Abstract
Voltage-gated ion channels respond to transmembrane electric fields through reorientations of the positively charged S4 helix within the voltage-sensing domain (VSD). Despite a wealth of structural and functional data, the details of this conformational change remain controversial. Recent electrophysiological evidence showed that equilibrium between the resting (Down) and activated (Up) conformations of KvAP-VSD from Aeropyrum pernix can be biased through reconstitution in lipids with or without phosphate groups. We investigated the structural transition between these functional states using site-directed spin labeling and EPR spectroscopic methods. Solvent accessibility and inter-helical distance determinations suggest that KvAP gates through S4 movements involving a ~3 Å upward tilt and simultaneous ~2 Å axial shift. This motion leads to large accessibly changes in the intracellular water-filled crevice and supports a novel model of gating that combines structural rearrangements and electric field remodeling.
Voltage-gated ion channels play a critical role in defining a wide variety of signaling processes throughout biology, controlling such basic cellular functions as electrical excitability, hormone secretion and osmotic balance. They assemble as functional tetramers, with a centrally located pore domain (helices S5 and S6) surrounded by four voltage-sensing domains (helices S1 to S4) that contain a set of highly conserved positive charges in the S4 helix1-3. Reorientation of these S4 gating charges within the transmembrane electric field trigger a transition from the resting (or “Down”) conformation to the active (or “Up”) conformation of the sensor, which is coupled to pore opening and an increase in ion conductance.
Crystal structures of a number of voltage dependent channels (KvAP, Kv1.2, NaVAb and NavRh)4-7, a cyclic nucleotide gated channel (MlotiK1)8 have demonstrated that the basic molecular architecture of all VSDs is based on a common scaffold. In VSDs, the four transmembrane segments form an anti-parallel helical bundle where some of the gating charges in S4 establish ion pair interactions with strategically placed countercharges in helices S1–S35-7. Defining the mechanism by which the movement of S4 leads to gating charge translocation and eventual pore opening requires detailed structural information of both Up and Down sensor conformations. However, all available VSD structures are currently thought to represent the Up conformation or a close conformational equivalent.
The main challenges to determine high resolution structural information in voltage dependent systems remain technical in nature: the difficulty associated with the simultaneous and vectorial application of electrical fields to a population of molecules. So far, these conditions have not been achieved experimentally in membrane-reconstituted systems. However, a plausible alternative is to develop approaches that shift the VSD conformational landscape so that in the absence of a bias potential the “Up” and “Down” conformations become biochemically stable. One option is to take advantage of the known role of lipid-protein interactions to change the energetic landscape of membrane-reconstituted voltage gated channels.
Electrophysiological studies have demonstrated that protein-lipid interactions play a critical role in the functional modulation of voltage-gated channels9-12. In Kv2.1 enzymatic hydrolysis of sphingomyelin to (phosphate-free) ceramide produce a leftward shift in the activation (G-V) curve, leading to a sharp increase in open probability near resting potentials9,11. Further, reconstitution of the prokaryotic K+ channel KvAP in membranes with progressively unavailable phosphodiester moieties lead to increasingly right-shifted activation curves, lower open probabilities and a reduction in voltage sensitivity10. Based on these results, it was suggested that the transition between Up and Down conformations might require gating charge interactions with surrounding phospholipids. More importantly, it appears that in the absence of phospholipids voltage sensors seem to be “trapped” in the Down conformation12.
Here, we set out to investigate the structural underpinnings of the lipid-dependent gating transitions in the prokaryotic K+ channel KvAP in differential lipid reconstitution by EPR spectroscopy. Solvent accessibility and inter-helical distance determinations showed that non phosphate-containing lipids triggered a reorientation of the S4 helix according to a novel Tilt-Shift model with a modest ~3 Å S4 displacement. This reorientation led to a large increase in intracellular water penetration within the VSD, which putatively refocused the electric field and rendered the VSD unresponsive to physiologically relevant changes in transmembrane voltage.
RESULTS
The conformation of KvAP VSD in DOTAP
Schmidt et al10 have shown that in the presence of non-phosphate lipids like 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP), the G-V curve of KvAP becomes shallower and shifts towards positive voltages. Fig. 1a shows a diagram depicting the relative positions of the KvAP G-V curve after reconstitution in phospholipid (POPE:POPG=3:1) and 80% non-phospholipid (DOTAP) bilayers (adapted from10). The shift in the voltage axis can be large enough (> ~120 mV) that even at 0 mV the channel is unable to open. KvAP reconstitution in 100% DOTAP membranes fully inhibits channel opening by putatively stabilizing the Down conformation of the sensor, as demonstrated by antibody binding and cysteine reactivity experiments12. These results led us to a relatively straightforward experimental strategy, where at 0 mV a VSD reconstituted in a PCPG (POPC:POPG=3:1) bilayer mostly populates the Up conformation and reconstitution in a DOTAP bilayer traps the sensor in a putative Down conformation (Fig. 1a, shaded areas).
Figure 1.
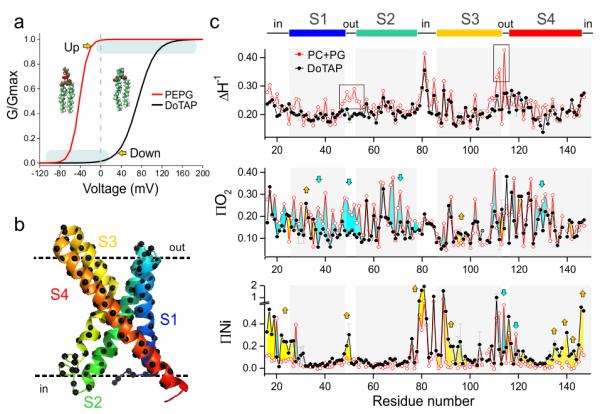
Functional and conformational changes induced by lipids. (a) Voltage dependence of KvAP was dramatically right shifted in the presence of non-phosphate lipids: 80% DOTAP with 20% POPE:POPG (black, Vh=73 mV, z=1.5) comparing to the phosphate lipids: 100% POPE:POPG = 3:1 (red, Vh=−42 mV, z=3.1). According to the simulated G-V curves, VSD of KvAP populates predominantly at activated (“Up”) state in phosphate lipids and at resting (“Down”) state in non-phosphate lipids at 0 mV, in the absence of asymmetric voltage under biochemical conditions. (b) Spin label was introduced to positions 16-147 on KvAP VSD one at a time for EPR spectroscopy studies in both phosphate lipid POPC:POPG=3:1 (PCPG) and non-phosphate lipid DOTAP. (c) Mobility ΔHo−1, oxygen accessibility ∏O2 and NiEDDA accessibility ∏NiEdda of KvAP VSD in PCPG (red) and DOTAP (black). 20 selected positions were repeated (n=3-5) and the standard deviation were shown in grey error bar. The grey regions represent the four putative transmembrane segments (S1, S2, S3 and S4) from the crystal structure. To facilitate comparison between the two conditions, we colored the area between the two data sets for both O2 and NiEdda accessibility profiles to highlight the degree and direction of the accessibility changes. Using the PCPG data (Up conformation) as reference, accessibility increases in the DOTAP-reconstituted sensor are colored in yellow, whereas accessibility reductions are shown as light blue areas. ∏NiEdda of S4 has a decreasing trend at the extracellular side and increasing trend at the intracellular side emphasized by the yellow arrows.
We pursued a spectroscopic approach to the study of lipid-dependent gating in KvAP by site-directed spin-labeling (SDSL) and electron paramagnetic resonance (EPR), using the known isolated VSD crystal structure (PDB 1ORS) as a reference4. EPR environmental parameters serve as powerful semi-quantitative constraints capable of characterizing overall conformation changes in liposome-reconstituted membrane proteins13. Indeed, we used EPR data to show that VSDs retain their overall Up conformation at 0 mV in PCPG liposomes, whether as part of a full-length channel or as an isolated domain14-16. A set of 132 single cysteine mutants (residues 16–147, Fig. 1b) of an isolated VSD construct (KvAP-VSD) reconstituted in DOTAP liposomes was used to characterize its putative Down conformation. This data set was evaluated in the context of previously published EPR data of KvAP-VSD in PCPG liposome, representing the Up conformation.
The environmental parameter profiles for probe mobility (ΔHo−1), lipid accessibility (to Oxygen, ∏O2) and aqueous accessibility (to NiEDDA, ∏NiEdda) in both Up and Down conformations are shown in Fig. 1c. There were noticeable changes in dynamics and solvent accessibility throughout the sensor. Our key observation was that the transition from the Up (PCPG) to the Down (DOTAP) conformation was accompanied by a large overall increase in ∏NiEdda towards the ends of each TM segment. This was particularly evident in regions that contribute to the lining of the intracellular crevasse (or inner crevice) in the sensor (blue arrows in Fig. 1c, bottom). At the same time, we saw essentially no changes in ∏NiEdda on S2 and the top part of S1, in agreement with the notion that S1 and S2 serve as anchor points for the structural rearrangements in S4. Furthermore, a recent solid-state NMR study characterizing the protein lipid interactions of KvAP in DOTAP or PCPG membranes17 found no changes in chemical shift (13C and 15Nε of indole group) in tryptophan 70 (on S2). This was consistent with our observations and the fact that S2 remains an anchoring point during gating. We noted that the above conformational changes were not a result of differences in KvAP-VSD aggregation behavior in either type of lipid (Supplementary Fig. 1a-d), or the physical properties of DOTAP membranes (Supplementary Fig. 1e-g). However, it must be noted that although slight differences in local dynamics between PCPG and DOTAP could affect the anisotropy values for the spin label (mobility), it shouldn’t dramatically affect the collision frequency between the spin label and paramagnetic reagent.
The large differences in water penetration between the Down (DOTAP) and Up (PCPG) sensor conformations could be best described by color mapping the ∏NiEdda values on the isolated KvAP-VSD crystal structure (Fig. 2a). The DOTAP-reconstituted sensor displayed a NiEdda-excluded area limited to a ~5 Å band positioned near the center of the bilayer. This aqueous exclusion region expands to almost ~15 Å in the PCPG-reconstituted sensor. In fact, the change in NiEdda penetration represented a shift in more than 3 turns of the S4 helix (~15 Å) from the intracellular opening of the crevice and about 1 turn of helix from adjacent residues at the top crevice.
Figure 2.
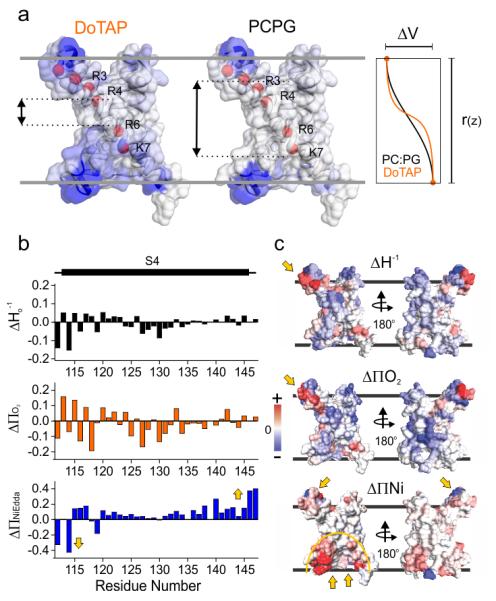
The lipid dependent conformation change. (a) Changes in water penetration leads to a refocusing of the transmembrane electric field. NiEdda data mapped onto the crystal structure of the isolated KvAP-VSD for DOTAP (left) and PCPG (center). The color spectrum is a linear scale between white (lowest NiEdda accessibility) and blue (highest). The red spheres depict the lowest two putative gating charges R133 (R6) and K136 (K7) in KvAP. Right, theoretical depiction of the voltage drop along KvAP-VSD after reconstitution in DOTAP (red) or PCPG (black). (b) The delta (DOTAP-PCPG) of ΔHo−1, ∏O2 and ∏NiEdda were plotted for the S4 region. There is clearly a tilt trend of a ∏NiEdda shown as a decrease at the top of S4 and increase of the bottom. (c) The delta of ΔHo−1, ∏O2 and ∏NiEdda were mapped onto the crystal structure of KvAP VSD. The bottom crevice has dramatic increase of ∏NiEdda accessibility accompanied by the slight increase from top crevice and decrease at S3-S4 loop region. There are obvious increases of ∏O2 at top of S4 (yellow arrow on Δ∏O2 map) suggesting the region getting closer into lipids.
The concept of focused electric fields in VSDs was initially proposed as a result of cysteine reactivity experiments18,19 and was indirectly estimated from fluorescence measurements20. In agreement with the present data, the hydration profile of KvAP in PCPG lipids by neutron scattering demonstrated that the extent of water penetration at both crevices21 was compatible with a focused electric field in the Up state of KvAP-VSD. We noted that these differences in NiEdda (water) penetration between the Up and Down conformations should lead to considerable changes in the internal dielectric constant along the sensor and refocusing the transmembrane electric field sensed by the gating charges in S4. Specifically, in the Down (DOTAP) conformation residues R133 and K136 (charges R6 and K7 in KvAP) would be exposed to a limited fraction of the field drop, but in the Up (PCPG) conformation would be placed well within the electric field. Having fewer charges within the electric field is likely to generate both a right shift and an apparent reduction in the slope of the G-V curve, as reported experimentally10. The theoretical change in transmembrane voltage along the VSD was depicted in the rightmost panel of Fig. 2a, where a shallower voltage profile is predicted in PCPG than in DOTAP-reconstituted sensors.
The nature of the S4 gating reorientation in KvAP
Fig. 2b showed a closer view of the EPR environmental parameters (ΔHo−1, ∏O2 and ∏NiEdda) in S4 and offers a perspective of the types of structural rearrangements in the VSD conformational landscape that take place during lipid-induced changes. The largest dynamic transitions (ΔΔHo−1) were found along the N-terminal half of S4, and these correlated well with overall changes in ∏O2. Most interestingly, the S4 Δ∏NiEdda profile was characterized by reductions in aqueous accessibility on its extracellular end, accompanied by an increase in accessibility towards its intracellular end. This was the same general pattern of solvent accessibility changes that has been observed on the extracellular end of S4 in various eukaryotic voltage-dependent channels (see ref. 22 for a review).
These results led to the consensus view that the transition from the Up to the Down conformation of the sensor must be accompanied by a downward movement of S423-26. However, the overall ∏NiEdda increase in the bottom cavity and the asymmetric ∏NiEdda change on S4 (Fig. 2) both appeared to be incompatible with a simple down movement of S4. Instead, the Δ∏NiEdda profile strongly suggested a movement where in addition to a relatively short downward shift, S4 undergoes a tilting motion (Supplementary Fig. 2a) that would allow increased ∏NiEdda penetration in both intra and extracellular crevices.
The above environmental changes were color mapped in the context of the KvAP-VSD crystal structure as shown in Fig. 2c. Three key observations emerged from these maps in the transition from the Up to the Down conformation. First, with the exception of the ends of S4, there was global reduction in local mobility. Second, as the opening of both VSD water crevices in DOTAP predicted a decrease the length of the hydrophobic center, the local oxygen concentration should decrease accordingly. This was consistent with the observed global reduction of ∏O2 around the sensor. The increase in ∏O2 observed on the lipid-facing regions on both ends of S4 is a combinational effect, since they would move further into the lipid environment. Third, the large increases in ∏NiEdda penetration on the intracellular face of the sensor mostly affected the S3-S4 half of the sensor, confirming the conformational stability of S1 and S2 during lipid-driven S4 movements.
To quantitatively characterize the lipid-driven structural rearrangements in KvAP, we carried out a series of double electron-electron resonance (DEER) measurements to determine average distances and distance distributions from a set of spin label pairs in both DOTAP and PCPG liposomes27. DEER-based distance determinations in liposomes is a notoriously challenging endeavor on account of sharply reduced sensitivity, increase in signal background and limitations in the accessible distance range28. To overcome these technical difficulties we took advantage of a newly characterized bi-functional spin label with reduced intrinsic dynamics (bis-SL Fig. 3a)29,30, and carried out the measurements at a higher microwave frequency (34 GHz, Q-band) (see Online Methods). The bis-SL pairs were designed to quantify the movement of S4 relative to the extracellular ends of S1 and S2, and the intracellular end of S2. We selected S1 and S2 as reference points since they were expected to undergo little or no conformational change upon S4 movement according to the EPR environmental parameters. The proper bi-SL attachment was confirmed by both mass spectrometry and CW-EPR spectra (Supplementary Fig. 3). All measured distances had a predominant distribution peak ranging from 22 to 38 Å (Fig. 4b, Supplementary Fig. 4a-c) and are self-consistent in regards to the basic geometric constrains expected from the crystal structure (Supplementary Fig. 4d).
Figure 3.
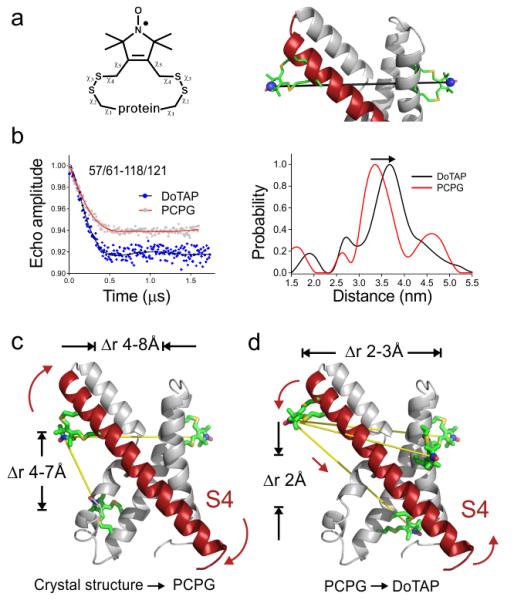
Quantitation of the conformation change by DEER distance measurement in lipids. (a) The scheme of the bi-functional spin label. Each distance between a pair of spin labels requires the introduction of four designed cysteine residues. (b) Representative raw DEER data (left) for a pair of distance measurement between 57/61 (bi-functional spin label attaching to position 57 and 61) and 118/121, and the distance distribution from Tikhonov regularization (right). The changes are small but confidently differentiable. (c) The absolute distances of 10 pairs in PCPG disagree with the crystal structure in a systematic way. It indicates that the crystal structure is over tilted 5~8 Å more than in PCPG. (d) The delta distances (DOTAP-PCPG) suggest a slight tilt and down movement of S4 at 2~3 Å. The distance change from PCPG to DOTAP is consistent in the direction of tilt of the S4.
Figure 4.
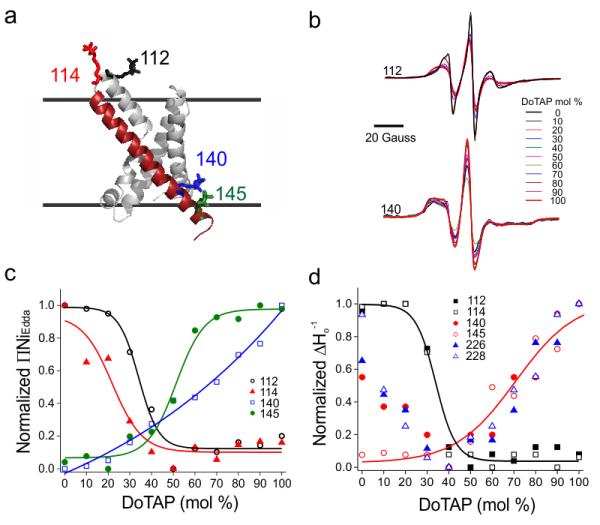
The lipid induced conformation change on KvAP VSD is titratable. (a) Spin labeled KvAP VSD proteins at four individual positions (112, 114, 140 and 145) were titrated with increments in the percentage of DOTAP. Residues 112 and 114 are on the extracellular side of S4 and residues 140 and 145 are on the intracellular side of S4. (b) Continuous wave EPR spectra of residues 112 and 145 upon DOTAP titration. The spectra were normalized by number of spins. As the percentage of DOTAP increases, the amplitudes are decreasing for residue 112 and increasing for residue 140. (c) The solvent accessibility (∏NiEdda) of S4 are decreasing for extracellular side (112, 114) and increasing for the intracellular side (140, 145). (d) The mobility ΔHo−1 change of extracellular side of S4 (112, 114), intracellular side of S4 (140, 145) and the pore (226, 228). The change on the intracellular side of S4 and the pore overlapped in the range of 40-100% DOTAP.
As an initial approach, we compared the measured distances in PCPG liposomes with those calculated from the crystal structure4 after individual incorporation of bis-SL pairs in silico. This structure is typically thought to represent the Up conformation of KvAP-VSD. To our surprise, we found large and systematic deviations between the experimental PCPG distances (Table I) and those predicted from the crystal structure. In PCPG the extracellular end of S4 appears to be 4~7 Å farther to the intracellular end of S2 than what the crystal structure would predict. At the same time, those labeled positions in S4 are 4~8 Å closer to the extracellular end of S1 and S2 in PCPG than in the crystal structure (Fig. 3c, Table I). These differences were beyond the uncertainties (~3 Å) of current DEER measurements. The apparent discrepancy strongly suggests that given the intrinsic flexibility of VSDs the present KvAP-VSD crystal structure appears to deviate from the physiologically-relevant Up conformation (in PCPG and at 0 mV). Although this should be considered a cautionary tale, these results are not surprising, for there is ample structural evidence pointing to the structural fragility of KvAP once it is extracted from the membrane31,32.
Table 1.
Inter-spin distances from KvAP-VSD in PCPG, DOTAP and the crystal structure. Data from double labeled Bi-SL labeled sensor.
| Double Cysteine Residue Number |
Distance(A) | Δ distance | Position | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Crystal | PCPG | DoTAP | PCPG- Crystal |
DoTAP- PCPG |
|||||
| Structure | Max | Dev | Max | Dev | |||||
| 39/43 | 118/121 | 33.1 | 27.9 | 5.6 | 30.8 | 4.8 | −5.2 | 2.9 | S1 Top |
| 121/125 | 28.8 | 20.9 | 2.2 | 23.6 | 3.1 | −7.9 | 2.7 | ||
| 40/44 | 118/121 | 38.1 | 33.7 | 3.0 | 34.2 | 2.9 | −4.4 | 0.5 | |
| 121/125 | 33.6 | 29.6 | 2.3 | 29.1 | 2.0 | −4.0 | −0.5 | ||
| 57/61 | 118/121 | 38.3 | 33.5 | 3.3 | 36.8 | 2.5 | −4.8 | 3.3 | S2 Top |
| 121/125 | 33.9 | 33.8 | 2.7 | 33.4 | 2.5 | −0.1 | −0.4 | ||
| 72/75 | 118/121 | 38.1 | 38.1 | 2.3 | 36.1 | 4.5 | 0 | −2.0 | S2 bottom |
| 121/125 | 32.4 | 33.1 | 3.0 | 32.1 | 2.6 | 0.7 | −1.1 | ||
| 74/77 | 118/121 | 22.2 | 26.4 | 5.4 | 25.9 | 3.8 | 4.2 | −0.5 | |
| 121/125 | 16.9 | 24.5 | 5.2 | 23.6 | 5.6 | 7.6 | −0.9 | ||
In contrast, the distance differences between DOTAP and PCPG reconstituted KvAP-VSD are more subtle, with maximal displacements around 3.3 Å (Table I). After mapping the differences onto the crystal structure (DOTAP-PCPG), there is a clear trend pointing to a systematic S4 movement: a ~3 Å tilt away from S1 and S2 together with a ~2 Å downward movement (Fig. 3d). The extent and direction of this tilt precisely coincides with proposed movements derived from the ∏NiEdda vectors (Supplementary Fig. 2a).
The conformational coupling between sensor and pore domain
To study the nature of the KvAP-VSD transitions along its conformational landscape we evaluated the dependence of the VSD structural changes in relation to the molar content of DOTAP in the membrane. We monitored aqueous accessibility and local dynamics changes from residues 112 and 114 at the extracellular end of S4, while positions 140 and 145 report movements on its intracellular end (Fig. 4a). Samples were reconstituted in mixtures of PCPG and DOTAP ranging from 0 to 100 mol % DOTAP and spectra were obtained at 10 mol % intervals (Fig. 4b).
Increasing DOTAP molar content leads to a sigmoid dependence in aqueous accessibility (Fig. 4c, left), where ∏NiEdda decreases on the extracellular side (residues 112 and 114) but gradually increases on the intracellular end of S4 (residues 140 and 145). This pattern is expected if S4 was moving downwards and tilting in relation to the plane of the bilayer. With the exception of position 140, DOTAP-dependent accessibility changes conform to a simple sigmoid dose-response curve, with midpoints at ~35, ~25 and ~50 mol % DOTAP for residues 112, 114 and 145, respectively. Similar lipid dependence was also observed for changes in local polarity from fluorescence spectroscopy determinations at position 111 (Supplementary Fig. 1d).
It was tempting to suggest that the increasing midpoints for the DOTAP dependence are a reflection of the sequential nature of the accessibility changes as the S4 segment moves downwards. In this scenario, as S4 reorients relatively to the membrane at increasing DOTAP contents, position 114 would be the first one to become membrane embedded, followed by position 112 and then position 145 emerged out of the membrane environment. The DOTAP dose-response curve for residue 140 was shallower and did not saturate, suggesting it did not fully emerge from its bilayer-buried position even at 100 mol % DOTAP.
Lipid-induced structural changes were conformationally coupled to the gating transitions in the pore domain, accounting for the observed differences in ion permeation in voltage-gated channels10,12. Electrophysiological data showed that at 0 mV KvAP experiences a sharp reduction in open probability when the fraction of DOTAP in the membrane was increased from 40 to 100 mol %10 (Supplementary Fig. 2b-d). Therefore, it was natural to ask whether the present set of structural changes in the isolated KvAP-VSD was somehow correlated to pore domain changes in full-length KvAP. We found that positions 226 and 228 (below the putative glycine hinge G220) showed large mobility differences as a function of DOTAP mol % (Fig. 4d). The DOTAP-dependent mobility changes in S6 took place in the same concentration range as those for the changes in open probability (Supplementary Fig. 2d). This pointed to a strong correlation between S6 mobility and open probability and supports the idea that residues 226 and 228 were useful reporters of the open-close equilibrium at the gate. Remarkably, the DOTAP dependence of ΔHo−1 at the intracellular end of S4 (140 and 145) overlapped with the lipid dependence of pore gate dynamics (positions 226 and 228) (Fig. 4d), in spite of their disparate location and spectral lineshape differences. This argued in favor of a concerted structural change linking the rearrangements at the intracellular end of S4 and the process of pore gate losing as the molar content of DOTAP increases in the bilayer. Of course, the present correlations between the S4 reorientations and dynamic changes along S6, although attractive, need to be taken with care. The S6 data derived from full-length KvAP, which pointed to additional complications in interpretation due to inter and intra subunit contacts, as well as protein-lipid interactions. The precise details of the conformational correlation between S4 and S6 movements were expected to be more complicated, but it was clear that the S4 C-terminus motion did indeed couple to changes in S6, setting the basis for electromechanical coupling in KvAP.
DISCUSSION
The mechanism of lipid-dependent gating in KvAP
A wealth of experimental data and computational simulations (see1,3,22 for reviews), along with several VSD structures4-7, have led to three fundamental mechanisms of S4 motion that aim to explain the origins of voltage dependence in VSDs. In the helical screw-sliding helix model, voltage sensing is the result of a choreographed reorientation where S4 undergoes a rotation and axial translation through discrete positions determined by the location of conserved countercharges33,34. A second model, based on the original description of the KvAP crystal structure, suggested that the sensor acts instead as a tethered hydrophobic cation4,31,35. Here, the S3-S4 helix-turn-helix hairpin (the “paddle”) would translocate large distances (15~20 Å) though the electric field drop across the lipid bilayer, triggering pore opening. Finally, it has been suggested that a refocusing of the electric field around the gating charges is electrically equivalent to mechanisms where the S4 helix moves, particularly in the presence of a focused electric field23,36. Though this mechanism has not been explicitly defined from a structural standpoint, it is expected that small S4 movements (~1-2 Å) can support relatively large charge movements.
We developed a simple approach to structurally compare explicitly-defined gating mechanisms on the basis of the present set of distance and accessibility data. We first calculated a reference Up model by driving the existing KvAP-VSD crystal structure (PDB 1ORS) through different conformations until it matched all available distance constraints determined in PCPG as shown in Table I and optimized with EPR environmental parameters37 (Supplementary Fig. 5a-d). This reference structure (Supplementary Fig. 5e) served as the baseline to evaluate the distance differences (Δr) with each of the three tested models. We assumed secondary structure integrity and rigid body motions to construct models of the Down conformation based on three conceptual gating models (Fig. 5a, left). The sliding helix-helix screw model was generated by modeling a ~5 Å downward S4 movement and a ~60° rotation according to its original description33,34. The paddle model was constructed after imposing a ~15 Å downward movement for the helix-turn-helix motif formed by the S4 and S3b helices around the putatively flexible Gly134 in S4. A third model, which we named the “Tilt-Shift” gating model, is calculated in the same way as the Up state model (see methods) with EPR environmental parameters and distance constraints. The Tilt-Shift model is characterized by a simple combination of a ~3 Å tilt and a ~2 Å downward shift along the S4 axis from Up state to Down state in KvAP-VSD (Supplementary Fig. 5f). Neither the paddle nor the sliding helix-helix screw models underwent data-driven minimization.
Figure 5.
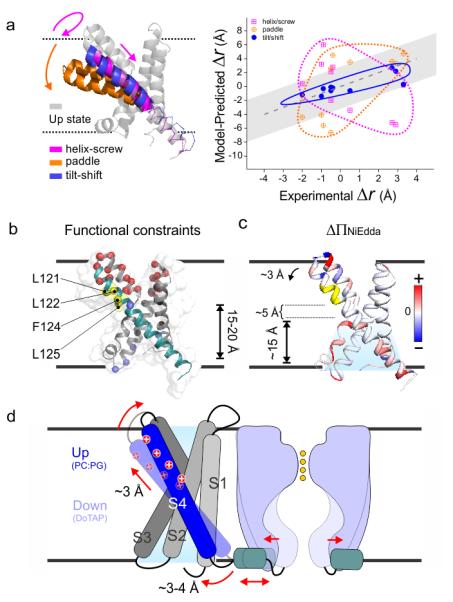
The Tilt-Shift model. (a) The distance change from Up state to Down state within KvAP VSD. (left) Up state VSD model (grey) was generated by MD simulation. The three Down states based on helix-screw (magenta), Paddle model (orange) and Tilt-Shift (blue) models. (right) The expected distance changes according to three models were plotted against experimental differences. The grey region indicated the uncertainty of measurement. Neither the helix-screw or the Paddle model is compatible with the experimental data, and only the Tilt-Shift model offer plausible explanation. (b) The alternative explanation to the existing data. (left) All the established state dependent accessibilities on KvAP focused on the region of 121-125. A Paddle model proposed a 15-20 Å downward movement of S4 to account for the their accessibility from the bottom. (c) Δ∏NiEdda was mapped on to the crystal structure. It shows the water penetration deep into the bottom crevice, where the region 121-125 is only ~ 5 Å above. Our observed ~ 3 Å Tilt-Shift of S4 should be sufficient to account for their accessibility from the bottom crevice. (d) Cartoon representation of the Tilt-Shift model on KvAP VSD. The ~25° tilt and ~2 Å down shift of the S4 could generate a 3~4 Å movement of the S4-S5 linker which seems to sufficient to open the pore and increases permeation of potassium current.
To qualitatively examine the types of S4 movements each model can support, the experimentally determined distance changes (Δr) for each of the spin label pairs in Table I were plotted against the calculated Δr from each model (Fig. 5a, right). A gray band in the background depicts the intrinsic uncertainty of the measurement. As the only model driven by the present dataset, the Tilt-Shift mechanism correlates favorably with the set of experimental distances. However, the large discrepancies seen with either the Helix Screw or the Paddle suggest that neither model could account for the type of S4 movement triggered by lipid interactions.
How many gating charges can be transferred under the proposed Tilt-Shift mechanism? Although the actual number of charges translocated per gating event remains unknown in KvAP, that number is expected to be between 1~3 eo net chargestransfer per sensor. In KvAP, the top two of the canonical four arginines (R117, R120, R123 and R126) are located on the extracellular side of the sensor, presumably establishing interactions with the membrane lipids. Since the S4 vertical movement in the Tilt-Shift model is around 2 Å with no obvious counter charge exchange for those arginines (based on MD simulation), some of the net charge transfer might be a direct result of the reorientation of these top charges. However, we propose that a larger portion of the charge transfer takes place as a consequence of the water penetration changes in local dielectric and subsequent refocusing of the electric field. This electric field compression or expansion would electrostatically influence R6 and K7 (Fig. 2), so that an upward movement of the electric field from the intracellular side would be equivalent to a downward movement of R6 and K7. Accordingly, we suggest the four arginines R1, R2, R3 and R4 on the top of S4 are the driving forces for the tilt toward the Down state driven by depolarization, but R6 and K7 are also important contributors to the gating charge transfer as a consequence of field refocusing.
Implications for the general mechanism of voltage sensing
A series of independent experimental approaches have been carried out to evaluate the nature and extent of the conformational changes in KvAP by identifying residues with state-dependent accessibilities to both sides of the sensor. Three approaches are worth highlighting: 1-Streptavidin-biotin trapping in a lipid bilayer under voltage clamp (with a 9-10 Å linker)31,35; 2- Radioactive labeling of inside out E. coli ghosts under voltage38; and 3-PEG5 mass tagging in PCPG and DOTAP liposomes at 0 mV12. Remarkably, and in spite of the diversity in methodology (including voltage vs. lipid gating), these experiments all converged onto a short region of S4 between residues 121-125 (Fig. 5b) as the only region of KvAP-VSD with conformation specific responses. Taking the isolated sensor crystal structure as a reference, this region is located about ~15 Å away from the intracellular membrane interface and it is accessible to intracellular side at Down state.
The canonical interpretation of these results, according to the paddle model, is that the S4 must traverse a large distance to account for this accessibility pattern31,35. So, how can we reconcile the rather modest DOTAP-driven S4 motion described above with the 15–20 Å motion proposed by biotin accessibility and mass tagging in KvAP? The Tilt-Shift model offers an alternative and plausible explanation for these results. We must first consider the extent of NiEdda penetration (Fig. 2) derived from the intracellular crevice opening as a result of S4 tilt and shift. Fig. 5c illustrates this point as the net increase of NiEdda penetration (red scale). In DOTAP, the aqueous solvent accesses up to ~15 Å through the opening of the intracellular crevice. This places residues backbone of 121-125 only about 5 Å above the upper penetration range of NiEdda. Considering that streptavidin-biotin trapping and mass tagging are based on reaching a kinetically absorbing state, is easy to suggest that the observed ~ 3 Å tilt and down shift should be sufficient to generate state dependent accessibilities for the region 121-125. Depending on the position of the hinge point for the S4 tilt, a ~3 Å displacement at the top of S4 could be amplified up to ~5 Å at the S4 bottom. The extent of this motion would be sufficient to gate the pore and is fully compatible with the ~4 Å displacement recently observed at the KvAP S4-S5 linker using LRET distance measurements39. The Tilt-Shift model, derived from lipid-dependent gating, correlates well with existing voltage-dependent gating data, although the exact mechanistic equivalency between the two stimuli remains to be determined.
Whether or not the Tilt-Shift gating mechanism is unique for KvAP remains an open question. Nevertheless, it is important to highlight the fact that of the currently available voltage sensing domain structures, the activated (Up) state of KvAP is somewhat of an outlier. KvAP has the sensor with the largest tilt of the S4 relative to the membrane normal (even after correcting for the S4 tilt with the present set of distances), it is the only sensor with a dramatic break in secondary structure (and bend) in S3, and as derived from its crystal structure, it also lacks an obvious candidate for a hydrophobic gasket or charge transfer center, typical of most other VSDs40,41. Nevertheless, the present data firmly points to a mechanism of lipid-driven gating consisting of a series of synergistic reorientations: S4 down movement, rotation, tilt, hydrophobic septum change and electric field refocusing. The relative prominence of these movements in other VSDs gating mechanisms will probably vary depending on the co-evolultion between the positive charges on S4 and the stabilizing countercharge partners in S1, S2 or S3. The details of the present model might also highlight the extent of the mechanistic diversity in the process of voltage sensing by VSDs.
Online Methods
Preparation, labeling and reconstitution of isolated voltage sensing domain and full length of KvAP
132 single cysteine mutants (residues 16-147) on KvAP VSD and 32 single cysteine mutants on KvAP full length (residues 225-256) were expressed and purified following published procedures4,14,15. Purified single cysteine mutants were labeled with a spin probe (1-oxyl-2,2,5,5,-tetramethylpyrrolidin-3-yl) methyl methanethiosulfonate (Toronto Research Chemicals) then reconstituted into POPC:POPG = 3:1 and DOTAP liposomes. The stock lipids in chloroform were dried under vacuum, and completely dissolved with 20 mM Tris pH 8.0, 150 mM NaCl, 60 mM β-OG and 10 mM DM at a lipid concentration of 10 mg/mL. Each spin labeled protein was mixed with 10 mg dissolved lipid at 1:250 (protein: lipid) molar ratio and dialyzed against 20 mM Tris pH 8.0 with 150 mM NaCl for three days. Fresh buffer was changed every 8~10 hours. The proteo-liposome was pellet down at 300,000 g for 20 min.
Ten quadruple-cysteine mutants (39/43-118/121, 39/43-121/125, 40/44-118/121, 40/44-121/125, 57/61-118/121, 57/61-121/125, 72/75-118/121, 72/75-121/125, 74/77-118/121 and 74/77-121/125) on isolated VSD were labeled with a bi-functional spin label (3,4-bis-(methanethiosulfonylmethyl)-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-1-yloxy radical, Toronto Research Chemicals) at a protein:label=1:20 ratio at room temperature for 30 min. Excess spin label was removed through size exclusion chromatography with a Superdex 200 column. The spin labeled protein was diluted with unlabeled isolated VSD WT and the dilution ratio is 1:20 for POPC:POPG reconstitution and 1:30 for DOTAP reconstitution. The spin labeled protein after dilution was mixed with appropriate amount of dissolved lipids and reconstitute in the same way as mentioned above.
Posphocholine lipids with a radical at head group (TEMPO-PC) and various positions along alkyl chain (16:0-5 Doxyl PC, 16:0-7 Doxyl PC, 16:0-10 Doxyl PC, 16:0-12 Doxyl PC, 16:14-5 Doxyl PC and 16:0-16 Doxyl PC) (Avanti Polar Lipids) were used to probe the oxygen and Ni[II] ethylenediaminediacetic acid (NiEdda) accessibilities inside liposomes (Supplementary Fig. 1). Spin labeled PC was mixed with POPC+POPG or DOTAP at 1:2000 molar ratio in chloroform. The mix was dried under vacuum, and hydrated with 20 mM Tris pH 8.0 and 150 mM NaCl. The mix was sonicated for 5 min and pellet down at 300,000 g for 20 min. The measurements were conducted with the gel phase pellet.
Fluorescence measurement
KvAP-VSD A111C was purified and individually labeled with either fluorescein 5-maleimide (excitationmax= 494 nm; emissionmax= 518 nm) or tetramethylrhodamine 5-maleimide (excitationmax= 544 nm; emissionmax= 572 nm) fluorescence dye. The individually labeled A11C mutant was mixed at donor: acceptor = 1:1 ratio, and reconstituted into POPC+POPG or DOTAP liposome in the same way as mentioned above. The fluorescence emission was recorded from 500 to 650 nm while excited at 494 nm in a PTI fluorimeter (PTI technology). The relative FRET intensity in 570-580 nm range indicates the closeness between donor and acceptor, as the relative aggregation level of KvAP-VSD.
Mass spectrometry
The spin labeled KvAP-VSD were precipitated with 5X (v/v) 100% acetone. The pellets were washed twice with DI water to remove ions and detergents. The pellet was completely dissolved in DMSO with 0.1% TFA at 100 μM concentration. The dissolved sample was directly injected into Agilent 6224 TOF-MS with 1290 UHPLC system without any prior column separation. Each sample was analyzed at four ionization voltages ranging from 50 V to 300 V. Data were acquired and analyzed with company software following standard protocols. Single predominant peak was obtained for each of purified spin labeled KvAP-VSD mutants.
EPR spectroscopy
Continuous-wave EPR spectroscopic measurements were carried on a Bruker EMX X-band spectrometer equipped with a dielectric resonator and a gas-permeable TPX plastic capillary at room temperature. Spectra were recorded at 2.0 mW incident power, 100 kHz modulation frequency, 1.0 Gauss modulation amplitude, 20.48 ms time constant, 40.96 ms conversion time and 120 Gauss scan width. The motion of the spin label at specific site was quantified by anisotropy of spectra shape with an empirical parameter as the inverse of peak-to-peak width of the central (mI = 0) resonance, which was conventionally termed as mobility (ΔHo−1)42. The mobility will be low for the region involving tertiary or quaternary contacts which can constrain the motion of spin label, but high for loop region where spin label has more freedom of motion. The accessibility (∏) of spin label to the paramagnetic relaxing reagent which can present predominantly in lipid (O2) or water (NiEdda) can be measured from power-saturation methods43, by the peak-to-peak amplitude of the central line of EPR spectra as a function of increasing incident microwave power. The measurements for each spin labeled mutant can be done after equilibration with air (21% O2) or N2 (0% O2) for oxygen accessibility (∏O2), or with 25 mM NiEdda + N2 or N2 (0 mM NiEdda) for NiEdda accessibility (∏Ni). High accessibility of the spin probe to O2 (∏O2) is suggestive of a lipid-exposed environment within membrane, whereas high NiEdda accessibility is a reflection of the probe being exposed to aqueous solvent outside membrane. Dynamic and structural information of KvAP-VSD inside the liposomes can be deduced by mobility and environmental accessibility at each residue.
Distance measurement in liposome is notoriously challenging primarily due to the crowding effect when protein was constrained onto the two-dimensional liposome. The non-specific intra-molecule spin-spin interactions can dominate the echo decay and mask off the desired inter-molecule spin-spin interaction, and increase noise by contributing the non-linear components into the baseline. The challenge was approached by the essential combination of three specific technical considerations: 1. Utilization of the bi-functional spin label. The spin label was able to attach to two adjacent cysteine residues simultaneously, which limit the spatial distribution of spin label thus boost up the echo oscillation above baseline. 2. Dilution of spin labeled protein with unlabeled protein up to 30 fold. 3. Measurement at higher frequency at Q-band (34 GHz). With combinational difficulties of decreased S/N by extensive dilution and increased noise by sample in liposome, only the measurements on Q-band pulse EPR spectrometer for ~20 hours are able to produce data with workable evolution time (1.5 – 2 μs) and S/N. Double electron-electron resonance (DEER) distance measurements were carried on a Bruker 580 pulsed EPR spectrometer operating at Q-band (34 GHz) with EN 5107D2 resonator at 83 K using an established four-pulse protocol44. Primary dipolar evolution was baseline-corrected and analyzed by Tikhonov regularization to determine distance distribution using the software DEER Analysis 2011 (available at website http://www.epr.ethz.ch/software/index) following standard protocol45.
Self-consistency of distances
The probabilities of distance distribution were normalized. Predominant single main peak (probability > 0.5, above the grey region in Supplementary Fig. 4b, 4c) was clearly resolved for all 10 pairs of distances in both liposomes among the range of 22 to 38 Å (Supplementary Fig. 4a), which lay in the most sensitive region of the pulsed EPR method. The reported average distances and the standard deviations were from single Gaussian fitted main peak. The width of the distribution are around 3~ 5 Å, which may not be well determined due to the limited evolution time and noisy background.
Due to the above mentioned technical difficulties, we first evaluated the reliability of distances from current measurements. Based on the existing crystal structure of KvAP VSD, we would expect certain pattern of distance variations based on the geometry of residues such as position and side chain orientation: 1. Positions 118/121 are further than 121/125 to all reference points on the crystal structure. We do observe all 10 distances measured with 118/121 (solid line, Supplementary Fig. 4b, 4c) are larger than the corresponding distances with 121/125 (dash line, Supplementary Fig. 4b, 4c) in both POPC+POPG and DOTAP liposomes. 2. Positions 40/44 are facing opposite to the S4, while 39/43 are facing closer to S4. We observe all 4 distances measured with 40/44 were larger than the corresponding one with 39/43. 3. Positions 72/75 are facing opposite S4 top, while 74/77 are facing toward. We observe all 4 distances measured with 72/75 are larger than the corresponding ones with 74/77. The measured distances are self-consistent with the geometric pattern expected from crystal structure.
Structure modeling
To resolve the distance discrepancy between experimental measurement in PCPG and expected values from crystal structure, we aim to refine the crystal structure model with experimental constrains to reach the Up state VSD by restrained molecular dynamic (RMD) simulation methods. To incorporate the accessibility restraints, we employed a previously developed method, pseudo-atom driven solvent accessibility refinement (PaDSAR) originally incorporated in CHARMM version c32a246. In brief, each residue of the protein was represented with two virtual particles (Supplementary Fig. 5a), backbone and the pre-assigned spin-probe pseudo-atoms (EP1 = buried, EP2 = water exposure and EP3 = lipid exposure). During the course of molecular dynamics (MD) calculations, restraint driving forces to spin-probe particles were governed by interactions with environment pseudo-atoms, backbone, O2 (OXY) and NiEdda (NIC). To incorporate the distance restraints, we measured a distance between pseudo-atoms (EPX), each of which is covalently cross-linked to the two alpha carbons of the corresponding labeled residues (Supplementary Fig. 5b). A dataset of ten distances determined in the mixed POPC+POPG (denoted as PCPG for simplification) and in DOTAP lipids were converted into distance restraints. Four distances between the top of S1 and the top of S4 segments and six distances were restrained between S2 and the top of S4. The distance restraints were imposed in the form of harmonic potential to force the distances between pseudo-atoms (EPX) toward a given value. Geometry of pseudo-atom EPX such as bond length, angle and torsion and force field parameters were defined in analogous ways as previous methods46 with the consideration of the crystal structure of bi-functional spin label on lysozyme47.
Structure refinement of the up state model
Structure refinements were performed by introducing distance and accessibility restraints in a stepwise fashion. In the first step, the KvAP-VSD crystal structure (PDB code:1ORS) was refined with the distance restraints only. The structure was restrained by a total of ten EPR distances determined in PCPG (Supplementary Table 1). Because of the limited distance information used in the calculation, our initial calculation started by fixing positions of the transmembrane (TM) segments, S1, S2, S3a and S3b, but allowing a rigid body motion of S4 to relax to a state that gave the best fit with the distance restraints. Then the position restraints were removed and the secondary structure constraints were employed to all the four TM segments instead. The resulting model was optimized with the solvent accessibility restraints using the PaDSAR method46. A total 108 residues were assigned to the pseudo-spin type from the profile plots of ΠO2, ΠNiEDDA and ΔHo−1 values determined in PCPG. The obtained model was subjected to the final RMD, in which all the distance and accessibility restraints used in the previous steps were introduced simultaneously. At this step, ten iterative cycles of RMD were performed with restraining the secondary structure of the four transmembrane helices during the calculation. The top ten structures that gave the best fit with experimental distances determined in PCPG were selected from MD trajectories and represent the final models of the up state and EPX-EPX distances are in good agreement with the experimental data. The averaged coordinates was used as the Up state VSD.
Structure refinement of the down state model
The modeling the down conformation is similar to that for the up model. The strategy employed in the calculation starts with the top ten structures that are refined with EPR distance and accessibility data from crystal structure. RMD runs of these structures were performed using the ten distance restraints derived from spin label distances determined in DOTAP. Due to the limited distance restraints and accuracy for the determination of conformational changes, we used the experimental observation (∏O2, ∏NiEDDA and ΔH o−1 profiles in PCPG and DOTAP) to direct conformational sampling. To reduce the number of degrees of freedom, the S2 position was kept fix and S1, S2, S3a, S3b and S4 can be moved as a rigid helix. In addition, the positions of residues 110, 118, 148 and 149 were restrained, allowing the S4 top, the S4 bottom and the S3b to tilt in a range from 2 to 8Å toward the membrane plane. However, the force constant of these positional restraints was decreased in the subsequence step, in which the PaDSAR run was employed to optimize the resulting models with the solvent accessibility data. Then, the obtained structures were subsequently subjected to RMD refinement with distance and accessibility restraints. In the last RMD run, we used the same set of the previous run, except all the positional restraints were eliminated. The final models that gave the best fit with the experimental distances determined in DOTAP were selected from MD trajectories (Supplementary Table 2). A superposition of ten selected structures gives the backbone RMSD of 1.6 Å (Supplementary Fig. 5e). A structure comparison between the up and down conformation by using the average structures results in the RMSD of 3.1 Å (Supplementary Fig. 5f).
Supplementary Material
Acknowledgements
We are thankful to H. Mchaourab and R. Stein for generous sharing of the Q-band spectrometer, assistance in data gathering and general insightful comments. We thank G. Meyer for the scripts to analyze power saturation data. We thank F. Bezanilla, B. Roux and members of the Perozo, Bezanilla and Roux labs for helpful discussions and invaluable experimental advice. This work was supported in part by US National Institutes of Health grants R01-GM57846, and U54-GM74946.
Footnotes
Author Contributions
Q.L. and E.P. designed the strategy, Q.L. and S.W. performed the experiments, P.T. performed the MD simulation, Q.L. analyzed the data, E.P. and Q.L. wrote the manuscript.
The authors declare no competing financial interests.
References
- 1.Bezanilla F. How membrane proteins sense voltage. Nat. Rev. Mol. Cell Biol. 2008;9:323–332. doi: 10.1038/nrm2376. [DOI] [PubMed] [Google Scholar]
- 2.Catterall WA. Ion channel voltage sensors: structure, function, and pathophysiology. Neuron. 2010;67:915–928. doi: 10.1016/j.neuron.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swartz KJ. Sensing voltage across lipid membranes. Nature. 2008;456:891–897. doi: 10.1038/nature07620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang Y, et al. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 5.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 6.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 2012;486:130–134. doi: 10.1038/nature11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clayton GM, Altieri S, Heginbotham L, Unger VM, Morais-Cabral JH. Structure of the transmembrane regions of a bacterial cyclic nucleotide-regulated channel. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1511–1515. doi: 10.1073/pnas.0711533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramu Y, Xu Y, Lu Z. Enzymatic activation of voltage-gated potassium channels. Nature. 2006;442:696–699. doi: 10.1038/nature04880. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt D, Jiang Q-X, MacKinnon R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature. 2006;444:775–779. doi: 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y, Ramu Y, Lu Z. Removal of phospho-head groups of membrane lipids immobilizes voltage sensors of K+ channels. Nature. 2008;451:826–829. doi: 10.1038/nature06618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng H, Liu W, Anderson LY, Jiang Q-X. Lipid-dependent gating of a voltage-gated potassium channel. Nat Commun. 2011;2:250. doi: 10.1038/ncomms1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McHaourab HS, Steed PR, Kazmier K. Toward the fourth dimension of membrane protein structure: insight into dynamics from spin-labeling EPR spectroscopy. Structure. 2011;19:1549–1561. doi: 10.1016/j.str.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuello LG, Cortes DM, Perozo E. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science. 2004;306:491–495. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- 15.Chakrapani S, Cuello LG, Cortes DM, Perozo E. Structural dynamics of an isolated voltage-sensor domain in a lipid bilayer. Structure. 2008;16:398–409. doi: 10.1016/j.str.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakrapani S, Sompornpisut P, Intharathep P, Roux B, Perozo E. The activated state of a sodium channel voltage sensor in a membrane environment. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5435–5440. doi: 10.1073/pnas.0914109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krepkiy D, Gawrisch K, Swartz KJ. Structural Interactions between Lipids, Water and S1-S4 Voltage-Sensing Domains. journal of molecular biology. 2012 doi: 10.1016/j.jmb.2012.07.015. doi:10.1016/j.jmb.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang N, Horn R. Evidence for voltage-dependent S4 movement in sodium channels. Neuron. 1995;15:213–218. doi: 10.1016/0896-6273(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 19.Ahern CA, Horn R. Focused electric field across the voltage sensor of potassium channels. Neuron. 2005;48:25–29. doi: 10.1016/j.neuron.2005.08.020. [DOI] [PubMed] [Google Scholar]
- 20.Asamoah OK, Wuskell JP, Loew LM, Bezanilla F. A fluorometric approach to local electric field measurements in a voltage-gated ion channel. Neuron. 2003;37:85–97. doi: 10.1016/s0896-6273(02)01126-1. [DOI] [PubMed] [Google Scholar]
- 21.Krepkiy D, et al. Structure and hydration of membranes embedded with voltage-sensing domains. Nature. 2009;462:473–479. doi: 10.1038/nature08542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tombola F, Pathak MM, Isacoff EY. How does voltage open an ion channel? Annu. Rev. Cell Dev. Biol. 2006;22:23–52. doi: 10.1146/annurev.cellbio.21.020404.145837. [DOI] [PubMed] [Google Scholar]
- 23.Yang N, George AL, Horn R. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 1996;16:113–122. doi: 10.1016/s0896-6273(00)80028-8. [DOI] [PubMed] [Google Scholar]
- 24.Larsson HP, Baker OS, Dhillon DS, Isacoff EY. Transmembrane movement of the shaker K+ channel S4. Neuron. 1996;16:387–397. doi: 10.1016/s0896-6273(00)80056-2. [DOI] [PubMed] [Google Scholar]
- 25.Mannuzzu LM, Moronne MM, Isacoff EY. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science. 1996;271:213–216. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- 26.Cha A, Bezanilla F. Characterizing voltage-dependent conformational changes in the Shaker K+ channel with fluorescence. Neuron. 1997;19:1127–1140. doi: 10.1016/s0896-6273(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 27.Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 28.Zou P, McHaourab HS. Increased sensitivity and extended range of distance measurements in spin-labeled membrane proteins: Q-band double electron-electron resonance and nanoscale bilayers. Biophys. J. 2010;98:L18–20. doi: 10.1016/j.bpj.2009.12.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fleissner MR, et al. Structure and dynamics of a conformationally constrained nitroxide side chain and applications in EPR spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16241–16246. doi: 10.1073/pnas.1111420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rayes RF, Kálai T, Hideg K, Geeves MA, Fajer PG. Dynamics of tropomyosin in muscle fibers as monitored by saturation transfer EPR of bi-functional probe. PLoS ONE. 2011;6:e21277. doi: 10.1371/journal.pone.0021277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang Y, Ruta V, Chen J, Lee A, MacKinnon R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature. 2003;423:42–48. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- 32.Lee S-Y, Lee A, Chen J, MacKinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Catterall WA. Molecular properties of voltage-sensitive sodium channels. Annu. Rev. Biochem. 1986;55:953–985. doi: 10.1146/annurev.bi.55.070186.004513. [DOI] [PubMed] [Google Scholar]
- 34.Guy HR, Seetharamulu P. Molecular model of the action potential sodium channel. Proc. Natl. Acad. Sci. U.S.A. 1986;83:508–512. doi: 10.1073/pnas.83.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruta V, Chen J, MacKinnon R. Calibrated measurement of gating-charge arginine displacement in the KvAP voltage-dependent K+ channel. Cell. 2005;123:463–475. doi: 10.1016/j.cell.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 36.Chanda B, Bezanilla F. A common pathway for charge transport through voltage-sensing domains. Neuron. 2008;57:345–351. doi: 10.1016/j.neuron.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 37.Sompornpisut P, Roux B, Perozo E. Structural refinement of membrane proteins by restrained molecular dynamics and solvent accessibility data. Biophys. J. 2008;95:5349–5361. doi: 10.1529/biophysj.108.142984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koag M-C, Papazian DM. Voltage-dependent conformational changes of KVAP S4 segment in bacterial membrane environment. Channels (Austin) 2009;3:356–365. doi: 10.4161/chan.3.5.9697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faure É, Starek G, McGuire H, Bernèche S, Blunck R. A limited 4 Å radial displacement of the S4-S5 linker is sufficient for internal gate closing in Kv channels. J. Biol. Chem. 2012;287:40091–40098. doi: 10.1074/jbc.M112.415497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lacroix JJ, Bezanilla F. Control of a final gating charge transition by a hydrophobic residue in the S2 segment of a K+ channel voltage sensor. Proc. Natl. Acad. Sci. U.S.A. 2011;108:6444–6449. doi: 10.1073/pnas.1103397108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 43.Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1994;91:1667–71. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeschke G. Distance measurements in the nanometer range by pulse EPR. Chemphyschem. 2002;3:927–32. doi: 10.1002/1439-7641(20021115)3:11<927::AID-CPHC927>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 45.Jeschke G, et al. DeerAnalysis2006 - a comprehensive software package for analyzing pulsed ELDOR data. Applied Magnetic Resonance. 2006;30:473–498. [Google Scholar]
- 46.Sompornpisut P, Roux B, Perozo E. Structural refinement of membrane proteins by restrained molecular dynamics and solvent accessibility data. Biophys J. 2008;95:5349–61. doi: 10.1529/biophysj.108.142984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fleissner MR, et al. Structure and dynamics of a conformationally constrained nitroxide side chain and applications in EPR spectroscopy. Proc Natl Acad Sci U S A. 2011;108:16241–6. doi: 10.1073/pnas.1111420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


