washed with pure water to remove salts and buffer. Membrane N-glycans were eluted with sequential addition of 10% acetonitrile, 20% acetonitrile, and 40% acetonitrile / 0.05% trifluoroacetic acid (v/v) in water. Samples were dried in vacuo.
Nano-LC/MS analysis and data processing
MS analysis was performed according to optimized procedures published by Hua et al.16 Briefly, aqueous glycan solutions were injected by autosampler onto a chip-mounted nano-LC column (Agilent Technologies), consisting of a 9 × 0.075 mm i.d. enrichment column and a 43 × 0.075 mm i.d. analytical column, both packed with 5 μm porous graphitized carbon as the stationary phase. A rapid glycan elution gradient was delivered to the analytical column at 0.4 μL/min using solutions of (A) 3.0% acetonitrile and 0.1% formic acid (v/v) in water, and (B) 90.0% acetonitrile and 0.1% formic acid (v/v) in water, ramping from 5% to 35.9% B over the course of 16.5 min. Remaining non-glycan compounds were flushed out with 100% B prior to re-equilibration.
MS spectra were acquired by a TOF MS detector (Model 6210, Agilent Technologies) in positive ionization mode over a mass range of m/z 600–2000 with an acquisition time of 1.5 seconds per spectrum. Mass correction was enabled using reference masses of m/z 622.029, 922.010, 1221.991, and 1521.971 (ESI-TOF Calibrant Mix G1969-85000, Agilent Technologies).
Raw LC/MS data was filtered with a signal-to-noise ratio of 5.0 and parsed into a series of extracted ion chromatograms using the Molecular Feature Extractor algorithm included in the MassHunter Qualitative Analysis software (Version B.04.00, Agilent Technologies). Using expected isotopic distribution and charge state information, extracted ion chromatograms were combined to create extracted compound chromatograms (ECCs) representing the summed signal from all ion species associated with a single compound (e.g. the doubly protonated ion, the triply protonated ion, and all associated isotopologues). Thus, each individual ECC peak could be taken to represent the total ion count associated with a single distinct compound.
Each ECC peak was matched by accurate mass to a comprehensive library of all possible complex, hybrid, and high mannose glycan compositions based on known biosynthetic pathways and glycosylation patterns.27, 28 Deconvoluted masses of each ECC peak were compared against theoretical glycan masses using a mass error tolerance of 20 ppm and a false discovery rate of 0.6%. As all samples originated from human cell lines, only glycan compositions containing hexose (Hex), N-acetylhexosamine (HexNAc), fucose (Fuc), and N-acetylneuraminic acid (NeuAc) were considered.
RESULTS AND DISCUSSION
Separation and quantitative profiling of cell membrane N-glycans
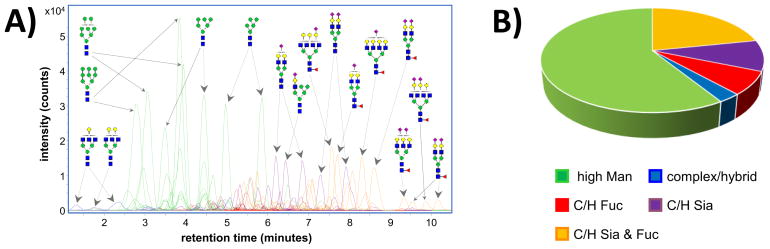
LC/MS-based glycan profiling provides a comprehensive look at the different glycan compositions and structures present on the cell membrane. On average, cell membrane glycan profiles yielded over 250 N-linked glycan compound peaks with over 100 distinct N-linked glycan compositions, spanning five orders of magnitude. Each of the identified compositions include two or more peaks corresponding to either structural and/or linkage isomers (regioisomers) or, in some cases, anomeric isomers. For example, Figure 1a shows chromatograms of cell membrane N-glycans identified on non-CD4 T-cells from human blood. From this data, the relative abundances of individual glycan compositions or structures were readily quantified, simply by integrating the ion counts associated with each peak and normalizing to the total (summed) ion count of all glycans detected in the sample. Where applicable, the results of duplicate analyses were averaged. Previous studies have already validated the quantitative reproducibility of the cell membrane glycan extraction procedure as well as the PGC nano-LC/MS platform, enabling rapid profiling and comparison of individual glycan abundances.16, 23
Figure 1.
a) Extracted compound chromatograms of cell membrane N-glycans identified on non-CD4 T-cells from human blood. Colors denote the biosynthetic class of each glycan. Glycan structures are putative, based on known biosynthetic pathways. b) Relative abundances of each glycan biosynthetic class on non-CD4 T-cell membranes.
In addition to individual glycan profiling, overarching differences in cell membrane glycosylation can be detected through glycan class profiling. In Figure 1a, the glycan compositional assignments have been used to sort glycan signals into different biosynthetic glycan classes, each represented by a different color — green for high mannose (high Man) glycans; blue for undecorated complex/hybrid (C/H) glycans; red for fucosylated complex/hybrid (C/H-F) glycans; purple for sialylated complex/hybrid (C/H-S) glycans; and orange for fucosylated-sialylated complex/hybrid (C/H-FS) glycans. Based on the summed ion counts of the individual glycan signals, relative abundances were assigned to each glycan class, creating a glycan class profile for the membrane N-glycans of each cell line (Figure 1b).
The ability of LC/MS-based glycomic analysis to detect unique cell membrane signatures at both the individual glycan level as well as the glycan class level has a number of potential clinical applications. To demonstrate this point, we hereby present a few clinically-relevant examples in which different cancer cell types are compared and differentiated.
Differentiation of cancerous and non-cancerous cells
When radiological imaging techniques detect the presence of tumors or lesions within a patient, doctors must determine whether these abnormal masses are malignant (cancerous), or benign (non-cancerous). While histological characteristics can often indicate whether a tumor is currently benign or cancerous, molecular markers can potentially increase accuracy, and might even indicate the prognosis and future inclinations of a tumor (e.g. likelihood of metastasizing).12, 15, 29, 30
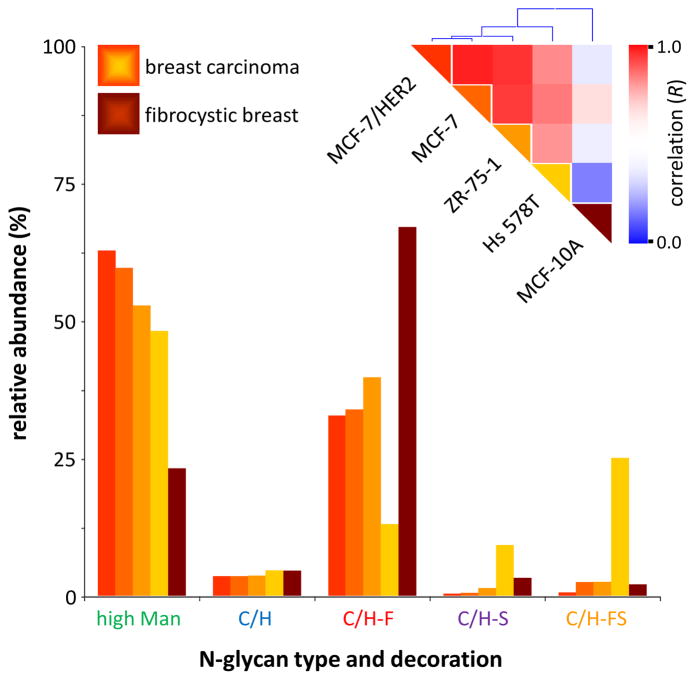
To demonstrate the utility of cell membrane glycan profiling towards tumor diagnosis, glycan class profiles (Figure 2) were compared amongst four cancerous breast cell lines (Hs-578T, ZR-75-1, MCF-7, and MCF-7/HER2) and one non-cancerous breast cell line (MCF-10A). All four cancerous cell lines originated from ductal breast carcinomas, whereas the non-cancerous MCF-10A cell line originated from fibrocystic breast tissue. Cursory inspection of the glycan class profiles reveals that all four breast carcinoma cell lines have much higher relative abundances (48–63%) of membrane high mannose glycosylation than the single non-cancerous fibrocystic breast cell line (23%). These results are consistent with previous research in which high mannose glycosylation is reported to increase during breast cancer.17
Figure 2.
Glycan class profiles of four cancerous breast cell lines (Hs-578T, ZR-75-1, MCF-7, and MCF-7/HER2) and one non-cancerous breast cell line (MCF-10A). Relative abundances are shown for each glycan biosynthetic class. Inset: Color-coded representation of the Pearson correlation coefficient (R) between each pair of cell lines, ranging from red (high correlation) to blue (low correlation), along with hierarchical clustering trees.
In addition to differentiating between cancerous and non-cancerous breast tissue, the glycan class profile (Figure 2) may also be used to differentiate amongst the four different breast carcinoma lines. For instance, cell line Hs-578T has much higher relative abundances of sialylated and fucosylated-sialylated complex/hybrid N-glycans compared to the other three breast carcinoma lines. Some of this increase may be associated with the presence of sialyl Lewis X (sLeX) or sialyl Lewis A (sLeA) epitopes, both of which are fucosylated-sialylated. Studies have shown that sLeX and/or sLeA are often found on the N-glycans of metastatic tumor cells, including carcinomas.12, 13 In clinical settings, direct detection of sLeX and/or sLeA on breast cancer carcinomas could help predict the metastatic potential of a tumor.
While informative at a macro level, the glycan class profiles shown in Figure 2 are ultimately just simplified representations of each cell’s membrane glycosylation, summed from hundreds of individual glycan signals. To more precisely quantify the similarities and differences between the different cell lines, these individual glycan signals were incorporated into pairwise Pearson correlations.31 Figure S1 shows a typical comparison between two breast cell lines: Hs-578T (cancerous) and MCF-10A (non-cancerous). Each point represents a different N-glycan, with the x-coordinate corresponding to the relative abundance of that glycan in Hs-578T cell membranes, and the y-coordinate corresponding to the relative abundance of that glycan in MCF-10A cell membranes. Based on these points, a best-fit line is drawn, and the Pearson correlation coefficient R is calculated. The low R-value of 0.217 indicates that the membrane glycosylation profiles of these two cell lines are quite different.
Pearson correlations were calculated for each possible pair of breast cell lines (Figure 2, inset), for a total of ten comparisons. In general, comparisons amongst cancerous breast cell lines show high correlation (0.700 < R < 0.932). Meanwhile, comparisons between cancerous and non-cancerous breast cell lines show low correlation (0.217 < R < 0.545). Based on these R-values, breast cell lines have been grouped into hierarchical clusters (Figure 2, inset). For example, closely-related cancerous breast cell lines MCF7 and MCF7/HER2 (R = 0.932) are grouped together into a single cluster. Meanwhile, non-cancerous breast cell line MCF-10A is grouped separately from all of the cancerous breast cell lines, providing rapid differentiation between non-cancerous and cancerous breast cells.
Identification of cancer cell origin and type
Once doctors identify a cancerous tumor in a patient, one of the first steps towards devising an effective treatment is to determine the primary origin of the cancer. However, in approximately 3% of patients, the primary cancer site is never found. Often, these cancers are poorly differentiated, lacking key morphological features that can be used to identify the origin of more developed cells.32–34 To make matters worse, poorly differentiated (high grade) cancers are typically the most aggressive, growing faster and spreading more rapidly than well-differentiated (low grade) cancers.35 Lymphomas, which originate from the lymphatic system, commonly exhibit poor differentiation.32, 36
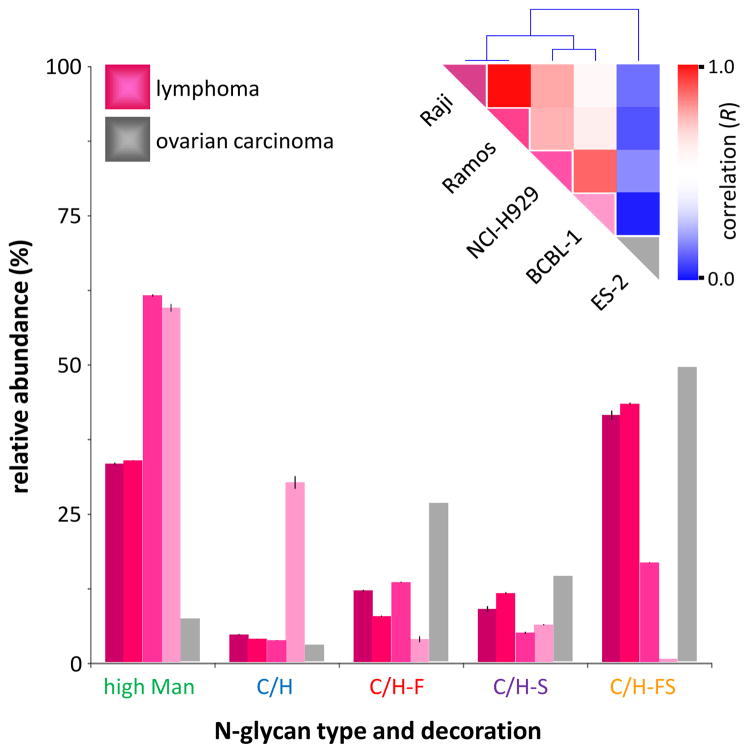
In Figure 3, four lymphoproliferative cancer cell lines (Raji, Ramos, NCI-H929, and BCBL-1) are compared with a single ovarian carcinoma cell line (ES-2). The Raji and Ramos cell lines originate from Burkitt’s lymphomas; the NCI-H929 from a multiple myeloma; the BCBL-1 from a primary effusion lymphoma; and the ES-2 from a poorly-differentiated ovarian clear cell carcinoma. As before, cursory inspection of the glycan class profiles (Figure 3) reveals stark differences between the different cell lines. For example, using just the relative abundances of the high mannose glycosylation, it is possible to separate the cell lines into three general groupings — the Raji and Ramos cell lines, at 33–34% high mannose glycosylation; the NCI-H929 and BCBL-1 cell lines, at 59–61% high mannose glycosylation; and the ES-2 cell line, at 7% high mannose glycosylation. These groupings are reflected in the Pearson correlations and hierarchical clustering (Figure 3, inset), in which the Raji and Ramos cell lines occupy one cluster, with very high correlation to each other; the NCI-H929 and BCBL-1 cell lines occupy another cluster, with a slightly less high correlation to each other; and the sole ovarian carcinoma ES-2 cell line is grouped apart from all of the lymphoma cell lines, with very low correlation to any of them.
Figure 3.
Glycan class profiles of four B-cell lymphoma cell lines (Raji, Ramos, NCI-H929, and BCBL-1) and one ovarian carcinoma cell line (ES-2). Relative abundances are shown for each glycan biosynthetic class. Inset: Color-coded representation of the Pearson correlation coefficient (R) between each pair of cell lines, ranging from red (high correlation) to blue (low correlation), along with hierarchical clustering trees.
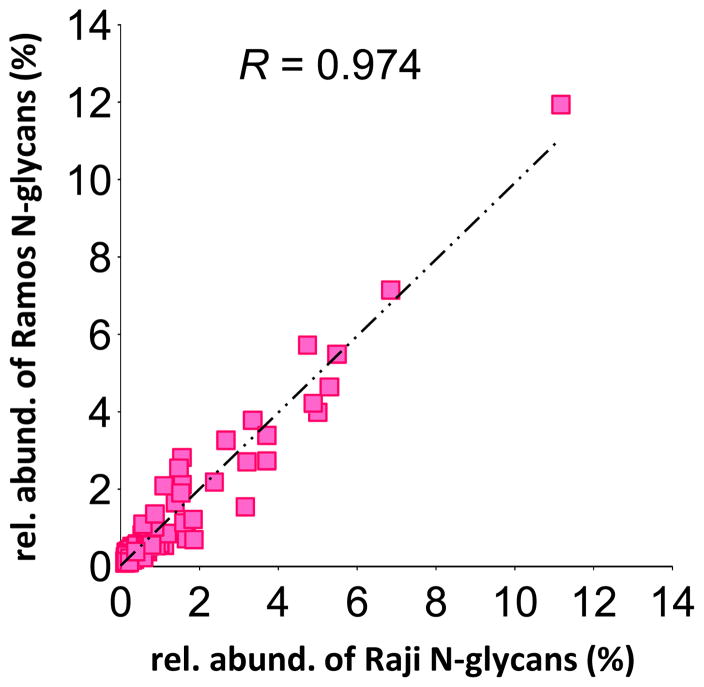
The incredibly high correlation of the Raji and Ramos cell lines (Figure 4) is quite significant biologically, because both of these cell lines derive from Burkitt’s lymphoma patients. However, the patients were different from each other in almost every other respect — Raji was isolated from an African in Nigeria, whereas Ramos was isolated from a Caucasian in the United States; Raji represents the endemic form of Burkitt’s lymphoma, whereas Ramos represents the spontaneous form; Raji was isolated in the early 1960s, whereas Ramos was isolated more than a decade later.37, 38 Yet, despite these differences, the individual glycan signals isolated from the Raji and Ramos cell membranes exhibit a startlingly high correlation (R = 0.974) — suggesting that, in at least some cases, glycosylation patterns can be linked to specific cancer variants even in spite of confounding genetic and environmental factors.
Figure 4.
Scatterplot showing the relative abundances of each glycan signal in the Raji vs Ramos B-cell lymphoma cell lines. Each point represents an individual glycan signal. Pearson correlation coefficient (R) is calculated based on the best-fit line.
Distinguishing between epithelial carcinomas from different organs
Identification of a primary cancer site enables doctors to focus diagnostic testing and treatment, significantly improving a patient’s chances for survival; however, radiological observations are often obfuscated by metastatic tumors. To make matters worse, carcinomas, which derive from a variety of epithelial tissues, often exhibit very similar histologic and biochemical characteristics, even when they originate from different organs. This similarity creates significant problems when doctors need to differentiate between primary and metastatic carcinomas.
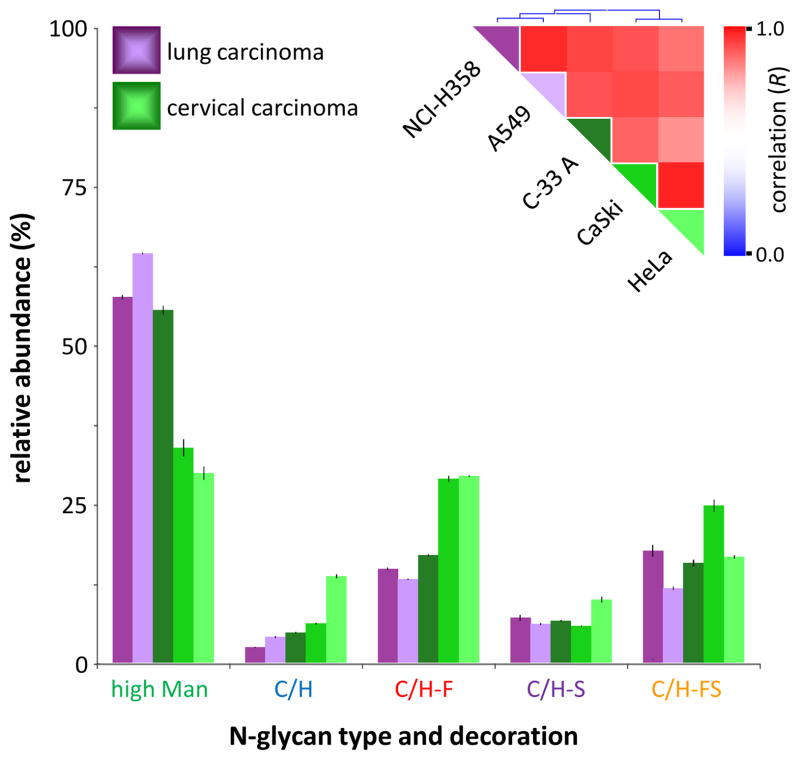
Cell membrane glycan profiling, however, may help determine the origin of a carcinoma. To demonstrate this concept, several cervical and lung carcinoma cell lines were selected for analysis. Cervical carcinomas commonly metastasize to the lung, forcing doctors to differentiate between primary lung carcinomas and metastatic cervical carcinomas prior to treatment.39 Therefore, in Figure 5, N-glycan profiles were compared amongst two lung carcinoma cell lines (NCI-H358 and A549) and three cervical carcinoma cell lines (C-33A, CaSki, and HeLa). Glycan class profiling as well as hierarchical clustering (Figure 5, inset) reveals two major clusters: one containing both lung carcinoma cell lines, and the other containing two of the three cervical carcinoma cell lines (CaSki and HeLa). The placement of the third cervical carcinoma cell line, C-33A, is more intriguing — clustering actually reveals that cervical carcinoma cell line C-33A is more closely related to lung carcinoma cell lines NCI-H358 and A549 than to its fellow cervical carcinoma cell lines CaSki and HeLa. C-33A is rather unique amongst cervical carcinoma cell lines in that it is completely negative for human papillomavirus (HPV) DNA, whereas most cervical carcinomas (and cell lines derived therefrom) harbor significant quantities of HPV DNA.40 For example, cervical carcinoma cell lines CaSki and HeLa are both HPV-positive and display highly correlated membrane glycosylation profiles (Figure S2), presumably due to mutual HPV-induced carcinogenesis. The cluster containing HPV-negative cervical carcinoma cell line C-33A and the two lung carcinoma cell lines, therefore, likely represents a grouping of cell lines for which carcinogenesis occurred due to largely genetic rather than environmental (e.g. viral) factors.
Figure 5.
Glycan class profiles of two lung carcinoma cell lines (NCI-H358 and A549) and three cervical carcinoma cell lines (C-33A, CaSki, and HeLa). Relative abundances are shown for each glycan biosynthetic class. Inset: Color-coded representation of the Pearson correlation coefficient (R) between each pair of cell lines, ranging from red (high correlation) to blue (low correlation), along with hierarchical clustering trees.
To distinguish amongst these individual cell lines, more detailed glycan profiling techniques were applied. Individual glycan species, identified and tracked by mass spectrometry, were employed as markers to differentiate amongst cell lines C-33A (cervical carcinoma), NCI-H358 (lung carcinoma), and A549 (lung carcinoma). Cervical carcinoma cell line C-33A was readily distinguished by elevated levels of glycans Man4GlcNAc2 and Man3GlcNAc2, which were present at 7-fold and 4-fold abundance, respectively, relative to the two lung carcinoma cell lines (Figure 6a). Lung carcinoma cell line NCI-H358, in turn, was readily distinguished from lung carcinoma cell line A549 by 6-fold elevated levels of glycans Man3GlcNAc2Fuc, Man3GlcNAc3Fuc, and Man3GlcNAc4Fuc (Figure 6b). Interestingly, all five marker glycans are biosynthetically related, each differing from the next by only one monosaccharide. Additionally, all represent relatively early steps in the glycan biosynthesis process, suggesting a lower degree of differentiation — one of the hallmarks of cancer glycosylation.2, 3
Figure 6.
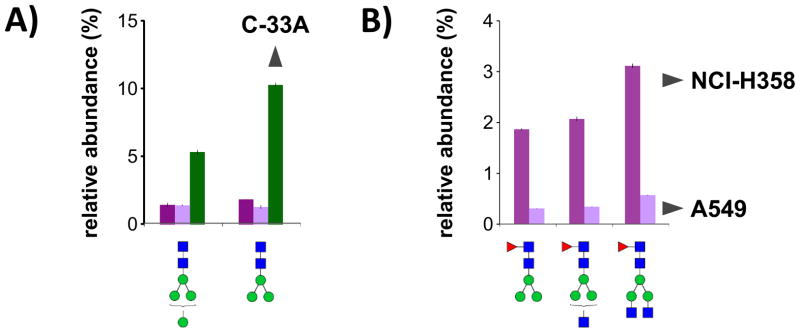
Relative abundances of biosynthetically-related glycans with high fold differences between carcinoma cell lines C-33A (cervical), NCI-H358 (lung), and A549 (lung). a) Increased levels of Man4GlcNAc2 and Man3GlcNAc2 distinguish C-33A from NCI-H358 and A549. b) Increased levels of Man3GlcNAc2Fuc, Man3GlcNAc3Fuc, and Man3GlcNAc4Fuc distinguish NCI-H358 from A549. Glycans are shown in order of biosynthesis, from left to right.
CONCLUSION
We have demonstrated for the first time direct analysis of cancer cell membrane N-glycans by nano-LC/MS, revealing stark differences in the glycosylation of different cell types. The platform is not only quantitatively reproducible, but also highly sensitive, with a large dynamic range.16 Additionally, porous graphitized carbon nano-LC provides structure-sensitive separation of isomeric glycans.41 Though structural elucidation was not pursued in the present study, the platform’s demonstrated structural separation capabilities may be easily coupled to tandem MS fragmentation for future glycan structure studies.31, 42, 43
The observation of such stark differences in the membrane N-glycosylation patterns of different cancer cells reveals new avenues for the differentiation and diagnosis of cancer type/origin. Previous research on cancer cell membrane glycosylation focused mainly on O-glycans, perhaps as a matter of convenience — antibodies against specific O-glycans (such as sLeX and sLeA) are widely available, whereas few if any antibodies exist that target specific N-glycans.12, 29, 30 However, recent studies have shown that the vast majority of cell membrane glycosylation is in fact N-linked, rather than O-linked.23 A shift of focus towards N-glycans, therefore, would provide a major boost in diagnostic biomarker candidates, especially now that a direct LC/MS analysis method has become available.
While the results of the present study provide a compelling argument for glycosylation-based cancer diagnosis, a crucial next step will be analysis of tumor samples from cancer patients. In our lab, preliminary experiments on patient T-cells (Figure 1) as well as microtomed epithelial tissue have already yielded glycan signals and profiles comparable to those of cultured cells, indicating the wide applicability of the membrane fractionation and LC/MS analysis procedures. Translation from cultured cell lines to patient tumors will provide an unprecedented glimpse of in vivo glycosylation, paving the way for a next-generation MS biopsy.
Supplementary Material
Acknowledgments
We are grateful for the support provided by the 2012 University-Institute Cooperation Program via the National Research Foundation of Korea; the Converging Research Center Program (2012K001505 for H. J. An) via the Ministry of Education, Science and Technology; and the National Institutes of Health (RO1GM049077 for C. B. Lebrilla).
Abbreviations
- PGC
porous graphitized carbon
- ECC
extracted compound chromatogram
- Hex
hexose
- HexNAc
N-acetylhexosamine
- Fuc
fucose
- NeuAc
N-acetylneuraminic acid
- Man
mannose
- C/H
undecorated complex/hybrid
- C/H-F
fucosylated complex/hybrid
- C/H-S
sialylated complex/hybrid
- C/H-FS
fucosylated-sialylated complex/hybrid
- sLeX
sialyl Lewis X
- sLeA
sialyl Lewis A
- HPV
human papillomavirus
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Sharon N, Lis H. Lectins as cell recognition molecules. Science. 1989;246(4927):227–234. doi: 10.1126/science.2552581. [DOI] [PubMed] [Google Scholar]
- 2.Dube DH, Bertozzi CR. Glycans in cancer and inflammation — potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4(6):477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 3.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5(7):526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 4.Rapoport E, Pendu J. Glycosylation alterations ofcells in late phase apoptosis from colon carcinomas. Glycobiology. 1999;9(12):1337–1345. doi: 10.1093/glycob/9.12.1337. [DOI] [PubMed] [Google Scholar]
- 5.Saito T, Miyoshi E, Sasai K, Nakano N, Eguchi H, Honke K, Taniguchi N. A Secreted Type of β1,6-N-Acetylglucosaminyltransferase V (GnT-V) Induces Tumor Angiogenesis without Mediation of Glycosylation. Journal of Biological Chemistry. 2002;277(19):17002–17008. doi: 10.1074/jbc.M200521200. [DOI] [PubMed] [Google Scholar]
- 6.Pili R, Chang J, Partis RA, Mueller RA, Chrest FJ, Passaniti A. The {alpha}-Glucosidase I Inhibitor Castanospermine Alters Endothelial Cell Glycosylation, Prevents Angiogenesis, and Inhibits Tumor Growth. Cancer Res. 1995;55(13):2920–2926. [PubMed] [Google Scholar]
- 7.Nita-Lazar M, Noonan V, Rebustini I, Walker J, Menko AS, Kukuruzinska MA. Overexpression of DPAGT1 Leads to Aberrant N-Glycosylation of E-Cadherin and Cellular Discohesion in Oral Cancer. Cancer Res. 2009;69(14):5673–5680. doi: 10.1158/0008-5472.CAN-08-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y-Y, Takahashi M, Gu J-G, Miyoshi E, Matsumoto A, Kitazume S, Taniguchi N. Functional roles of N-glycans in cell signaling and cell adhesion in cancer. Cancer Science. 2008;99(7):1304–1310. doi: 10.1111/j.1349-7006.2008.00839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaheri A, Mosher DF. High molecular weight, cell surface-associated glycoprotein (fibronectin) lost in maglinant transformation. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 1978;516(1):1–25. doi: 10.1016/0304-419x(78)90002-1. [DOI] [PubMed] [Google Scholar]
- 10.The Role of Integrins in Tumorigenesis and Metastasis. Cancer Investigation. 1998;16(5):329–344. doi: 10.3109/07357909809084653. [DOI] [PubMed] [Google Scholar]
- 11.Casey R, Oegema T, Jr, Skubitz K, Pambuccian S, Grindle S, Skubitz AN. Cell membrane glycosylation mediates the adhesion, migration, and invasion of ovarian carcinoma cells. Clin Exp Metastasis. 2003;20(2):143–152. doi: 10.1023/a:1022670501667. [DOI] [PubMed] [Google Scholar]
- 12.Ohyama C, Tsuboi S, Fukuda M. Dual roles of sialyl Lewis X oligosaccharides in tumor metastasis and rejection by natural killer cells. EMBO J. 1999;18(6):1516–1525. doi: 10.1093/emboj/18.6.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takada A, Ohmori K, Yoneda T, Tsuyuoka K, Hasegawa A, Kiso M, Kannagi R. Contribution of Carbohydrate Antigens Sialyl Lewis A and Sialyl Lewis X to Adhesion of Human Cancer Cells to Vascular Endothelium. Cancer Research. 1993;53(2):354–361. [PubMed] [Google Scholar]
- 14.Kartner N, Riordan; Ling V. Cell surface P-glycoprotein associated with multidrug resistance in mammalian cell lines. Science. 1983;221(4617):1285–1288. doi: 10.1126/science.6137059. [DOI] [PubMed] [Google Scholar]
- 15.Bell DR, Gerlach JH, Kartner N, Buick RN, Ling V. Detection of P-glycoprotein in ovarian cancer: a molecular marker associated with multidrug resistance. Journal of Clinical Oncology. 1985;3(3):311–5. doi: 10.1200/JCO.1985.3.3.311. [DOI] [PubMed] [Google Scholar]
- 16.Hua S, Williams CC, Dimapasoc LM, Ro GS, Ozcan S, Miyamoto S, Lebrilla CB, An HJ, Leiserowitz GS. Isomer-specific chromatographic profiling yields highly sensitive and specific potential N-glycan biomarkers for epithelial ovarian cancer. Journal of Chromatography A. 2013;1279(0):58–67. doi: 10.1016/j.chroma.2012.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Leoz MLA, Young LJT, An HJ, Kronewitter SR, Kim J, Miyamoto S, Borowsky AD, Chew HK, Lebrilla CB. High-Mannose Glycans are Elevated during Breast Cancer Progression. Molecular & Cellular Proteomics. 2011;10(1) doi: 10.1074/mcp.M110.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hua S, An HJ, Ozcan S, Ro GS, Soares S, DeVere-White R, Lebrilla CB. Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst. 2011;136(18):3663–3671. doi: 10.1039/c1an15093f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua S, Lebrilla C, An HJ. Application of nano-LC-based glycomics towards biomarker discovery. Bioanalysis. 2011;3(22):2573–2585. doi: 10.4155/bio.11.263. [DOI] [PubMed] [Google Scholar]
- 20.Naka R, Kamoda S, Ishizuka A, Kinoshita M, Kakehi K. Analysis of Total N-Glycans in Cell Membrane Fractions of Cancer Cells Using a Combination of Serotonin Affinity Chromatography and Normal Phase Chromatography. Journal of Proteome Research. 2005;5(1):88–97. doi: 10.1021/pr0502976. [DOI] [PubMed] [Google Scholar]
- 21.Pilobello KT, Slawek DE, Mahal LK. A ratiometric lectin microarray approach to analysis of the dynamic mammalian glycome. Proceedings of the National Academy of Sciences. 2007;104(28):11534–11539. doi: 10.1073/pnas.0704954104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuno A, Kato Y, Matsuda A, Kaneko MK, Ito H, Amano K, Chiba Y, Narimatsu H, Hirabayashi J. Focused Differential Glycan Analysis with the Platform Antibody-assisted Lectin Profiling for Glycan-related Biomarker Verification. Molecular & Cellular Proteomics. 2009;8(1):99–108. doi: 10.1074/mcp.M800308-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.An HJ, Gip P, Kim J, Wu S, Park KW, McVaugh CT, Schaffer DV, Bertozzi CR, Lebrilla CB. Extensive Determination of Glycan Heterogeneity Reveals an Unusual Abundance of High Mannose Glycans in Enriched Plasma Membranes of Human Embryonic Stem Cells. Molecular & Cellular Proteomics. 2012;11(4) doi: 10.1074/mcp.M111.010660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baum LG. Developing a Taste for Sweets. Immunity. 2002;16(1):5–8. doi: 10.1016/s1074-7613(02)00265-0. [DOI] [PubMed] [Google Scholar]
- 25.Hua S, An HJ. Glycoscience aids in biomarker discovery. Biochemistry and Molecular Biology Reports. 2012;45(6):323–330. doi: 10.5483/bmbrep.2012.45.6.132. [DOI] [PubMed] [Google Scholar]
- 26.Kronewitter SR, de Leoz MLA, Peacock KS, McBride KR, An HJ, Miyamoto S, Leiserowitz GS, Lebrilla CB. Human Serum Processing and Analysis Methods for Rapid and Reproducible N-Glycan Mass Profiling. Journal of Proteome Research. 2010;9(10):4952–4959. doi: 10.1021/pr100202a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kronewitter SR, An HJ, Leoz MLd, Lebrilla CB, Miyamoto S, Leiserowitz GS. The development of retrosynthetic glycan libraries to profile and classify the human serum N-linked glycome. PROTEOMICS. 2009;9(11):2986–2994. doi: 10.1002/pmic.200800760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hua S, Jeong HN, Dimapasoc LM, Kang I, Han C, Choi J-S, Lebrilla CB, An HJ. Isomer-Specific LC/MS and LC/MS/MS Profiling of the Mouse Serum N-Glycome Revealing a Number of Novel Sialylated N-Glycans. Analytical Chemistry. 2013;85(9):4636–4643. doi: 10.1021/ac400195h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamori S, Kameyama M, Imaoka S, Furukawa H, Ishikawa O, Sasaki Y, Kabuto T, Iwanaga T, Matsushita Y, Irimura T. Increased Expression of Sialyl Lewisx Antigen Correlates with Poor Survival in Patients with Colorectal Carcinoma: Clinicopathological and Immunohistochemical Study. Cancer Research. 1993;53(15):3632–3637. [PubMed] [Google Scholar]
- 30.Jørgensen T, Berner A, Kaalhus O, Tveter KJ, Danielsen HE, Bryne M. Up-Regulation of the Oligosaccharide Sialyl LewisX: A New Prognostic Parameter in Metastatic Prostate Cancer. Cancer Research. 1995;55(9):1817–1819. [PubMed] [Google Scholar]
- 31.Oh MJ, Hua S, Kim BJ, Jeong HN, Jeong SH, Grimm R, Yoo JS, An HJ. Analytical platform for glycomic characterization of recombinant erythropoietin biotherapeutics and biosimilars by MS. Bioanalysis. 2013;5(5):545–559. doi: 10.4155/bio.12.327. [DOI] [PubMed] [Google Scholar]
- 32.Greco FA, Oien K, Erlander M, Osborne R, Varadhachary G, Bridgewater J, Cohen D, Wasan H. Cancer of unknown primary: progress in the search for improved and rapid diagnosis leading toward superior patient outcomes. Annals of Oncology. 2012;23(2):298–304. doi: 10.1093/annonc/mdr306. [DOI] [PubMed] [Google Scholar]
- 33.Pavlidis N, Pentheroudakis G. Cancer of unknown primary site: 20 questions to be answered. Annals of Oncology. 2010;21(suppl 7):vii303–vii307. doi: 10.1093/annonc/mdq278. [DOI] [PubMed] [Google Scholar]
- 34.Oien KA. Pathologic Evaluation of Unknown Primary Cancer. Seminars in Oncology. 2009;36(1):8–37. doi: 10.1053/j.seminoncol.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 35.Keiser J, Bergsland E, Nakakura E. The Diagnosis and Management of Neuroendocrine Carcinoma of Unknown Primary. NEUROENDOCRINE TUMOR. 2012:37. [Google Scholar]
- 36.Fizazi K, Greco FA, Pavlidis N, Pentheroudakis G Group ObotEGW. Cancers of unknown primary site: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of Oncology. 2011;22(suppl 6):vi64–vi68. doi: 10.1093/annonc/mdr389. [DOI] [PubMed] [Google Scholar]
- 37.Epstein MA, Achong BG, Barr YM, Zajac B, Henle G, Henle W. Morphological and Virological Investigations on Cultured Burkitt Tumor Lymphoblasts (Strain Raji) Journal of the National Cancer Institute. 1966;37(4):547–559. [PubMed] [Google Scholar]
- 38.Klein G, Giovanella B, Westman A, Stehlin JS, Mumford D. An EBV-Genome-Negative Cell Line Established from an American Burkitt Lymphoma; Receptor Characteristics. EBV Infectibility and Permanent Conversion into EBV-Positive Sublines by in vitro Infection. Intervirology. 1975;5(6):319–334. doi: 10.1159/000149930. [DOI] [PubMed] [Google Scholar]
- 39.Friedlander M, Grogan M. Guidelines for the Treatment of Recurrent and Metastatic Cervical Cancer. The Oncologist. 2002;7(4):342–347. [PubMed] [Google Scholar]
- 40.Yee C, Krishnan-Hewlett I, Baker C, Schlegel R, Howley P. Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. The American journal of pathology. 1985;119(3):361. [PMC free article] [PubMed] [Google Scholar]
- 41.Wu S, Tao N, German JB, Grimm R, Lebrilla CB. Development of an Annotated Library of Neutral Human Milk Oligosaccharides. Journal of Proteome Research. 2010;9(8):4138–4151. doi: 10.1021/pr100362f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hua S, Nwosu C, Strum J, Seipert R, An H, Zivkovic A, German JB, Lebrilla C. Site-specific protein glycosylation analysis with glycan isomer differentiation. Anal Bioanal Chem. 2012;403(5):1291–1302. doi: 10.1007/s00216-011-5109-x. [DOI] [PubMed] [Google Scholar]
- 43.Hua S, Hu CY, Kim BJ, Totten SM, Oh MJ, Yun N, Nwosu CC, Yoo JS, Lebrilla CB, An HJ. Glyco-analytical multispecific proteolysis (Glyco-AMP): A simple method for detailed and quantitative glycoproteomic characterization. Journal of Proteome Research. 2013 doi: 10.1021/pr400442y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.