Abstract
Neoadjuvant chemotherapy (NAC) induces a pathologic complete response (pCR) in approximately 30% of patients with triple-negative breast cancers (TNBC). In patients lacking a pCR, NAC selects a subpopulation of chemotherapy-resistant tumor cells. To understand the molecular underpinnings driving treatment-resistant TNBCs, we performed comprehensive molecular analyses on the residual disease (RD) of 74 clinically-defined TNBCs after NAC including next-generation sequencing (NGS) on 20 matched pre-treatment biopsies. Combined NGS and digital RNA expression analysis identified diverse molecular lesions and pathway activation in drug-resistant tumor cells. Ninety percent of the tumors contained a genetic alteration potentially treatable with a currently available targeted therapy. Thus, profiling residual TNBCs after NAC identifies targetable molecular lesions in the chemotherapy-resistant component of the tumor which may mirror micro-metastases destined to recur clinically. These data can guide biomarker-driven adjuvant studies targeting these micro-metastases to improve the outcome of patients with TNBC who do not respond completely to NAC.
Keywords: next-generation sequencing, breast cancer, neoadjuvant chemotherapy
Introduction
Neoadjuvant chemotherapy (NAC) is used increasingly in patients with triple-negative breast cancer (TNBC), a subtype lacking expression of estrogen receptor (ER), progesterone receptor (PR) or human epidermal growth factor-2 (HER2) amplification. The goals of NAC are to increase likelihood of breast-conserving surgery and to eliminate clinically silent micro-metastases. Approximately 30% of TNBC patients who receive NAC achieve a pathological complete response (pCR). These patients have a favorable recurrence-free (RFS) and overall survival (OS)(1-3). The remaining patients with residual viable cancer in the breast or lymph nodes exhibit high rates of metastatic recurrence and an overall poor long term outcome(1-3).
Immunohistochemistry (IHC) of the proliferation marker Ki67 in the post-NAC residual disease (RD) has been shown to correlate with patient outcome (4-6). Previous studies showing the prognostic ability of Ki67 after NAC included all subtypes of breast cancer (i.e. HER2-enriched, luminal A, luminal B and basal-like), which also offer prognostic information (7-9). We have recently shown that these subtypes differ vastly in their post-NAC Ki67 scores, confounding the prognostic utility of Ki67 in this setting (10), and has been confirmed by other investigators(9). Furthermore, Ki67 scoring is difficult to standardize among clinical laboratories and many studies have defined different ‘cutoffs’ for patient stratification, ranging from 14-50%(4-6). Finally, the Ki67 scoring of the post-NAC residual tumor is not actionable as it does not identify a pathogenic driver of the tumor and, as such, a drug target and rational treatment decision.
Intuition suggests that tumor cells remaining after NAC contain the cancer cell population intrinsically resistant to chemotherapy. These tumor cells likely mirror the micro-metastatic component of the disease that is ultimately responsible for distant metastases, and is unlikely to be highly sensitive to further chemotherapy once clinical metastases become evident. The standard of care for patients with TNBC who have RD after NAC is observation as therapies that would be effective in reducing recurrences are unknown. Thus, we molecularly-profiled the RD remaining after NAC in a cohort of 111 TNBCs (including gene expression analysis of 89 tumors and NGS of 80 tumors, 74 of which were TNBC) to identify lesions that could be therapeutically targeted in adjuvant trials.
Results
Ki67 does not predict clinical outcome in TNBCs
Since TNBC is a heterogeneous subtype of breast cancer(11), we determined if Ki67 could predict patient outcome within this clinical subtype by scoring Ki67 in the RD of a cohort of 111 TNBCs after NAC. Patient demographics are listed in Supplementary data, Table 1. Molecular subtyping based on gene expression using the PAM50 centroids(7) revealed a predominance of tumors with basal-like gene expression (Supplementary data, Figure 1A). After adjusting for 7 tumors that exhibited HER2 amplification (see below), 70% of TNBCs were basal-like, which is similar to previously published rates in larger datasets (12). Basal-like status was associated with a trend toward worse RFS and OS (Log-rank p=0.12 and p=0.058, respectively; Supplementary data, Figure 1B). As we have previously demonstrated(10), the Ki67 score in the RD varied significantly among molecular subtypes within this TNBC cohort, but was not prognostic (Supplementary data, Figure 1C-D). Ki67 staining decreased significantly in response to chemotherapy (p<0.0001, paired-t-test; Supplementary data, Figure 1E), but this change was not different among the molecular subtypes (Supplementary data, Figure 1F). Tumor cellularity was significantly decreased between the pre and post-NAC samples (paired t-test p<0.0001) (Supplementary data, Figure 1G). Node status at surgery (an established prognostic marker), but not a change in Ki67, was predictive of both RFS and OS, although this effect appeared to be confined only to post-menopausal women (Supplementary data, Fig 2). These data suggest that the underlying molecular subtype may confound the prognostic ability of Ki67 score in the RD after NAC.
Genomic alterations in drug-resistant residual cancers after NAC
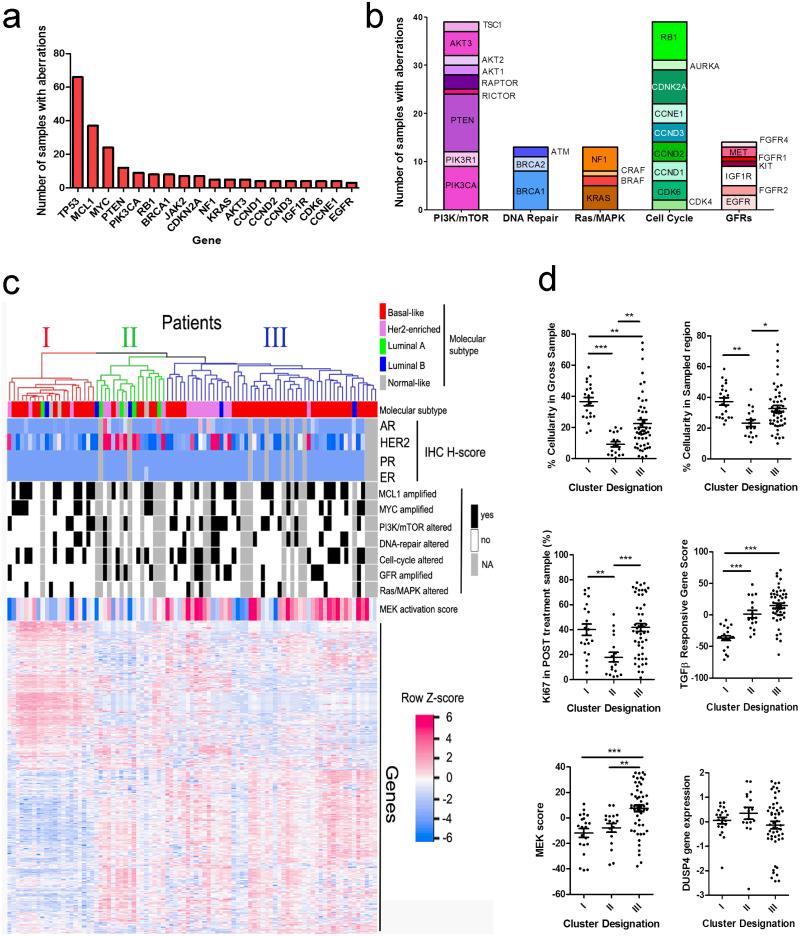
To identify targetable molecular lesions present in breast cancers after NAC, we performed targeted next-generation sequencing (NGS) of 3,320 exons of 182 oncogenes and tumor suppressors plus 37 introns of 14 genes frequently rearranged in cancer (Supplementary data, Table 2), including intervallic targets throughout the genome for copy number alteration (CNA) analysis. NGS analysis was attempted in 85 formalin-fixed, paraffin-embedded (FFPE) cancers with sufficiently high tumor cellularity (>20%); 81 (95%) were successfully analyzed. Mean depth of coverage across all samples was 609X (range: 131-1215). Six samples lacked sufficient depth of coverage (<200X) to make calls in CNAs with confidence. Seven tumors harbored ERBB2 amplification (confirmed by FISH) and were excluded from further analysis. All remaining post-NAC tumors were ER- and PR-negative by IHC. Thus, 74 tumors had evaluable NGS data, 68 of which also had CNA data. No obvious differences between the NGS-evaluable population and the entire cohort were observed in terms of outcome or clinical characteristics. NGS analysis revealed a diversity of lesions, many of which were present in less than 5% of samples (Figure 1A and Supplementary data, Table 3).
Figure 1. Targetable alterations and pathways in TNBCs after NAC.
A) Most common recurrently altered genes detected by NGS, representing amplifications, deletions, rearrangements and known somatic mutations. B) Organization and representation of altered genes (n=81 tumors) into 5 functional and targetable pathways. A total of 118 genomic alterations were identified across 81 tumors (1.5 alterations/tumor). C) Integrated molecular analysis of residual tumors, using unsupervised clustering based on gene expression patterns (NanoString). D) Scatterplots depicting the differences among the clusters identified in (C) for cellularity in the entire FFPE block cross section; cellularity in the sampled (macro-dissected) hotspot; Ki67 score; TGF-β response signature; MEK signature; and DUSP4 gene expression. *p<0.05; ** p<0.01; ***p<0.001.
Alterations in TP53 were identified in 72/81 (89%), which is similar to other studies of basal-like or TNBC including The Cancer Genome Atlas (TCGA) dataset (~85%)(13, 14). The next most common alterations included MCL1 (54%) and MYC (35%) gene amplifications. MYC amplifications were detected primarily in basal-like tumors (42% basal vs. 10% all others; Fisher’s exact p=0.018) and with a similar frequency as in the basal-like cohort in the TCGA (Supplementary data, Table 4). Compared to basal-like primary tumors in the TCGA, we detected a higher frequency of MCL1 amplifications (54% in post-NAC TNBC vs. 19% in TCGA basal-like tumors; p=0.0006), PTEN deletions or mutations (trend, p=0.0697) and JAK2 amplifications (trend, p=0.08) in the RD. Amplifications in CDK6 and CCND1-3 were collectively enriched as well (24% in post-NAC TNBC vs. 10% in TCGA basal-like tumors). This difference suggests that these alterations are present at higher frequency in chemotherapy-treated TNBCs, and may play a role in de novo or acquired therapeutic resistance. However, it is important to note that these comparisons of copy-number alterations with the TCGA data are made between platforms (NGS versus Affymetrix SNP arrays), and thus some variation in calling rates and detection of alterations may be platform-specific.
Identified alterations were categorized into several key pathway or functional groups: cell cycle alterations (amplifications in CDK4, CDK6, CCND1-3, CCNE1 or AURKA and loss of CDKN2A, CDKN2B or RB1); PI3K/mTOR alterations (amplifications of AKT1-3, PIK3CA, RAPTOR, RICTOR, loss or mutation of PTEN, truncations or nonsense mutations in TSC1, amplifications or mutations in PIK3CA or PIK3R1); growth factor receptor (GFR) amplifications (EGFR, MET, KIT, FGFR1-2 and 4 or IGF1R); Ras/MAPK alterations (amplifications/gains of KRAS, BRAF, RAF1, or truncations of NF1); or DNA repair alterations (truncations, loss or mutations of BRCA1, BRCA2, or mutations in ATM) (Figure 1B). Importantly, over 90% of patients had alterations in at least one of these clinically targetable pathways.
Gene expression analysis
NanoString gene expression analysis was performed in 104/111 samples; 89 samples (86%) passed quality control metrics (Supplementary data, Table 5). Sixty-five samples were analyzed by both NGS and NanoString. Overall, 450 transcripts were quantified. These 450 transcripts were selected based on their inclusion in published gene expression signatures or based on their association with the post-NAC Ki67 score we reported recently (15). Specifically, we included the PAM50 genes (7), a signature of MEK activation (16), a signature of TGFβ activation (17) and genes we have previously shown to correlate with the post-NAC Ki67 score (10). These signatures were selected based on our previous studies demonstrating association of DUSP4 loss with the MEK activation signature, and with the enrichment of TGFβ inducible genes after NAC (15, 18). There was excellent concordance between gene expression and CNAs or mutations in cases where both were assessed (Supplementary data, Figure 3). Gene expression data were utilized to predict the molecular subtype using the PAM50 centroids. Of the 89 samples, 10 were predicted to be of the luminal subtype. These samples were confirmed ER- and PR-negative by IHC. Gene expression analysis confirmed low ESR1 and PGR expression for all samples, with the exception of two outliers for ESR1 mRNA expression (basal-like and luminal B respectively, both ER- by IHC, data not shown). Neither of these samples was included in the NGS analysis. This phenomenon has been noted and discussed elsewhere (19), and one possible explanation to the presence of ER/PR negative samples with luminal-like gene expression patterns as the ‘Luminal Androgen Receptor’ subtype which expresses the AR hormone receptor (11).
Visualization of expression patterns identified distinct gene signatures that did not correlate with the breast cancer molecular subtype or pathway alterations identified by NGS, but appeared to correlate with the MEK signature score (Figure 1C). To explore these patterns, we identified the three most prominent gene expression clusters (Clusters I-III, Figure 1C) by hierarchical clustering. These clusters contained core gene sets of 84, 6, and 30 genes, respectively expressed within each cluster (Supplementary data, Table 6). Tumor cellularity in the gross specimen appeared to be a defining factor of these clusters, where cluster II represented the most paucicellular tumors with the lowest Ki67 staining (Figure 1D). Importantly, the cellularity was less of a defining feature after considering the cellularity of the ‘hotspot’ regions that were macro-dissected for gene expression analysis. Thus, the gene expression patterns may be influenced by tumor sampling but could also be reflective of the underlying microenvironment resulting from a strong anti-tumor effect from neoadjuvant therapy. Clusters I and III appeared more similar in terms of cellularity and Ki67 staining (Figure 1D). However, Cluster I had a distinct lack of expression of TGFβ-responsive genes. Cluster III had a high MEK signature score. This cluster contained a group of tumors with low expression of DUSP4, a negative-regulator of the MAPK pathway, offering at least one possible mechanism of MEK activation (Figure 1D). Importantly, survival of the patients comprising these clusters was not significantly different (data not shown).
Bioinformatic exploration of these genesets with the Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb) suggested that Cluster I was driven primarily by luminal-like breast expression patterns (Supplementary Data, Table 7), despite a lack of ER, PR, or AR IHC staining in these tumors. Furthermore, several overlapping gene signatures suggested that tri-methylated H3K27 (H3K27me3) genes were highly expressed in this cluster. H3K27me3 maintains epigenetic silencing of developmental genes in stem cells, leaving them poised for expression upon differentiation (20). Thus, we speculate Cluster I is composed of more differentiated tumors, with high Ki67 and low TGFβ and MEK activity. In contrast, Cluster III-expressed genes overlapped with invasive signatures across many types of cancer, including signatures of poorly-differentiated cancers, suggesting that this cluster reflects tumors maintained in a less differentiated state and toward a higher stem-cell like hierarchy. The expression patterns in cluster III, reflective of high MEK and TGF-β activation (including tumors with DUSP4 loss), are consistent with stem-like phenotypes induced by these pathways, as we have previously demonstrated (15, 18, 21).
Selection of oncogenic alterations by chemotherapy
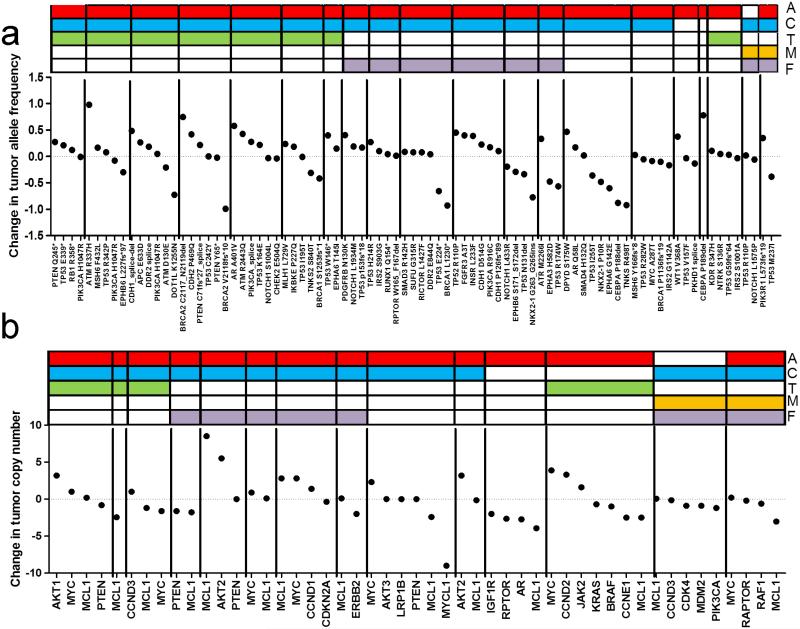
In order to quantify enrichment of alterations during NAC, we analyzed 20 matched pre-treatment biopsies by NGS. We detected gain (not detected in pre-treatment while detected in post-treatment sample) and enrichment (detected in pre-treatment but increased in post-treatment sample) in mutational allele frequencies and copy number estimations in 41 patient-specific alterations (Supplementary data, Figure 4). Many of these enrichments and gains occurred in genes comprising cell cycle regulators and PIK3CA/mTOR pathway genes. Although the number of alterations in GFRs and DNA repair genes were low in this subset, significant gains and enrichments were noted in these pathways as well.
Some paired samples demonstrated gains or enrichments across several lesions, suggesting a difference in regional sampling or tumor purity between the pre- and post-therapy specimen. To accommodate these variations, we normalized each sample to its estimated tumor purity (see methods) to calculate a fold change in allele or copy number frequency across each tumor pair. This produced an expected pattern of normal distribution around zero of changes in alterations as a result of NAC, assuming that most alterations should not be selected for or against by chemotherapy (Figure 2A-B). When analyzed by this method, several alterations were highly enriched relative to other within-sample alterations. These included two mutations in ATM; R337H and R2443Q, TP53 T253fs*11, a CDH1 splice site deletion, KDM6A L214fs*, AR A401V, and DPYD S175W. When examining CNAs in tumor pairs, we found that copy numbers of AKT and CCND family members were increased in 3 of 4 tumors each. Although copy number of MYC and MCL1 were enriched in several cases following NAC, this effect was not consistent in all tumor pairs. Furthermore, there was no clear concordance of case-specific enrichment with the therapeutic agents utilized for NAC. However, since the frequency of MCL1 amplifications was higher in this post-NAC cohort relative to primary tumors in the TCGA, this discordance suggests MCL1 amplification may be associated with de novo resistance to chemotherapy, but is not enriched further upon treatment.
Figure 2. Quantitative changes in gene alterations in TNBC tumor pairs before and after NAC.
A) Change in allele frequency of known and likely somatic mutations during NAC, adjusted for tumor purity assessment. Each segment (n=20) represents a patient. Type of neoadjuvant chemotherapy is depicted with a different color at the top; A: Adriamycin, C: cyclophosphamide, T: taxane, M: methotrexate, F: fluorouracil. B) Change in copy-number during NAC, adjusted for tumor purity assessment.
Co-amplification of MYC and MCL1 in the RD of TNBC
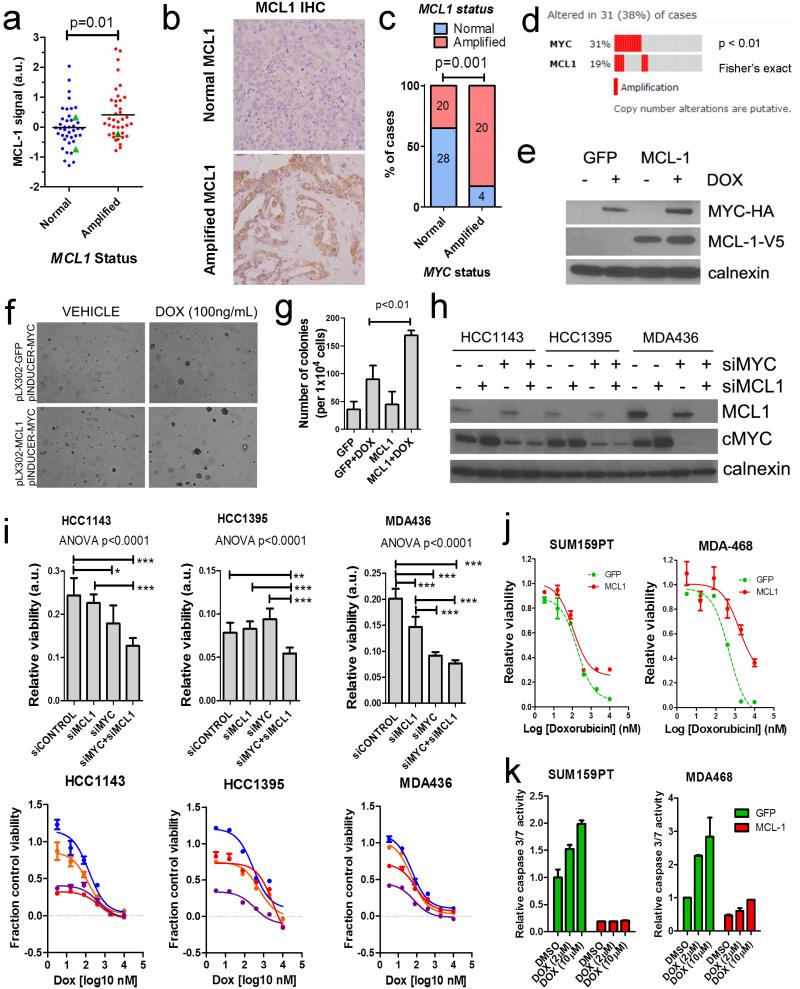
The anti-apoptosis MCL1 protein is dynamically regulated during cell cycle progression and shows rapid turnover rates in cancer cells (22). To determine whether MCL1 CNAs contribute to higher protein levels in breast cancer, we performed IHC for MCL1 on tissue microarrays (TMAs) of this cohort. MCL1 amplification was significantly associated with increased protein expression (p=0.01; Figure 3A-B). However, MCL1 amplification does not appear to be the sole factor in modulating protein expression in breast cancer, as several samples showed high MCL-1 protein levels by IHC in the absence of CNAs. We also detected 3 frameshift or nonsense mutations in FBXW7, the E3 ubiquitin ligase responsible for targeting MCL-1 (and MYC) for proteasome-mediated degradation (23). However, presence of these mutations was not associated with higher protein levels of MCL-1 (Figure 3A).
Figure 3. Co-amplification and interaction of MYC and MCL1 in TNBCs.
A) MCL-1 IHC score as quantified on TMAs of TNBCs after NAC. Signal intensity (a.u: arbitrary units) was normalized for tumor area and number of nuclei. FBXW7-mutant patients are shown as green triangles. B) Example images of high and low MCL-1 expressing tumors by IHC. C) Co-amplification of MCL1 and MYC in residual breast tumors assessed in this study. Absolute numbers of tumors are shown in bars (p=0.001, Fisher’s exact test). D) Co-amplification of MCL1 and MYC in primary basal-like breast tumors in the TCGA(13, 38). E) Western blot of MCF10A cells expressing pLX302-GFP (control) or pLX302-MCL-1 (V5-tagged) and pINDUCER22-MYC (HA tagged) ± doxycycline treatment. F) Soft agar colony formation assay of MCF10A cells in (E) ± doxycycline where indicated. G) Quantification of colonies from (F). Each bar represents the mean colony number of triplicate wells ± SD. H) siRNA knockdown of MYC and MCL-1 in HCC1143 (MYC-amplified, MCL1-gain), HCC1395 (MYC-amplified), and MDA-436 (MYC-amplified and MCL1-amplified) cells (13). I) Baseline viability (upper panels) and response to dose titration of 72 h of doxorubicin (lower panels) of cell lines after siRNA knockdown. Viability curves are shown as relative to siCONTROL, DMSO treated controls. J) Viability curves of cells transduced with MCL-1 or GFP control and treated for 48 h with a dose titration of doxorubicin. Viability was measured with CellTiter-Glo (Promega). K) Caspase cleavage in cells from (J) after 5 h doxorubicin at the indicated doses. Caspase 3/7 cleavage was measured with Caspase-Glo (Promega).
We detected a high degree of concordance between CNAs in both MYC and MCL1. MCL1 expression has been shown to facilitate MYC-induced lung cancers and leukemogenesis(24-26), although this interaction has not been shown in breast cancer. Indeed, 83% of MYC amplified tumors also showed CNAs at MCL1 (p=0.001; Figure 3C). Co-occurence of MYC and MCL1 amplification was not associated with altered prognosis (RFS or OS) compared to patients with amplification of either gene alone in this dataset (data not shown). This co-occurrence was also present in the basal-like breast cancers in the TCGA (p<0.01; Figure 3D), although the frequency of MCL1 alterations was lower.
To test whether MCL1 overexpression facilitates MYC-induced transformation in breast cells, we stably expressed MCL1 or GFP (control) in MCF10A cells and then transduced the cells with a doxycycline-inducible MYC vector (Figure 3E). While MYC induction induced sporadic transformation of MCF10A ascertained by anchorage-independent growth assays, concurrent overexpression of MCL1 markedly increased MCF10A colony formation (Figure 3F-G). Furthermore, in TNBC cell lines demonstrating gains or amplifications in MYC and/or MCL1 including HCC1143 (MYC-amplified, MCL1-gains), HCC1395 (MYC-amplified), and MDA-436 (MYC-amplified and MCL1-amplified) (13), siRNA knockdown of MYC and MCL1 reduced cell viability (Figure 3H-I). siRNA-targeting of MYC and MCL1 did not alter the relative sensitivity to doxorubicin, a commonly utilized chemotherapeutic agent in the neoadjuvant setting. However, knockdown of these oncogenes increased the fractional kill at lower doses of doxorubicin relative to non-targeting siRNA (siCONTROL) treated cells (Figure 3I). Furthermore, lentiviral-mediated overexpression of MCL-1 increased resistance to doxorubicin and docetaxel (Figure 3J and Supplementary data, Figure 5A-B). Resistance to doxorubicin was mediated in part by decreased baseline and doxorubicin-mediated apoptosis (Figure 3K). Thus, MYC and MCL1 enhance cell fitness, and MCL1 additionally protects TNBCs from chemotherapy-induced apoptosis.
Molecular alterations in the RD after NAC correlate with patient outcome
Next, we explored the prognostic impact of genomic alterations and gene expression signatures identified by NGS and NanoString analysis, respectively (16, 18). Gene-specific alterations occurring in at least 8 (>10%) analyzed tumors were tested for prognostic impact (RFS and OS) by the likelihood ratio test (Table 1). Alterations in pathways, gene expression signature scores and clinical variables were also tested. Of note, JAK2 amplification was strongly associated with poor RFS (p=0.006; HR=3.36), while BRCA1 truncations/mutations and JAK2 amplification predicted poor OS (p=0.041; HR=2.5, and p=0.002; HR 4.16, respectively). PTEN alterations were a favorable prognostic factor for OS (p=0.03; HR=0.14). MYC amplifications also demonstrated a trend toward worse OS (P=0.084; HR=1.78). When alterations were categorized into functional pathways, DNA repair alterations were weakly associated with poor OS (P=0.09; HR=1.89). Interestingly, a high MEK activation score (16) predicted poor RFS and OS (P=0.059; HR=1.758 and P=0.013; HR=2.264, respectively). These data offer insights into molecular alterations that may predict the natural history of TNBC after NAC.
Table 1.
Prognostic ability of clinical factors and molecular alterations
| Recurrence-free survival |
Overall survival |
|||||||
|---|---|---|---|---|---|---|---|---|
| Clinical (All patients)* | n | events | HR | p- value |
n | events | HR | p- value |
| Ki67 score > 15 | 105 | 60 | 0.915 | 0.772 | 108 | 52 | 1.179 | 0.623 |
| Node positive | 104 | 60 | 1.664 | 0.068 | 107 | 52 | 1.534 | 0.147 |
| Age > 50 | 105 | 60 | 0.932 | 0.790 | 108 | 52 | 0.872 | 0.630 |
| Post-menopausal | 105 | 60 | 0.681 | 0.138 | 108 | 52 | 0.711 | 0.221 |
| Neoadjuvant taxane | 105 | 60 | 1.255 | 0.390 | 108 | 52 | 1.157 | 0.625 |
| Genes (TNBC only) | ||||||||
| MCL1 = amp | 64 | 40 | 0.620 | 0.130 | 67 | 36 | 0.620 | 0.140 |
| MYC = amp | 64 | 40 | 1.370 | 0.400 | 67 | 36 | 1.780 | 0.084 |
| PIK3CA = amp/mut | 64 | 40 | 0.530 | 0.380 | 67 | 36 | 0.300 | 0.210 |
| BRCA1 = trunc/mut | 64 | 40 | 1.700 | 0.260 | 67 | 36 | 2.500 | 0.041 |
| RB1 = del/trunc | 64 | 40 | 0.910 | 0.850 | 67 | 36 | 0.760 | 0.660 |
| PTEN = del/trunc/mut | 64 | 40 | 0.570 | 0.280 | 67 | 36 | 0.140 | 0.030 |
| JAK2= amp | 64 | 40 | 3.360 | 0.006 | 67 | 36 | 4.160 | 0.002 |
| Pathways (TNBC only) | ||||||||
| Cell cycle altered | 64 | 40 | 0.580 | 0.100 | 67 | 36 | 0.560 | 0.110 |
| DNA repair altered | 64 | 40 | 1.570 | 0.253 | 67 | 36 | 1.890 | 0.090 |
| PI3K/mTOR altered | 64 | 40 | 0.813 | 0.540 | 67 | 36 | 0.660 | 0.280 |
| Ras/MAPK altered | 64 | 40 | 1.000 | 0.980 | 67 | 36 | 0.860 | 0.755 |
| GFR amplified | 64 | 40 | 1.030 | 0.920 | 67 | 36 | 1.220 | 0.600 |
| Gene signatures (TNBC only) | ||||||||
| MEK signature = int/high | 79 | 42 | 2.120 | 0.035 | 81 | 37 | 3.170 | 0.004 |
| TGFβ signature = int/high | 79 | 42 | 1.250 | 0.500 | 81 | 37 | 1.570 | 0.207 |
Clinical data analysis of clinically defined TNBC includes 7 samples later identified as ERBB2 amplified by NGS.
p≤0.05 (significant) or p≤0.1 (statistical trend) defined in bold.
Prognostic interaction of MEK activation and MYC amplification
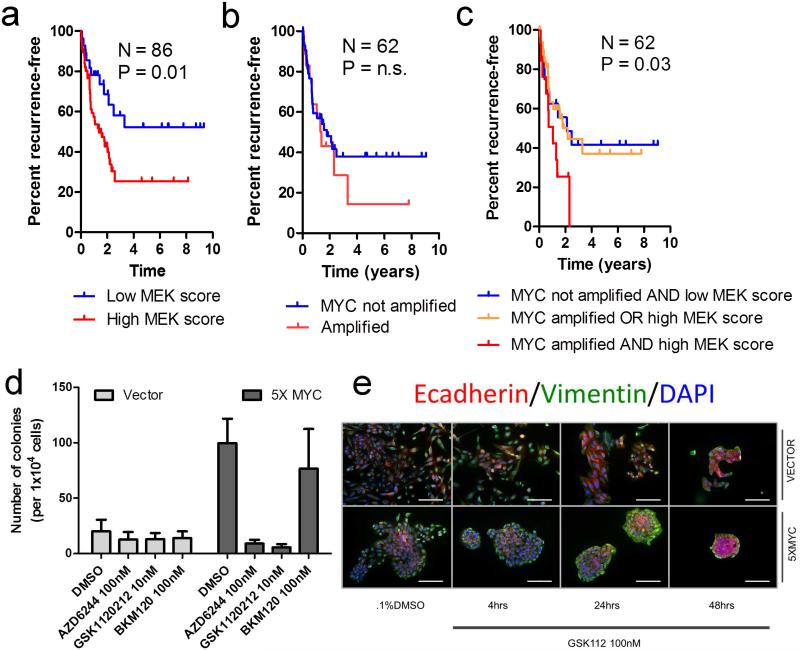
MYC amplifications in the RD trended toward an association with poor OS, while high MEK transcriptional output was a risk factor for reduced RFS and OS. Importantly, the Ras/MAPK pathway has been shown to cooperate with the MYC oncogene (27, 28). Thus, we tested the possibility that these perturbations may interact with one another. When the interaction term was tested by Cox proportional hazards analysis, a significant interaction was noted for RFS but not for OS (P=0.03 and p=0.83, respectively). Kaplan-Meier analysis confirmed this association (Figure 4A-C).
Figure 4. Interaction of MYC amplification with MEK pathway activity correlates with poor prognosis in TNBCs.
A) Kaplan Meier (KM) analysis of RFS in patients with a high MEK transcriptional signature(16) (highest 66%) vs. all others (lowest 33%). B) KM analysis of RFS in MYC-amplified patients versus those with normal MYC copy number. C) Combined KM analysis of patients with a high MEK transcriptional signature and MYC amplification versus those with either or neither alteration. D) Quantification of 3-week soft agar colony formation assays using MCF10A cells stably-transduced with MYC (5X MYC) vs. vector control, plated in the presence or absence of a single dose of AZD6244/selumetinib, GSK1120212/trametinib or the pan-PI3K inhibitor BKM120 at the indicated concentrations. Bars represent the mean colony number ± SD of 3 replicates. E) Immunofluorescence of E-cadherin, vimentin and DAPI in cells from (D) grown on chamber slides and treated with 100 nM GSK1120212/trametinib. Scale bars represent 50 μm.
The effect of the interaction between MYC and MEK activation on patient outcome suggested a mechanistic interaction linking these pathways to tumor progression. This cooperation has been demonstrated in experimental models, where MEK stabilizes MYC expression (28-35). For example, c-Myc overexpression in transgenic mice results in spontaneous breast tumors which activate MEK through the generation of KRAS mutations (31). To test this interaction on a molecular level, we utilized MCF10A cells stably transduced with MYC (5XMYC)(36). Stable expression of MYC induced the formation of anchorage-independent MCF10A colonies. However, treatment with a single dose of a MEK1/2 inhibitor (GSK1120212/trametinib or AZD6244/selumetinib) completely abolished the ability of MYC to induce MCF10A colonies (Figure 4D and Supplementary data, Figure 6 A-B). This effect was MEK-specific, as treatment with the pan-PI3K inhibitor BKM120 had no effect on MCF10A anchorage-independent growth. Treatment of 5XMYC cells with a MEK inhibitor resulted in the formation of polarized normal acini, as observed by immunofluorescence for basal (CK5, vimentin), luminal (CK8, e-cadherin), and a tight-junction marker (ZO-1) (Figure 4E, and Supplementary data Figure 6C-D). These data suggest that MYC cooperates with Ras/MAPK to drive anchorage-independent growth in breast cancer.
Molecular profiling for rational selection of adjuvant targeted therapies
Despite the high likelihood of recurrence, the current standard of care for patients with TNBC who do not achieve a pCR after NAC is watchful waiting. Patients who recur with metastatic cancer are less likely to exhibit prolonged responses to conventional anti-cancer therapy, since the micrometastases that generated these clinical recurrences have already been exposed to chemotherapy in the neoadjuvant setting. This unmet need suggests the identification of actionable molecular targets in the RD could, in turn, be explored in adjuvant trials after NAC and mastectomy. Since the spectrum of alterations present in such tumors is highly diverse, we integrated existing preclinical and clinical data into an ‘actionability’ table of rational therapies for the identified alterations (Table 2 and Supplementary data, Table 6).
Table 2.
Actionability of lesions identified in at least three post-NAC specimens
| Gene Symbol |
# altered | Category | Potential therapy |
|---|---|---|---|
| TP53 | 73 | D | Prognostic (poor, potentially sensitive to WEE1 inhibitors e.g. MK1775) |
| MCL1 | 40 | C | Resistance to anti-tubulins e.g.paclitaxel, MCL1 inhibitor in development |
| MYC | 24 | C | Aurora kinase inhibitors e.g. MLN8237, AMG 900; possible sensitivity to CDK inhibitors |
| PIK3CA | 13 | B | PI3 kinase, mTOR inhibitors e.g. Everolimus, Temsirolimus, others |
| PTEN | 12 | B | PI3K/mTOR inhibitors e.g. GSK2636771, Everolimus, Temsirolimus, others |
| BRCA1 | 9 | B | PARP Inhibitors e.g. Olaparib, CEP-9722, Rucaparib, others |
| RB1 | 9 | D | Prognostic |
| JAK2 | 8 | D | JAK2 inhibitors e.g. Ruxolitinib, others |
| ERBB2 | 7 | A | Herceptin, Lapatinib and others |
|
CDKN2A/
B |
7 | E | CDK4/6 inhibitors e.g. PD0332991, LEE011, P276-00 |
| NF1 | 5 | C | MAPK/PI3K/mTOR inhibitors e.g. MSC1936369B, Everolimus, Temsirolimus, others |
| AKT3 | 5 | C | AKT inhibitors e.g. MK2206, PI3K/mTOR inhibitors e.g. Everolimus, Temsirolimus |
| KRAS | 5 | A | Resistance to cetuximab, MEK inhibitors e.g. MEK162 |
| CCND1 | 5 | C | CDK4/6 inhibitors e.g. PD0332991, LEE011, P276-00 |
| CCND3 | 4 | C | CDK inhibitors, Kinetin riboside |
| CCNE1 | 4 | C | CDK2/4/6 inhibitors e.g. ABT-888, PD0332991, LEE011, P276-00 |
| CCND2 | 4 | C | CDK inhibitors, Kinetin riboside |
| CDK6 | 4 | C | CDK4/6 inhibitors e.g. PD0332991, LEE011, P276-00 |
| IGF1R | 4 | C | IGF1-R Inhibitors e.g. AMG-479, BMS-754808, MK-0646, IMC A12, others |
| LRP1B | 3 | E | Biologically relevant, presently no known targeted therapies |
| PIK3R1 | 3 | C | PI3K-PATHWAY INHIBITORS |
| ATM | 3 | C | PARP Inhibitors e.g. Olaparib, CEP-9722, Rucaparib |
| BRCA2 | 3 | B | PARP Inhibitors e.g. Olaparib, CEP-9722, Rucaparib, others |
| EGFR | 3 | A | Cetuximab, Panitumumab and others |
| FBXW7 | 3 | C | Resistance to anti-tubulins, potential sensitivity to PI3K/mTOR inhibitors |
| CDK4 | 3 | C | CDK4/6 inhibitors e.g. PD0332991, LEE011, P276-00 |
| RPTOR | 3 | E | Biologically relevant, possible sensitivity to mTORC1 and mTORC2 inhibitors |
| Category A: Approved/standard alterations that predict sensitivity or resistance to approved/standard therapies | |||
| Category B: Alterations that are inclusion or exclusion criteria for specific experimental therapies | |||
| Category C: Alterations with limited evidence that predict sensitivity or resistance to standard or experimental therapies | |||
| Category D: Alterations with prognostic or diagnostic utility | |||
| Category E: Alterations with clear biological significance in cancer (i.e. driver mutations) without clear clinical implications to date | |||
Of note, we detected two tumors (2.5%) with presumed loss of function alterations in TSC1: a truncation of an intergenic region between intron 8 and the 3’UTR contained in the 23rd intron (Supplementary data, Figure 7) which was detected in matched pre- and post-NAC samples in one patient, and a nonsense mutation (TSC1_Q516*, 20% allele frequency) in another patient (Supplementary data, Table 2). Loss of TSC1 function has been proposed as a basis for significant clinical responses in metastatic bladder cancer to the TORC1 inhibitor everolimus (37), and may also represent a therapeutically actionable target in patients with breast cancer. Importantly, loss of function TSC1 deletions, truncations or nonsense mutations were not identified in the TCGA breast cancer study (13, 38).
Discussion
Herein, we have described the genomic landscape of drug-resistant tumor cells remaining in the breast of patients with TNBC after anti-cancer chemotherapy. We also performed serial analysis to detect changes in CNAs and mutations before and after NAC. These data provide insights into genomic alterations which may predict de novo or acquired resistance to standard anti-cancer therapies in TNBC and could inform on the effective use of rational molecularly targeted agents in adjuvant trials. In an effort to not confound our results with tumors with variable residual cancer burden (RCB), we focused on a cohort of cancers with significant macroscopic residual disease after NAC. Indeed, this represents a cohort with a particularly poor prognosis (median survival ~18 months).
Several molecular insights were gained through this analysis. We showed that the Ki67 score after NAC does not provide prognostic information in patients with TNBC. Further, we confirmed our previous report demonstrating that Ki67 in the RD is intimately related to the underlying molecular subtype (15). We also found frequent co-amplification of MCL1 and MYC which conferred an advantage in anchorage-independent growth. Importantly, these co-amplifications were more frequent in this study compared to those previously reported in primary basal-like breast tumors. Amplification of the MYC oncogene coinciding with gene expression signatures of MEK activity identified a group of patients with very poor prognosis. Further, MEK inhibitors potently inhibited 3D growth of MYC-overexpressing cells, suggesting a role for MEK inhibitors in MYC-amplified breast cancers.
We also detected a higher frequency of several potentially targetable alterations in this cohort of post-treatment TNBCs compared to basal-like primary breast cancers in the TCGA. These included PTEN alterations (PI3K and AKT inhibitors), amplifications of JAK2 (ruxolinib or tofacitinib), CDK6, CCND1, CCND2, CCND3 (CDK4/6 inhibitors) and IGF1R (dalotuzumab). Importantly, several patients’ tumors showed an enrichment of AKT family CNAs and cyclin D family CNAs after NAC, suggesting an association of these alterations with resistance to chemotherapy. TSC1 truncations and mutations were also identified. These alterations have been associated with high sensitivity to the TORC1 inhibitor everolimus in other tumor types (37), suggesting they generate tumor dependence on the TOR pathway.
Overall, this analysis provides new information on the molecular alterations present in chemotherapy-resistant tumor cells within TNBCs. As supported by the poor outcome of patients with TNBC that recur with metastatic disease after an incomplete response to NAC, we surmise these persistent tumor cells are resistant to conventional cytotoxic chemotherapies without the addition of novel agents targeting these oncogenic pathways. Further, these data suggest that molecular analysis of TNBCs not achieving a pCR to NAC should be performed routinely in order to stratify patients according to this information to rational adjuvant trials with molecularly targeted agents.
Materials and methods
Patients and tumor specimens
One-hundred-eleven (111) surgically-resected tumor samples were from patients with triple negative breast cancer diagnosed and treated with neoadjuvant chemotherapy at the Instituto Nacional de Enfermedades Neoplásicas in Lima, Perú. Clinical and pathological data were retrieved from medical records under an institutionally approved protocol (INEN 10-018). Tumors were determined as triple negative if they were negative for estrogen receptor, progesterone receptor and HER2 overexpression measured by IHC. A subset of cases was subjected to HER2 FISH to resolve discrepant findings between the HER2 IHC results and the PAM50 subtype assignment. The results were further verified by comparison with the NGS results. The diagnostic biopsy (pre-NAC) was obtained for NGS analysis in a subset (n=21) of these patients.
Immunohistochemistry
Antigen retrieval for Ki67 was performed using HpH Buffer (pH 9.0) in a decloaking chamber (Biocare Medical, Concord, MA). The Ki67 antibody (m7240; DAKO, Denmark) was utilized at a 1:75 dilution overnight. Visualization was performed using the 4 Plus Detection System (Biocare) and DAB (DAKO) as the chromogen. The section was scanned at 100x magnification and the area containing the highest number of positive cells was selected. Positive and negative tumor cells were manually counted at 400x; the percentage of positive cells was calculated with at least 700 viable cells.
Antigen retrieval for ER and PR were performed using citrate buffer (pH 6) in a decloaking chamber (Biocare Medical, Concord, MA). The ER (6F11; Vector Labs, Burlingame, CA) and PR (PgR636; DAKO, Carpenteria, CA) antibodies were utilized at 1:200 and 1:50 dilutions, respectively, for a 1-h incubation. Visualization for both antibodies was performed using the Envision Detection System (DAKO, Carpenteria, CA) and DAB (DAKO, Carpenteria, CA) as the chromogen. The percentage of invasive tumor cells with nuclear staining and the average intensity of all positively staining tumor cells in the in the section were manually counted per CAP/ASCO guidelines(39).
Antigen retrieval for HER2 was performed using HpH Buffer (pH 9.0) in a decloaking chamber (Biocare Medical, Concord, MA). The HER2 antibody (#2242; Cell Signaling, Beverly, MA) was utilized at a 1:200 dilution overnight. Visualization was performed using the Envision Detection System (DAKO) and DAB (DAKO) as the chromogen. The percentage of invasive tumor cells with membranous staining at the highest intensity level was manually assessed and recorded per CAP/ASCO guidelines(40).
Antigen retrieval for MCL-1 was performed in citrate buffer (pH 6.0) under pressure for 15 min; endogenous peroxidase activity was blocked by incubating with 3% H2O2 for 10 min. The sections were incubated with Mcl-1 antibody (Santa Cruz, sc-819) at 1:800 dilution overnight at 4° Celsius and developed by using DAB substrate (Vector Laboratories, Burlingame, CA). Automated slide scanning and scoring were performed at the Vanderbilt Epithelial Biology Center Imaging Resource. Images were captured and quantitated using the Ariol SL-50 automated microscope system (Leica Microsystems, San Jose, CA) at 20X. Selected areas at original resolution are displayed. Immunoreactivity intensity scores were determined in areas of residual tumor cells selected by expert breast pathologists (JMG and MGK) and averaged for redundant tissue cores.
HER2 fluorescence in situ hybridization
FISH for detection of amplification of HER2 was performed using the PathVysion HER-2 DNA Probe Kit (PathVysion Kit, Abbott Molecular, Des Planes, IL) utilizing the Vysis LSI HER-2/neu 17q11.2-12 SpectrumOrange TM and Vysis CEP 17 17p11.1-q11.1 SpectrumGreen Alpha Satellite DNA probes. Images were visualized on a Fluorescence Olympus BX60 Microscope and analyzed using the Genus™ for Genetic Image Analysis software, version 3.6. The ratio of HER2 to CEP 17 signals was recorded and reported as an average ratio per CAP/ASCO guidelines (40).
Immunofluorescence
Immunofluorescence staining was performed as described previously(41). All primary and secondary antibodies were diluted in 12% Fraction V BSA (RPI-Cat#A30075). The following antibodies and dilutions were used: ZO-1 (Life Technologies-Cat#617300) 1:200, E-Cadherin (BD-Cat#610182) 1:200, Vimentin (Covance-Cat#PCK-594P) 1:500, CK5 (Covance-Cat#PRB-160P-100) 1:500, CK8 (RDI Fitzgerald-Cat#20R-CP004) 1:500. Secondary goat antibodies were highly cross-adsorbed and used at 1:200 (Molecular Probes/Life Technologies). Chamber slides were briefly rinsed in PBS and fixed in 4% paraformaldehyde with 0.1% Triton-X100 for 20 min. Cells were rinsed 3 times with PBS and primary antibodies were applied overnight. Secondary antibodies were incubated for 20 min and rinsed 4 times in PBS then mounted in SlowFade +DAPI (Molecular Probes/Life Technologies).
Tissue microarrays
Triplicate 0.6-mm cores were punched from post-NAC FFPE tumor blocks from the patient’s surgical resection specimen using the Beecher Manual Tissue Arrayer MTA-1 (Beecher Scientific, Beecher, IL). Three arrays containing approximately 100 cores per array were generated.
Gene expression analyses
RNA was isolated from FFPE tumor blocks by macro-dissecting tumor-rich regions from 3-6 10-μm sections. RNA was purified using the RNEasy FFPE kits (Qiagen). Gene expression analysis was performed by NanoString as previously described(10). Raw transcript counts were subtracted from background (negative input control). Normalization of raw transcript counts was performed by dividing by the geometric mean of 7 housekeeper-control genes; NPAS1, NAGA, POLR1B, CD40, WAS, B2M, and TUBB. Housekeeper-normalized transcript counts were log2 transformed and data were row z-score standardized prior to further analysis. PAM50 analysis was performed using the PAM50 qRT-PCR centroids(7) on the normalized log2 gene expression data prior to z-standardization.
Sequencing and primary sequence data analysis
Eighty-five (85) post-NAC tumors and 26 pre-NAC biopsies were submitted for NGS at Foundation Medicine Inc. A 4-μm of hematoxylin and eosin stained slide was reviewed by an expert pathologist to ensure 1) a sample volume of ≥1mm3, 2) nucleated cellularity ≥80% or ≥30,000 cells, and 3) that ≥20% of the nucleated in the sample are derived from the tumor. DNA was extracted from 40-μm of unstained FFPE sections, typically 4x10 μm sections by digestion in a Proteinase K buffer for 12-24 h and then purified with the Promega® Maxwell 16 Tissue LEV DNA kit; 50-200 ng of dsDNA in 50-100 uL water in microTUBEs were fragmented to ~200 bp by sonication (3 min, 10% Duty, Intensity=5, 200 Cycles/Burst ; Covaris E210) before purification with a 1.8X volume of AMPure XP Beads (Agencourt). SPRI purification and subsequent library construction with the NEBNext kits (E6040S, NEB), containing mixes for end repair, dA addition and ligation, are performed in 96-well plates (Eppendorf) on a Bravo Benchbot (Agilent) using the “with-bead” protocol 43 to maximize reproducibility and library yield. Indexed (6-bp barcodes) sequencing libraries are PCR amplified with HiFi™ (Kapa) for 10 cycles, 1.8X SPRI purified and quantified by qPCR (Kapa SYBR® Fast) and sized on a LabChip® GX (Caliper); size selection is not performed. Paired end sequencing (49×49 cycles) was performed using the HiSeq2000 (Illumina). Sequence data from genomic DNA were mapped to the reference human genome (hg19) using the BWA aligner(42). PCR duplicate read removal and sequence metric collection was performed using Picard (http://picard.sourceforge.net) and SAMtools (43). Local alignment optimization was performed using GATK(44).
Genomic alteration detection
Base substitution detection was performed using a Bayesian methodology, which allows detection of somatic mutations at a low mutation allele frequency (MAF) and increased sensitivity for mutations at hotspot sites(45) through the incorporation of tissue-specific prior expectations: P(Mutation present| Read data "R") = P(Frequency of mutation "F" > 0|R) ∞ 1 – P(R|F = 0)P(F = 0), where P(R|F) is evaluated with a multinomial distribution of the observed allele counts using empirically observed error rates and P(F = 0) is the prior expectation of mutation in the tumor type. To detect indels, de novo local assembly in each targeted exon was performed using the de-Bruijn approach(46). Candidate calls are filtered using a series of quality metrics, including strand bias, read location bias and a custom database of sequencing artifacts derived from normal controls. Germline alterations are identified and filtered using dbSNP (version 135, http://www.ncbi.nlm.nih.gov/projects/SNP/), 1000genomes (http://www.1000genomes.org/), and subsequently annotated for known and likely somatic mutations using the COSMIC database (version 62, http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/). Detection of CNAs was performed by obtaining a log-ratio profile of the sample by normalizing the sequence coverage obtained at all exons against a process-matched normal control. The profile is segmented and interpreted using allele frequencies of ~1,800 additional genome-wide SNPs to estimate tumor purity and copy number based on established methods(47-49) by fitting parameters of the equation, where and lrseg, Cseg are the log-ratios and copy numbers at each segment and sample purity, respectively. Focal amplifications are called at segments with ≥6 copies and homozygous deletions at 0 copies, in samples with tumor cell purity >20%.
To normalize for tumor content between pre-and post-NAC matched samples, allele frequencies or copy number estimations were divided by fractional tumor purity to calculate normalized allele frequency or copy number for the individual sample. The pre-NAC sample normalized frequency/copy number was subtracted from the post-NAC sample normalized frequency or copy number to calculate the absolute change in allele frequency or copy number.
TSC1 deletion verification
PCR primers amplifying across the predicted breakpoint in intron 23 (F: ACCCAATCTCACCAAGGTCC; R: CACCTTTTCTCGCTGAAAGCA, product: 100bp) and spanning the truncation from intron 23-intron 8 (F: AACAACTCTCCAAGAGTATCCGCAG R: TGGTGCGAAAAGAGCTGTTGCTT, product: 170bp) were utilized for PCR detection of the truncated allele.
Cell culture
MCF10A cells were cultured in DMEM/F12 nutrient mix with 5% horse serum (GIBCO), 20 ng/mL EGF, 0.5 mg/mL hydrocortisone, 100 ng/mL cholera toxin and 10 μg/mL insulin (all from Sigma). HCC1143 and HCC1395 were cultured in RPMI + 10% fetal bovine serum (FBS; GIBCO); MDA-436 and MDA-468 were cultured in DMEM + 10% FBS. SUM159PT cells were cultured in DMEM + 5% FBS and 0.5 μg/mL hydrocortisone. All cells were cultured at 37°C in at 5% CO2. MCF10A cells were purchased from ATCC. All TNBC cell lines (HCC1143, HCC1395, MDA-436, MDA-468, and SUM159PT) were obtained from the sources described in (11) and were confirmed by DNA fingerprinting (Cell Line Genetics, Madison WI) on 3/24/11 to consist of a single cell line and to match the DNA fingerprint on file at ATCC. Frozen stocks archived shortly after fingerprinting from the confirmed cell lines were utilized in these studies.
Chemicals
GSK1120212 (trametinib), AZD6244 (selumetinib), and BKM-120 were purchased from Selleckchem, dissolved in DMSO and utilized at dilutions resulting in a final concentration of <0.1% DMSO in all studies in vitro. Doxorubicin was purchased from Sigma and was solubilized in DMSO at a concentration of 100 mM.
siRNA knockdown
siRNA knockdown was performed as previously described (21). Cells were transfected with 20 nM siCONTROL (non-targeting siRNA), 10nM siMYC + 10 nM siCONTROL, 10 nM siMCL1 + 10 nM siCONTROL, or 10 nM siMYC + 10 nM siMCL1. Constructs for siMYC and siMCL1 were purchased from Ambion (s9129 and s8583, respectively).
MCL-1 overexpression
Cells were transduced with lentiviral particles derived from 293FT cells transfected with pLX302-MCL1 or GFP. GFP and MCL1 vectors were purchased from Thermo Scientific (Open Biosystems).
Soft agar colony formation assays
These assays were carried out in 6- or 12-well dishes using 5×104 or 1×104 cells, respectively. A single cell suspension in 0.4% agarose in 1X media was layered on top of a bottom layer of 0.8% agarose in 1X media in the presence of inhibitors or DMSO (control). Fresh 1X media (no drug) was applied to cells every 3-4 days to protect against dehydration. Colonies measuring >80 μm were counted after 2-3 weeks on a Gelcount Scanner (Oxford Optronix).
Immunoblotting
Immunoblotting was carried out as described43. Antibodies used for immunoblotting were: p-ERK1/2 (p-T202/Y204; #9101), ERK1/2 (#9102), p-AKT (p-S473; #9271), AKT (# 9272), HA-tag (#2367), calnexin (#2433) (all from Cell Signaling), and V5-tag (Invitrogen R960-25).
Statistical analysis
Statistics were performed where indicated using R(50) or GraphPad Prism (GraphPad Software, San Diego, CA). p<0.05 was considered statistically significant and p<0.1 was considered a statistical trend.
Supplementary Material
Statement of Significance.
This study demonstrates the spectrum of genomic alterations present in residual triple-negative breast cancer (TNBC) after neoadjuvant chemotherapy (NAC). Since TNBCs that do not achieve a complete response after NAC are likely to recur as metastatic disease at variable times after surgery, these alterations may guide the selection of targeted therapies immediately after mastectomy before these metastases become evident.
Footnotes
Notes:
Financial support: This work was supported by the NIH Breast Cancer Specialized Program of Research Excellence (SPORE) grant P50 CA98131, Vanderbilt-Ingram Cancer Center Support Grant P30 CA68485 and Core Services Grants CA14837, CA105436, and CA68485, The Lee Jeans Translational Breast Cancer Research Program (to CLA), Susan G. Komen for the Cure Foundation grants SAC100013 (CLA) and SAC110030 (JAP), Komen Career Catalyst grant KG100677 (RSC) and Komen Post-Doctoral award PDF12229712 (JMB), Department of Defense Post-Doctoral Fellowship award W81XWH-12-1-0026 (CDY) and Idea Award BC120793 (RSC), and NIH R01CA143126 (RSC).
Potential conflicts of interest: KW, JSR, PJS, RY, MC, VAM, and GP are paid employees and/or stockholders of Foundation Medicine.
Other Notes: None
References
- 1.Guarneri V, Broglio K, Kau SW, Cristofanilli M, Buzdar AU, Valero V, et al. Prognostic value of pathologic complete response after primary chemotherapy in relation to hormone receptor status and other factors. J Clin Oncol. 2006;24:1037–44. doi: 10.1200/JCO.2005.02.6914. [DOI] [PubMed] [Google Scholar]
- 2.Kuerer HM, Newman LA, Smith TL, Ames FC, Hunt KK, Dhingra K, et al. Clinical course of breast cancer patients with complete pathologic primary tumor and axillary lymph node response to doxorubicin-based neoadjuvant chemotherapy. J Clin Oncol. 1999;17:460–9. doi: 10.1200/JCO.1999.17.2.460. [DOI] [PubMed] [Google Scholar]
- 3.Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–81. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 4.Guarneri V, Piacentini F, Ficarra G, Frassoldati A, D'Amico R, Giovannelli S, et al. A prognostic model based on nodal status and Ki-67 predicts the risk of recurrence and death in breast cancer patients with residual disease after preoperative chemotherapy. Ann Oncol. 2009;20:1193–8. doi: 10.1093/annonc/mdn761. [DOI] [PubMed] [Google Scholar]
- 5.Jones RL, Salter J, A'Hern R, Nerurkar A, Parton M, Reis-Filho JS, et al. The prognostic significance of Ki67 before and after neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat. 2009;116:53–68. doi: 10.1007/s10549-008-0081-7. [DOI] [PubMed] [Google Scholar]
- 6.Masuda H, Masuda N, Kodama Y, Ogawa M, Karita M, Yamamura J, et al. Predictive factors for the effectiveness of neoadjuvant chemotherapy and prognosis in triple-negative breast cancer patients. Cancer Chemother Pharmacol. doi: 10.1007/s00280-010-1371-4. [DOI] [PubMed] [Google Scholar]
- 7.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 9.von Minckwitz G, Schmitt WD, Loibl S, Muller BM, Blohmer JU, Sinn BV, et al. Ki67 Measured after Neoadjuvant Chemotherapy for Primary Breast Cancer. Clin Cancer Res. 2013;19:4521–31. doi: 10.1158/1078-0432.CCR-12-3628. [DOI] [PubMed] [Google Scholar]
- 10.Balko JM, Cook RS, Vaught DB, Kuba MG, Miller TW, Bhola NE, et al. Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance. Nat Med. 2012 doi: 10.1038/nm.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rody A, Karn T, Liedtke C, Pusztai L, Ruckhaeberle E, Hanker L, et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011;13:R97. doi: 10.1186/bcr3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–9. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balko JM, Cook RS, Vaught DB, Kuba MG, Miller TW, Bhola NE, et al. Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance. Nat Med. 2012;18:1052–9. doi: 10.1038/nm.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A. 2009;106:4519–24. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Padua D, Zhang XH, Wang Q, Nadal C, Gerald WL, Gomis RR, et al. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133:66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhola NE, Balko JM, Dugger TC, Kuba MG, Sanchez V, Sanders M, et al. TGF-beta inhibition enhances chemotherapy action against triple-negative breast cancer. J Clin Invest. 2013;123:1348–58. doi: 10.1172/JCI65416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist. 2013;18:123–33. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapman-Rothe N, Curry E, Zeller C, Liber D, Stronach E, Gabra H, et al. Chromatin H3K27me3/H3K4me3 histone marks define gene sets in high-grade serous ovarian cancer that distinguish malignant, tumour-sustaining and chemo-resistant ovarian tumour cells. Oncogene. 2012 doi: 10.1038/onc.2012.477. [DOI] [PubMed] [Google Scholar]
- 21.Balko JM, Schwarz LJ, Bhola NE, Kurupi R, Owens P, Miller TW, et al. Activation of MAPK pathways due to DUSP4 loss promotes cancer stem cell-like phenotypes in basal-like breast cancer. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-13-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ertel F, Nguyen M, Roulston A, Shore GC. Programming cancer cells for high expression levels of Mcl1. EMBO Rep. 2013;14:328–36. doi: 10.1038/embor.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–9. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allen TD, Zhu CQ, Jones KD, Yanagawa N, Tsao MS, Bishop JM. Interaction between MYC and MCL1 in the genesis and outcome of non-small-cell lung cancer. Cancer Res. 2011;71:2212–21. doi: 10.1158/0008-5472.CAN-10-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beverly LJ, Varmus HE. MYC-induced myeloid leukemogenesis is accelerated by all six members of the antiapoptotic BCL family. Oncogene. 2009;28:1274–9. doi: 10.1038/onc.2008.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang Z, Luo H, Payton JE, Cain J, Ley TJ, Opferman JT, et al. Mcl1 haploinsufficiency protects mice from Myc-induced acute myeloid leukemia. J Clin Invest. 2010;120:2109–18. doi: 10.1172/JCI39964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–21. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–14. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han G, Wang Y, Bi W. C-Myc overexpression promotes osteosarcoma cell invasion via activation of MEK-ERK pathway. Oncol Res. 2012;20:149–56. doi: 10.3727/096504012x13522227232237. [DOI] [PubMed] [Google Scholar]
- 31.Hollern DP, Yuwanita I, Andrechek ER. A mouse model with T58A mutations in Myc reduces the dependence on KRas mutations and has similarities to claudin-low human breast cancer. Oncogene. 2013;32:1296–304. doi: 10.1038/onc.2012.142. [DOI] [PubMed] [Google Scholar]
- 32.Liu M, Casimiro MC, Wang C, Shirley LA, Jiao X, Katiyar S, et al. p21CIP1 attenuates Ras- and c-Myc-dependent breast tumor epithelial mesenchymal transition and cancer stem cell-like gene expression in vivo. Proc Natl Acad Sci U S A. 2009;106:19035–9. doi: 10.1073/pnas.0910009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marampon F, Ciccarelli C, Zani BM. Down-regulation of c-Myc following MEK/ERK inhibition halts the expression of malignant phenotype in rhabdomyosarcoma and in non muscle-derived human tumors. Mol Cancer. 2006;5:31. doi: 10.1186/1476-4598-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Podsypanina K, Politi K, Beverly LJ, Varmus HE. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci U S A. 2008;105:5242–7. doi: 10.1073/pnas.0801197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serra JM, Gutierrez A, Alemany R, Navarro M, Ros T, Saus C, et al. Inhibition of c-Myc down-regulation by sustained extracellular signal-regulated kinase activation prevents the antimetabolite methotrexate- and gemcitabine-induced differentiation in non-small-cell lung cancer cells. Mol Pharmacol. 2008;73:1679–87. doi: 10.1124/mol.107.043372. [DOI] [PubMed] [Google Scholar]
- 36.Horiuchi D, Kusdra L, Huskey NE, Chandriani S, Lenburg ME, Gonzalez-Angulo AM, et al. MYC pathway activation in triple-negative breast cancer is synthetic lethal with CDK inhibition. J Exp Med. 2012;209:679–96. doi: 10.1084/jem.20111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Comprehensive molecular portraits of human breast tumours Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch Pathol Lab Med. 2010;134:907–22. doi: 10.5858/134.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131:18–43. doi: 10.5858/2007-131-18-ASOCCO. [DOI] [PubMed] [Google Scholar]
- 41.Owens P, Pickup MW, Novitskiy SV, Chytil A, Gorska AE, Aakre ME, et al. Disruption of bone morphogenetic protein receptor 2 (BMPR2) in mammary tumors promotes metastases through cell autonomous and paracrine mediators. Proc Natl Acad Sci U S A. 2012;109:2814–9. doi: 10.1073/pnas.1101139108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Compeau PE, Pevzner PA, Tesler G. How to apply de Bruijn graphs to genome assembly. Nat Biotechnol. 2011;29:987–91. doi: 10.1038/nbt.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Loo P, Nordgard SH, Lingjaerde OC, Russnes HG, Rye IH, Sun W, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107:16910–5. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yau C, Mouradov D, Jorissen RN, Colella S, Mirza G, Steers G, et al. A statistical approach for detecting genomic aberrations in heterogeneous tumor samples from single nucleotide polymorphism genotyping data. Genome Biol. 2010;11:R92. doi: 10.1186/gb-2010-11-9-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Team RC. R Foundation for Statistical Computing. Vienna, Austria: 2012. R: A language and environment for statistical computing. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.