Abstract
Orchestration of many processes relying on intracellular signal transduction is recognized to require the generation of hydrogen peroxide as a second messenger, yet relatively few molecular details of how this oxidant acts to regulate protein function are currently understood. This review describes emerging chemical tools and approaches that can be applied to study protein oxidation in biological systems, with a particular emphasis on a key player in protein redox regulation, cysteine sulfenic acid. While sulfenic acids (within purified proteins or simple mixtures) are detectable by physical approaches like X-ray crystallography, nuclear magnetic resonance and mass spectrometry, the propensity of these moieties to undergo further modification in complex biological systems has necessitated the development of chemical probes, reporter groups and analytical approaches to allow for their selective detection and quantification. Provided is an overview of techniques that are currently available for the study of sulfenic acids, and some of the biologically meaningful data that have been collected using such approaches.
Keywords: sulfenic acid, redox regulation, redox signaling, mass spectrometry, chemical probes
I. Introduction
Since the emergence of life on Earth, biological systems have relied on redox reactions to fuel metabolism and growth. Under aerobic conditions, these reactions are often associated with the production of reactive oxygen species (ROS). These species include both radical and non-radical species with varying reactivities toward protein functional groups. Examples of ROS include superoxide (O2·−), hydrogen peroxide (H2O2), hydroxyl radical (OH·), organic hydroperoxides (ROOH), hypohalous acids (e.g., HOCl and HOBr) and many other species. The controlled and localized production of ROS (e.g., H2O2) is now widely acknowledged as an integral regulatory component of signaling, metabolism and epigenetics under physiological conditions (Finkel, 2011; Sena & Chandel, 2012; Wallace & Fan, 2010). Infections and a range of dietary or environmental stressors, however, induce acute or chronic accumulation of these species in humans leading to a wide range of pathologies such as aging, cancer, diabetes, cardiovascular diseases, and others. Further emphasizing the dual beneficial and harmful effects of ROS, our own bodies employ ROS to fight against disease and repair injured tissues (e.g., immune response to infection and wound healing) (Bryan et al., 2012). In clinical applications, radiation and chemotherapies, which rely on the generation of ROS, are used in the fight against cancer. Thus, ROS are tightly associated with both normal physiology and disease pathology and investigations of the ROS targets, the products of oxidation, and the timing of oxidation processes are critical to disease prevention, diagnosis and therapy.
Oxidative regulation of biological processes by ROS has gained increasing attention over the years and has been refueled by the development of chemical reagents and technologies that now enable the investigation of these processes. Currently, the field of redox biology revolves around the following three major areas of investigation: (i) identification of protein targets of oxidation and the nature of oxidative modification, (ii) identification of the precise sites of modification in proteins and quantification of the extent of oxidation, and (iii) study of the functional consequences of oxidation on the individual protein activity and, on a larger scale, on the regulation of metabolic, signaling and epigenetic networks. These issues are highly complex and require multiple approaches. For example, there are currently more than 35 reported types of oxidative modifications in proteins targeting ~13 different amino acid residues (Madian & Regnier, 2010). This review will focus primarily on classical and emerging methods for detection of sulfenic acid (also known as S-hydroxycysteine, abbreviated here as CySOH), the critical intermediate oxoform in oxidative pathways that lead to formation of disulfides, sulfenamides and higher order sulfinic or sulfonic acid species (Fig. 1).
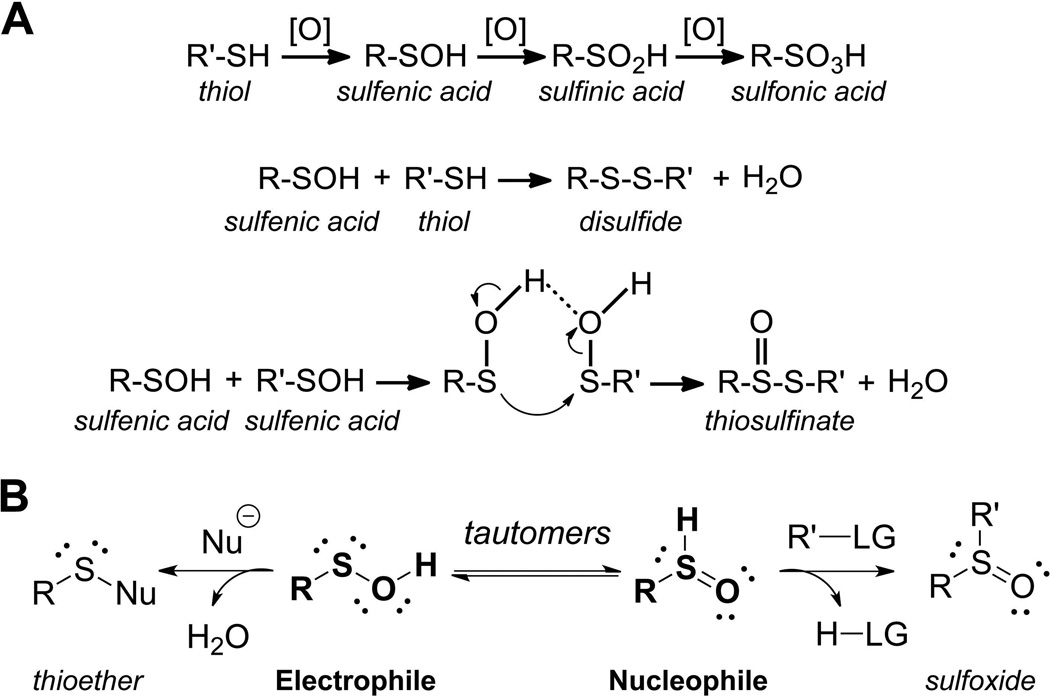
FIGURE 1.
Reactions mediated by R-SOH. A. Products of cysteine oxidation generated by further oxidation of R-SOH (CySOH) to R-SO2H/R-SO3H or condensation reactions to generate disulfides and thiosulfinates. B. R-SOH tautomeric equilibrium; R-SOH electrophilic properties distinguish this species from R-SH.
Mechanistically, cysteine can be modified to -SOH (sulfenic state) through its reaction with hydrogen peroxide (H2O2), organic hydroperoxides (ROOH), peroxynitrous acid (ONOOH) and peroxynitrite anion (ONOO−) (cumulatively referred to as [O] in Fig. 1) (Poole, Karplus & Claiborne, 2004; Kettenhofen & Wood, 2010). Over the past decade, oxidative modification of cysteine to sulfenic acid has emerged as an important post-translational modification in proteins and was shown to play a pivotal role in regulating protein functions under both physiological and oxidative stress conditions (Poole & Nelson, 2008; Wani et al., 2011b; Leonard & Carroll, 2011; Roos & Messens, 2011). The large-scale identification of cysteinyl sites which selectively undergo modifications by intracellular oxidants and electrophiles is an area that has only relatively recently been made possible by new chemical tools and approaches. These methods have provided considerable substantiation of the view that all cysteines within proteins are not equivalent in their reactivity toward oxidants or electrophiles. Rather, at low pH (<6), only a restricted subset of cysteines in proteins are reactive toward electrophiles such as the widely used iodoacetamide (Wu, Kwon & Rhee, 1998). Profiles gathered by mass spectrometry of iodoacetamide- versus maleimide-reactive cysteines within cellular proteins have further verified that the difference in the mechanisms through which these reagents work (i.e., through SN2 reaction or Michael addition, respectively) is reflected in the partially overlapping, yet considerably distinct, reactivity patterns with these two alkylating agents (Dennehy et al., 2006). Proteomic analysis of hydroxynonenal (HNE) Michael adducts also identified a distinct subset of sites susceptible to modification by this electrophile (Codreanu et al., 2009). In another study, reactivity of cysteinyl residues toward iodoacetamide at two different concentrations was assessed in order to detect the rare cysteinyl residues within cellular proteomes that are hyperreactive toward this reagent (Weerapana et al., 2010). The sites identified using this approach were enriched in protein cysteinyl residues involved in nucleophilic and reductive catalysis as well as sites of oxidative modification. As further elaborated below, redox proteomics studies to identify protein oxidation sites have become increasingly available to reveal products of cysteinyl oxidation either under reversible redox regulatory control or, alternatively, as a result of reversible or irreversible oxidative damage.
Structural and bioinformatics analyses of sulfenic acid sites in proteins have provided some indications of features within protein microenvironments that promote reactivity of cysteines toward oxidants and/or enhance stability of CySOH within proteins. Early information from crystal structures of the protein environment surrounding the sulfenic acid site from two unrelated cysteine-based peroxidases, the flavoprotein NADH peroxidase and the peroxiredoxin PrxVI, was used with other data to suggest that the stabilization of this moiety was strongly influenced by (i) the lack of solvent accessibility, (ii) the lack of a proximal thiol group, and (iii) the presence of a H-bonding group nearby (Claiborne et al., 1999). A later bioinformatic study focused on sequence and electrostatic features around CySOH modifiable sites from a diverse set of ~50 proteins (Salsbury et al., 2008). Conclusions from this study were that (i) polar but uncharged residues, and particularly Thr, were overrepresented in the structures near SOH sites and can exert a large influence on the Cys thiol (and presumably CySOH) pKa; (ii) three of the four charged residues were underrepresented in sites near the CySOH; and (iii) solvent accessibility did not appear to be a key distinguishing feature of these sites overall. It should be noted, when interpreting these findings, that the amino acids surrounding the sites used for this analysis were identified from protein structures with the Cys of interest in the fully reduced form (CySH), which would not account for any perturbations in structure upon oxidation to CySOH. The proteins used in this dataset are also likely to be quite heterogeneous with respect to their reactivities toward oxidants that generate the CySOH, and their ability to stabilize the SOH once formed. In another report, thermodynamic studies of thiol sulfenylation showed that while the oxidation of CyS− is driven by polar residues (e.g., Arg 127 in human peroxiredoxin), CySH oxidation is favored by a more hydrophobic environment (Billiet et al., 2012).
Once formed, the metastable CySOH species can be further transformed into more stable products through condensation and oxidation reactions (Fig. 1a). These products include disulfides and sulfenamides (the condensation product of sulfenic acid and amine or amide nitrogens), sulfinic and sulfonic acids, thiosulfinates and other species (Jeong et al., 2011). In cells, some of these products are readily reversed to yield the reduced thiol form via the action of cellular reductants like thioredoxin, glutaredoxin and glutathione (Salmeen et al., 2003). Hyperoxidation to sulfinic or sulfonic acid may occur if sulfenic acids encounter high oxidant concentrations, a process which is typically irreversible in the cell. However, an exception to this is now recognized. A specialized repair protein, sulfiredoxin, can rescue at least a subset of cysteine-based peroxidases (peroxiredoxins) in which hyperoxidation to sulfinic acid occurs (Biteau, Labarre & Toledano, 2003; Jönsson et al., 2008a; Jönsson et al., 2008b). In cells, nearly 5% of protein cysteines were reported to exist in sulfinic or sulfonic acid states (Hamann et al., 2002), which is in the range of the estimated phosphoprotein content. Considering that sulfinic and sulfonic acids represent only two of the multiple oxidation products, it becomes quite clear that regulation by oxidative processes may far exceed the regulation mediated by phosphorylation events. The versatility of CySOH reactivity comes from its dual function as electrophile and nucleophile, as noted in examples of its chemistry described below (Fig. 1b).
Thus CySOH is both a product and a critical intermediate of oxidative processes. The advantages of focusing on this species for detection of oxidative processes are: (i) knowledge is gained regarding the reactive site where the oxidation chemistry is initiated (which is not the case if both Cys of a disulfide are simultaneously detected), (ii) the presence of this species most likely indicates an active process (unlike the relative stability that a detected disulfide might exhibit), and (iii) from a technical perspective, it should represent a simpler subset of sites before divergence into multiple oxidation products. The scarcity and often transience of CySOH in proteins makes its detection challenging, however. Given the significance of this oxidative modification in proteins, various analytical methods and probes have been developed to label, identify, and quantify these species using both targeted and discovery approaches. The methods described here take advantage of the dual electrophilic and nucleophilic properties of CySOH discussed above and shown in Fig. 1b.
II. Direct Detection of Sulfenic Acids
The typically unstable nature of sulfenic acids formed in proteins and the often “spectrally silent” changes that occur upon oxidation of the thiol (or thiolate) form of Cys to sulfenic acid has made the detection and analysis of this species a challenge. For this reason, much of the effort devoted to their detection has relied on more indirect means, e.g., by chemical trapping with sulfenic acid-reactive probes, by their selective reduction using arsenite, or by generation of more stable oxidized products that allow one to infer the intermediacy of the sulfenic acid form. However, in cases of stabilized sulfenic acids and proteins amenable to high resolution structural studies, X-ray crystallography and NMR analyses of proteins of interest have offered significant insights into the sulfenic acids present in a select group of proteins. Mass spectrometric analyses without first derivatizing the sulfenic acid can also provide evidence for this redox state, although not without considering the additional oxidation products and adducts that could form during the analyses. Use of these direct biophysical methods for investigations of a number of protein sulfenic acids is discussed in this section.
A. Sulfenic acids detected by X-ray crystallography
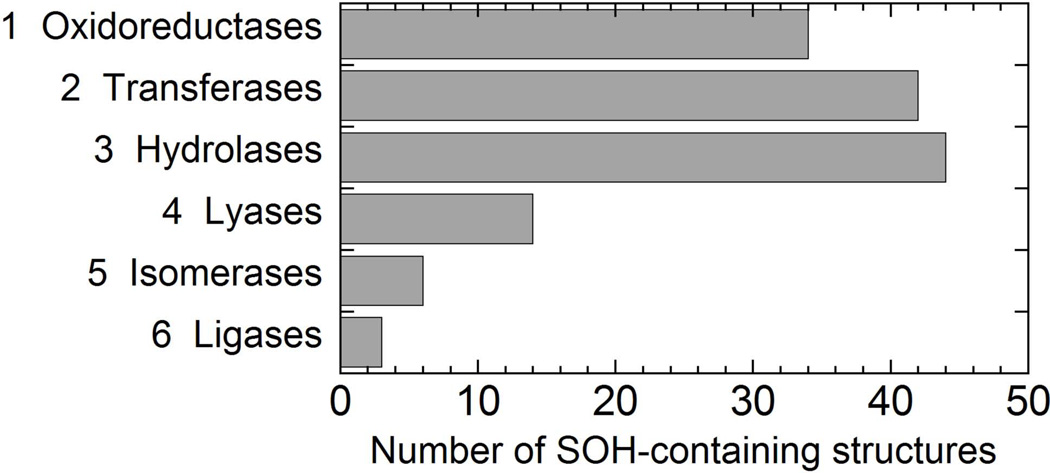
Early efforts to capture crystallographic evidence for a sulfenic acid in the oxidized form of enterococcal NADH peroxidase, a cysteine-based peroxidase with a highly stabilized sulfenic acid formed at the active site proximal to the flavin (Poole & Claiborne, 1989), produced high quality crystallographic information regarding the hyperoxidized, sulfonic acid form (Stehle, Claiborne & Schulz, 1993), but the sulfenic acid form remained difficult to capture for X-ray analysis until cryo conditions were used (PDB ID 1JOA) (Yeh, Claiborne & Hol, 1996). Unfortunately, with X-ray crystallographic analyses, even if the folded, crystallized protein is highly resistant to air oxidation, the use of synchrotron radiation which generates highly reactive hydroxyl radicals will sometimes promote non-physiological cysteine oxidation (Xu & Chance, 2005). Another cysteine-based peroxidase belonging to the peroxiredoxin family, human PrxVI, was apparently more amenable to crystallization and stability of the naturally oxidized, sulfenic acid form (PDB ID 1PRX), requiring no unusual experimental conditions to capture it in the crystal (Choi et al., 1998). Since 1998, the number of structures deposited in the Protein Data Bank (PDB) has grown by more than 10-fold (to over 87,000 at the end of 2012), and among these, many more examples of CySOH have been recognized. There are now (up to the end of 2012) over 400 protein structures in the PDB which include an annotated S-hydroxy or S-oxycysteine (designated CSO or CSX, respectively), only a subset of which are known or suspected to play catalytic or regulatory roles. Filtering these at the “95% sequence similarity” level, 198 proteins remain, of which 143 are enzymes. Shown in Fig. 2 are the numbers of CSO- or CSX-containing structures classified by function according to the Enzyme Classification system. It is clear that CySOH formation is not only observed in oxidoreductases, but also significantly in transferases and hydrolases and to a lesser extent in other functional classes (Fig. 2). A selected set of these protein structures is summarized in Table 1. In fact, the oxidation of Cys residues before or during crystallographic analysis is likely to be far more prevalent than this suggests as such additional density for the oxygen associated with the sulfur in the CySOH is often of partial occupancy and is not necessarily interpreted or reported as a CySOH.
FIGURE 2.
Number of sulfenic acid-containing enzyme structures in the PDB as of Dec. 25, 2012, categorized by function. The PDB included 198 protein structures (filtered at 95% similar) containing CSO (S-hydroxycysteine) or CSX (S-oxycysteine) residues; of these, 143 were enzymes.
Table 1.
Representative proteins whose X-ray crystal structures contain CySOH, emphasizing the oxidoreductasesa
| PDB ID |
Protein Name | Species | CySOH Site |
Date | Enzymatic (or other) function |
Reference |
|---|---|---|---|---|---|---|
| 1JOA | NADH Peroxidase | Enterococcus faecalis | 42 | 1996 | Oxidoreductase | (Yeh, Claiborne & Hol, 1996) |
| 1GSN | Glutathione reductase | Homo sapiens | 63 | 1998 | Oxidoreductase | (Becker et al., 1998) |
| 1PRX | Peroxiredoxin, Prx6 subfamily | Homo sapiens | 47 | 1998 | Oxidoreductase | (Choi, et al., 1998) |
| 1XVW | Peroxiredoxin, AhpE | Mycobacterium tuberculosis | 45 | 2005 | Oxidoreductase | (Li et al., 2005) |
| 1XVQ | Peroxiredoxin, Tpx subfamily | Mycobacterium tuberculosis | 80 | 2006 | Oxidoreductase | (Rho et al., 2006) |
| 2ZCT | Peroxiredoxin, Prx6 subfamily | Aeropyrum pernix | 50 | 2008 | Oxidoreductase | (Nakamura et al., 2008) |
| 3TJJ | Peroxiredoxin PrxIV | Homo sapiens | 124 | 2011 | Oxidoreductase | (Cao et al., 2011) |
| 1DNU | Myeloperoxidase | Homo sapiens | 150 | 2001 | Oxidoreductase | (Blair-Johnson, Fiedler & Fenna, 2001) |
| 1K3I | Galactose oxidase precursor | Fusarium sp. | 228 | 2001 | Oxidoreductase | (Firbank et al., 2001) |
| 1ME8 | Inosine monophosphate dehydrogenase | Tritrichomonas foetus | 319 | 2002 | Oxidoreductase | (Prosise, Wu & Luecke, 2002) |
| 1OZU | Cu-Zn superoxide dismutase | Homo sapiens | 111 | 2003 | Oxidoreductase | (Elam et al., 2003) |
| 1JOX | Glyceraldehyde 3-phosphate dehydrogenase | Oryctolagus cuniculus | 149 | 2003 | Oxidoreductase | (Cowan-Jacob et al., 2003) |
| 2VYN | Glyceraldehyde 3-phosphate dehydrogenase | Rattus norvegicus | 75 | 2009 | Oxidoreductase | (Frayne et al., 2009) |
| 1Y1F | Formylglycine-generating enzyme | Homo sapiens | 336 | 2005 | Oxidoreductase | (Dierks et al., 2005) |
| 1YQW | Ni-Fe hydrogenase | Desulfovibrio fructosovorans | 543 | 2005 | Oxidoreductase | (Volbeda et al., 2005) |
| 1WUI | Ni-Fe hydrogenase | Desulfovibrio vulgaris | 84 | 2005 | Oxidoreductase | (Ogata et al., 2005) |
| 2CVO | Putative N-acetyl-gamma-glutamyl-phosphate reductase | Oryza sativa | 145 | 2005 | Oxidoreductase | (Nonaka et al., 2005) |
| 2D1S | Luciferase | Luciola cruciata | 64 | 2006 | Oxidoreductase | (Nakatsu et al., 2006) |
| 3CDU | NADH oxidase | Lactobacillus sanfranciscensis | 42 | 2006 | Oxidoreductase | (Lountos et al., 2006) |
| 2QTZ | Methionine sulfoxide reductase | Homo sapiens | 421 | 2007 | Oxidoreductase | (Wolthers et al., 2007) |
| 3BQG | Methionine sulfoxide reductase MsrA | Neisseria meningitides | 207 | 2008 | Oxidoreductase | (Ranaivoson et al., 2008) |
| 3B4Y | Glucose-6-phosphate dehydrogenase FGD1 | Mycobacterium tuberculosis | 95 | 2008 | Oxidoreductase | (Bashiri et al., 2008) |
| 3BB0 | Vanadium apochloroperoxida se | Curvularia inaequalis | 69 | 2008 | Oxidoreductase | (de Macedo-Ribeiro et al., 2008) |
| 2WSD | CotA laccase | Bacillus subtilis | 35 | 2008 | Oxidoreductase | (Durao et al., 2008) |
| 3FE5 | 3-hydroxyanthranilat e 3,4-dioxygenase | Bos taurus | 134 | 2009 | Oxidoreductase | (Dilovic et al., 2009) |
| 3ULK | Ketol-acid reductoisomerase | Escherichia coli | 20 | 2012 | Oxidoreductase | (Wong et al., 2012) |
| 1I9T | RNA triphosphatase of mRNA capping enzyme | Mus musculus | 126 | 2001 | Hydrolase | (Changela et al., 2001) |
| 1OET | PTP1B | Homo sapiens | 215 | 2003 | Hydrolase | (van Montfort, et al., 2003) |
| 1YML | CDC 25B phosphatases | Homo sapiens | 473 | 2005 | Hydrolase | (Buhrman et al., 2005) |
| 2BMX | Aminopeptidase P | Escherichia coli | 249 | 2006 | Hydrolase | (Nakamura, et al., 2008) |
| 2HKP | Ulp1 SUMO protease | Saccharomyces cerevisiae | 603 | 2008 | Hydrolase | (Xu et al., 2008) |
| 2VRN | DJ1 family member | Deinococcus radiodurans | 115 | 2008 | Hydrolase | (Fioravanti et al., 2008) |
| 3C6B | S-formylglutathione hydrolase | Saccharomyces cerevisiae | 60 | 2008 | Hydrolase | (Legler et al., 2008) |
| 2ZZD | Thiocyanate hydrolase | Thiobacillus thioparus | 133 | 2009 | Hydrolase | (Arakawa et al., 2009) |
| 2FN6 | Sortase A | Streptococcus pyogenes | 208 | 2009 | Hydrolase | (Race et al., 2009) |
| 4AZ4 | Cobalt peptide deformylase | Escherichia coli | 129 | 2012 | Hydrolase | (Strianese et al., 2012) |
| 3A2Z | Gsp amidase | Escherichia coli | 59 | 2010 | Hydrolase & Ligase | (Chiang et al., 2010) |
| 1IRE | Nitrile hydratase | Pseudonocardia thermophila | 113 | 2001 | Lyase | (Miyanaga et al., 2001) |
| 3HRM | SarZ | Staphylococcus aureus | 13 | 2009 | Transcriptional regulator | (Poor et al., 2009) |
| 3T8R | CymR | Staphylococcus aureus | 25 | 2012 | Transcriptional regulator | (Ji et al., 2012) |
| 2W9Y | CE-FAR-7 lipid binding protein | Caenorhabditis elegans | 9 | 2009 | Lipid Transport | (Jordanova et al., 2009) |
| 2VEC | Yhak “bicupin” | Escherichia coli | 122 | 2009 | Unknown | (Gurmu et al., 2009) |
Included in the table are all of the protein structures from the pdb (filtered at 95% “similar”) returned in searches using the terms “sulfenic,” “sulphenic,” “sulfenate,” or “sulphenate,” excluding unpublished work. All additional oxidoreductases returned in searches for “CSO” (S-hydroxycysteine) or “CSX” (S-oxycysteine) (again filtered at 95% similar) are also included. Proteins are sorted initially by enzymatic function, and secondarily by date, except for the local grouping of closely related proteins where observed.
B. NMR studies to detect sulfenic acids
NMR using proteins labeled with [3-13C]-cysteine has been used to monitor changes in cysteine protonation state (which enables NMR-based pKa analyses) or redox state in proteins exhibiting high quality NMR spectra (Jeng, Holmgren & Dyson, 1995; Wilson et al., 1995; Wishart & Sykes, 1994). Pioneering studies with the sulfenic acid form of the NADH peroxidase discussed above provided the first NMR-based analysis of this redox form of a protein (Crane, Vervoort & Claiborne, 1997). Assisted with assignments of the various forms by chemical shift predictions made by the C-13 NMR Module of ChemIntosh (SoftShell International), the Cys42 sulfenic acid was found to correspond to a chemical shift of 41.3 ppm (comparable to a predicted value of 39.6 ppm), higher than the 30.8 ppm observed for the reduced, thiolate form of the enzyme (predicted at 28.5 ppm). Moreover, oxidatively-inactivated NADH peroxidase, reflecting sulfinic and/or sulfonic acid formation at Cys42 (Poole & Claiborne, 1989), gave a 13C chemical shift of 57.0 ppm; because the expected chemical shifts of Cys sulfinic and sulfonic acids are so similar (predicted values of 58.8 and 56.0 ppm, respectively), this 57 ppm species could not be unambiguously assigned to either oxidation state. It must also be taken into account that the secondary structure and overall protein microenvironment exert additional influences on the actual 13Cβ chemical shifts observed in a given protein (Wishart & Sykes, 1994). This work also provided strong support for the presence of the Cys42 sulfenic acid of NADH peroxidase in its sulfenate (R-SO−) form, rather than O-protonated (R-SOH) or S-protonated sulfoxide (R–S(=O)H forms (Crane, Vervoort & Claiborne, 1997) (Fig. 1B). NMR as well as crystallographic evidence using small molecules to investigate, at higher resolution than possible with proteins, the bond lengths between the O and S of stabilized sulfenic acids has also supported the predominance of the sulfenic acid (as opposed to sulfoxide) form in these molecules (Crane, Vervoort & Claiborne, 1997). Taken together, the results with NADH peroxidase nicely documented the progressive oxidation of the sulfur from thiolate (30.8 ppm), to sulfenate (41.3 ppm), then sulfinic and/or sulfonic acid states (57.0 ppm), reflecting the increasing deshielding of the sulfur atom with oxidation.
C. Direct observation of sulfenic acids by mass spectrometry
Mass spectrometry is in principle an excellent way to detect oxidation of a Cys thiol group to a sulfenic acid, which increases the mass of a protein by 16 amu (one oxygen atom). The main consideration must be, however, the extent to which adventitious oxidation or conversion to other species may be occurring during sample workup and/or analysis. For example, air oxidation (particularly in the presence of trace metals) can lead to sulfenic acid formation in vitro (Kim & Raines, 1994; Rehder & Borges, 2010a) , as well as loss of CySOH through hyperoxidation to sulfinic or sulfonic acids (Ellis & Poole, 1997b; Fuangthong & Helmann, 2002). Anaerobic conditions and/or rapid analysis of each sample can help mitigate such problems (Ellis & Poole, 1997b; Poole et al., 2007). Denaturants present during sample workup or analysis can also have a detrimental effect on sulfenic acid stability. Because of this, the preparation of samples using non-denaturing volatile buffers in the absence, or minimal presence, of acetonitrile are important factors in stabilizing CySOH for MS analysis. In select cases, the CySOH detection can, however, be achieved under denaturing conditions, such as in the presence of formic acid (0.1% in water) (Qian et al., 2012). It should also be noted that any thiol-containing molecules present in the sample will rapidly attack to condense with accessible sulfenic acids to form disulfide bonds, so these must be avoided if such sulfenic acids are to be stabilized. Considerable evidence has also emerged over the last decade that sulfenamides can form, quite readily in some proteins (e.g., in PTP1B and B. subtilis OhrR), as reversible condensation products of sulfenic acids and proximal nucleophilic nitrogens (Lee, Soonsanga & Helmann, 2007; Salmeen, et al., 2003; van Montfort et al., 2003). The desolvation of the analyte molecules during electrospray analysis may well promote formation of this species (Qian, et al., 2012). Unfortunately, the mass signature for sulfenamide formation is a loss of 2 Da relative to the unmodified protein, the same mass change that is observed when disulfide bonds are formed, making clear identification of such a species as the sulfenamide more challenging by MS alone and in the absence of MS/MS analysis. Depending on the ionization properties of specific proteins, the small 2 Da mass change can also be quite difficult to demonstrate unequivocally. We should also note that there are other oxidative modifications (besides sulfenic acid formation) that can lead to a +16 amu mass change: (i) hydroxylation of lysine, tryptophan, proline, and aspartate; (ii) oxidation of methionine to sulfoxide; (iii) formation of 3,4-dihydroxy-phenylalanine (DOPA) from tyrosine; (iv) oxidation of histidine to oxohistidine; and, (v) oxidation of proline to γ-glutamyl semialdehyde (Hovorka et al., 2002; Amici et al., 1989). Therefore, one should not simply rely only on the MS analysis to assess formation of CySOH. Mutagenesis, chemical labeling and MS/MS studies should accompany this as has been used to demonstrate the presence of sulfenic acids in a number of proteins (Boschi-Muller et al., 2000; Fuangthong & Helmann, 2002; Poole, et al., 2007; Qian et al., 2011; Qian, et al., 2012; Rehder & Borges, 2010a) and peptides (Rehder & Borges, 2010a; Shetty & Neubert, 2009; Shetty, Spellman & Neubert, 2007).
III. Chemical Approaches for Detection of Oxidized Thiols and/or Sulfenic Acids
Many proteins are not amenable to the crystallographic or NMR-based analyses described above that can directly detect sulfenic acids. Direct analysis by mass spectrometry is also limited by the inherent instability of this redox species under aerobic and/or denaturing conditions. Moreover, there is a need to have available multiple complementary tools to detect and study this often transient redox form in order to better understand its properties within the context of the specific proteins of interest.
Before turning to chemical modification methods to identify proteins which form oxidized Cys modifications and especially CySOH in cells and tissues, it is important to note that a genetic method has also been introduced for identifying such proteins as an alternative to chemical labeling. Given studies of the sensitivity of the yeast transcription regulator Yap1 toward disulfide bond formation through a CySOH intermediate (Delaunay et al., 2002), Wood’s group developed a genetically engineered protein probe (Yap1-cCRD) with reactivity toward CySOH in a range of proteins (Takanishi, Ma & Wood, 2007; Takanishi & Wood, 2011). The protein contains a histidine tag for enrichment and a CySOH reactive thiol in the C-terminus (Cys598). When expressed in target cells, Yap1-cCRD reacts with endogenous CySOH proteins resulting in disulfide-linked proteins that can then be pulled down along with Yap1-cCRD using His-tag affinity columns, reduced to release the cross-linked protein, separated by SDS-PAGE, in-gel digested and then analyzed by mass spectrometry. Using this approach the authors were able to identify ~ 30 presumed CySOH proteins in Escherichia coli (Takanishi, Ma & Wood, 2007) and Saccharomyces cerevisiae (Takanishi & Wood, 2011). The drawbacks of the Yap1-based strategy are the relative low efficiency and the somewhat high risk of false identifications (e.g., disulfide bond formation during sample processing, and co-elution of non-CySOH proteins due to complex protein-protein interactions that may or may not involve disulfide bond formation).
With the recent expansion in the availability of tools for the analysis of oxidative posttranslational modifications, sulfenic acids can be targeted selectively for analysis, or be an intended or unintended part of redox proteomics analyses conducted to identify oxidation targets in cells, depending on the approach used. Described below are some of the approaches and reagents currently available for detecting thiol oxidation and, more specifically, sulfenic acid formation within proteins.
A. Methods relying on reversal of cysteine oxidation by general or specific reductants
Approaches undertaken to identify proteins, or at higher resolution specific sites within proteins, which undergo modifications on thiol groups must at the outset be tailored to minimize the loss or “scrambling” of the modifications present in the sample. Efficient trapping of thiol groups to prevent further reaction is thus an essential part of any successful protocol to identify thiol modifications present in biological samples. Often overlooked are sulfenic acid groups which, if unprotected, will be prone to hyperoxidation, disulfide bond formation with thiol groups, or potentially other chemical fates during the workup of samples, and especially upon denaturation and/or digestion of the proteins. A common initial step is to block free thiol groups with an alkylating agent like N-ethylmaleimide (NEM); depending on the intended analysis, dimedone could also be included for alkylation of sulfenic acids. Both of these reagents have the advantage of being cell permeable, so that the initial blocking steps can be done prior to cell lysis, although one can never be certain that all redox modifications are trapped in precisely the same state as they were in the untreated cells. Acidic conditions are generally preferred for the alkylation conditions in order to slow thiol-disulfide exchange, although at very high alkylating agent concentrations this may be less important (Hansen & Winther, 2009). Use of an alternative alkylating agent such as iodoacetate or iodoacetamide is also possible, although these reagents become less reactive when acidic conditions are used and most thiol groups are protonated. Full alkylation is not always possible in the absence of denaturation, but denaturants are not always part of protocols for such analyses.
If a detectable group or affinity handle like biotin is included on the initial alkylating agent, then the analytical procedure takes advantage of the fact that thiol modifications present in the protein(s) under analysis will typically block alkylation at that site. In these cases, the loss of thiol reactivity due to the modification can then be detected as a decrease in the incorporation of the alkylating agent into that protein or site (Sethuraman et al., 2004). This allows for detection of both reversible and irreversible modifications including alkylations, but does not report on the type of modification present (Chouchani et al., 2011).
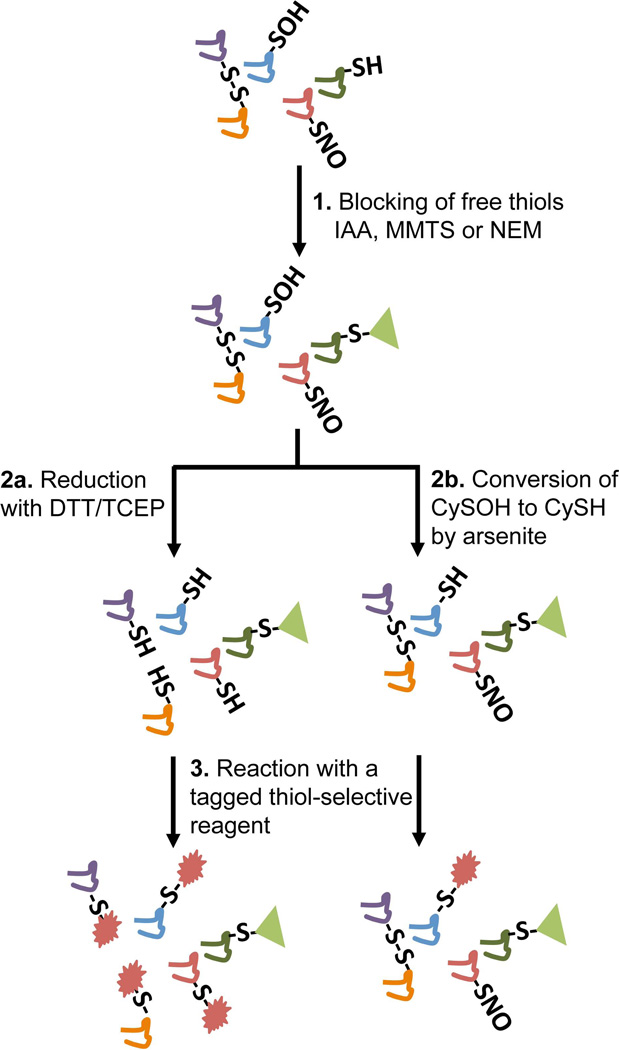
A more sensitive and in some cases selective approach to detect reversible thiol modification is to block thiols in the first step with an unlabeled alkylating agent (e.g., NEM), then reduce or reverse the thiol modification to “unmask” new thiol groups that can then be alkylated with a detectable thiol probe (Fig. 3). Reduction by either DTT or TCEP prior to the second alkylation provides a comprehensive way to detect oxidation of thiols to various reversible products including disulfides, sulfenic acids and S-nitrosothiols (Fig. 3, path a).
FIGURE 3.
The tag-switch approach to detect CySOH, S-nitrosothiol, and disulfide-containing proteins together (path 2a) and CySOH selectively (path 2b). The thiol-selective reagent in step 3 can be tagged with biotin, fluorescent dyes, His-tag, and others depending on the downstream detection method (Western blot, enrichment/MS, 2D-DIGE, etc).
More selective uses of this general approach have also been used. The “biotin switch” approach was introduced by Jaffrey and Snyder for detection of S-nitrosated cysteinyl (CySNO) groups (Jaffrey et al., 2001; Jaffrey & Snyder, 2001); following a thiol blocking step, ascorbate is added to selectively reduce CySNO groups, then the nascent thiol groups are labeled with a detectable thiol-directed reagent like N-[6-(biotinamido)hexyl]-3'-(2'-pyridyldithio)propionamide (HPDP-biotin) (Chouchani, et al., 2011). In a modification to the standard assay introduced by Hogg and Gladwin, addition of copper to the ascorbate in the reduction step increases the sensitivity without loss of selectivity, improving the ability to detect CySNO in protein samples (Wang et al., 2008). On the basis of the biotin switch approach for detecting CySNO, Eaton and colleagues introduced a related biotin switch approach where the reductant used was arsenite rather than ascorbate in order to selectively reduce CySOH rather than CySNO (Saurin et al., 2004) (Fig. 3, path b). Upon alkylation of these nascent thiols with biotin-maleimide and subsequent affinity capture, gel separation and digestion steps, a set of 17 proteins was identified by mass spectrometry from among the biotinylated proteins isolated from rat heart tissues previously perfused with H2O2. Unfortunately, two of the proteins thus identified have no cysteine residues at all and may have remained bound to other labeled proteins during the affinity capture procedure (Saurin, et al., 2004), suggesting a need for higher stringency washes of the bound material before elution from the streptavidin-agarose beads. The lability of the sulfenic acid moiety also raises concerns about the effectiveness and sensitivity of this assay in detecting protein sulfenic acids.
B. Chemical cross-reactivity of sulfenic acids with reagents used in detecting other protein posttranslational modifications
Many of the approaches for detecting protein oxidations, and particularly thiol oxidations, rely on the use of reagents intended to specifically target known functional groups. The assumptions of specificity or selectivity are not always completely valid, however. For example, NEM, iodoacetamide and methyl methanethiosulfonates are often used as thiol-specific blocking agents, but all exhibit some degree of reactivity toward sulfenic acids (Poole & Ellis, 2002), a cross-reactivity which can skew results in unexpected ways. Because it generates a disulfide rather than irreversible alkylated product, MMTS can also induce intra- and intermolecular disulfide bonds in proteins that were not present at the outset, leading to the conclusion that this reagent should be avoided as a thiol-blocking agent (Hansen & Winther, 2009).
Another under-recognized cross-reactivity exhibited by sulfenic acids is with 2,4-dinitrophenylhydrazine (DNPH), the reagent that is used to detect carbonylation in oxidatively stressed biological samples (Dalle-Donne et al., 2009). Thus, a significant component of the aldehydes and ketones purportedly detected in the large number of publications that use this approach may in fact be cysteine sulfenic acids which are likely to be formed under much milder oxidative conditions than would be expected to introduce carbonyl groups into proteins. Another point of concern is with the ascorbate used for selectively reducing S-nitrosothiols; at least one report indicates that ascorbate can also act as a reductant for sulfenic acids to support turnover of several peroxiredoxins with their peroxide substrates (Monteiro et al., 2007), raising the possibility that sulfenic acid crossreactivity contributes to the signal observed in some S-nitrosothiol targeting experiments.
One additional type of cross-reactivity with reagents typically used to assess thiols in proteins must also be considered when sulfenic acids may be present in a given sample. Aromatic disulfides have been noted to react with sulfenic acids (Torchinsky, 1981), and this reactivity was confirmed in later studies which demonstrated that 5,5’-dithiobis(2-nitrobenzoic acid) (DTNB), a reagent often used to assess thiol contents in proteins, does indeed react with protein sulfenic acids, likely yielding the thiosulfinate product (Poole & Ellis, 2002). Assessment of the DTNB reactivity with sulfenic acids is complicated, however, by the even greater reactivity of sulfenic acids toward the 2-nitro-5-thiobenzoate (typically referred to as TNB) that is released upon reaction of DTNB with thiols or sulfenic acids. These chemistries emphasize the care that must be taken when thiol contents are evaluated using aromatic disulfides like DTNB and 4,4’-dithiodipyridine (Hansen & Winther, 2009) in samples that may contain sulfenic acids.
C. Dimedone alkylation and UV/Vis spectral approaches to detect sulfenic acids
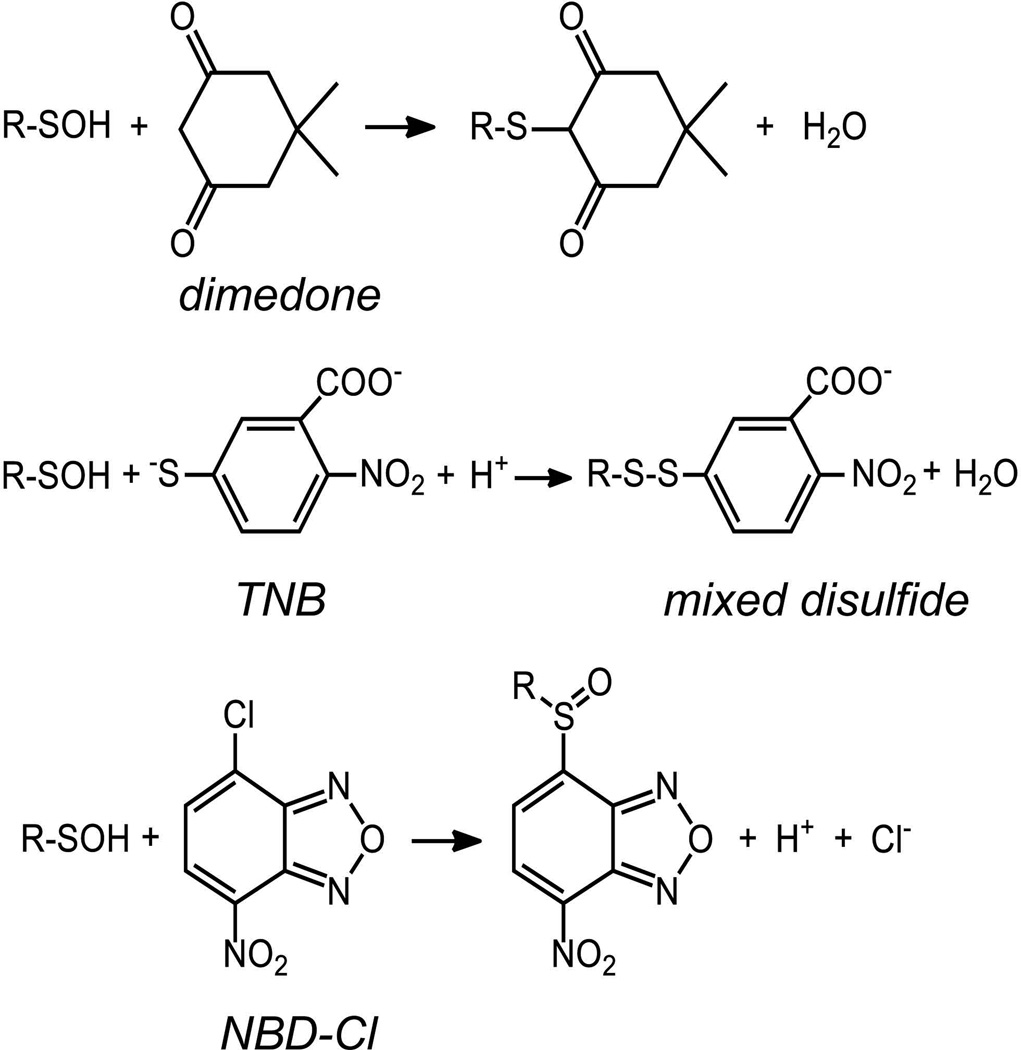
The unique reactivity of sulfenic acids toward nucleophiles such as 5,5-dimethyl-1,3-cyclohexanedione (dimedone), benzylamine, and thiol groups was the primary way in which this species was originally identified within reversibly oxidized, inactivated forms of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and papain (Benitez & Allison, 1974; Lin, Armstrong & Gaucher, 1975; Allison, 1976). Dimedone, the most widely used reagent for sulfenic acid trapping, reacts with sulfenic acids to generate a covalent thioether linkage that is not reversed by reductants like DTT or TCEP (Fig. 4). Importantly, dimedone was shown to be unreactive toward functional groups such as S-nitrosothiol, methionine sulfoxide, aldehydes and amines in aqueous solution and neutral pH (Poole et al., 2005). In the 1970s, this reagent could be purchased in radioactive form, giving the investigator a way to quantitatively evaluate the level of incorporation of the reagent into the protein sample. In the last several decades, however, a radioactive version of this reagent has not been available. In the absence of the detectable radioisotopically-labeled dimedone, mass spectrometry can be used to detect the incorporation of the dimedone into proteins of interest based on the mass increase of 138 amu imparted by dimedone modification (Willett & Copley, 1996; Ellis & Poole, 1997b; Percival et al., 1999; Carballal et al., 2003; Conway, Poole & Hutson, 2004). More recently, detection of dimedone incorporation into proteins has also become possible using antibodies which recognize this moiety (Seo & Carroll, 2009; Maller, Schroder & Eaton, 2011). As described below, several groups have also developed new sets of chemical probes to directly trap and detect sulfenic acids based on the 1,3-dicarbonyl core of dimedone. In order to evaluate sulfenic acids within proteins before such reagents became available, however, several spectroscopic methods for evaluating sulfenic acid formation within pure proteins were developed, as described next.
FIGURE 4.
The reaction products of R-SOH (CySOH) reaction with dimedone, TNB and NBD-Cl.
An important approach to quantify sulfenic acids in proteins relies on their propensity to generate disulfide bonds with thiol groups. In particular, the bright yellow, small thiol-containing molecule TNB, described above as the product of DTNB reduction, reacts stoichiometrically with sulfenic acids and is well suited for this purpose (Poole & Claiborne, 1989; Ellis & Poole, 1997a; Poole & Ellis, 2002; Poole, 2008; Rehder & Borges, 2010b). As long as the TNB solution is freshly prepared, is protected as much as possible from air oxidation, and does not include any residual DTNB or DTT, the loss of the strong 412 nm absorbance of TNB upon addition of a sulfenic acid-containing sample (if the sulfenic acid is sufficiently exposed) correlates directly with the sulfenic acid content. Moreover, it is possible to isolate the protein product(s) of this reaction through ultrafiltration, and then titrate with DTT in order to observe the increase in 412 nm absorbance as the TNB is released from the protein (Fig. 5). This TNB-based analytical approach has been used to quantitatively establish the presence of sulfenic acid in oxidized forms of NADH peroxidase (Poole & Claiborne, 1989) and the bacterial peroxiredoxin AhpC (Ellis & Poole, 1997a; Poole & Ellis, 2002), as well as in oxidized human serum albumin (Turell et al., 2008) and more complex biological samples (Rehder & Borges, 2010b).
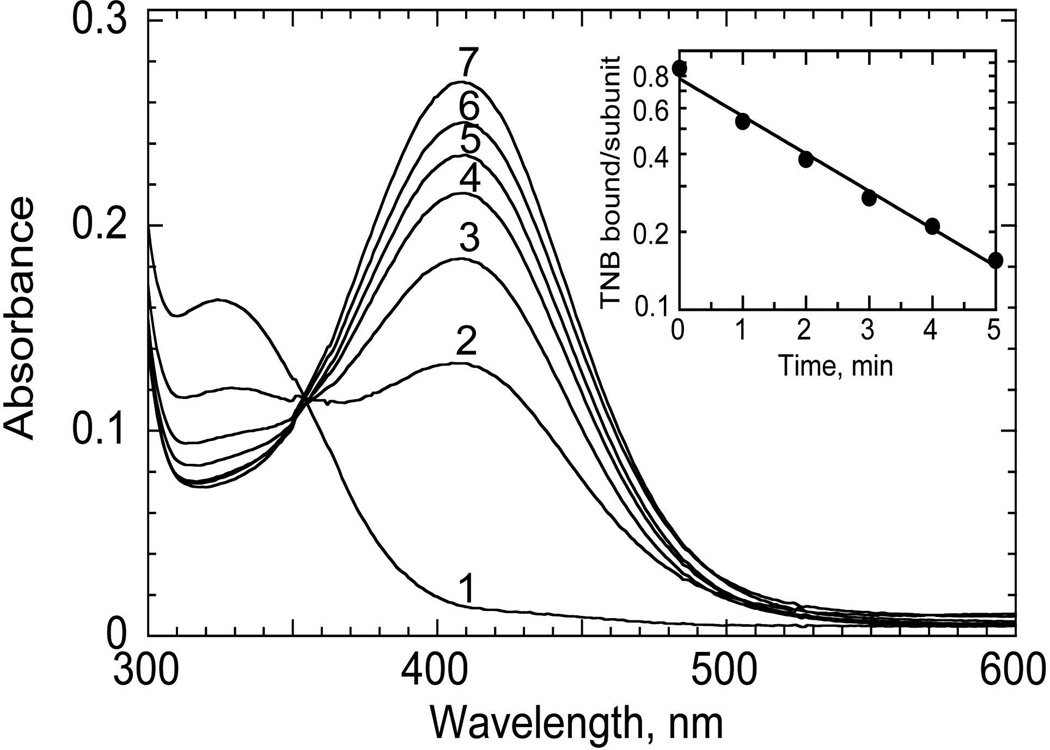
FIGURE 5.
Reduction of TNB-labeled C165S AhpC by dithiothreitol (DTT). The mutant enzyme (20 nmol in 0.6 mL) pretreated with 1 equivalent of H2O2 followed by 20 equivalents of TNB was washed free of excess TNB and treated with a 10-fold excess of DTT. Spectra shown are before (1) and after addition of DTT for 1, 2, 3, 4, 5, and 30 min (spectra 2–7, respectively). (Inset) Semilogarithmic plot of the change in absorbance, converted into units of TNB/subunit, versus time. Reprinted with permission from Ellis HR, Poole LB. 1997. Biochemistry 36:13349–13356. Copyright 1997 American Chemical Society.
In an effort to establish additional criteria that could be used to directly demonstrate sulfenic acid formation upon peroxide-mediated oxidation of the C165S mutant of bacterial AhpC, a method was developed that used the electrophilic reagent NBD chloride (7-chloro-2-nitrobenzo-2-oxa-1,3-diazole) to detect sulfenic acids (Ellis & Poole, 1997b). This reagent reacts nearly as readily with sulfenic acids as with thiol groups at neutral pH, and the products of this reaction in the two cases are spectrally distinguishable. While the NBD-thiol adduct, a thioether, absorbs maximally at ~420 nm, the adduct with sulfenic acid (either a sulfenate ester or sulfoxide) absorbs maximally at ~347 nm, very similar to the unreacted reagent (Fig. 6a). The NBD adduct with the thiol group is also fluorescent, whereas the adduct with sulfenic acid is not (Ellis & Poole, 1997b). Ultimate proof of these products was obtained using mass spectrometry, where the NBD adduct with the sulfenic acid was shown to be 16 amu larger than the comparable adduct with the thiol group (Fig. 6b & c). Reactivity of NBD chloride toward other amino acid side chains can occasionally cause problems in these analyses, as well, although the adducts with amino or tyrosyl groups are typically formed only at higher pH and are readily distinguished by their distinct spectral properties (i.e., λmax values are 382 and 480 nm for NBD adducts with tyrosines and amines, respectively) (Aboderin & Boedefeld, 1976; Ghosh & Whitehouse, 1968; Miki, 1985). As a possible advantage of this reagent over dimedone, the NBD adducts with either thiol or sulfenic acid are reducible by DTT, allowing for the recovery of the reduced form of the protein after analysis. Since its introduction, this approach has been used to detect the presence of sulfenic acids in a number of proteins (Denu & Tanner, 1998; Ma, Takanishi & Wood, 2007; Fuangthong & Helmann, 2002; Carballal, et al., 2003; Griffiths, King & Cooney, 2002; Silva et al., 2008). With both of the spectral approaches to label sulfenic acids described in this section, modification with TNB or NBD, pretreatment with dimedone blocks the incorporation of the chromophore, adding another layer of proof to the experimental evidence for sulfenic acid formation in proteins of interest (Ellis & Poole, 1997b).
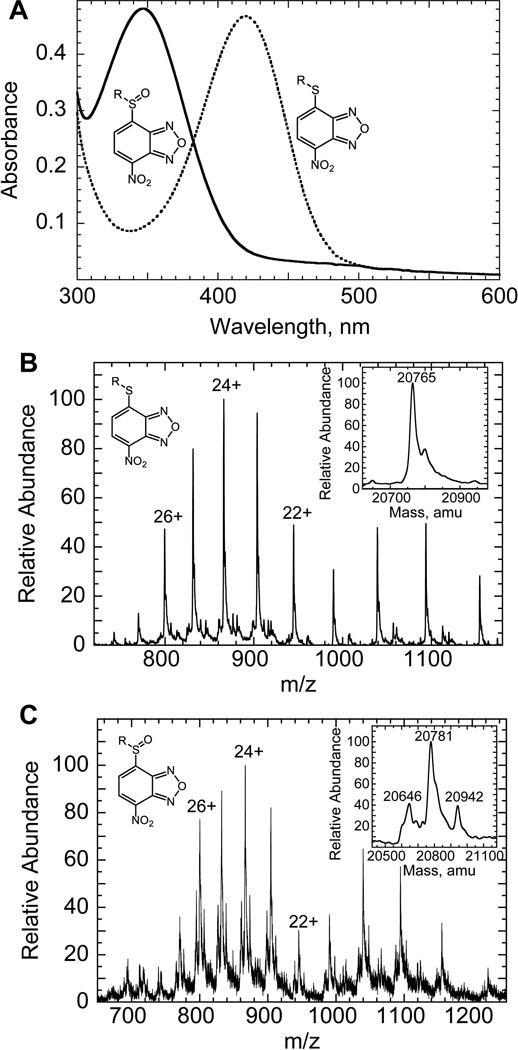
FIGURE 6.
Reaction of CySOH with NBD-Cl. A. Spectroscopic characteristics of NBD-modified AhpC mutants. The Cys-S(O)-NBD conjugate was generated by incubating 30 nmol of C165S AhpC with 1 equiv of H2O2 under anaerobic conditions for 15 min, followed by incubation with 2 equiv of NBD-Cl (0.6 mL total volume) for 30 min in the standard buffer containing 50 mM KCl (solid line). Modified proteins were concentrated by ultrafiltration and rediluted into standard buffer three times prior to absorbance measurements. The Cys-S-NBD conjugate was generated under conditions similar to the Cys-S(O)-NBD conjugate, but without pretreatment with H2O2 (dotted line). The rate of formation of the Cys-S(O)-NBD conjugate was a little slower than that of the Cys-S-NBD conjugate, with spectral changes half complete by about 5 and 2 1/2 min, respectively. B and C. Electrospray mass spectra for NBD-modified products of reduced and oxidized C165S AhpC. NBD-modified C165S proteins (7–10 nmol) generated with or without pretreatment by 1 eq of H2O2 were concentrated by ultrafitration and rediluted five times with deionized water, generating final samples of about 400 µL containing 50% HPLC-grade acetonitrile and 1% formic acid. The capillary temperature was 80 °C, and the spray voltage was 3.5 kV. The mass spectrometer was calibrated in the high-MW range using myoglobin. Each panel shows the full ESI-MS pattern of peaks with several of the peak charges labeled; the insets show a deconvolution of the series of peaks to give the final mass value(s). Panel B shows the mass spectrum obtained from a sample of the Cys-S-NBD conjugate of C165S; Panel C shows that for the Cys-S(O)-NBD conjugate. The two additional products detected in the Cys-S(O)-NBD sample correspond to over-oxidized protein which was not labeled by NBD (20,645.5 +/− 3.3) and a small amount of Cys-S(O)-NBD product that was labeled with a second NBD (20,942.2 +/− 1.8). Note that this latter product was present in amounts too small to be detected spectrally (Panel A). Adapted with permission from Ellis HR, Poole LB. 1997. Biochemistry 36:15013–15018. Copyright 1997 American Chemical Society.
D. Development of proteomics-friendly approaches to detect sulfenic acids
The methods described above often work well with purified proteins for detection and quantitation of sulfenic acid formation, but they are of limited value when the samples of interest are more complex and broader, proteomics-level analyses are desired. For example, NBD chloride reacts with both thiol and sulfenic acid groups, requiring either differential mass spectrometry or spectroscopic analyses to distinguish the products, and the presence of more than a few additional Cys residues in a given protein to be analyzed can cause significant problems in such analyses. Further, both TNB and NBD adducts are labile in the presence of reductants or can migrate to different locations when attacked by other residues either intra- or intermolecularly. Dimedone is a better reagent in that it forms a stable alkylated species, but it offers no useful affinity handle or detectable group to facilitate proteomic-level analyses.
By far the most important tool in proteomics-level analyses is mass spectrometry, and we describe below the utility of this technology to studies of the oxidized cysteine proteome. Moreover, chemical probes based on the 1,3-cyclohexadione reactive core of dimedone have been developed recently by several groups, and the different types of probes currently available and their utility for analyzing the sulfenic acid proteome are summarized below.
1. Mass spectrometry approaches for analyzing cysteine and oxidized cysteine proteomes
Mass spectrometry has emerged as a critical tool in the analysis of biological systems, allowing for both the high-throughput monitoring of temporal changes in proteins and their post-translational modifications in response to extracellular cues, as well as in-depth investigations of selected biochemical processes. Its application for the monitoring of protein oxidation was recognized early in the development of proteomics workflows. Steps like reduction and blocking by alkylation of cysteine residues in proteins, and inclusion of potential methionine oxidation within the database search algorithms are, with few exceptions, present in all proteomics workflows.
Early work to quantitatively evaluate proteins across entire proteomes took advantage of the distinct reactivity of protein thiol groups and their high prevalence in proteins in order to analyze the relative abundance of specific proteins across a range of conditions. The first isotope-coded affinity tags (ICAT) developed by Aebersold’s group contained a thiol reactive group (e.g., an iodoacetyl group), a linker carrying stable isotopes (with heavy and light versions to distinguish between paired samples) and an affinity tag for enrichment of labeled peptides (Gygi et al., 1999). The frequency of cysteine residues in proteins (~ 90% of the proteins contain at least one cysteine residue, with an average of 11 cysteines/protein) ensured a high coverage of the proteome. It also allowed for the detection of low abundance proteins by reducing sample complexity (e.g., in the yeast proteome, ~10% of all tryptic peptides are cysteine-containing peptides) (Gygi, et al., 1999). This early strategy of protein quantification has been modified over the years to include cleavable linkers (acid-cleavable or enzymatic), other enrichment or detection tags (e.g., fluorescent dyes), and a range of reactive groups for assessing protein or peptide abundance that extend beyond targeting of cysteinyl residues (e.g., amine reactive reagents such as iTRAQ or TMT). While the relative change in protein abundance is an important factor in controlling the level of activity of many proteins, it has become increasingly important to evaluate proteomes for their functional qualities. One way to address this need is to apply activity-based protein profiling (ABPP) which provides a readout of targeted enzymatic or binding activities under selected conditions (Cravatt, Wright & Kozarich, 2008). Proteomic methods are also increasingly available to identify particular posttranslational modifications across a wide range of proteins. It is within the context of this need for more and better defined information that the field of redox proteomics has developed.
As described above, ICAT reagents targeting free thiols can be used to distinguish between (at least) two samples, and such reagents were soon applied to differentially label reduced versus reversibly oxidized cysteines to provide a readout of “redox status” (OxICAT) (Leichert et al., 2008). Based on the approaches described in Section IIIA above, two different isotope-coded reagents could be incorporated in the first thiol blocking step and the second labeling step after reduction by DTT or TCEP (or more targeted protein reductants like thioredoxin) to release the thiol groups from those modifications (disulfides, S-nitrosocysteine or sulfenic acid modifications) that are susceptible to the reductant used. As described above, a variation on this theme included simply blocking the free thiols initially present with a thiol reactive reagent (typically iodoacetamide, NEM or MMTS), reduction of the targeted oxidized species to free thiols (e.g., using ascorbate or arsenite for CySNO or CySOH reduction, respectively) and labeling of resulting thiols with a tagged thiol reactive group (e.g., biotin-maleimide, biotin-iodoacetamide, or HPDP-biotin) (Fig. 3). With this approach, only the targeted set of proteins are labeled for affinity capture and further analysis by Western blots or mass spectrometry. As is always the case with such approaches, the selectivity and labeling efficiency of the chosen chemical reagents toward thiols and the modified targets are key to the accuracy of this analysis. The caveats pertaining to cross-reactivity of thiol-blocking reagents or selective reduction of cysteine oxoforms were discussed in Section IIIA.
In order to more specifically target the sulfenic acid proteome for analysis by mass spectrometry and other techniques, a number of new reagents have been developed based on dimedone which allow for the incorporation of detectable or affinity tags into proteins at sulfenic acid sites, as described in the next section.
2. New reagents for labeling proteins containing sulfenic acids
Given the caveats of the indirect biotin switch strategies described above, recent work from our groups as well as others (Poole, et al., 2005; Charles et al., 2007; Poole, et al., 2007; Reddie et al., 2008; Leonard, Reddie & Carroll, 2009; Qian, et al., 2011; Qian, et al., 2012; Truong et al., 2011) has focused on the more direct identification of CySOH proteins and the cysteine modification sites using methods based on sulfenic acid-selective chemical labeling and enrichment followed by Western blot analysis (e.g., to monitor the CySOH status in known proteins) or MS (in discovery or targeted types of analyses). The first readily detectable reagents with selectivity toward sulfenic acids included fluorescent groups (methylcoumarin or isatoic acid) linked to 1,3-cyclohexadione (dimedone lacking the dimethyl group on the ring) (Poole, et al., 2005; Klomsiri et al., 2010). Studies with these reagents and dimedone confirmed the selectivity of such 1,3-dicarbonyl reagents toward CySOH relative to other reactive groups and modifications in proteins. Several years later new reagents were developed which incorporated either rhodamine or fluorescein as the detectable group (Poole, et al., 2007), or biotin attached through a linker to 1,3-cyclohexadione (Poole, et al., 2007) or dimedone (Charles, et al., 2007) for subsequent affinity enrichment of labeled proteins or peptides (Fig. 7). In a similar vein, reagents with azide or alkyne groups attached to the 1,3-cyclohexadione core were also introduced, allowing incorporation of a detectable group or affinity handle subsequent to the labeling step using Staudinger ligation or click chemistry approaches (Reddie, et al., 2008; Seo & Carroll, 2011; Leonard, Reddie & Carroll, 2009) (Fig. 7). These new tools enabled the identification of new sets of oxidation-sensitive proteins not previously possible (Charles, et al., 2007; Leonard, Reddie & Carroll, 2009; Reddie, et al., 2008; Wani, et al., 2011b), although there is still a need for improvements in the capacity for these probes and approaches to effectively identify specific reactive cysteinyl groups in oxidatively-modified proteins (as described in more detail below).
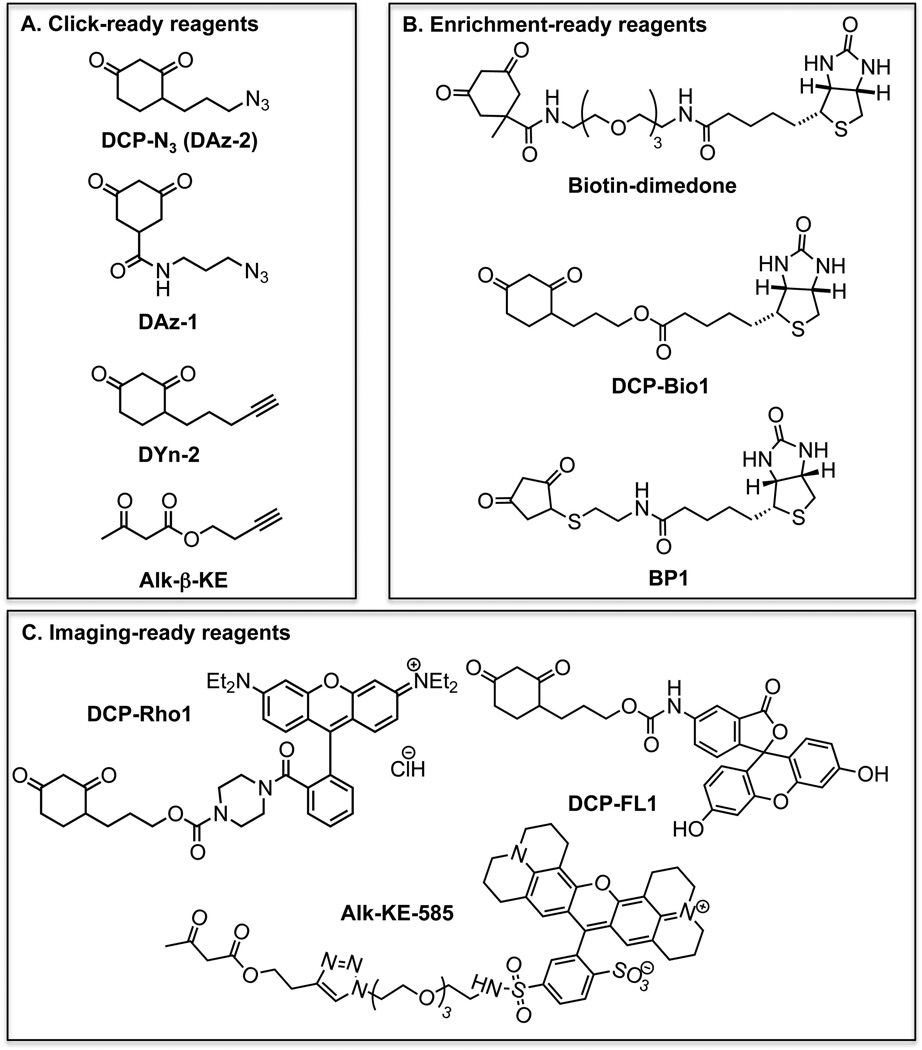
FIGURE 7.
Nucleophilic chemical probes for selective labeling of CySOH species. A. Alkyne and azide-linked reagents for tag addition post-labeling using the click reaction. B. Biotin-linked reagents for enrichment and/or Western blot detection. C. Reagents tagged with fluorescent dyes for imaging application.
Improvements in synthesis and compatibility with MS using 1,3-dicarbonyl reagents (BP1 and Alk-β-KE)
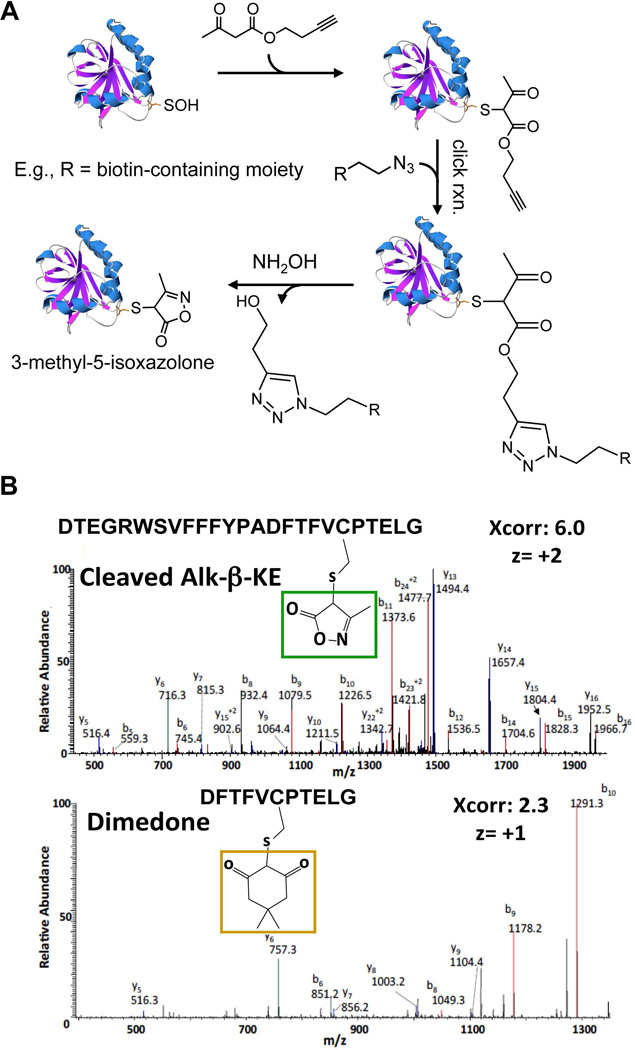
While proven to be selective in labeling CySOH proteins, the attachment of biotin, alkyne/azide or fluorescent dyes to the reactive 1,3-cyclohexanedione moiety requires complex synthesis schemes that result in an overall low yield (Charles, et al., 2007; Leonard, Reddie & Carroll, 2009; Poole, et al., 2007). The detailed mechanism on how dimedone reacts with CySOH has not been extensively studied, but the reaction has been proposed to proceed through a 1,4-addition or direct nucleophilic substitution (Reddie & Carroll, 2008). These scenarios imply that other 1,3-diketone containing compounds may potentially react with CySOH. This hypothesis has led to synthesis of ethylthio-1,3-cyclopentanedione, its biotin conjugate (BP1), and linear alkyne 1,3-β-ketoester (e.g., Alk-β-KE) derivatives (structures shown in Fig. 7) (Qian, et al., 2011; Qian, et al., 2012). Thorough reactivity studies using recombinant proteins and cell lysates proved these probes to be selective toward CySOH and showed distinct pH dependent reactivity profiles with CySOH (Fig. 7). In particular, there was a significant increase in the reactivity of ethylthio-1,3-cyclopentanedione and Alk-β-KE toward CySOH at lower and higher pH, respectively, while at physiological pH the reactivity was comparable with dimedone. The use of low pH during cell lysis to achieve CySOH labeling has the added advantages of quenching ROS-inducing reactions and limiting post-lysis oxidation. The potential for side-reactions of Alk-β-KE with amines (e.g., lysine) was raised in a recent review article (Paulsen & Carroll, 2013). Data in Qian, et al. (2012) do not support this concern regarding cross-reactivity. The Alk-β-KE was tested with recombinant AhpC, a protein that contains numerous lysine and arginine groups in addition to the amino-terminal group. There was only one adduct formed, which the MS/MS analysis showed to be at the CySOH site. Further selectivity studies using cell lysates showed no evidence of labeling with the Alk-β-KE in the presence of a reducing agent (e.g., TCEP), further proving the selectivity of this probe toward CySOH and lack of reaction with amines or other amino acids. The reference cited by Paulsen and Carroll describes the reactivity of a 1,3-diketone (not β-ketoester) with a reactive amine in an engineered catalytic antibody that was specifically raised against the 1,3-diketone hapten (Zhu, et al., 2009). Attachment of fluorophores, biotin, alkyne or azide tags to either the 1,3-cyclopentanedione or the β-ketoester can be achieved with high yield and in just a few chemical steps. In the case of Alk-β-KE, azide-containing tags can be added through the robust click reaction and can be subsequently removed using hydroxylamine to yield the smaller tag 3-methyl-5-isoxazolone (Fig. 8a). This is particularly useful during procedures that involve enrichment followed by Western blot or MS detection. In fact, the resulting 3-methyl-5-isoxazolone tag was shown to improve the fragmentation pattern of labeled peptides in MS/MS analysis relative to dimedone labeled peptides (Fig. 8b). Introduction of an acid-labile tag has also been used to improve MS properties of labeled CySOH products (Truong, et al., 2011).
FIGURE 8.
Reaction of CySOH in proteins with Alk-β-KE. A. Labeled proteins can be conjugated with reporter tags using click-reaction. The reporter tag can then be removed using NH2OH. The protein structure shown is C165S AhpC and was generated using Swiss PDB Viewer 4.0.1 based on the PDB entry 3EMP. B. Positive ion LC-MS/MS spectra of C46 containing peptides in C165S AhpC labeled by 3-methyl-5-isoxazolone after NH2OH cleavage (top), and dimedone (bottom). The series of b and y ions confirm the sequence of the AhpC peptide and C46 modification.
Improvements in quantification using isotope-coded dimedone derivatives
Isotope-coded dimedone and 2-iododimedone (ICDID) were used with the goal of labeling CySOH and free thiol groups at the same cysteine site in different molecules of a target protein; the extent of CySOH formation was quantified relative to the thiol content in the GAPDH and yeast Gpx3 proteins used to test this approach (Seo & Carroll, 2011). In another approach, isotope-labeled heavy d6-DAz-2 and light DAz-2 were used to label CySOH proteins at different concentrations of H2O2 (Truong, et al., 2011). The labeled proteins were then combined and conjugated with the acid-cleavable linker attached to biotin, alkyneYn-ACL, for LC-MS based quantitative analysis.
Improvements in selectivity toward specific CySOH proteins or groups of proteins
Strategies to target cyclic 1,3-diketones to protein classes of interest have recently been described (Leonard et al., 2011). The 1,3-diketones are derivatized with a module that directs binding to the protein class of interest (e.g., protein tyrosine phosphatases), and with a reporter tag to enable detection of labeled proteins.
Potential for new chemical approaches
As discussed in the Introduction, the -SOH in proteins can have both electrophilic and nucleophilic properties (Fig. 1b). The labeling strategies described here exploit primarily the electrophilic properties of protein sulfenic acids and rely upon the addition of activated carbon nucleophiles to yield thioether adducts. Early work shows, however, that electrophiles such as 3-cyclohexene-1-carboxylate, tetrahydrophthalimide and dihydropyran could potentially react with protein sulfenic acids (e.g., in GAPDH) to give sulfoxide adducts (Benitez & Allison, 1974). The thermally concerted reaction of sulfenic acids with alkenes to generate sulfoxides could develop into new chemical approaches to target CySOH in a mechanistically distinct manner that does not involve anionic species and relies on the nucleophilic properties of the sulfenic acid. It must be considered, however, that the selectivity toward CySOH may be difficult to achieve with electrophilic reagents, given the nucleophilic properties of thiols and other functional groups.
3. Approaches to detect chemically-trapped sulfenic acids in proteins
As described above, selective probes of CySOH have been designed to introduce a detectable group (e.g., a fluorophore, biotin, or alkyne or azide “handle”) into the reactive sites of interest for further analyses. Downstream analytical techniques vary depending on the specific reagent used and the research question to be addressed. Approaches used can be tailored for analyzing purified proteins, mixtures of proteins, or even cellular samples, and both targeted and global analyses of oxidation-sensitive CySOH can be conducted, as described briefly below and summarized in Fig. 9. For further methodological details and advice, the reader may wish to consult additional literature on the subject (Poole & Nelson, 2008; Poole, 2008; Klomsiri, et al., 2010; Nelson et al., 2010; Leonard & Carroll, 2011).
FIGURE 9.
Workflow examples for identification and detection of CySOH proteins.
Labeling Approaches
Initial labeling of the CySOH present in a given sample can be conducted through incubation of the CySOH-targeted probes with individual or mixtures of proteins; moreover, probes can be added either subsequent to addition of an oxidant or exposure to conditions of interest, or simultaneously with the treatment of interest (Conway, Poole & Hutson, 2004; Poole, et al., 2007). Intact cells in culture or even tissues or organs can also be exposed to CySOH-directed reagents in cases where these reagents are cell permeable, as is true of the parental reagent dimedone, as well as DCP-Bio1, DCP-Rho1, the DAz and DYn reagents, BP1 and Alk-β-KE (Charles, et al., 2007; Kaplan et al., 2011; Klomsiri, et al., 2010; Leonard & Carroll, 2011; Nelson, et al., 2010; Qian, et al., 2011; Qian, et al., 2012). Given a choice between DAz (azide-labeled) and DYn (alkyne-labeled) reagents, there has been evidence with other types of probes that the alkyne derivatives may be better choices (Speers & Cravatt, 2004). Moreover, inorganic azide can react with sulfenic acids (Allison, 1976), adding a degree of uncertainty as to whether or not organic azides are in fact inert toward CySOH.
As an alternative to incubations with intact cells and tissues, reagents can be added to lysis buffers for labeling at the time of lysis, although care must be taken to minimize adventitious oxidation (see below). Labeled samples are then processed in various ways, depending on the type of reagent used and the intended analytical procedures. For most, if not all, subsequent analyses of labeled proteins, free probe must be removed using precipitation with organic solvents, gel filtration chromatography or other means; reactivity of these probes with non-CySOH sites under physiological conditions is minimal, but this is not necessarily the case at pH extremes, high temperature or other sample workup procedures. Moreover, when biotin-linked reagents are used, free probe will interfere with binding of the biotinylated proteins to beads during affinity capture.
If intact cells or tissues are to be labeled, then the toxicity of the reagent and its propensity to perturb the normal biological processes of the cell or tissue must also be considered and minimized. For example, the parental reagent dimedone can exhibit toxicity when applied to cells at low mM concentrations (Qian, et al., 2012), suggesting that caution should be exercised with all such CySOH-trapping reagents if incubated with cells or tissues for extended periods of time.
Affinity capture and analysis of biotinylated CySOH proteins
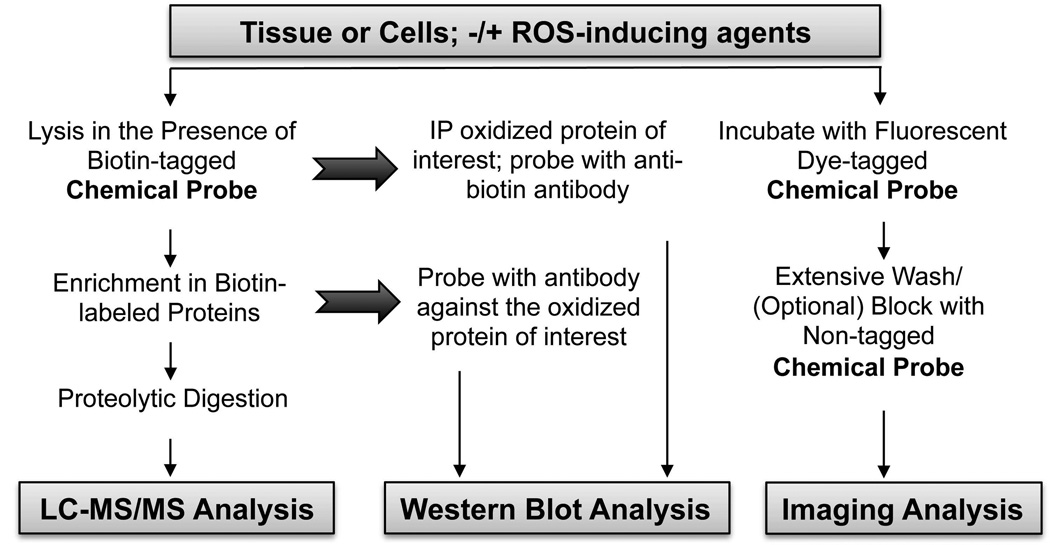
Most popular with investigators is the use of biotinylated probes, applied in biotinylated form to the samples (e.g., DCP-Bio1, biotin-dimedone, or BP1) or generated following attachment of this affinity handle to alkyne- or azide-linked CySOH reagents (Charles, et al., 2007; Leonard, Reddie & Carroll, 2009; Poole, et al., 2007; Qian, et al., 2011; Qian, et al., 2012; Reddie, et al., 2008; Seo & Carroll, 2011). This allows for use of the highly efficient affinity capture procedures using avidin or streptavidin beads to enrich for labeled proteins before subsequent analytical steps, or alternatively for the use of blotting procedures to detect the biotinylation associated with the proteins in the sample (Fig. 9). For samples undergoing affinity capture procedures, “sticky” proteins can be removed through a pre-clearing step with non-avidin beads; then the samples are incubated with streptavidin beads, ideally washed extensively and with stringent conditions to remove loosely associated proteins (including a DTT wash in order to dissociate proteins bound only through disulfide bonds to other CySOH containing proteins), and eluted in a manner compatible with the next steps in the analysis. Typically, such analytical procedures include one dimensional or two dimensional polyacrylamide gel electrophoresis, Western blotting or proteolytic digestions, and/or mass spectrometry. If one has a list of candidate proteins to be tested, the enriched material can be subjected to Western blots to identify the oxidized proteins in the sample; this approach is useful for obtaining time course data after cell treatments to get a sense of the amounts and time dependence of the oxidation of specific proteins of interest (Michalek et al., 2007; Nelson, et al., 2010; Wani, et al., 2011b; Kaplan, et al., 2011; Crump et al., 2012). To increase the confidence that specific proteins identified by this procedure are indeed oxidized, a complementary immunoprecipitation approach is also included whereby the specific protein of interest is first immunoprecipitated, then resolved on an SDS gel and blotted for the presence of biotin using an anti-biotin antibody or an HRP-streptavidin conjugate for detection of the label (Nelson, et al., 2010; Wani, et al., 2011b; Crump, et al., 2012).
Global detection of CySOH formation across proteomes
Often there is a mixture of proteins within the samples to be analyzed and more global approaches are of interest. In these cases one or two-dimensional gel electrophoresis can be followed either by detection of fluorophores using gel imaging procedures, or detection of the biotin moiety through the blotting procedures described above (Poole, et al., 2007; Wani, et al., 2011b). Total biotin content within labeled samples can also be subjected to quantitative analysis with kits designed for this purpose (Klomsiri, et al., 2010; Nelson, et al., 2010; Wani, et al., 2011b).
Mass spectrometry to identify labeled CySOH proteins and reactive sites
Of great interest to the field, as introduced above, is the use of mass spectrometry to identify proteins or peptides containing CySOH, although the current state of the technology is not yet optimized for high-throughput identification of the peptides containing the labeled sites within complex mixtures. Ideally, the labeled peptides can be enriched by affinity purification after protein or gel samples are digested, then eluted and identified through MS/MS analyses; thus far such analyses have had only marginal success, however, due to the difficulty in generating unambiguous results from complex samples (Nelson, et al., 2010; Qian, et al., 2012). Most of the studies have therefore relied on enrichment only of the labeled, undigested proteins, then MS (or LC-MS/MS) analysis of any peptides generated by digestion of these proteins, allowing for identification of the oxidized proteins, but not the sites of labeling within these proteins (Charles, et al., 2007; Leonard, Reddie & Carroll, 2009; Reddie, et al., 2008). One advantage of this less direct identification of oxidized proteins is the potentially higher confidence of assignments based on observation of multiple peptides from a putative oxidized protein. However, without clear identification of the labeled peptide, all assignments of CySOH proteins must be considered tentative.
Imaging of samples to detect CySOH formation in cells and tissues
In addition to molecular analyses, there has been some use of imaging analyses to identify where protein oxidation occurs within treated or untreated cells, and to get a sense of the levels of protein oxidation provoked within cells or tissues upon treatment with reagents of interest (Charles, et al., 2007; Kaplan, et al., 2011; Paulsen et al., 2011). Where fluorescent probes are used, the fluorophores can either be part of the probe when the labeling is performed (e.g., DCP-FL1 or DCP-Rho1), or they can be introduced into the azide or alkyne derivatives (Fig. 7). Biotin is also detectable using fluorescently-tagged avidin conjugates or immunohistochemistry. An application was also developed to observe CySOH formation within blood vessels of ischemic and nonischemic muscle tissues after injection of the muscle with DCP-Bio1 (Kaplan, et al., 2011). Recent studies using DCP-Rho1 with cultured cells show great promise for enabling analyses of protein oxidation associated with growth factor-stimulated signal transduction pathways within cells (L.B. Poole, L.W. Daniel and colleagues, manuscript in preparation).
IV. Case Studies in Sulfenic Acid Detection and Analysis
To accommodate both Western blot and MS studies, the first efforts were directed at attaching fluorescent dyes or a biotin “handle” to 1,3-cyclohexanedione to generate reagents amenable to enrichment and/or detection by Western blot and MS methods (Poole, et al., 2005; Charles, et al., 2007; Poole, et al., 2007; Klomsiri, et al., 2010). The first proteomics analysis using a biotin-dimedone strategy was reported by the Eaton group (Charles, et al., 2007). In this study rat hearts were perfused first with the biotin-dimedone derivative, followed by perfusion with H2O2 to induce CySOH formation. The biotin-labeled proteins in the tissue homogenate were further enriched by avidin chromatography, separated by SDS-PAGE, in-gel digested and identified by LC-MS analysis. Similar to the group’s earlier findings (Saurin, et al., 2004), twenty-four proteins were identified as oxidized to CySOH in response to H2O2 treatment. As mentioned above, in this as well as other studies described here, the actual identification of cysteine modification sites has not been accomplished. This poses a significant issue as the identification of proteins without the identification of cysteine modification sites can introduce false positives as result of multiple factors. These include complex protein-protein interactions (e.g., a protein that interacts with the labeled protein may be pulled down and identified, yet this will not be a labeled redox-active protein). While stringent washing procedures during the sample preparation described earlier (Wani, et al., 2011b; Klomsiri, et al., 2010) can alleviate some of these issues, it is imperative that we continue to improve chemical reagents, workflows, and detection methods to achieve sensitivity and selectivity required for identification of CySOH sites in proteins.
Similarly, other proteomics studies using azide-based dimedone probes to target sulfenic acid-modified proteins in cells have been reported. In these studies, cells were incubated with the dimedone probe, and the biotin tag was introduced at a later step using the Staudinger ligation (Leonard, Reddie & Carroll, 2009; Reddie, et al., 2008). More than 175 new proteins were identified as putative CySOH proteins. In these studies, the CySOH modification sites in these proteins were assigned based on previously published literature where the sites were identified using recombinant proteins and/or systematic mutagenesis of cysteine sites.
In our laboratories, we have used DCP-Bio1 and DCP-Rho1 to identify and/or detect localized protein oxidation in a number of systems in order to better understand how to use these reagents to effectively obtain important information about biological oxidations, as summarized next.
A. Strategies for labeling of samples with CySOH-directed chemical probes in intact cells and tissues
Many of the reagents described here are cell membrane permeable, allowing for their use with intact cells and tissues. Alternative procedures to label biological samples involve the trapping of modified proteins at distinct times after perturbation by including the reagent in the lysis buffer. Neither labeling approach avoids concerns that artifactual oxidation and/or labeling can occur. While artifactual protein oxidation can be caused by the lysis procedure itself, it is also well established that labeling of intact cells diverts the normal biology as proteins become chemically modified at their oxidized cysteinyl sites, in some cases triggering much higher oxidation levels due to the treatment itself or by perturbing the relay of signaling or metabolic events (Crump, et al., 2012; Kaplan, et al., 2011; Michalek, et al., 2007; Qian, et al., 2012). In order to minimize artifactual protein oxidation caused by lysis to obtain a “snapshot” of protein oxidation status at the time of lysis, procedures have been developed and sentinel proteins established to evaluate and ameliorate the artifactual oxidation provoked by lysis, as described further in the next section (Klomsiri, et al., 2010).
In designing a labeling protocol, the relative timing of the ROS-inducing stimulation of cells/tissue versus chemical probe addition for trapping the ensuing CySOH species is an important variable. The reagent, if cell permeable, can be preincubated with the cells or tissues prior to induction of intracellular ROS by receptor agonists, allowing for trapping of CySOH species as they are formed. One argument for preincubation with the probe is that this allows more accumulation of the trapped CySOH species as they cycle potentially multiple times between reduced and oxidized species during incubation. As alluded to above, the counterargument is that preincubation with the probe leads to covalent labeling of CySOH proteins which in turn will alter the kinetics and perhaps function of these proteins, leading in some cases to increased ROS and stimulus-independent (non-physiological) CySOH formation. This is in particular an issue when chemical probes are used in concentrations higher than 1 mM and for extended periods of time. Our protocols for labeling have largely favored the addition of probe either at the time of lysis, or for only 5 to 10 min prior to lysis, to minimize diversion of the normal biological processes. We have demonstrated, through labeling of cell lysates, that even transiently generated CySOH species can be captured by reagents like DCP-Bio1 at 1 or 5 mM in spite of their propensity to form intraprotein disulfide bonds (e.g., human PrxI through PrxIV as well as the phosphatases PTEN and SHP-2 were labeled in TNFα-treated HEK-293 cells) (Nelson, et al., 2010).
B. Evaluation of dimedone-based probe reactivities with protein CySOH and development of approaches to minimize and evaluate artifactual oxidation
With a set of known CySOH-generating proteins and an expanding arsenal of reagents available to us, we have conducted experiments to better understand the reactivities of the proteins and probes and to tailor our procedures to minimize adventitious oxidation that may occur during sample workup. Using CySOH forms of (i) the bacterial peroxiredoxin AhpC (the C165S mutant, where Cys46-SOH is stabilized), (ii) the C84,94S mutant of R-specific free methionine sulfoxide redutase (fRMsr) from E. coli, (iii) a truncated, mutated form of E coli OxyR known as “C4A-RD”, and (iv) papain, kinetics of labeling and the effects of pH were analyzed with reagents of interest (Klomsiri, et al., 2010; Poole, et al., 2007). Briefly, oxidized proteins incubated with 1 mM DCP-Bio1 were labeled at rates ranging from 0.003 (for AhpC) to 1.65 (for papain) min−1 at pH 7 and 25 °C (given as pseudo-first order rates at this concentration of DCP-Bio1). As tested for fRMsr, rates with DCP-Bio1 were not different between pH 5.5 and 8.0, reflecting the relative pH insensitivity exhibited by dimedone (Klomsiri, et al., 2010). Other, more recently developed reagents do exhibit differences in reactivity with changes in pH, however. For example, labeling of AhpC with the cyclopentadione reagent BP1 is increased at lower pH values (Qian, et al., 2011), whereas the linear 1,3-dicarbonyl reagent Alk-β-KE increased in reactivity with CySOH at higher pH values (Qian, et al., 2012). While the varying pH dependence of reactivity is not well understood, it is not surprising that different (folded) proteins react with these reagents at different rates, as their protein microenvironments and variable accessibility will have a strong effect on rates of reaction with these alkylating agents.
Non-native oxidation processes occurring during labeling and workup of samples are always a concern when one seeks information on the redox status of proteins within their natural biological settings. We therefore developed a protocol to minimize such adventitious oxidation during lysis. Briefly, both NEM and iodoacetamide at 10 mM were included in the lysis buffer to trap free thiols, as were catalase to rapidly scavenge H2O2 and DTPA to chelate metals (Klomsiri, et al., 2010). Given its high reactivity with H2O2 to form CySOH, the His-tagged, C208S mutant of C4A-RD OxyR was employed as a reporter of postlysis Cys oxidation, allowing for sensitive detection of adventitious oxidation occurring in the sample. Pre-biotinylated bacterial proteins (AhpC or fRMsr) were also added to samples during workup to control for loss of biotinylated proteins during affinity capture and analytical procedures (Klomsiri, et al., 2010; Nelson, et al., 2010). Finally, it was demonstrated that the extent of incorporation of DCP-Bio1 into cellular proteins is strongly dependent on the protein concentration (Klomsiri, et al., 2010). This effect can be an important factor when different samples, which may contain different concentrations of protein, are labeled in order to compare extent of labeling in subsequent analyses.
C. PDGF-Induced CySOH Formation in the Akt2 Isoform of NIH 3T3 Cells
Inhibitory Akt2 oxidation at Cys124 was shown to occur in cellular proteins using DCP-Bio1 upon platelet-derived growth factor (PDGF)-stimulated proliferative and migratory signaling in NIH 3T3 cells (Wani, et al., 2011b; Wani et al., 2011a). After stimulation with PDGF, cells were lysed in the presence of DCP-Bio1. Biotin incorporation into cellular proteins increased within 5 min of PDGF stimulation and a number of DCP-Bio1 labeled proteins were identified by MS. These included PLCγ1 and Akt2, both well-known mediators of PDGF signaling. Redox sensitivity of PLCγ1 was also reported earlier in PC12 cells stimulated with dopamine (Kim et al., 2002). Western blot analysis of streptavidin-agarose enriched samples from PDGF-stimulated cells lysed in the presence of DCP-Bio1 showed increased PLCγ1 oxidation with PDGF stimulation compared with unstimulated control. With regard to Akt2, the peptide identified by MS contained a cysteine site (Cys124) labeled by DCP-Bio1. Interestingly, Cys124 is situated in the linker region connecting the pleckstrin homology (PH) domain to the catalytic kinase domain of Akt2 and is not conserved in the other Akt isoforms. Follow-up studies (Fig. 10) of the selective oxidation of Akt2 and its functional consequence in a variety of biological systems (fibroblasts, myoblasts/ myotubes, adipocytes) showed that: (i) oxidation initiated at Cys124 was inhibitory to Akt2 kinase activity; (ii) ~50% of phosphorylated Akt2 was in oxidized and inactive state in cells stimulated with PDGF; this process was reversible - kinase activity could be restored by removal of PDGF or treatment of oxidized protein with reducing agents; (iii) the mechanism of inhibition of Akt2 kinase activity with oxidation included formation of multiple disulfides involving cysteine residues 60 and 77 in the PH domain, 124 in the linker region, and 297 and 311 in the kinase domain; (iv) mutation of Cys124 decreased significantly the redox sensitivity of Akt2; and (v) redox regulation of Akt2 was linked to physiological outputs such as glucose uptake, cell migration, and cell cycle (Wani, et al., 2011b; Wani et al., 2011a).
FIGURE 10.
Selective inhibition of Akt2 activity by oxidation during PDGF signaling. A. Summary diagram of Akt2 activity assay. GSK3 phosphorylation was used as a measure of Akt2 kinase activity. B. WT Akt2 and Cys124Ser Akt2 proteins immunoprecipitated from lysates of PDGF-stimulated (20 ng/mL, 10 min) NIH 3T3 cells were oxidized in vitro with H2O2 before kinase activity assays. Quantification of the kinase activity shows a higher degree of inhibition of WT Akt2 activity by oxidation (p < 0.001) relative to Cys124Ser Akt2 (p = 0.029). C. Isoform-specific inactivation of Akt2 kinase with oxidation. Kinase activities of recombinant purified active Akt1, Akt2, and Akt3 were assayed before and after oxidation with H2O2D. Akt2 oxidation is PDGF-dependent and reversible, decreasing with prolonged PDGF stimulation. Lysates from PDGF-stimulated NIH 3T3 cells were used to capture DCP-Bio1–labeled proteins and probed for Akt2. Two additional controls were used in which cells were treated with PDGF alone for the first 30 min and then incubated further for 30 and 90 min without PDGF. Additionally, biotinylated AhpC was added to the normalized lysates as an internal control before affinity capture and probed for AhpC. Reprinted from Wani R, Qian J, Yin L, Bechtold E, King SB, Poole LB, Paek E, Tsang AW, Furdui CM. 2011. Proc Natl Acad Sci U S A 108:10550–10555.
D. Localized CySOH formation in proteins critical to VEGF-stimulated endothelial migration and angiogenesis
Over the past several years, pioneering work by Ushio-Fukai and colleagues has highlighted the importance of localized CySOH formation in proteins which modulate processes important to endothelial cell proliferation and migration (Oshikawa et al., 2010; Kaplan et al., 2011). Imaging of cells and Western blotting of oxidized proteins after DCP-Bio1 labeling were used to demonstrate CySOH formation in IQGAP, a VEGF receptor (VEGFR2) binding scaffold protein involved in ROS-dependent EC migration and post-ischemic angiogenesis. IQGAP oxidation was associated with the lamellipodial leading edge where it colocalizes with NADPH oxidase (Fig. 11). CySOH formation was demonstrated in both cultured endothelial cells, where VEGF responses could be monitored, as well as small blood vessels of the ischemic hindlimb in mice where levels of oxidation were much greater than in the non-ischemic tissue. Oxidation responses to VEGF signaling within lipid rafts were also observed in endothelial cells, where engineered expression of lipid raft-associated extracellular superoxide dismutase (ecSOD) stimulated both protein tyrosine phosphatase oxidation (for both PTP1B, as shown in Fig. 11, and DEP1) and angiogenesis in the hindlimb ischemia model. As both DEP1 and PTP1B negatively regulate tyrosine phosphorylation in VEGFR2, this localized oxidation in lipid raft microdomains is of critical importance to the proliferative and migratory responses of endothelial cells in angiogenesis and wound repair.
FIGURE 11.
Localized CySOH in VEGF signaling. A. Role of CySOH in endothelial cell migration and angiogenesis. Upon activation of VEGFR2 by VEGF, IQGAP1 translocates to the leading edge where it forms VEGFR2/IQGAP1/NADPH oxidase signaling complex in migrating EC. ROS derived from this complex leads to localized CySOH formation of IQGAP1/VEGFR2 at the leading edge, which may contribute to ROS-dependent directional EC migration. Thus, protein CySOH formation may play an important role in VEGF-induced redox signaling involved in endothelial repair in response to injury and postnatal angiogenesis. Supporting data for the model are included in panels B, C, and D. B. and C. Protein CySOH formation co-localizes with IQGAP1 at the leading edge during VEGF-stimulated EC migration. HUVECs stimulated with VEGF were double-labeled with anti-IQGAP1 antibody (red) and CySOH probe DCP-Bio1 (green). White arrows point to the leading edge where CySOH and IQGAP1 localize. In B (lower panel), cells were incubated with 10 mM dimedone together with the CySOH probe to show the specificity of DCP-Bio1 labelling. In C, IQGAP1-depleted cell (white dotted line) and IQGAP1-expressing cell in IQGAP1 siRNA-transfected HUVECs are shown. Small white arrows point out that depletion of IQGAP1 using siRNA reduces CySOH formation at lamellipodial plasma membranes. Bar scale indicates 10 µm. D. VEGF stimulation increases CySOH formation of IQGAP1 in HUVEC in a redox-dependent manner. HUVECs were pre-treated with or without thiol antioxidant N-acetyl cysteine (NAC, 20 mM for 1 hr) and then stimulated with VEGF (50 ng/ml) for indicated times. Lysates containing biotin-labeled CySOH trapping reagent were immunoprecipitated with anti-IQGAP1 antibody, followed by immunoblotting with anti-biotin antibody. Dimedone was added to the lysate to compete with the DCP-Bio1 binding to the CySOH-containing proteins. E. Model for role of ecSOD-derived H2O2 in VEGFR2 signaling linked to angiogenesis. Extracellular H2O2 generated by ecSOD localized at caveolae/lipid rafts via its heparin-binding domain induces oxidative inactivation of DEP1 and PTP1B in these microdomains, thereby promoting VEGF-induced VEGFR2 phosphorylation, which may contribute to EC migration and proliferation in vitro as well as angiogenesis in vivoF. ecSOD induces inactivation and oxidation of PTP1B localized in caveolae/lipid rafts. (Upper) PTP1B activity in caveolae/lipid rafts and non-caveolae/lipid rafts in Ad.LacZ and Ad.ecSOD-infected HUVECs were measured using pNPP as a substrate after IP with anti-PTP1B antibody. The values were expressed as a ratio to Ad.LacZ infected PTP activity (defined as 1.0) in each fraction (n=3) *p<0.05. (Lower) Ad.LacZ or Ad.ecSOD-infected HUVECs were extracted in the presence of DCP-Bio1. After sucrose gradient centrifugation, DCP-Bio1 labeled proteins from caveolae/lipid rafts and non-caveolae/lipid rafts were affinity captured with streptavidin beads, followed by immunoblotting with anti-PTP1B antibody. Similar results were obtained for DEP-1. Panels A through D are reprinted with permission from Kaplan N, Urao N, Furuta E, Kim SJ, Razvi M, Nakamura Y, McKinney RD, Poole LB, Fukai T, Ushio-Fukai M. 2011. Free Radic Res 45:1124–1135. Copyright 2011 Informa UK Ltd. Panels E and F are reprinted from Oshikawa J, Urao N, Kim HW, Kaplan N, Razvi M, McKinney R, Poole LB, Fukai T, Ushio-Fukai M. 2010. PLoS One 5:e10189.
V. Future Directions
Regulation of many enzymes, transcriptional regulators and, in a broader view, cell signaling processes is now recognized to be highly dependent on oxidative events that are only now beginning to come to light. The new reagents and procedures combined with computational approaches that are increasingly available bode well for the future where oxidation effects will be able to be integrated with expanding knowledge of other posttranslational modifications that act to shape the metabolic, signaling and epigenetics states of biological systems. However, while we and others have successfully applied these new reagents and MS methods to identify and quantify CySOH modifications in recombinant or overexpressed proteins, it is important to note that global mapping of the sites of CySOH modification directly in cells under the conditions of endogenous protein expression has not yet been accomplished. With continued efforts to improve the reactivity of CySOH-directed probes, as well as the ionization and fragmentation properties of the resulting protein and peptide adducts, the potential for an enhanced mechanistic understanding of redox regulation and signaling is great and may well lead to the development and implementation of new therapeutic approaches to combat disease.
Acknowledgements
The authors thank J.A. Reisz for assistance with figures. This work was supported by grants from the National Institutes of Health to L.B.P. (R33 CA126659 and R01 GM050389) and C.M.F. (R01 CA136810),
Reference list
- Aboderin AA, Boedefeld E. Reaction of chicken egg white lysozyme with 7-chloro-4-nitrobenz-2-oxa-1,3-diazole. II. Sites of modification. Biochim Biophys Acta. 1976;420:177–186. doi: 10.1016/0005-2795(76)90356-1. [DOI] [PubMed] [Google Scholar]
- Allison WS. Formation and reactions of sulfenic acids in proteins. Acc. Chem. Res. 1976;9:293–299. [Google Scholar]
- Amici A, Levine RL, Tsai L, Stadtman ER. Conversion of amino acid residues in proteins and amino acid homopolymers to carbonyl derivatives by metal-catalyzed oxidation reactions. J Biol Chem. 1989;264:3341–3346. [PubMed] [Google Scholar]
- Arakawa T, Kawano Y, Katayama Y, Nakayama H, Dohmae N, Yohda M, Odaka M. Structural basis for catalytic activation of thiocyanate hydrolase involving metal-ligated cysteine modification. J Am Chem Soc. 2009;131:14838–14843. doi: 10.1021/ja903979s. [DOI] [PubMed] [Google Scholar]
- Bashiri G, Squire CJ, Moreland NJ, Baker EN. Crystal structures of F420-dependent glucose-6-phosphate dehydrogenase FGD1 involved in the activation of the anti-tuberculosis drug candidate PA-824 reveal the basis of coenzyme and substrate binding. J Biol Chem. 2008;283:17531–17541. doi: 10.1074/jbc.M801854200. [DOI] [PubMed] [Google Scholar]
- Becker K, Savvides SN, Keese M, Schirmer RH, Karplus PA. Enzyme inactivation through sulfhydryl oxidation by physiologic NO-carriers. Nat Struct Biol. 1998;5:267–271. doi: 10.1038/nsb0498-267. [DOI] [PubMed] [Google Scholar]
- Benitez LV, Allison WS. The inactivation of the acyl phosphatase activity catalyzed by the sulfenic acid form of glyceraldehyde 3-phosphate dehydrogenase by dimedone and olefins. J Biol Chem. 1974;249:6234–6243. [PubMed] [Google Scholar]
- Billiet L, Geerlings P, Messens J, Roos G. The thermodynamics of thiol sulfenylation. Free Radic Biol Med. 2012;52:1473–1485. doi: 10.1016/j.freeradbiomed.2011.12.029. [DOI] [PubMed] [Google Scholar]
- Biteau B, Labarre J, Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425:980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- Blair-Johnson M, Fiedler T, Fenna R. Human myeloperoxidase: structure of a cyanide complex and its interaction with bromide and thiocyanate substrates at 1.9 Å resolution. Biochemistry. 2001;40:13990–13997. doi: 10.1021/bi0111808. [DOI] [PubMed] [Google Scholar]
- Boschi-Muller S, Azza S, Sanglier-Cianferani, Talfournier F, Van Dorsselear A, Branlant G. A sulfenic acid enzyme intermediate is involved in the catalytic mechanism of peptide methionine sulfoxide reductase from Escherichia coli . J. Biol. Chem. 2000;275:35908–35913. doi: 10.1074/jbc.M006137200. [DOI] [PubMed] [Google Scholar]
- Bryan N, Ahswin H, Smart N, Bayon Y, Wohlert S, Hunt JA. Reactive oxygen species (ROS)--a family of fate deciding molecules pivotal in constructive inflammation and wound healing. Eur Cell Mater. 2012;24:249–265. doi: 10.22203/ecm.v024a18. [DOI] [PubMed] [Google Scholar]
- Buhrman G, Parker B, Sohn J, Rudolph J, Mattos C. Structural mechanism of oxidative regulation of the phosphatase Cdc25B via an intramolecular disulfide bond. Biochemistry. 2005;44:5307–5316. doi: 10.1021/bi047449f. [DOI] [PubMed] [Google Scholar]
- Cao Z, Tavender TJ, Roszak AW, Cogdell RJ, Bulleid NJ. Crystal structure of reduced and of oxidized peroxiredoxin IV enzyme reveals a stable oxidized decamer and a non-disulfide-bonded intermediate in the catalytic cycle. J Biol Chem. 2011;286:42257–42266. doi: 10.1074/jbc.M111.298810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballal S, Radi R, Kirk MC, Barnes S, Freeman BA, Alvarez B. Sulfenic acid formation in human serum albumin by hydrogen peroxide and peroxynitrite. Biochemistry. 2003;42:9906–9914. doi: 10.1021/bi027434m. [DOI] [PubMed] [Google Scholar]
- Changela A, Ho CK, Martins A, Shuman S, Mondragon A. Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme. Embo J. 2001;20:2575–2586. doi: 10.1093/emboj/20.10.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles RL, Schröder E, May G, Free P, Gaffney PR, Wait R, Begum S, Heads RJ, Eaton P. Protein sulfenation as a redox sensor: proteomics studies using a novel biotinylated dimedone analogue. Mol Cell Proteomics. 2007;6:1473–1484. doi: 10.1074/mcp.M700065-MCP200. [DOI] [PubMed] [Google Scholar]
- Chiang BY, Chen TC, Pai CH, Chou CC, Chen HH, Ko TP, Hsu WH, Chang CY, Wu WF, Wang AH, Lin CH. Protein S-thiolation by Glutathionylspermidine (Gsp): the role of Escherichia coli Gsp synthetASE/amidase in redox regulation. J Biol Chem. 2010;285:25345–25353. doi: 10.1074/jbc.M110.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HJ, Kang SW, Yang CH, Rhee SG, Ryu SE. Crystal structure of a novel human peroxidase enzyme at 2.0 Å resolution. Nat Struct Biol. 1998;5:400–406. doi: 10.1038/nsb0598-400. [DOI] [PubMed] [Google Scholar]
- Chouchani ET, James AM, Fearnley IM, Lilley KS, Murphy MP. Proteomic approaches to the characterization of protein thiol modification. Curr Opin Chem Biol. 2011;15:120–128. doi: 10.1016/j.cbpa.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claiborne A, Yeh JI, Mallett TC, Luba J, Crane EJ, 3rd, Charrier V, Parsonage D. Protein-sulfenic acids: diverse roles for an unlikely player in enzyme catalysis and redox regulation. Biochemistry. 1999;38:15407–15416. doi: 10.1021/bi992025k. [DOI] [PubMed] [Google Scholar]
- Codreanu SG, Zhang B, Sobecki SM, Billheimer DD, Liebler DC. Global analysis of protein damage by the lipid electrophile 4-hydroxy-2-nonenal. Mol Cell Proteomics. 2009;8:670–680. doi: 10.1074/mcp.M800070-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway ME, Poole LB, Hutson SM. Roles for cysteine residues in the regulatory CXXC motif of human mitochondrial branched chain aminotransferase enzyme. Biochemistry. 2004;43:7356–7364. doi: 10.1021/bi0498050. [DOI] [PubMed] [Google Scholar]
- Cowan-Jacob SW, Kaufmann M, Anselmo AN, Stark W, Grutter MG. Structure of rabbit-muscle glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2003;59:2218–2227. doi: 10.1107/s0907444903020493. [DOI] [PubMed] [Google Scholar]
- Crane EJ, 3rd, Vervoort J, Claiborne A. 13C NMR analysis of the cysteine-sulfenic acid redox center of enterococcal NADH peroxidase. Biochemistry. 1997;36:8611–8618. doi: 10.1021/bi9707990. [DOI] [PubMed] [Google Scholar]
- Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- Crump KE, Juneau DG, Poole LB, Haas KM, Grayson JM. The reversible formation of cysteine sulfenic acid promotes B-cell activation and proliferation. Eur J Immunol. 2012;42:2152–2164. doi: 10.1002/eji.201142289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle-Donne I, Carini M, Orioli M, Vistoli G, Regazzoni L, Colombo G, Rossi R, Milzani A, Aldini G. Protein carbonylation: 2,4-dinitrophenylhydrazine reacts with both aldehydes/ketones and sulfenic acids. Free Radic Biol Med. 2009;46:1411–1419. doi: 10.1016/j.freeradbiomed.2009.02.024. [DOI] [PubMed] [Google Scholar]
- de Macedo-Ribeiro S, Renirie R, Wever R, Messerschmidt A. Crystal structure of a trapped phosphate intermediate in vanadium apochloroperoxidase catalyzing a dephosphorylation reaction. Biochemistry. 2008;47:929–934. doi: 10.1021/bi7018628. [DOI] [PubMed] [Google Scholar]
- Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- Dennehy MK, Richards KA, Wernke GR, Shyr Y, Liebler DC. Cytosolic and nuclear protein targets of thiol-reactive electrophiles. Chem Res Toxicol. 2006;19:20–29. doi: 10.1021/tx050312l. [DOI] [PubMed] [Google Scholar]
- Denu JM, Tanner KG. Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: Evidence for a sulfenic acid intermediate and implications for redox regulation. Biochemistry. 1998;37:5633–5642. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- Dierks T, Dickmanns A, Preusser-Kunze A, Schmidt B, Mariappan M, von Figura K, Ficner R, Rudolph MG. Molecular basis for multiple sulfatase deficiency and mechanism for formylglycine generation of the human formylglycine-generating enzyme. Cell. 2005;121:541–552. doi: 10.1016/j.cell.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Dilovic I, Gliubich F, Malpeli G, Zanotti G, Matkovic-Calogovic D. Crystal structure of bovine 3-hydroxyanthranilate 3,4-dioxygenase. Biopolymers. 2009;91:1189–1195. doi: 10.1002/bip.21167. [DOI] [PubMed] [Google Scholar]
- Durao P, Chen Z, Silva CS, Soares CM, Pereira MM, Todorovic S, Hildebrandt P, Bento I, Lindley PF, Martins LO. Proximal mutations at the type 1 copper site of CotA laccase: spectroscopic, redox, kinetic and structural characterization of I494A and L386A mutants. Biochem J. 2008;412:339–346. doi: 10.1042/BJ20080166. [DOI] [PubMed] [Google Scholar]
- Elam JS, Taylor AB, Strange R, Antonyuk S, Doucette PA, Rodriguez JA, Hasnain SS, Hayward LJ, Valentine JS, Yeates TO, Hart PJ. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat Struct Biol. 2003;10:461–467. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- Ellis HR, Poole LB. Roles for the two cysteine residues of AhpC in catalysis of peroxide reduction by alkyl hydroperoxide reductase from Salmonella typhimurium . Biochemistry. 1997a;36:13349–13356. doi: 10.1021/bi9713658. [DOI] [PubMed] [Google Scholar]
- Ellis HR, Poole LB. Novel application of 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole to identify cysteine sulfenic acid in the AhpC component of alkyl hydroperoxide reductase. Biochemistry. 1997b;36:15013–15018. doi: 10.1021/bi972191x. [DOI] [PubMed] [Google Scholar]
- Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioravanti E, Dura MA, Lascoux D, Micossi E, Franzetti B, McSweeney S. Structure of the stress response protein DR1199 from Deinococcus radiodurans: a member of the DJ-1 superfamily. Biochemistry. 2008;47:11581–11589. doi: 10.1021/bi800882v. [DOI] [PubMed] [Google Scholar]
- Firbank SJ, Rogers MS, Wilmot CM, Dooley DM, Halcrow MA, Knowles PF, McPherson MJ, Phillips SE. Crystal structure of the precursor of galactose oxidase: an unusual self-processing enzyme. Proc Natl Acad Sci U S A. 2001;98:12932–12937. doi: 10.1073/pnas.231463798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayne J, Taylor A, Cameron G, Hadfield AT. Structure of insoluble rat sperm glyceraldehyde-3-phosphate dehydrogenase (GAPDH) via heterotetramer formation with Escherichia coli GAPDH reveals target for contraceptive design. J Biol Chem. 2009;284:22703–22712. doi: 10.1074/jbc.M109.004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuangthong M, Helmann JD. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc. Natl. Acad. Sci USA. 2002;99:6690–6695. doi: 10.1073/pnas.102483199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh PB, Whitehouse MW. 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole: a new fluorigenic reagent for amino acids and other amines. Biochem J. 1968;108:155–156. doi: 10.1042/bj1080155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths SW, King J, Cooney CL. The reactivity and oxidation pathway of cysteine 232 in recombinant human alpha 1-antitrypsin. J Biol Chem. 2002;277:25486–25492. doi: 10.1074/jbc.M203089200. [DOI] [PubMed] [Google Scholar]
- Gurmu D, Lu J, Johnson KA, Nordlund P, Holmgren A, Erlandsen H. The crystal structure of the protein YhaK from Escherichia coli reveals a new subclass of redox sensitive enterobacterial bicupins. Proteins. 2009;74:18–31. doi: 10.1002/prot.22128. [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- Hamann M, Zhang T, Hendrich S, Thomas JA. Quantitation of protein sulfinic and sulfonic acid, irreversibly oxidized protein cysteine sites in cellular proteins. Methods Enzymol. 2002;348:146–156. doi: 10.1016/s0076-6879(02)48634-x. [DOI] [PubMed] [Google Scholar]
- Hansen JM, Go YM, Jones DP. Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu Rev Pharmacol Toxicol. 2006;46:215–234. doi: 10.1146/annurev.pharmtox.46.120604.141122. [DOI] [PubMed] [Google Scholar]
- Hansen RE, Winther JR. An introduction to methods for analyzing thiols and disulfides: Reactions, reagents, and practical considerations. Anal Biochem. 2009;394:147–158. doi: 10.1016/j.ab.2009.07.051. [DOI] [PubMed] [Google Scholar]
- Hovorka SW, Biesiada H, Williams TD, Huhmer A, Schoneich C. High sensitivity of Zn2+ insulin to metal-catalyzed oxidation: detection of 2-oxo-histidine by tandem mass spectrometry. Pharm Res. 2002;19:530–537. doi: 10.1023/a:1015164200431. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001. 2001:PL1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- Jeng MF, Holmgren A, Dyson HJ. Proton sharing between cysteine thiols in Escherichia coli thioredoxin: implications for the mechanism of protein disulfide reduction. Biochemistry. 1995;34:10101–10105. doi: 10.1021/bi00032a001. [DOI] [PubMed] [Google Scholar]
- Jeong J, Jung Y, Na S, Jeong J, Lee E, Kim MS, Choi S, Shin DH, Paek E, Lee HY, Lee KJ. Novel oxidative modifications in redox-active cysteine residues. Mol Cell Proteomics. 2011;10:M110 000513. doi: 10.1074/mcp.M110.000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Zhang L, Sun F, Deng X, Liang H, Bae T, He C. Staphylococcus aureus CymR is a new thiol-based oxidation-sensing regulator of stress resistance and oxidative response. J Biol Chem. 2012;287:21102–21109. doi: 10.1074/jbc.M112.359737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson TJ, Murray MS, Johnson LC, Lowther WT. Reduction of cysteine sulfinic acid in peroxiredoxin by sulfiredoxin proceeds directly through a sulfinic phosphoryl ester intermediate. J Biol Chem. 2008a;283:23846–23851. doi: 10.1074/jbc.M803244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson TJ, Tsang AW, Lowther WT, Furdui CM. Identification of intact protein thiosulfinate intermediate in the reduction of cysteine sulfinic Acid in peroxiredoxin by human sulfiredoxin. J Biol Chem. 2008b;283:22890–22894. doi: 10.1074/jbc.C800124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordanova R, Groves MR, Kostova E, Woltersdorf C, Liebau E, Tucker PA. Fatty acid- and retinoid-binding proteins have distinct binding pockets for the two types of cargo. J Biol Chem. 2009;284:35818–35826. doi: 10.1074/jbc.M109.022731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Urao N, Furuta E, Kim SJ, Razvi M, Nakamura Y, McKinney RD, Poole LB, Fukai T, Ushio-Fukai M. Localized cysteine sulfenic acid formation by vascular endothelial growth factor: role in endothelial cell migration and angiogenesis. Free Radic Res. 2011;45:1124–1135. doi: 10.3109/10715762.2011.602073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenhofen NJ, Wood MJ. Formation, reactivity, and detection of protein sulfenic acids. Chem Res Toxicol. 2010;23:1633–1646. doi: 10.1021/tx100237w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Raines RT. A misfolded but active dimer of bovine seminal ribonuclease. Eur J Biochem. 1994;224:109–114. doi: 10.1111/j.1432-1033.1994.tb20001.x. [DOI] [PubMed] [Google Scholar]
- Kim JR, Kwon KS, Yoon HW, Lee SR, Rhee SG. Oxidation of proteinaceous cysteine residues by dopamine-derived H2O2 in PC12 cells. Archives of Biochemistry and Biophysics. 2002;397:414–423. doi: 10.1006/abbi.2001.2691. [DOI] [PubMed] [Google Scholar]
- Klomsiri C, Nelson KJ, Bechtold E, Soito L, Johnson LC, Lowther WT, Ryu SE, King SB, Furdui CM, Poole LB. Use of dimedone-based chemical probes for sulfenic acid detection: evaluation of conditions affecting probe incorporation into redox-sensitive proteins. Methods Enzymol. 2010;473:77–94. doi: 10.1016/S0076-6879(10)73003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Soonsanga S, Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci U S A. 2007;104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legler PM, Kumaran D, Swaminathan S, Studier FW, Millard CB. Structural characterization and reversal of the natural organophosphate resistance of a D-type esterase, Saccharomyces cerevisiae S-formylglutathione hydrolase. Biochemistry. 2008;47:9592–9601. doi: 10.1021/bi8010016. [DOI] [PubMed] [Google Scholar]
- Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard SE, Carroll KS. Chemical 'omics' approaches for understanding protein cysteine oxidation in biology. Curr Opin Chem Biol. 2011;15:88–102. doi: 10.1016/j.cbpa.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Leonard SE, Garcia FJ, Goodsell DS, Carroll KS. Redox-based probes for protein tyrosine phosphatases. Angew Chem Int Ed Engl. 2011;50:4423–4427. doi: 10.1002/anie.201007871. [DOI] [PubMed] [Google Scholar]
- Leonard SE, Reddie KG, Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem Biol. 2009;4:783–799. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- Li S, Peterson NA, Kim MY, Kim CY, Hung LW, Yu M, Lekin T, Segelke BW, Lott JS, Baker EN. Crystal structure of AhpE from Mycobacterium tuberculosis, a 1-Cys peroxiredoxin. J Mol Biol. 2005;346:1035–1046. doi: 10.1016/j.jmb.2004.12.046. [DOI] [PubMed] [Google Scholar]
- Lin WS, Armstrong DA, Gaucher GM. Formation and repair of papain sulfenic acid. Can J Biochem. 1975;53:298–307. doi: 10.1139/o75-042. [DOI] [PubMed] [Google Scholar]
- Lountos GT, Jiang R, Wellborn WB, Thaler TL, Bommarius AS, Orville AM. The crystal structure of NAD(P)H oxidase from Lactobacillus sanfranciscensis: insights into the conversion of O2 into two water molecules by the flavoenzyme. Biochemistry. 2006;45:9648–9659. doi: 10.1021/bi060692p. [DOI] [PubMed] [Google Scholar]
- Ma LH, Takanishi CL, Wood MJ. Molecular mechanism of oxidative stress perception by the Orp1 protein. J Biol Chem. 2007;282:31429–31436. doi: 10.1074/jbc.M705953200. [DOI] [PubMed] [Google Scholar]
- Madian AG, Regnier FE. Proteomic identification of carbonylated proteins and their oxidation sites. J Proteome Res. 2010;9:3766–3780. doi: 10.1021/pr1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller C, Schroder E, Eaton P. Glyceraldehyde 3-phosphate dehydrogenase is unlikely to mediate hydrogen peroxide signaling: studies with a novel anti-dimedone sulfenic acid antibody. Antioxid Redox Signal. 2011;14:49–60. doi: 10.1089/ars.2010.3149. [DOI] [PubMed] [Google Scholar]
- Michalek RD, Nelson KJ, Holbrook BC, Yi JS, Stridiron D, Daniel LW, Fetrow JS, King SB, Poole LB, Grayson JM. The requirement of reversible cysteine sulfenic acid formation for T cell activation and function. J Immunol. 2007;179:6456–6467. doi: 10.4049/jimmunol.179.10.6456. [DOI] [PubMed] [Google Scholar]
- Miki M. Chemical modification of tropomyosin with NBD-chloride. J Biochem. 1985;97:1067–1072. doi: 10.1093/oxfordjournals.jbchem.a135149. [DOI] [PubMed] [Google Scholar]
- Miyanaga A, Fushinobu S, Ito K, Wakagi T. Crystal structure of cobalt-containing nitrile hydratase. Biochem Biophys Res Commun. 2001;288:1169–1174. doi: 10.1006/bbrc.2001.5897. [DOI] [PubMed] [Google Scholar]
- Monteiro G, Horta BB, Pimenta DC, Augusto O, Netto LE. Reduction of 1-Cys peroxiredoxins by ascorbate changes the thiol-specific antioxidant paradigm, revealing another function of vitamin C. Proc Natl Acad Sci U S A. 2007;104:4886–4891. doi: 10.1073/pnas.0700481104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Yamamoto T, Abe M, Matsumura H, Hagihara Y, Goto T, Yamaguchi T, Inoue T. Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate. Proc Natl Acad Sci U S A. 2008;105:6238–6242. doi: 10.1073/pnas.0709822105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu T, Ichiyama S, Hiratake J, Saldanha A, Kobashi N, Sakata K, Kato H. Structural basis for the spectral difference in luciferase bioluminescence. Nature. 2006;440:372–376. doi: 10.1038/nature04542. [DOI] [PubMed] [Google Scholar]
- Nelson KJ, Klomsiri C, Codreanu SG, Soito L, Liebler DC, Rogers LC, Daniel LW, Poole LB. Use of dimedone-based chemical probes for sulfenic acid detection; methods to visualize and identify labeled proteins. Methods Enzymol. 2010;473:95–115. doi: 10.1016/S0076-6879(10)73004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka T, Kita A, Miura-Ohnuma J, Katoh E, Inagaki N, Yamazaki T, Miki K. Crystal structure of putative N-acetyl-gamma-glutamyl-phosphate reductase (AK071544) from rice (Oryza sativa) Proteins. 2005;61:1137–1140. doi: 10.1002/prot.20679. [DOI] [PubMed] [Google Scholar]
- Ogata H, Hirota S, Nakahara A, Komori H, Shibata N, Kato T, Kano K, Higuchi Y. Activation process of [NiFe] hydrogenase elucidated by high-resolution X-ray analyses: conversion of the ready to the unready state. Structure. 2005;13:1635–1642. doi: 10.1016/j.str.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Oshikawa J, Urao N, Kim HW, Kaplan N, Razvi M, McKinney R, Poole LB, Fukai T, Ushio-Fukai M. Extracellular SOD-derived H2O2 promotes VEGF signaling in caveolae/lipid rafts and post-ischemic angiogenesis in mice. PLoS One. 2010;5:e10189. doi: 10.1371/journal.pone.0010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen CE, Carroll KS. Cysteine-Mediated Redox Signaling: Chemistry, Biology, and Tools for Discovery. Chem Rev. 2013 doi: 10.1021/cr300163e. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, Carroll KS. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat Chem Biol. 2011;8:57–64. doi: 10.1038/nchembio.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival MD, Ouellet M, Campagnolo C, Claveau D, Li C. Inhibition of cathepsin K by nitric oxide donors: evidence for the formation of mixed disulfides and a sulfenic acid. Biochemistry. 1999;38:13574–13583. doi: 10.1021/bi991028u. [DOI] [PubMed] [Google Scholar]
- Poole LB. Measurement of protein sulfenic acid content. Curr Protoc Toxicol Unit 17. 2008;2:1–21. doi: 10.1002/0471140856.tx1702s38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole LB, Claiborne A. The non-flavin redox center of the streptococcal NADH peroxidase. II. Evidence for a stabilized cysteine-sulfenic acid. J Biol Chem. 1989;264:12330–12338. [PubMed] [Google Scholar]
- Poole LB, Ellis HR. Identification of cysteine sulfenic acid in AhpC of alkyl hydroperoxide reductase. Methods Enzymol. 2002;348:122–136. doi: 10.1016/s0076-6879(02)48632-6. [DOI] [PubMed] [Google Scholar]
- Poole LB, Karplus PA, Claiborne A. Protein sulfenic acids in redox signaling. Annu. Rev. Pharmacol Toxicol. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- Poole LB, Klomsiri C, Knaggs SA, Furdui CM, Nelson KJ, Thomas MJ, Fetrow JS, Daniel LW, King SB. Fluorescent and affinity-based tools to detect cysteine sulfenic acid formation in proteins. Bioconjug Chem. 2007;18:2004–2017. doi: 10.1021/bc700257a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole LB, Nelson KJ. Discovering mechanisms of signaling-mediated cysteine oxidation. Curr Opin Chem Biol. 2008;12:18–24. doi: 10.1016/j.cbpa.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole LB, Zeng BB, Knaggs SA, Yakubu M, King SB. Synthesis of chemical probes to map sulfenic acid modifications on proteins. Bioconjug Chem. 2005;16:1624–1628. doi: 10.1021/bc050257s. [DOI] [PubMed] [Google Scholar]
- Poor CB, Chen PR, Duguid E, Rice PA, He C. Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus . J Biol Chem. 2009;284:23517–23524. doi: 10.1074/jbc.M109.015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosise GL, Wu JZ, Luecke H. Crystal structure of Tritrichomonas foetus inosine monophosphate dehydrogenase in complex with the inhibitor ribavirin monophosphate reveals a catalysis-dependent ion-binding site. J Biol Chem. 2002;277:50654–50659. doi: 10.1074/jbc.M208330200. [DOI] [PubMed] [Google Scholar]
- Qian J, Klomsiri C, Wright MW, King SB, Tsang AW, Poole LB, Furdui CM. Simple synthesis of 1,3-cyclopentanedione derived probes for labeling sulfenic acid proteins. Chem Commun (Camb) 2011;47:9203–9205. doi: 10.1039/c1cc12127h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Wani R, Klomsiri C, Poole LB, Tsang AW, Furdui CM. A simple and effective strategy for labeling cysteine sulfenic acid in proteins by utilization of beta-ketoesters as cleavable probes. Chem Commun (Camb) 2012;48:4091–4093. doi: 10.1039/c2cc17868k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race PR, Bentley ML, Melvin JA, Crow A, Hughes RK, Smith WD, Sessions RB, Kehoe MA, McCafferty DG, Banfield MJ. Crystal structure of Streptococcus pyogenes sortase A: implications for sortase mechanism. J Biol Chem. 2009;284:6924–6933. doi: 10.1074/jbc.M805406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranaivoson FM, Antoine M, Kauffmann B, Boschi-Muller S, Aubry A, Branlant G, Favier F. A structural analysis of the catalytic mechanism of methionine sulfoxide reductase A from Neisseria meningitidis . J Mol Biol. 2008;377:268–280. doi: 10.1016/j.jmb.2008.01.021. [DOI] [PubMed] [Google Scholar]
- Reddie KG, Carroll KS. Expanding the functional diversity of proteins through cysteine oxidation. Curr Opin Chem Biol. 2008;12:746–754. doi: 10.1016/j.cbpa.2008.07.028. [DOI] [PubMed] [Google Scholar]
- Reddie KG, Seo YH, Muse Iii WB, Leonard SE, Carroll KS. A chemical approach for detecting sulfenic acid-modified proteins in living cells. Mol Biosyst. 2008;4:521–531. doi: 10.1039/b719986d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehder DS, Borges CR. Cysteine sulfenic acid as an intermediate in disulfide bond formation and nonenzymatic protein folding. Biochemistry. 2010a;49:7748–7755. doi: 10.1021/bi1008694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehder DS, Borges CR. Possibilities and pitfalls in quantifying the extent of cysteine sulfenic acid modification of specific proteins within complex biofluids. BMC Biochem. 2010b;11:25. doi: 10.1186/1471-2091-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho BS, Hung LW, Holton JM, Vigil D, Kim SI, Park MS, Terwilliger TC, Pedelacq JD. Functional and structural characterization of a thiol peroxidase from Mycobacterium tuberculosis . J Mol Biol. 2006;361:850–863. doi: 10.1016/j.jmb.2006.05.076. [DOI] [PubMed] [Google Scholar]
- Roos G, Messens J. Protein sulfenic acid formation: from cellular damage to redox regulation. Free Radic Biol Med. 2011;51:314–326. doi: 10.1016/j.freeradbiomed.2011.04.031. [DOI] [PubMed] [Google Scholar]
- Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- Salsbury FR, Jr, Knutson ST, Poole LB, Fetrow JS. Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid. Protein Sci. 2008;17:299–312. doi: 10.1110/ps.073096508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AT, Neubert H, Brennan JP, Eaton P. Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proc Natl Acad Sci U S A. 2004;101:17982–17987. doi: 10.1073/pnas.0404762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YH, Carroll KS. Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc Natl Acad Sci U S A. 2009;106:16163–16168. doi: 10.1073/pnas.0903015106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YH, Carroll KS. Quantification of protein sulfenic acid modifications using isotope-coded dimedone and iododimedone. Angewandte Chemie. 2011;50:1342–1345. doi: 10.1002/anie.201007175. [DOI] [PubMed] [Google Scholar]
- Sethuraman M, McComb ME, Heibeck T, Costello CE, Cohen RA. Isotope-coded affinity tag approach to identify and quantify oxidant-sensitive protein thiols. Mol Cell Proteomics. 2004;3:273–278. doi: 10.1074/mcp.T300011-MCP200. [DOI] [PubMed] [Google Scholar]
- Shetty V, Neubert TA. Characterization of novel oxidation products of cysteine in an active site motif peptide of PTP1B. J Am Soc Mass Spectrom. 2009;20:1540–1548. doi: 10.1016/j.jasms.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty V, Spellman DS, Neubert TA. Characterization by tandem mass spectrometry of stable cysteine sulfenic acid in a cysteine switch peptide of matrix metalloproteinases. J Am Soc Mass Spectrom. 2007;18:1544–1551. doi: 10.1016/j.jasms.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva GM, Netto LE, Discola KF, Piassa-Filho GM, Pimenta DC, Barcena JA, Demasi M. Role of glutaredoxin 2 and cytosolic thioredoxins in cysteinyl-based redox modification of the 20S proteasome. Febs J. 2008;275:2942–2955. doi: 10.1111/j.1742-4658.2008.06441.x. [DOI] [PubMed] [Google Scholar]
- Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Stehle T, Claiborne A, Schulz GE. NADH binding site and catalysis of NADH peroxidase. Eur J Biochem. 1993;211:221–226. doi: 10.1111/j.1432-1033.1993.tb19889.x. [DOI] [PubMed] [Google Scholar]
- Strianese M, Palm GJ, Milione S, Kuhl O, Hinrichs W, Pellecchia C. A FRET enzyme-based probe for monitoring hydrogen sulfide. Inorg Chem. 2012;51:11220–11222. doi: 10.1021/ic301363d. [DOI] [PubMed] [Google Scholar]
- Takanishi CL, Ma LH, Wood MJ. A genetically encoded probe for cysteine sulfenic acid protein modification in vivo. Biochemistry. 2007;46:14725–14732. doi: 10.1021/bi701625s. [DOI] [PubMed] [Google Scholar]
- Takanishi CL, Wood MJ. A genetically encoded probe for the identification of proteins that form sulfenic acid in response to H2O2 in Saccharomyces cerevisiae . Journal of proteome research. 2011;10:2715–2724. doi: 10.1021/pr1009542. [DOI] [PubMed] [Google Scholar]
- Torchinsky YM. Properties of SH groups. In: Metzler D, editor. Sulfur in Proteins. Oxford, England: Pergamon; 1981. [Google Scholar]
- Truong TH, Garcia FJ, Seo YH, Carroll KS. Isotope-coded chemical reporter and acid-cleavable affinity reagents for monitoring protein sulfenic acids. Bioorg Med Chem Lett. 2011;21:5015–5020. doi: 10.1016/j.bmcl.2011.04.115. [DOI] [PubMed] [Google Scholar]
- Turell L, Botti H, Carballal S, Ferrer-Sueta G, Souza JM, Duran R, Freeman BA, Radi R, Alvarez B. Reactivity of sulfenic acid in human serum albumin. Biochemistry. 2008;47:358–367. doi: 10.1021/bi701520y. [DOI] [PubMed] [Google Scholar]
- van Montfort RL, Congreve M, Tisi D, Carr R, Jhoti H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature. 2003;423:773–777. doi: 10.1038/nature01681. [DOI] [PubMed] [Google Scholar]
- Volbeda A, Martin L, Cavazza C, Matho M, Faber BW, Roseboom W, Albracht SP, Garcin E, Rousset M, Fontecilla-Camps JC. Structural differences between the ready and unready oxidized states of [NiFe] hydrogenases. J Biol Inorg Chem. 2005;10:239–249. doi: 10.1007/s00775-005-0632-x. [DOI] [PubMed] [Google Scholar]
- Wallace DC, Fan W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010;10:12–31. doi: 10.1016/j.mito.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kettenhofen NJ, Shiva S, Hogg N, Gladwin MT. Copper dependence of the biotin switch assay: modified assay for measuring cellular and blood nitrosated proteins. Free Radic Biol Med. 2008;44:1362–1372. doi: 10.1016/j.freeradbiomed.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani R, Bharathi NS, Field J, Tsang AW, Furdui CM. Oxidation of Akt2 kinase promotes cell migration and regulates G1-S transition in the cell cycle. Cell Cycle. 2011a;10:3263–3268. doi: 10.4161/cc.10.19.17738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani R, Qian J, Yin L, Bechtold E, King SB, Poole LB, Paek E, Tsang AW, Furdui CM. Isoform-specific regulation of Akt by PDGF-induced reactive oxygen species. Proc Natl Acad Sci U S A. 2011b;108:10550–10555. doi: 10.1073/pnas.1011665108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett WS, Copley SD. Identification and localization of a stable sulfenic acid in peroxide-treated tetrachlorohydroquinone dehalogenase using electrospray mass spectrometry. Chem Biol. 1996;3:851–857. doi: 10.1016/s1074-5521(96)90071-x. [DOI] [PubMed] [Google Scholar]
- Wilson NA, Barbar E, Fuchs JA, Woodward C. Aspartic acid 26 in reduced Escherichia coli thioredoxin has a pKa > 9. Biochemistry. 1995;34:8931–8939. doi: 10.1021/bi00028a001. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- Wolthers KR, Lou X, Toogood HS, Leys D, Scrutton NS. Mechanism of coenzyme binding to human methionine synthase reductase revealed through the crystal structure of the FNR-like module and isothermal titration calorimetry. Biochemistry. 2007;46:11833–11844. doi: 10.1021/bi701209p. [DOI] [PubMed] [Google Scholar]
- Wong SH, Lonhienne TG, Winzor DJ, Schenk G, Guddat LW. Bacterial and plant ketol-acid reductoisomerases have different mechanisms of induced fit during the catalytic cycle. J Mol Biol. 2012;424:168–179. doi: 10.1016/j.jmb.2012.09.018. [DOI] [PubMed] [Google Scholar]
- Wu Y, Kwon KS, Rhee SG. Probing cellular protein targets of H2O2 with fluorescein-conjugated iodoacetamide and antibodies to fluorescein. FEBS Lett. 1998;440:111–115. doi: 10.1016/s0014-5793(98)01415-x. [DOI] [PubMed] [Google Scholar]
- Xu G, Chance MR. Radiolytic modification of sulfur-containing amino acid residues in model peptides: fundamental studies for protein footprinting. Anal Chem. 2005;77:2437–2449. doi: 10.1021/ac0484629. [DOI] [PubMed] [Google Scholar]
- Xu Z, Lam LS, Lam LH, Chau SF, Ng TB, Au SW. Molecular basis of the redox regulation of SUMO proteases: a protective mechanism of intermolecular disulfide linkage against irreversible sulfhydryl oxidation. Faseb J. 2008;22:127–137. doi: 10.1096/fj.06-7871com. [DOI] [PubMed] [Google Scholar]
- Yeh JI, Claiborne A, Hol WG. Structure of the native cysteine-sulfenic acid redox center of enterococcal NADH peroxidase refined at 2.8 Å resolution. Biochemistry. 1996;35:9951–9957. doi: 10.1021/bi961037s. [DOI] [PubMed] [Google Scholar]
- Zhu X, Tanaka F, Lerner RA, Barbas CF3rd, Wilson IA. Direct observation of an enamine intermediate in amine catalysis. J Am Chem Soc. 2009;131(51):18206–18207. doi: 10.1021/ja907271a. [DOI] [PMC free article] [PubMed] [Google Scholar]