Abstract
Although CYP101D1 and P450cam catayze the same reaction at a similar rate and share strikingly similar active site architectures, there are significance functional differences. CYP101D1 thus provides an opportunity to probe what structural and functional features must be shared and what can differ yet maintain high catalytic efficiency. Crystal structures of the cyanide complex of wild type CYP101D1 and it active site mutants, D259N and T260A, have been solved. The conformational changes in CYP101D1 upon cyanide binding are very similar to P450cam indicating a similar mechanism for proton delivery during oxygen activation using solvent assisted proton transfer. The D259N-CN− complex shows a perturbed solvent structure compared to wild type which is similar to what was observed in the oxy-complex of the corresonding D251N mutant in P450cam. As in P450cam the T260A mutant is highly uncoupled while the D259N gives barely detectable activity. Despite these similarities, CYP101D1 is able to use the P450cam redox partners while P450cam cannot use the CYP101D1 redox partners. Thus the strict requirement of P450cam for its own redox partner is relaxed in CYP101D1. Differences in the local environment of the essential Asp (Asp259 in CYP101D1) provides a strucutral basis for understanding these functional differences.
Cytochromes P450 catalyze a variety of monooxygenase reactions that require electron transfer from redox partners. P450cam, a soluble bacterial cytochrome from Pseudomonas putida that hydroxylates camphor, has served as the primary model to address questions on the detailed mechanisms of electron transfer and O2 activation. The entire P450cam system consists of an iron-sulfur ferredoxin, putdiaredoxin (Pdx), that transfers electrons from the FAD protein, putidaredoxin reductase (Pdr), to P450cam. P450cam is unusual in having a strict and specific requirement for Pdx as an electron donor, which suggests that the specificity for Pdx is related to the Pdx-induced structural change required for oxygen activation.1–3 The recent P450cam-Pdx crystal and NMR structures4,5 indicate that the effector role of Pdx is to shift P450cam toward the open conformation, which enables the establishment of a water-mediated H-bonded network that is required for proton-coupled electron transfer and oxygen activation.5 Pdx binding has been proposed to free the critical residue, Asp251, from salt bridging interactions with Arg186 and Lys178 in order to serve its catalytic function6,7 in shuttling protons to dioxygen.
CYP101D1 is a close homolog to P450cam and catalyzes exactly the same camphor hydroxylation reaction, uses a similar ferredoxin (Arx) and ferredoxin reductase (Adr), but exhibits subtle and possibly important differences.8 Adjacent to the heme iron ligand, Cys357, is Leu358 in P450cam while this residue is Ala in CYP101D1. Leu358 plays a role in binding of the P450cam redox partner, putidaredoxin (Pdx).9 On the opposite side of the heme about 15 – 20 Å away the critical residue Asp251 in P450cam forms strong ion pairs with Arg186 and Lys178. In CYP101D1 a Gly replaces Lys178. Thus, the local electrostatic environment and ion pairing is substantially different in CYP101D1. Another important difference relates to the effect of K+ ions on substrate binding and spin state. Potassium ions promote both susbtrate binding and high-spin P450cam.10 P450cam has a K+ binding site near Tyr96 whose side chain OH group H-bonds with the camphor carbonyl oxygen of camphor so K+ helps to stabilize key substrate-protein interactions.11 CYP101D1 lacks this K+ binding site and K+ has only a modest effect on the fraction of high-spin CYP101D1.8 The addition of 200 mM KCl to the CYP101D1-camphor complex increases the spin state from 30% to only 40%.8 In our recent study aimed at comparing these two enzymes, these sites were systematically mutated in P450cam to the corresponding residues in CYP101D1.12 The data show that although individually the mutants have little effect on activity or structure, in combination there is a major drop in enzyme activity due the mutants being locked in the low-spin state thus preventing electron transfer from the P450cam redox partner, Pdx. Overall these results illustrate the delicate balance between the open and closed conformational states13 as well as a strong structural connection between the Pdx binding site (Leu358) and the region around Asp251 that undergoes a large structural change in the open/closed transition in P450cam. The results along with some structural and biochemical studies on CYP101D1 indicate that these complex interactions between spin-state, redox partner binding, and conformational changes might be less stringent or different in CYP101D1 primarily because this P450 is predominantly low-spin even in the presence of substrate14 but still is as active as P450cam. This opens up some interesting questions on how two such similar P450s can behave so differently and what, if any, the connection is between these various levels of conformational dynamics, activity, and biological function. Here we present the crystal structures of cyanide and camphor bound complexes of wild type CYP101D1 and its active site mutants, D259N and T260A, and enzyme activities. Since CYP101D1 is so similar to P450cam, a comparison of the two with respect to redox partner specificity has also been addressed to better understand whether the effector/allosteric role of redox partner binding is a general property of P450s or is unique to P450cam.
Experimental procedures
Protein expression and purification
The E. coli codon optimized genes encoding full-length ArR, Arx and CYP101D1 were sub cloned into the vector pET28a (Novagen Inc.) An N-terminal 6XHis tag was incorporated for both CYP101D1 and PdR. Arx gene was subcloned without the 6XHis tag. All proteins were expressed in Escherichia coli strain BL21(DE3).
For expression of wild type/ mutant CYP101D1, ArR and Arx, cells were grown at 37 °C in 1 liter flasks of Luria-Bertani broth (LB) medium containing the appropriate antibiotic until the optical density at 600nm reached 0.6–0.8. Recombinant protein production was induced with 1 mM isopropyl 1-thio-D galactopyranoside for overnight at 25°C following which cells were harvested by centrifugation and re-suspended in respective lysis buffers for further purification as described below.
For CYP101D1 (and its mutants), pink cell pellets were re-suspended in 50 mM potassium phosphate, pH 7.4, 150mM NaCl and then lysed by sonication at 4°C. The crude extracts were centrifuged at 16000 rpm for 1 hr and supernatant was loaded on to a Ni- IMAC column (BIO-RAD). The column was washed with lysis buffer containing 10mM imidazole and the target protein was eluted with elution buffer (50mM potassium phosphate, pH 7.4, 300 mM imidazole). The pooled fractions were concentrated and then buffer-exchanged into 50mM potassium phosphate and incubated for 4–5 hours with high purity thrombin (bovine) at room temperature (approximately 10 units of thrombin per mg of CYP101D1) to cleave the His tag. The incubated mixture was reloaded into Ni-IMAC and flow through was collected and concentrated and buffer exchanged with 20mM Tris, pH 8.0, 1 mM DTT. The protein was further purified on a Hi Trap Q column (GE Healthcare) using a linear gradient of 0–1M NaCl in 20mM Tris, pH 8.0, 1mM DTT. Gel filtration chromatography on a Superdex-75 (GE Healthcare) column was used to obtain highly pure CYP101D1 using buffer 20mM Tris, pH 8.0, 1 mM DTT.
ArR was purified using Ni-IMAC column as described for CYP101D1 using the same buffer composition. The elute from the Ni column was concentrated and loaded on Superdex 75 as described in the protocol for CYP101D1.
For Arx purification, the brown cell pellet was resuspended in 50mM potassium phosphate, pH 7.4, 1mM DTT. After sonication and high-speed centrifugation, the supernatant was loaded on DEAE sepharose fast flow (GE Healthcare). The column was washed and protein was eluted with a gradient of 50–500mM NaCl in 50mM potassium phosphate, pH 7.4, 1 mM DTT. The fractions containing Arx were pooled together and further purified using Superdex 75 (GE Healthcare) using buffer 20mM Tris, pH 8, 150mM NaCl, 1mM DTT.
Pdr and Pdx were expressed and purified using previously established protocols.15,16
Spectroscopic Studies
All UV-visible spectroscopy was performed using a Cary 3 spectrophotometer. CYP101D1 content was measured using an extinction coefficient of 107 mM−1 cm−1 at 418 nm 17. Concentrations of Arx and ArR were calculated using extinction coefficients of 9.3 mM−1 cm−1 at 414 nm and 10.0 mM−1 cm−1 at 458 nm, respectively 17. Concentrations of Pdx and Pdr were calculated using extinction coefficients of 5.9 mM−1cm−1 at 455 nm and 11.0 mM−1 cm−1 at 454 nm, respectively.18,19
Enzyme Assays
Cytochrome P450 hydroxylation activity was determined in the complete system of three proteins (CYP101D1, ArR, Arx) by measuring rates of camphor-dependent NADH oxidation at 25 °C following previously established protocols.20 The concentration of proteins and substrates used ensured that the rates measured were under steady state saturating conditions. Briefly, the reaction mixture of 1.2 mL contained 0.5 μM ArR, 5 μM Arx, and 0.5 μM P450 in 50 mM Tris, pH 7.4. The rate of NADH oxidation was measured by monitoring the absorbance change at 340 nm using 6.22 mM−1 cm−1. The reaction was initiated by first adding NADH (200 μM final concentration) in the absence of camphor to obtain the background rate. Substrate-dependent NADH oxidation was then assayed in the presence of 200 μM camphor and was calculated as the difference between the measured rate and the rate of nonspecific NADH oxidation in the absence of camphor (background). The assay was repeated with a 1mM final concentration of camphor. After the completion of the reaction (followed by UV-Vis), the reaction mixture was subjected organic extraction with dichloromethylene for product formation analysis using GC-MS according to previously established protocol.21
Crystallization of wild type CYP101D1 and mutants
All protein (wild type or mutant CYP101D1) crystals were grown at room temperature using the hanging drop vapor diffusion method in 100 mM Tris-HCl buffer, pH 8.2, 1.4–1.6M ammonium sulphate as the reservoir solution. The initial droplets contained 2 μL of protein solution at a concentration of ~30–35 mg/mL, and 2 μL of the reservoir solution and was equilibrated against a 500 μL of the reservoir solution. To generate camphor bound crystals, individual CYP101D1 crystals were soaked in reservoir solution containing saturated camphor and 20% glycerol (as cryoprotectant) for 1–2 hrs and flash cooled with liquid nitrogen. To generate the cyanide and camphor bound complex of CYP101D1 and mutants, the same protocol for generating the camphor bound complex was used and the reservoir solution in addition contained 50–60mM KCN. For substrate free conditions, crystals were soaked in reservoir solution containing 20% glycerol for 15–20 minutes and then flash frozen.
All data were collected remotely using the Stanford Synchrotron Radiation Lightsource (SSRL) beamline as indicated in Table 1. Data were indexed, integrated, and scaled with either MOSFLM,22 XDS23 or HKL2000.24 Molecular replacement calculations were carried out with Phaser25 through the CCP4i26 graphic interface using ferric camphor-bound wild type CYP101D1 (PDB code 3LXI) as a search model. Further structure refinements were performed using Phenix.refine.27 Table 1 lists data collection and refinement statistics.
Table 1.
Crystallographic data collection and refinement statistics
| Data set | CYP101D1 bound to camphor | CYP101D1 bound to camphor and cyanide | CYP101D1 substrate free glycerol bound | D259N CYP101D1 bound to camphor | D259N CYP101D1 bound to camphor and cyanide | T260A CYP101D1 bound to camphor |
|---|---|---|---|---|---|---|
| PDB Accesion Code | 4C9K | 4C9L | 4C9M | 4C9N | 49CO | 4C9P |
| Data collection | ||||||
| Space group | P6422 | P6422 | P6422 | P6422 | P6422 | P6422 |
| Resolution (Å) | 2.18 | 1.81 | 1.80 | 2.30 | 1.98 | 1.80 |
| Radiation source | SSRL 7-1 | SSRL 7-1 | SSRL 7-1 | SSRL 7-1 | SSRL 7-1 | SSRL 11-1 |
| Completeness (%) | 99.2 (99.2) | 99.7 (99.3) | 99.9 (100) | 100 (100) | 100 (100) | 99.8 (98.6) |
| No of unique reflections | 69832 (9954) | 121806 (5915) | 121820 (5966) | 59543 (8519) | 94411 (4631) | 123065 (17526) |
| Redundancy | 14.2 (14.3) | 9.4 (8.0) | 10.6 (10.1) | 20.5 (16.8) | 22.7 (14.5) | 16.2 (14.9) |
| Rsym or Rmerge | 0.085 (0.49) | 0.076 (0.38) | 0.075 (0.26) | 0.211 (1.06) | 0.169 (0.742) | 0.084 (1.07) |
| Rpim | 0.023 (0.133) | 0.036 (0.126) | 0.034 (0.126) | 0.034 (0.126) | 0.047 (0.260) | 0.021 (0.28) |
| I/σ(I) | 28.5 (7.0) | 16.4 (4.4) | 18.5 (6.2) | 15.0 (4.2) | 30.15 (3.8) | 22.3 (3.0) |
| Refinement | ||||||
| Resolution (Å) | 2.18 | 1.80 | 1.80 | 2.20 | 1.97 | 1.79 |
| B factor (mean) (Å2) | 30.81 | 23.67 | 23.20 | 35.65 | 23.16 | 30.02 |
| R work | 0.175 | 0.152 | 0.154 | 0.176 | 0.157 | 0.168 |
| R free | 0.218 | 0.182 | 0.188 | 0.208 | 0.185 | 0.193 |
| r.m.s.d bonds (Å) | 0.007 | 0.011 | 0.01 | 0.008 | 0.010 | 0.007 |
| r.m.s.d angles (°) | 1.155 | 1.402 | 1.353 | 1.153 | 1.317 | 1.192 |
| No of atoms | ||||||
| Protein | 6496 | 6496 | 6496 | 6504 | 6504 | 6476 |
| Ligand/Ions | 130 | 110 | 92 | 130 | 110 | 92 |
| Water | 543 | 977 | 927 | 485 | 819 | 782 |
Values for highest resolution shell are in parenthesis. Since Rmerge tends to increase with high multiplicity, Rpim, which is a multiplicity corrected version, has also been reported here 38.
Calculation of Salt Bridge Stability
In order to compare the strength of salt bridges involving a conserved Asp-Arg pair (Asp251-Arg186 in P450cam and Asp259-Arg188 in CYP101D1), the procedures used by Lounnas and Wade28 adapted from Hendsch and Tidor29 and the program Delphi30 were used. Each crystal structure was protonated and energy minimized using Amber31 and Delphi calculations were carried using full Amber charges. The stability of the salt bridge is given as ΔGtot = ΔGsol + ΔGprt + ΔGbrd where ΔGsol is the solvation energy, ΔGprt is the interaction of the side chains with the protein, and ΔGbrd is the bridging interaction energy between side chains. For these calculations charges were either turned off (set to zero) or left on. ΔGsolv is the sum of grid energies for the Asp in water, Arg in water, Asp with charges on in the protein with the rest of the protein charges off, and the same calculations with the Arg side chain. ΔGprt is the sum of grid energies for all charges on, both side chain charges on and protein charges off, and both side chains with charges off and protein charges on. ΔGbrd is the sum of grid energies of the Asp with charges on and the protein charges off, the Arg with charges on and the protein charges off, and both side chain charges on and the protein charges off. The internal and external dielectric constant were set to 4.0 and 80.0, respectively, at a salt concentration of 0.1M. The grid size was 0.5 Å per grid an only a single calculation was carried out.
Results
Crystal structures of substrate-free and –bound
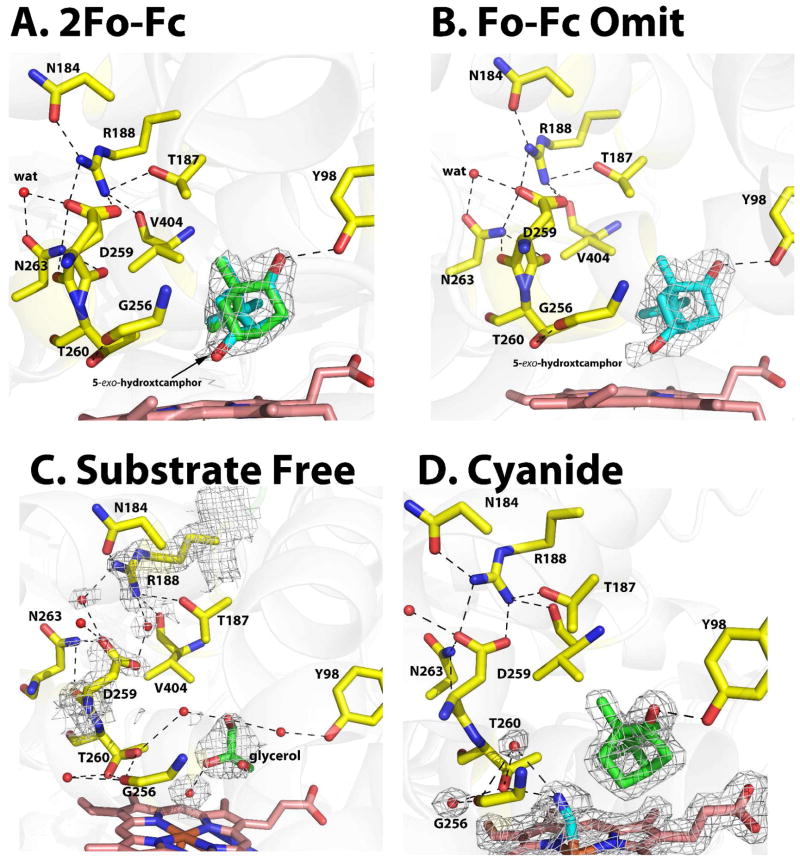
These structures already have been solved8 and here we present two new observations. Although the active sites of CYP101D1 and P450cam are nearly identical, one notable difference is that there is extra density extending from the C5 carbon of camphor which we interpret as the product, 5-exo-hydroxycamphor (Fig. 1A and B). This extra density could be due to a nonproductive binding mode of camphor with the carbonyl oxygen oriented toward the iron. This, however, is very unlikely since the extra desnity clearly is attached to a SP3 carbon and not SP2 planar which would be expected for the carbonyl oxyen atom. Moreover, if the camphor were partially misoriented then we would expect to see residual Fo-Fc difference density around the carbonyl oxygen atom which we do not. The best model has a 50:50 mix of substrate and 5-exo-hydroxycamphor suggesting that x-ray driven reduction of the crystals results in substrate hydroxylation. We have never observed this type of behavior with P450cam. The only time we have seen product in P450cam crystals is when the reduced-oxy complex receives high doses of x-rays or in crystals of the P450cam-Pdx complex. Thus CYP101D1 is more susceptible to x-ray driven O2 activation and substrate hydroxylation.
Figure 1.
Structures of the wild type CYP101D1 active site with camphor bound (A and B), in the absence of substrate (C), and the camphor-bound cyanide complex (D). Panel A shows the 2Fo-Fc map contoured at 1.0σ while panel B shows the omit Fo-Fc map contoured at 3.0σ. Electron density extending from C5 clearly indicates hydroxycamphor at about 50% occupancy. Hydroxycamphor is cyan and camphor is green. The substrate free sructure (Panel C) with glycerol repalcing the substrate shows that the ion pair between Asp259 and Arg188 has been disrupted. The cyanide complex (panel D) shows the 2Fo-Fc omit electron density contoured at 1.0σ. Water molecules are represented as red spheres. Electrostatic interactions are represented as black dashed lines.
Another possibly important difference involves Asp259 which is the essential Asp251 in P450cam. In P450cam the peptide carbonyl of Asp251 adopts an odd conformation and points perpendicular to the I helix axis rather than parallel as in a normal helix. In CYP101D1 the Asp259 peptide adopts two conformations in molecule A (Fig. 1A) of the two molecules in the asymmetric unit while in molecule B there is only one conformation. It thus appears that the peptide of Asp259 is more relaxed or flexible in CYP101D1 compared to corresponding Asp251 in P450cam. The salt bridge interactions between Asp259 and Arg188 in CYP101D1 appear weaker compared to the corresponding interactions in P450cam (residues in P450cam are Asp251 and Arg186). Unlike Asp251 in P450cam, Asp259 in CYP101D1 must compete with Asn184, Thr187 and Val404 all of which are close enough for electrostatic interactions with Arg188. Most importantly, the Asp251-Lys178 ion pair found in P450cam is missing in CYP101D1 since the Lys178 in P450cam corresponds to Gly180 in CYP101D1. As a result, water takes the place of the missing Lys side chain (Fig. 1A). We also estimated the stability of the Asp259-Arg188 ion pair using a computational approach employed earlier for P450cam.28 As a control we calculated the stability of the Asp251-Arg186 ion pair in P450cam to be −8.6kcal/mol which compares well with −8.44kcal/mol from the published value.28 In CYP101D1 the stability of this ion pair is calculated to be −5.9kcal/mol.
We attempted to obtain the substrate-free structure in hopes of capturing CYP101D1 in the open conformation. However, as in previous studies8 substrate-free CYP101D1 crystallized in the closed form and like the previous studies a solvent molecule is in the active site. In our case this solvent molecule is gycerol while in the earlier structure dioxane is bound.8 Although CYP101D1 remains closed there clearly is some loosening of the structure in regions important for function. In the substrate bound state Asp259 ion pairs with Arg188 which is analogous to the Asp251-Arg186 pair in P450cam. However, in the closed glycerol-bound structure the Asp259-Arg188 interaction is disrupted and water moves in to bridge between Asp259 and Arg188 (Fig. 1C). When P450cam adopts the open conformation the Asp251 ion pairs with neighboring residues are broken and Arg186 adopts a totally different orientation.
CYP101D1-Cyanide Complex
We chose to examine ferric-cyanide complexes since cyanide has been shown to be an excellent mimic of oxy-P450cam.32 Both O2 and cyanide binding result in an opening up of the I helix which enables two water molecules to move into the active site and establish an H-bonded network with O2/CN− that is thought to be important for O2 activation. The main question we address here is whether or not the same changes take place in CYP101D1.
The conformational changes in CYP101D1 upon cyanide binding are very similar to P450cam (Figure 1D). The cyanide molecule points toward Thr260, which rotates to form a hydrogen bond with the distal nitrogen atom of CN−. This further results in a widening of the groove formed by the stretch of residues encompassing Gly256 to Thr260. Overall, cyanide binding to CYP101D1 involves (a) breaking of the Thr260-Gly256 H-bond (b) movement of the Asp259 peptide to the orientation facing Asn263, and (c) opening of the I helix to accommodate two new water molecules in the active site, each H bonded to both Thr260 and Gly256 (Figure 1B). Overall the structure indicates a similar mechanism for proton delivery during oxygen activation in CYP101D1 and P450cam using solvent assisted proton transfer.
D251N Mutant Structures
In P450cam, the conserved Asp251 is crucial for shuttling protons from solvent molecules to the iron bound oxygen molecule.6,7,33 Structural studies on the oxygen complex of mutant D251N in P450cam showed that the Gly248-Thr252 H bond did not break and as a result the catalytically important water molecules could not enter the active site33 resulting in about a 100-fold decrease in activity.6,7 In our study, we have made the corresponding mutation D259N in CYP101D1 and solved the camphor-bound bound structure to 2.2 Å (Table 1). The peptide of Asn259 residue can adopt two alternate orientations as observed for the wild type enzyme (Fig. 2A). In molecule A, we observe the presence of some product in the hydroxycamphor density similar to the wild type enzyme. However, the contribution from hydroxycamphor is much less compared to the wild type enzyme. In D259N mutant molecule A, the occupancy for hydroxycamphor is ~30% where as in molecule B, it was only, ~15–20%. The H bonding network involving the side chains of Asn259 and Arg188 are altered with the loss of the salt bridge interactions. Arg188 moves away and new water molecules participate in the H bonding network. In the D259N-CN− complex the I helix adopts a similar open conformation as in the wild type CN− structure that includes two new water molecules (Fig. 2B). However, these waters are positioned quite differently. In the wild type structure one of the new waters can directly H-bond with CN− while in D259N this water is about 4.6Å from the CN−. The second new water molecule can also only H bond with Gly256 and not with Thr260. Part of the reason is that Thr260 adopts a different rotamer conformation in D259N which blocks the water from approaching much closer to CN−. These differences probably result from the I helix moving slightly closer to the CN− in D259N than in wild type. In addition, the Asp259-Arg188 ion pair in wild type more rigidly holds the structure in place while in D259N the side chain adopts a new rotamer conformation leaving Asn259 with no direct protein interactions.
Figure 2.
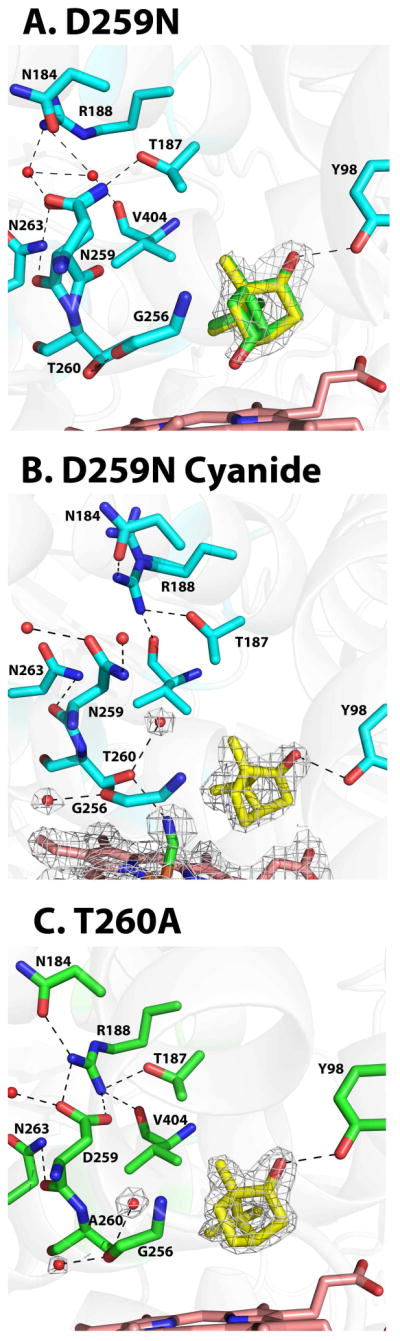
Structures of CYP101D1 D259N mutant. A) Camphor-bound structure showing the 2Fo-Fc electron density map contoured at 1.0σ. The occupancy of hydoxycamphor, ≈30%, which is lower than wild type. Hydroxycamphor is green and camphor is yellow. B) Cyanide complex showing the 2Fo-Fc electron density map contoured at 1.0σ. C) T260A mutant structure shown the 2Fo-Fc electron density map contoured at 1.0σ. There is no indication of hydroxycamphor.
T260A Mutant Structure
Another key residue in P450cam is Thr251. The T251A mutant is highly uncoupled33–35 and is thought to play an important role in the H-bonding requirements for O2 activation by possibly helping to stabilize the hydroperoxy intermediate.33 We made the corresponding T260A mutation in CYP101D1 and solved the structure of the camphor bound complex to 1.8 Å. We attempted to solve the cyanide complex as well but the electron density for the cyanide was weak indicating very low occupancy binding. Therefore, we did not further pursue this structure. Similar to the P450cam,34 in the camphor bound T260A mutant of CYP101D1, the I helix opens up thus enabling the two water molecules to enter the active site that are present only in the O2 or CN− wild type structures (Fig. 2C). We did not observe any hydroxycamphor density for this mutant in the active site for either molecule in the asymmetric subunit. The H bonding network involving the side chains of Arg188 and Asp259 are similar to the wild type enzyme.
Steady State Assays
Previous NADH oxidation assays carried showed that CYP101D1 has high turnover rates, coupling efficiency and affinity for camphor (Kd ~ 9μM).8 We performed activity assays with the wild type and mutant enzymes to test the effects of the mutations D259N and T260A on enzyme activity. Consistent with the earlier studies, the wild type CYP101D1 has high turnover rates comparable to P450cam and the reaction is highly coupled (Table 2). The presence or absence of the N term 6X His tag did not have any significant effect on enzyme activity. The mutant T260A retained significant (~50%) NADH turnover rates compared to the wild type but the reaction is highly uncoupled. On the other hand, the D259N lowers activity to the extent that no significant NADH turnover or product formation could be observed.
Table 2.
NADH turnover rates
| P450 type | Redox partner | NADH turnover (min−1) | Coupling efficiency (%) |
|---|---|---|---|
| WT CYP101D1 | Arr/Arx | 950 ± 42 | 95 |
| T260A CYP101D1 | Arr/Arx | 522 ± 20 | NA |
| D259N CYP101D1 | Arr/Arx | NA | NA |
| WT CYP101D1 | Pdr/Pdx | 40 ± 12 | 69 |
| WT P450cam | Pdr/Pdx | 930 ± 28 | 95 |
| WT P450cam | Arr/Arx | NA | NA |
The reaction mixture contained 0.5 μM Arr, 5 μM Arx, and 0.5 μM P450 in 50 mM Tris, pH 7.4 The value for P450cam are taken from our previous work.12 The rate of NADH oxidation for CYP101D1 in presence of either 200 μM or 1mM D-camphor at room temperature was measured by monitoring the absorbance change at 340 nm using 6.22 mM−1 cm−1. No significant difference in rates were observed at the two camphor concentrations. NA means no significant turnover or product formation was observed.
Redox Partner Selectivity
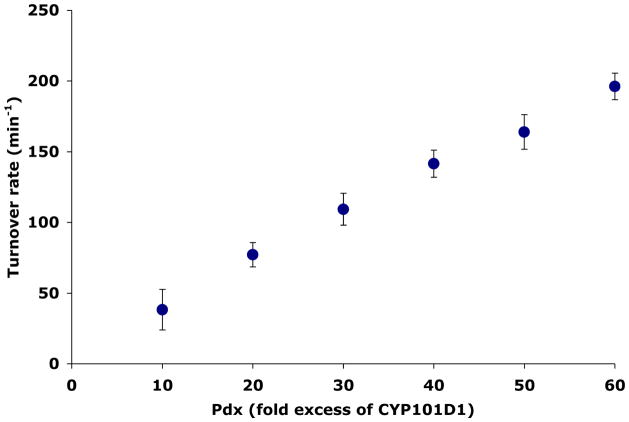
Since the recent P450cam-Pdx crystal structure5 strongly implicates Pdx binding as being critical to structural changes involving Asp251 required for proton coupled electron transfer, we next examined the question of redox partner selectivity in CYP101D1. The supporting proteins for CYP101D1, the Arx ferredoxin and ArR ferredoxin reductase, are very similar in structure to the P450cam counterparts, Pdx and Pdr, although residues at the docking interfaces are substantially different.8 We measured activity of the wild type CYP101D1 using the P450cam redox partners, Pdr and Pdx. As shown in Fig. 3 there is a linear increase in activity with increasing Pdx concentration but does not saturate although the reaction is significantly coupled (Table 2). This is consistent with relatively robust electron transfer reaction but poor binding of Pdx to CYP101D1 and that full saturation is not achieved even at the highest concentration of Pdx used, 60μM. This shows that CYP101D1 is not as selective for its redox partner as P450cam which further suggests that Arx may not serve a major effector role in CYP101D1 as opposed to Pdx which plays a critical effector role in P450cam. As expected we failed to observe any significant activity with P450cam using ArR and Arx, the CYP101D1 redox partners.
Figure 3.
NADH turnover rates for CYP101D1 with the redox partners of P450cam Pdr and Pdx. The rate increases linearly upon increasing the Pdx concentration.
Discussion
CYP101D1 was the first P450 to be found that catalyzes exactly the same reaction as P450cam using very similar supporting redox proteins.14,17 CYP101D1 thus provides an opportunity to probe what structural and functional features must be shared and which can differ yet maintain high catalytic efficiency. One of the more interesting and possibly unusual properties of P450cam is its strict requirement for its own redox partner, Pdx. In addition, camphor shifts the conformational equilibrium of P450cam completely from the open low-spin state to the closed high-spin state.13 CYP101D1 shares neither of these properties. Earlier studies8 as well as our own more recent work shows that camphor shifts CYP101D1 to at most 40–45% high-spin yet CYP101D1 is as active and as tightly coupled as P450cam. In addition, the CYP101D1 is not specific for its own ferredoxin electron donor, Arx, since Pdx can substitute for Arx. This suggest that specific protein-protein interactions may not play as important an effector role in CYP101D1 as in P450cam.
We first address the question of incomplete spin shift in CYP101D1. With P450cam it now appears that the high-spin substrate bound state is associated with the closed conformation while the low-spin state is associated with the open state.13 The closed to open switch in P450cam results in a large structural change involving a significant repositioning of the F and G helices which effectively exposes the active site to bulk solvent.13 So far there is no structure of an open CYP101D1 but there is an open structure of CYP101D2.36 CYP101D1 and CYP101D2 share 62% sequence identity.36 Open CYP101D2 has camphor bound but in a non-productive orientation where the substrate carbonyl oxygen H-bonds with the water ligand coordinated to the heme iron and thus is a substrate bound low-spin complex. If CYP101D1 also can form a substrate-bound low-spin complex then reduction of the low-spin form would result in dissociation of the water ligand, which then might allow the camphor to reorient into a productive position. The main difference between P450cam and CYP101D1 then would be that substrate bound low-spin CYP101D1 can be reduced but not low-spin P450cam and, perhaps, P450cam cannot form a low-spin substrate bound complex.
To address the question on the effector role of redox partner binding, we first needed to check if the key active site residues important in proton coupled electron transfer in P450cam are also critical in CYP101D1. As expected, the answer is yes. Both Thr260 and Asp259 are important. The various CYP101D1-CN− structures also show that CN− induces very much the same changes in CYP101D1 as it does in P450cam even to the extent that the same “catalytic” waters enter the active site to provide part of the proton delivery machinery. The D259N-CN− structure is particularly insightful since here the catalytic waters are not properly placed which also is the effect of this mutation on P450cam. With P450cam the D251N mutant has much reduced activity but just enough to measure about a 5-fold increase in the kinetic solvent isotope effect37 which strongly argues that Asp251 plays an important role in proton delivery to dioxygen. This view is consistent with our current CYP101D1 results and shows that the proton delivery machinery operates the same in both P450s.
It also has been proposed that Asp251 in P450cam is intimately tied to the effector role of Pdx.5 In the closed conformation Asp251 is tied up with ion pairs to Lys178 and Arg186 so it is not possible for Asp251 to participate in a proton shuttle network. However, when Pdx binds these salt bridges are broken suggesting that one of the main effector roles of Pdx is to free Asp251 to perform its role in proton coupled electron transfer. Asp259 in CYP101D1 also is tied up with Arg188 but the residue corresponding to Lys178 in P450cam, which also ion pairs with D251, is a glycine in CYP101D1. In addition, we found that although substrate-free CYP101D1 with a glycerol molecule in the active site is in the closed conformation, the Asp259-Arg188 ion pair has been broken and replaced by water molecules that bridge between these two residues. It thus is easier to free Asp259 in CYP101D1 to serve its role in proton delivery while P450cam is more firmly tied down in the closed conformation and requires Pdx to provide additional stabilization of the open form which frees Asp251 from its salt bridges. In sharp contrast, CYP101D1 does not require this extra “help” since the Asp259-Arg188 interaction is more readily disrupted. One final observation indicating that it is easier for CYP101D1 to adopt the active conformation is that product is observed in electron density maps in CYP101D1 but not in P450cam. If O2 activation requires proton coupled electron transfer, then x-ray induced reduction must be accompanied by proton transfer to dixoygen in order to hydroxylate the substrate. The tightly closed conformation in P450cam prevents ready access of bulk solvent protons to the active site so even upon reduction of the heme iron by x-rays, product does not form. However, since the active site entry in CYP101D1 is looser, solvent protons can enter more easily and product is observed.
An equally challenging question to address is the biological advantage/disadvantage for a tightly controlled system like P450cam vs. a more promiscuous system like CYP101D1. P450cam is encoded on a plasmid and is part of a tightly regulated operon so that the P450cam proteins are produced only when needed. When camphor is the only or primary carbon source, it would be critical to efficiently utilize reducing equivalents for only camphor oxidation. Without such selectivity any low potential reductant could turnover P450cam resulting in wasted reducing equivalents not utilized for camphor oxidation in addition to the generation of toxic peroxide and superoxide. Selectivity is achieved by tightly coupling substrate binding to spin-state, redox partner binding, and proton coupled electron transfer. In contrast, CYP101D1 is one of several P450s in Novosphingobium aromaticivorans so camphor oxidation by CYP101D1 is unlikely to be critical for using camphor as a carbon source since one of the other several P450s could serve this function. In addition, Novosphingobium aromaticivorans has only one ferredoxin, Arx, that services several other P450s which is somewhat reminiscent of mammalian liver P450s where several promiscuous P450s are supported by a single reductase. In these cases an additional level of control requiring an effector role for the reductase is not required. It is tempting to speculate that the CYP101D1 is representative of a simpler P450 system where fewer levels of control are required since CYP101D1 is not an essential component in the oxidative assimilation of a specific carbon source and thus is not critical to the organisms survival. Nature, however, required a higher level of control with P450cam-like systems where the P450 is an essential component in energy transduction and thus must be more tightly regulated and controlled.
Acknowledgments
This work was supported by NIH grant GM33688 and NIH/NCI Institutional Training Grant Fellowship T32CA009054
This paper involves research carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. We also thank Dr. Huiying Li and Dr. Irina Sevrioukova for helpful discussions and advise.
Abbreviations
- CYP
Cytochrome P450
- Pdx
Putidaredoxin
- Pdr
Putidaredoxin reductase
- ArR
ferredoxin reductase
- Arx
[2Fe-2S] ferredoxin
- NADH
Nicotinamide adenine dinucleotide hydride
- GC-MS
Gas chromatography-mass spectrometry
Footnotes
Coordinates and structure factors have been deposited in the Protein Data Base under accession numbers 4C9K, 4C9L, 4C9M, 4C9N, 49CO, 4C9P
References
- 1.Tyson CA, Lipscomb JD, Gunsalus IC. The role of putidaredoxin and P450 cam in methylene hydroxylation. The Journal of biological chemistry. 1972;247:5777–5784. [PubMed] [Google Scholar]
- 2.Lipscomb JD, Sligar SG, Namtvedt MJ, Gunsalus IC. Autooxidation and hydroxylation reactions of oxygenated cytochrome P-450cam. J Biol Chem. 1976;251:1116–1124. [PubMed] [Google Scholar]
- 3.Nagano S, Shimada H, Tarui A, Hishiki T, Kimata-Ariga Y, Egawa T, Suematsu M, Park SY, Adachi S-i, Shiro Y, Ishimura Y. Infrared spectroscopic and mutational studies on putidaredox-induced conformational changes in ferrous CO-P450cam. Biochemistry. 2003;42:14507–14514. doi: 10.1021/bi035410p. [DOI] [PubMed] [Google Scholar]
- 4.Hiruma Y, Hass MA, Kikui Y, Liu WM, Olmez B, Skinner SP, Blok A, Kloosterman A, Koteishi H, Lohr F, Schwalbe H, Nojiri M, Ubbink M. The Structure of the Cytochrome P450cam-Putidaredoxin Complex Determined by Paramagnetic NMR Spectroscopy and Crystallography. J Mol Biol. 2013 doi: 10.1016/j.jmb.2013.07.006. in press. [DOI] [PubMed] [Google Scholar]
- 5.Tripathi S, Li H, Poulos TL. Structural basis for effector control and redox partner recognition in cytochrome P450. Science (New York, NY. 2013;340:1227–1230. doi: 10.1126/science.1235797. [DOI] [PubMed] [Google Scholar]
- 6.Gerber NC, Sligar SG. A role for Asp-251 in cytochrome P-450cam oxygen activation. J Biol Chem. 1994;269:4260–4266. [PubMed] [Google Scholar]
- 7.Gerber NS, Sligar SG. Catalytic mechanism of cytochrome P450 - evidence for a distal charge relay system. J Amer Chem Soc. 1992;174:725–735. [Google Scholar]
- 8.Yang W, Bell SG, Wang H, Zhou W, Hoskins N, Dale A, Bartlam M, Wong LL, Rao Z. Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444. J Biol Chem. 2010;285:27372–27384. doi: 10.1074/jbc.M110.118349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagano S, Tosha T, Ishimori K, Morishima I, Poulos TL. Crystal structure of the cytochrome P450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding. J Biol Chem. 2004;279:42844–42849. doi: 10.1074/jbc.M404217200. [DOI] [PubMed] [Google Scholar]
- 10.Lange R, Debey P. Spin transition of camphor-bound cytochrome P-450. 1 local paH and electrostatic interactions. Eur J Biochem. 1979;94:485–489. doi: 10.1111/j.1432-1033.1979.tb12916.x. [DOI] [PubMed] [Google Scholar]
- 11.Poulos TL, Finzel BC, Howard AJ. High-resolution crystal structure of cytochrome P450cam. J Mol Biol. 1987;195:687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 12.Batabyal D, Li H, Poulos TL. Synergistic Effects of Mutations in Cytochrome P450cam Designed To Mimic CYP101D1. Biochemistry. 2013;52:5396–5402. doi: 10.1021/bi400676d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee YT, Wilson RF, Rupniewski I, Goodin DB. P450cam visits an open conformation in the absence of substrate. Biochemistry. 2010;49:3412–3419. doi: 10.1021/bi100183g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell SG, Wong LL. P450 enzymes from the bacterium Novosphingobium aromaticivorans. Biochrm Biophys Res Comm. 2007;360:666–672. doi: 10.1016/j.bbrc.2007.06.119. [DOI] [PubMed] [Google Scholar]
- 15.Sevrioukova IF, Poulos TL. Putidaredoxin reductase, a new function for an old protein. J Biol Chem. 2002;277:25831–25839. doi: 10.1074/jbc.M201110200. [DOI] [PubMed] [Google Scholar]
- 16.Sevrioukova I, Gracia C, Li H, Bhaskar B, Poulos TL. Crystal structure of putidaredoxin, the [2Fe-2S] component of the P450cam monooxygenase system from Pseudomonas putida. J Molec Biol. 2003;333:377–392. doi: 10.1016/j.jmb.2003.08.028. [DOI] [PubMed] [Google Scholar]
- 17.Bell SG, Dale A, Rees NH, Wong LL. A cytochrome P450 class I electron transfer system from Novosphingobium aromaticivorans. Appl Microbiol Biotechnol. 2010;86:163–175. doi: 10.1007/s00253-009-2234-y. [DOI] [PubMed] [Google Scholar]
- 18.Kuznetsov VY, Blair E, Farmer PJ, Poulos TL, Pifferitti A, Sevrioukova IF. The putidaredoxin reductase-putidaredoxin electron transfer complex: theoretical and experimental studies. J Biol Chem. 2005;280:16135–16142. doi: 10.1074/jbc.M500771200. [DOI] [PubMed] [Google Scholar]
- 19.Gunsalus IC, Wagner GC. Bacterial P-450cam methylene monooxygenase components: cytochrome m, putidaredoxin, and putidaredoxin reductase. Methods Enzymol. 1978;52:166–188. doi: 10.1016/s0076-6879(78)52019-3. [DOI] [PubMed] [Google Scholar]
- 20.Sevrioukova I, Hazzard JT, Tollin G, Poulos TL. Laser flash induced electron transfer in P450cam monoxygenase: putidaredoxin reductase-putidaredoxin interaction. Biochemistry. 2001;40:10592–10600. doi: 10.1021/bi010874d. [DOI] [PubMed] [Google Scholar]
- 21.Churbanova IY, Poulos TL, Sevrioukova IF. Production and characterization of a functional putidaredoxin reductase-putidaredoxin covalent complex. Biochemistry. 2010;49:58–67. doi: 10.1021/bi901872s. [DOI] [PubMed] [Google Scholar]
- 22.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr. 2011;D67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lounnas V, Wade RC. Exceptionally stable salt bridges in cytochrome P450cam have functional roles. Biochemistry. 1997;36:5402–5417. doi: 10.1021/bi9622940. [DOI] [PubMed] [Google Scholar]
- 29.Hendsch ZS, Tidor B. Do salt bridges stabilize proteins? A continuum electrostatic analysis. Protein Sci. 1994;3:211–226. doi: 10.1002/pro.5560030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilson MK, Honig BH. Calculation of electrostatic potentials in an enzyme active site. Nature. 1987;330:84–86. doi: 10.1038/330084a0. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case D. Development and testing of a general Amber force field. J Amer Chem Soc. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 32.Fedorov R, Ghosh DK, Schlichting I. Crystal structures of cyanide complexes of P450cam and the oxygenase domain of inducible nitric oxide synthase-structural models of the short-lived oxygen complexes. Arch Biochem Biophys. 2003;409:25–31. doi: 10.1016/s0003-9861(02)00555-6. [DOI] [PubMed] [Google Scholar]
- 33.Nagano S, Poulos TL. Crystallographic study on the dioxygen complex of wild-type and mutant cytochrome P450cam - Implications for the dioxygen activation mechanism. J Biol Chem. 2005;280:31659–31663. doi: 10.1074/jbc.M505261200. [DOI] [PubMed] [Google Scholar]
- 34.Raag R, Martinis SA, Sligar SG, Poulos TL. Crystal structure of the cytochrome P-450CAM active site mutant Thr252Ala. Biochemistry. 1991;30:11420–11429. doi: 10.1021/bi00112a008. [DOI] [PubMed] [Google Scholar]
- 35.Kimata Y, Shimada H, Hirose T, Ishimura Y. Role of Thr-252 in cytochrome P450cam: a study with unnatural amino acid mutagenesis. Biochem Biophys Res Commun. 1995;208:96–102. doi: 10.1006/bbrc.1995.1310. [DOI] [PubMed] [Google Scholar]
- 36.Yang W, Bell SG, Wang H, Zhou W, Bartlam M, Wong LL, Rao Z. The structure of CYP101D2 unveils a potential path for substrate entry into the active site. Biochem J. 2011;433:85–93. doi: 10.1042/BJ20101017. [DOI] [PubMed] [Google Scholar]
- 37.Vidakovic M, Sligar SG, Li H, Poulos TL. Understanding the role of the essential Asp251 in cytochrome p450cam using site-directed mutagenesis, crystallography, and kinetic solvent isotope effect. Biochemistry. 1998;37:9211–9219. doi: 10.1021/bi980189f. [DOI] [PubMed] [Google Scholar]
- 38.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science (New York, NY. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]