Abstract
Mutations in zinc-finger in cerebellum 3 (ZIC3) result in heterotaxy or isolated congenital heart disease (CHD). The majority of reported mutations cluster in zinc-finger domains. We previously demonstrated that many of these lead to aberrant ZIC3 subcellular trafficking. A relative paucity of N- and C-terminal mutations has, however, prevented similar analyses in these regions. Notably, an N-terminal polyalanine expansion was recently identified in a patient with VACTERL, suggesting a potentially distinct function for this domain. Here, we report ZIC3 sequencing results from 440 unrelated patients with heterotaxy and CHD, the largest cohort yet examined. Variants were identified in 5.2% of sporadic male cases. This rate exceeds previous estimates of 1% and has important clinical implications for genetic testing and risk-based counseling. Eight of 11 were novel, including 5 N-terminal variants. Subsequent functional analyses included 4 additional reported but untested variants. Aberrant cytoplasmic localization and decreased luciferase transactivation were observed for all zinc-finger variants, but not for downstream or in-frame upstream variants, including both analyzed polyalanine expansions. Collectively, these results expand the ZIC3 mutational spectrum, support a higher than expected prevalence in sporadic cases, and suggest alternative functions for terminal mutations, highlighting a need for further study of these domains.
Keywords: ZIC3, left-right patterning, symmetry, heterotaxy, congenital heart disease
INTRODUCTION
Bilateral symmetry is broken early in development following establishment of the anterior-posterior and dorsal-ventral body axes. Embryos of some species begin to manifest left-right (L-R) asymmetries as early as initial cleavage stages, establishing molecular gradients and initiating signaling cascades that foreshadow additional L-R pattering processes at the ciliated embryonic node (reviewed in Vandenberg and Levin, 2010). Failure to properly establish L-R asymmetry results in heterotaxy, a condition of significant morbidity and mortality associated with abnormal positioning and disrupted morphogenesis of the thoraco-abdominal viscera. Much of the clinical severity results from complex and heterogeneous heart malformations that are classically accompanied by abnormalities in spleen position and number, malrotation of the developing gut tube, and/or situs anomalies of the liver and stomach. The overall genetics of heterotaxy are complex and have been reviewed in detail elsewhere (Sutherland and Ware, 2009); however, both de novo and familial mutations have been reported, including pedigrees suggestive of autosomal recessive, autosomal dominant, and X-linked inheritance patterns (Vitale, et al., 2001; Belmont, et al., 2004; Zhu, et al., 2006).
The clinical significance of the zinc-finger in cerebellum 3 gene (ZIC3; MIM# 300265), the first human heterotaxy gene to be described (Gebbia, et al., 1997) and the only known genetic cause of X-linked heterotaxy (MIM# 306955), is well documented (Gebbia, et al., 1997; Megarbane, et al., 2000; Ware, et al., 2004; Tzschach, et al., 2006; Ware, et al., 2006b; Chhin, et al., 2007; De Luca, et al., 2010; Wessels, et al., 2010; Chung, et al., 2011; D’Alessandro, et al., 2011; Ma, et al., 2012). Estimates from our initial analyses of 194 individuals with heterotaxy and heterotaxy spectrum CHD suggested that 75% of pedigrees consistent with X-linked inheritance and 1% of sporadic heterotaxy cases are attributable to ZIC3 mutations (Ware, et al., 2004). Nevertheless, variable expressivity is common, resulting in a wide spectrum of clinical manifestations ranging from situs abnormalities and isolated heart defects to more complex diagnoses involving additional midline, gastrointestinal, urogenital, and/or central nervous system anomalies (Casey, et al., 1993; Gebbia, et al., 1997; Ware, et al., 2004; De Luca, et al., 2010; Wessels, et al., 2010; Chung, et al., 2011; D’Alessandro, et al., 2011; Ma, et al., 2012). Such phenotypic complexity is mirrored in mice deficient for Zic3, which exhibit a wide variety of congenital abnormalities including heterotaxy, vertebral and rib anomalies, neural tube defects, and cerebellar hypoplasia (Purandare, et al., 2002; Ware, et al., 2006a; Jiang, et al., 2013; Sutherland, et al., 2013).
Recent reports additionally associate ZIC3 mutations with VACTERL, a constellation of defects (vertebral, anal, cardiac, tracheo-esophageal, renal, radial, and limb) demonstrating phenotypic overlap with heterotaxy (Wessels, et al., 2010; Chung, et al., 2011). A polyalanine tract expansion in one of these patients (Wessels, et al., 2010) is particularly intriguing as similar tract expansions have been associated with a variety of human genetic disorders, including holoprosencephaly caused by mutations in ZIC2 (Messaed and Rouleau, 2009).
Both the complexity of ZIC3-associated phenotypes and its broad expression during embryogenesis (Nagai, et al., 1997; McMahon and Merzdorf, 2010; Quinn, et al., 2012) are indicative of diversity in ZIC3 developmental function, including recognized roles in neural and neural crest development (Nakata, et al., 1997; Klootwijk, et al., 2000), limb bud digitation (Quinn, et al., 2012), cardiac morphogenesis, and L-R patterning (Kitaguchi, et al., 2000; Purandare, et al., 2002; Ware, et al., 2004; Ware, et al., 2006b; Ware, et al., 2006a; Zhu, et al., 2007b; Zhu, et al., 2007a; Jiang, et al., 2013; Sutherland, et al., 2013). Studies utilizing Zic3-null mice suggest that Zic3 is required for progression into and through gastrulation (Ware, et al., 2006a; Cast, et al., 2012) and that it acts upstream of Nodal signaling at the embryonic node (Purandare, et al., 2002; Ware, et al., 2006b).
The ZIC3 gene encodes a highly conserved zinc-finger protein belonging to the GLI superfamily of transcription factors (Mizugishi, et al., 2001; Sakai-Kato, et al., 2008). The most highly conserved elements of the GLI superfamily members are the zinc-finger DNA-binding domains (Herman and El-Hodiri, 2002; Aruga, et al., 2006). Five of these domains encompass over a third of the ZIC3 amino acid sequence and collectively comprise a DNA-binding domain that facilitates not only transcriptional activation of target genes, but also binding of transcriptional cofactors and subcellular trafficking of the ZIC3 protein (Mizugishi, et al., 2001; Ware, et al., 2004; Bedard, et al., 2007; Hatayama, et al., 2008; Zhu, et al., 2008; Lim, et al., 2010). Interestingly, relatively few ZIC3 polymorphisms have been reported in 1000 genomes (1KG, http://browser.1000genomes.org/) and NHLBI Exome Sequencing Project (ESP, http://evs.gs.washington.edu/EVS/) databases, potentially reflecting high levels of sequence conservation throughout the entire coding sequence (Ware, et al., 2004).
In this study, we report results from ZIC3 mutation screening in 440 unrelated patients with heterotaxy and isolated heterotaxy-spectrum CHD and perform in vitro functional testing of 15 ZIC3 variants. These studies represent not only the first major sequencing effort to include coverage of the recently identified fourth exon of ZIC3 (Bedard, et al., 2011), but also the first functional assessment of expansions of the ZIC3 polyalanine tract. Our sequencing results increase the total number of reported ZIC3 variants from 23 to 31 (Stenson, et al., 2003) and notably include the most N-terminal (Asp6GlufsX32) and C-terminal (Ala447Gly) variants yet identified. Collectively, these data expand the known ZIC3 mutation spectrum, particularly in N-terminal encoding sequences, and reveal a higher than anticipated incidence in patients with sporadic heterotaxy (3.8% vs. 1%), particularly in affected males. Furthermore, our functional analyses confirm the importance of the zinc-finger regions for both ZIC3 localization and transcriptional target activation and suggest the existence of alternative functional consequences for variants in N- and C-terminal regions, including those in the polyalanine tract.
MATERIALS AND METHODS
Patient recruitment and phenotypic classification
Informed consent was obtained from all study participants according to protocols approved by the institutional review board at Cincinnati Children’s Hospital Medical Center (CCHMC). Using our previously described classification system (Ware, et al., 2004), each participant was categorized as having situs inversus, heterotaxy, or isolated heterotaxy spectrum CHD based on phenotypic information collected from patient histories and chart review. Heterotaxy was considered to be “familial” if the presenting pedigree met one or more of the following criteria: 1) autosomal dominant, autosomal recessive, or X-linked inheritance; 2) more than one family member with heterotaxy or laterality disorder regardless of degree of relationship; 3) heterotaxy in the proband and a first degree relative with isolated CHD; and/or 4) heterotaxy in the proband and a first degree relative with situs-related defect(s), including but not limited to gut malrotation, isolated dextrocardia, bilateral superior vena cava, or right-sided stomach. Heterotaxy identified in pedigrees not meeting one or more of these criteria was classified as “sporadic”.
Variant detection
Genomic DNA was prepared from participant blood samples following standard protocols. DNA was PCR-amplified with oligonucleotide primers designed to encompass all ZIC3 coding regions and splice junctions (RefSeq: NM_003413.3), including the recently identified fourth exon (Bedard, et al., 2011). Sequencing primer sequences were as follows: exon 1aF: 5′-TCCTATCCCTCTGCAGGAGAC-3′ and exon 1aR 5′-ACTCGCGCGTTGAGTTGAAGG-3′; exon 1bF 5′-GAATCCCTTCGGGGACTCAAC-3′ and exon 1bR 5′-CAGCTCCTGCTTGATAGGCTG-3′; exon 1cF 5′-CAGTTTCCTAACTACAGCCCC-3′ and exon 1cR 5′-GTGGATGGTCACTGACAGCGC-3′; exon 2F 5′-GCTGCTTGCCTCTGAGAAAC-3′ and exon 2R 5′-ACGTGGAAGACAGTGGGTTG-3′, exon 3F 5′-GCTCTTGTTTTTGCTTGCAC-3′ and exon 3R 5′-CATTTCCATCTGATTGGTCTC-3′; and exon 4F 5′-AAGAGGAAATGTGGCCTGTTT-3′ and exon 4R 5′-CACTTCAAGGTAACAGACATCCA-3′. All amplifications were performed at 57°C as 20μL reactions containing 20ng genomic DNA, 10μL Fast Master Mix (Roche) and 10pmol of each primer. PCR-amplified products were treated with exonuclease 1 and shrimp alkaline phosphatase prior to sequencing. Bi-directional sequencing was performed using BigDye Terminator v3.1 on an ABI 3730XL DNA analyzer and mutation analysis was completed with Mutation Surveyor v4.0.5 (SoftGenetics Inc.). Detected variants were then queried against the Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk/) and nomenclature was assigned to novel changes according to Human Genome Variation Society (HGVS, http://www.hgvs.org/) guidelines.
Variant interpretation
Pathogenicity of each ZIC3 variant was interpreted according to clinical molecular diagnostic laboratory standards, taking into account type of mutation, bioinformatic prediction of protein function (if applicable), prevalence in control populations (1KG, ESP public variant databases), segregation with disease phenotype, and functional analyses.
Expression constructs and mutagenesis
Methods for generating an N-terminal hemagglutinin (HA)-tagged, wild-type ZIC3 expression construct (HA-ZIC3) have been previously described (Ware, et al., 2004). This plasmid contains the entire human ZIC3-A ORF subcloned into a pHM6 expression vector (Roche). Mutations were introduced into the wild-type ZIC3 sequence via Quik-Change site-directed mutagenesis (Agilent) and confirmed by Sanger sequencing. Primers used to generate each mutant ZIC3 construct are listed in Supp. Table S1.
Cell culture
NIH/3T3 cells were obtained from the American Type Culture Collection. Cells were grown in high glucose DMEM (HyClone) supplemented with 10% newborn calf serum (HyClone) using a Forma Series II 3110 Water-Jacketed CO2 Incubator (Thermo Scientific) maintained at 37°C and 5% CO2.
Luciferase assays
Luciferase assays were conducted as previously described (Ware, et al., 2004). NIH/3T3 cells were seeded onto 9.6cm2 6-well plates (BD-Falcon) at 2 X 105 cells/mlone day prior to transfection with FuGENE HD transfection reagent (Promega) following manufacturer’s protocols. A total of 2μg of DNA and a DNA:FuGENE HD ratio of 1:3 were used for each transfection. Wild-type or mutant HA-ZIC3 constructs were co-transfected with an SV40 firefly luciferase reporter (pGL3-SV40, Promega), and a Renilla luciferase reporter (pRL-TK, Promega) to control for transfection efficiency. A promoterless luciferase vector (pGL3-Basic, Promega) was used for normalization. Cells were lysed 24 h after transfection and luciferase activities were determined using the Dual Luciferase Reporter Assay System (Promega) and a Glomax 96 Microplate Luminometer (Promega). Firefly luciferase activities were normalized to Renilla luciferase activity. Fold activations for each ZIC3 mutant were calculated with respect to wild-type values. Results for each construct represent average values obtained from a minimum of 3 independent experiments repeated in triplicate. Statistical analyses were completed using GraphPad Instat v. 3.
Immunocytochemistry and subcellular localization
NIH/3T3 cells were seeded onto 15mm diameter circular glass coverslips (Electron Microscopy Sciences) at 1 X 105 cells/ml one day prior to transfection with either wild-type or mutant HA-ZIC3. For each transfection, 2μg of DNA were transfected at a 1:3 ratio with FuGENE HD transfection reagent (Promega). After 24 h, transfected cells were washed in PBS and fixed for 20 minutes at room temperature in 4% paraformaldehyde/PBS. Following 3 washes in PBS and 3 additional washes in 0.1% BSA/PBS, cells were blocked for 1 h at room temperature with 50% normal donkey serum/0.3% Triton X-100/PBS. Cells were subsequently incubated for 1 h at room temperature in a 1:200 dilution of anti-HA rabbit polyclonal antibody (NB600-363, Novus Biologicals), washed 3 times in 0.1% BSA/PBS, incubated for 1 h in a 1:200 dilution of Alexa Fluor 488 goat anti-rabbit IgG (A-11008, Molecular Probes), washed 3 more times in 0.1% BSA/PBS, and mounted using Vectashield Hardset with DAPI (Vector Labs). Slides were imaged using an Eclipse E400 fluorescence microscope equipped with a DXM1200F digital camera and ACT-1 v. 2.62 imaging software (Nikon). Merged images were generated in Adobe Photoshop CS5 extended v. 12.0×32 and subsequently imported to Image J v. 1.45s (rsbweb.nih.gov/ij/) for analysis. Cells were scored as having either “nuclear”, “cytoplasmic”, or “nuclear and cytoplasmic” ZIC3 expression. At least 250 cells across a minimum of 3 independent experiments were scored for each ZIC3 construct.
RESULTS
Identification of ZIC3 variants
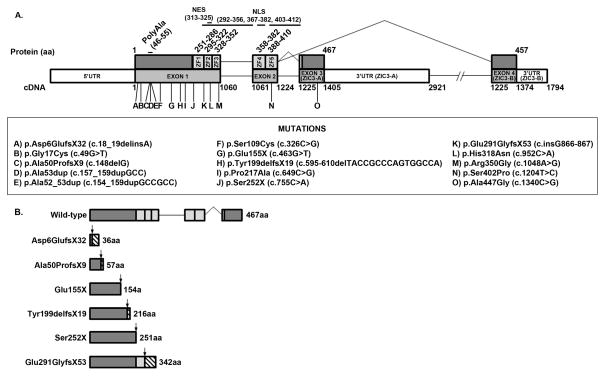
ZIC3 coding and splice junction regions were fully sequenced in 440 unrelated patients with heterotaxy or isolated heterotaxy-spectrum CHD. Demographics of the cohort are summarized in Table 1. A total of 11 ZIC3 variants were identified in 15 patients, including 8 that were novel (Table 2). Collectively, the identified variants encompass 7 point mutations (2 nonsense, 5 missense), 2 indels (1 deletion, 1 deletion-insertion), and 2 duplications (including 1 polyalanine expansion). Figure 1A depicts amino acid and cDNA locations for each variant. Truncations and their predicted effects on ZIC3 protein structure are also shown (Figure 1B). In all but one pedigree, ZIC3 sequencing was performed on the proband. The sole exception was an asymptomatic mother (Patient 12) whose DNA was sequenced in the absence of an available sample from her son with heterotaxy. Sequencing identified a heterozygous C to A transversion resulting in a nonsense mutation that prematurely truncates the ZIC3 protein at a highly conserved serine 252 residue bordering the first zinc-finger domain (Ser252X). The severe phenotype of the proband and the carrier status of his mother imply likely X-linked transmission of the variant allele. Pathogenicity of the variant is also supported by the identification of a nearby nonsense mutation (Gln249X) in multiple individuals of a family with comparably complex CHD and situs abnormalities (Ware, et al., 2004).
Table 1.
Heterotaxy and CHD cohort demographics
| Racial and Ethnic Categories | Totals (%)(n=440) |
|---|---|
| American Indian/Alaskan | 1 (0.23) |
| Asian | 14 (3.18) |
| Black or African American | 52 (11.82) |
| Caucasian | 222 (50.45) |
| Hispanic / Latino | 77 (17.50) |
| Native Hawaiian / Other Pacific Islander | 1 (0.23) |
| Mixed | 23 (5.23) |
| Other1 | 6 (1.36) |
| Unknown | 44 (10.00) |
| Totals (%) | 440 (100.00) |
| Gender | |
|---|---|
| Females | Males |
| 0 | 1 |
| 5 | 9 |
| 23 | 29 |
| 93 | 129 |
| 26 | 51 |
| 0 | 1 |
| 9 | 14 |
| 2 | 4 |
| 23 | 21 |
| 181 (41.14) | 259 (58.86) |
| Inheritance | ||
|---|---|---|
| Familial | Sporadic | Unknown |
| 0 | 1 | 0 |
| 1 | 10 | 3 |
| 7 | 30 | 15 |
| 32 | 126 | 64 |
| 8 | 57 | 12 |
| 0 | 1 | 0 |
| 6 | 17 | 0 |
| 0 | 6 | 0 |
| 5 | 16 | 23 |
| 59 (13.41) | 264 (60.00) | 117 (26.59) |
Includes Arabic, Sephardic Jewish, and Sri-Lankan racial categories
Table 2.
Summary of ZIC3 variants detected in heterotaxy cohort
| Patient | Amino Acid Change | Nucleotide Change | Exon | Gender | Ethnicity / Race | Inheritance | Phenotype | Detailed Phenotype |
|---|---|---|---|---|---|---|---|---|
| 1 | p.Asp6GlufsX32 | c.18_19delinsA | 1 | M | Caucasian / African American | Familial; maternal half-brother with asplenia (d. age 7 years) | Heterotaxy | Gut malrotation, asplenia, ASD, CAVC, PA |
| 2 | p.Gly17Cys | c.49G>T | 1 | M | Caucasian | Sporadic | Isolated CHD | HLHS, MS with hypoplastic MV, dysplastic PV, PS, l- TGA, DILV, ASD, congenital coronary anomaly |
| 3 | p.Gly17Cys | c.49G>T | 1 | M | Caucasian | Unknown | Heterotaxy | Esophageal atresia |
| 4 | p.Gly17Cys | c.49G>T | 1 | F | Unknown | Sporadic; sister and mother mutation positive, presumed normal phenotype | Isolated CHD | Williams Syndrome, dextrocardia, d- TGA, TA, severely hypoplastic RV, mild hypoplasia of ascending Ao |
| 5 | p.Gly17Cys | c.49G>T | 1 | M | Caucasian | Sporadic | Heterotaxy | Abd SI, dextrocardia, single ventricle, PA, TA, PAPVR, dilated Ao, R Ao arch, MPA w/o connection, ASD, VSD |
| 6 | p.Ala53dup | c.157_159dupGCC | 1 | M | Hispanic or Latino | Sporadic | Isolated CHD | PS, d-TGA, VSD |
| 7 | p.Ser109Cys | c.326C>G | 1 | M | African American | Sporadic | Isolated CHD | d-TGA, PDA |
| 8 | p.Glu155X | c.463G>T | 1 | M | Caucasian | Sporadic | Heterotaxy | Midline liver, dextrocardia, CAVC, PA, Ao arch abnormalities, MGA, DORV, TAPVR, SVC abnormalities |
| 9 | p.Tyr199delfsX19 | c.delTACCGCCCA GTGGCCA595-610 | 1 | M | Unknown | Sporadic | Heterotaxy | Dextrocardia, VSD, ventriculomegaly, no stomach |
| 10 | p.Pro217Ala | c.649C>G | 1 | M | Hispanic or Latino | Sporadic | Isolated CHD | Ventricular inversion, hypoplastic LV & MV, d-TGA, DORV, VSD |
| 11 | p.Pro217Ala | c.649C>G | 1 | F | African American | Familial; maternally inherited; mutation- positive sister with CHDs; stillborn brother, cause unknown | Heterotaxy | Gut malrotation, polysplenia, midline liver, interrupted IVC |
| 12 | p.Ser252X | c.755C>A | 1 | F | Caucasian | Sporadic | Unaffected carrier mother; sequenced in lieu of sample from affected son with heterotaxy | Abd SI, asplenia, CAVC, PA |
| 13 | p.Glu291GlyfsX53 | c.866dupG | 1 | M | Caucasian | Familial; 2 sibs with “Ivemark”, 1 with heart defect | Heterotaxy | Abd SI, asplenia, CAVC, PA, d-TGA |
| 14 | p.His318Asn | c.952C>A | 1 | F | Caucasian | Familial; brother with isomerism/asplenia, mat. uncle with complex CHD | VACTERL -like | L hip dysplasia, VSD |
| 15 | p.Ala447Gly | c.1340C>G | 3 | F | Caucasian | Sporadic | Isolated CHD | PA, VSD, ASD |
RefSeq: NP_003404.1, NM_003413.3
Abbreviations: Abd SI = abdominal situs inversus; Ao = aorta; ASD = atrial septal defect; CAVC = complete atrioventricular canal; CHD = congenital heart disease; d-TGA = dextro-transposition of the great arteries; DILV = double inlet left ventricle; DORV = double outlet right ventricle; HCM = hypertrophic cardiomyopathy, HLHS = hypoplastic left heart syndrome; IVC = inferior vena cava; l-TGA = levo-transposition of the great arteries; LV = left ventricle; MGA = malposed great arteries; MPA = main pulmonary artery; MS = mitral stenosis; MV = mitral valve; PA = pulmonary atresia; PAPVR = partial anomalous pulmonary venous return; PDA = patent ductus arteriosus; PS = pulmonic stenosis; PV = pulmonary valve; RAA = RV = right ventricle; SI = situs inversus; SVC = superior vena cava; TA = tricuspid atresia; TAPVR = total anomalous pulmonary venous return; VACTERL = vertebral, anomalies, anal atresia, cardiovascular anomalies, tracheoesophageal fistula, renal/radial anomalies, limb defects; VSD = ventricular septal defect
Figure 1. ZIC3 structure.
(A) Wild-type human ZIC3, RefSeq: NP_003404.1, NM_003413.3. Locations of ZIC3 variants are indicated. cDNA numbering begins at the A (position +1) of the ATG initiation codon (codon 1). ZF = zinc-finger domain. (B) Predicted protein structures and amino acid lengths for wild-type and truncating ZIC3 variants. Arrows represent mutation sites. Hatched bars indicate altered out-of-frame amino acid sequences.
Familial disease was associated with 4 of the 11 ZIC3 variants. Two of these encode truncated proteins. The first, an out-of-frame indel at the 5′ end of the ZIC3 gene (Asp6GlufsX32), was associated with heterotaxy in a male proband (Patient 1) and his maternal half-brother. The resulting protein is severely truncated at 36 amino acids and lacks all known functional domains. The second variant, an out-of-frame duplication of a single G nucleotide near the 3′ end of exon 1, was identified in a male proband with heterotaxy and a family history of 2 siblings with Ivemark syndrome, one of whom also had CHD (Patient 13). This variant was predicted to encode a 342 amino acid protein missing all but the first zinc-finger domain (Glu291GlyfsX53). Notably, a nonsense mutation resulting in loss of the same functional regions (Gln294X) was previously reported in the context of familial heterotaxy (Gebbia, et al., 1997).
The 2 remaining familial variants identified in our cohort were missense. The first, an N-terminal variant replacing a semi-conserved proline with an alanine residue (Pro217Ala), was identified in a female proband with heterotaxy (Patient 11) and was inherited from her asymptomatic mother. The family history was significant for a stillborn brother of unknown cause and a variant-positive sister with CHD. The second identified missense mutation replaces a highly conserved histidine residue with an asparagine in the second zinc-finger domain (His318Asn) and was found in a female proband with isolated CHD (Patient 14). The family history was exceptional for a brother with situs abnormalities and a maternal uncle with complex CHD, both of whom are deceased with unknown mutation status. A nearby and similarly conserved zinc-finger 2 missense mutation (Thr323Met) was previously identified in over 10 related individuals with heterotaxy and related anomalies (Gebbia, et al., 1997).
A total of 8 ZIC3 variants were identified in individuals with sporadic disease, including the aforementioned Ser252X nonsense mutation in Patient 12. These variants ranged from single amino acid substitutions to more complex genetic abnormalities. Five were point mutations, including 4 missense variants found in 3 males and a single female with isolated CHD. Each was found either upstream or downstream of the zinc-finger domains. Two (Gly17Cys and Ala447Gly) affected highly conserved amino acids, while 2 (Ser109Cys and Pro217Ala) altered amino acids incompletely conserved among species. Although Ser109Cys and Ala447Gly are novel, both Gly17Cys and Pro217Ala were previously reported in patients with isolated CHD (Ware, et al., 2004; De Luca, et al., 2010). In our cohort, Gly17Cys and Pro217Ala variants were also identified in 3 patients with classic heterotaxy (Patients 3, 5, and 11). The remaining sporadic point mutation, a novel nonsense mutation encoding a 154 amino acid protein truncated upstream of the zinc-finger regions (Glu155X), was identified in a male patient with heterotaxy (Patient 8). Additionally, an out-of-frame deletion removing 16bp midway through exon 1 (Tyr199delfsX19), was detected in a male patient with heterotaxy (Patient 9). The resulting 216 amino acid protein lacks all 5 zinc-finger domains. Finally, a male patient with isolated CHD (Patient 6) was found to carry a single alanine residue expansion (Ala53dup) in the ZIC3 polyalanine tract. A similar single alanine expansion was recently identified in a male patient with isolated CHD (d-TGA) (D’Alessandro, et al., 2013), while larger expansions of 2 and 3 alanine residues were previously identified in a male patient with VACTERL association (Wessels, et al., 2010) and a female patient with heterotaxy, respectively (D’Alessandro, et al., 2013).
In summary, we have identified 11 ZIC3 variants in a cohort of 440 patients with heterotaxy and CHD, including 8 that were novel, 5 of which fall in the N-terminal domain. Detection of variants in 10/264 (3.8%) sporadic cases, including 8/154 (5.2%) males and 2/111 (1.8%) females, indicated a higher than expected yield patients with sporadic disease, based on past estimates of 1% (Ware, et al., 2004). Our analyses additionally verify previous associations of ZIC3 mutations with isolated CHD (Megarbane, et al., 2000; Ware, et al., 2004; D’Alessandro, et al., 2013) and reinforce existing evidence for pathogenicity of ZIC3 deficiency in a subset of carrier females (Gebbia, et al., 1997; Ware, et al., 2004; De Luca, et al., 2010; Chung, et al., 2011; Ma, et al., 2012).
ZIC3 variants alter luciferase reporter gene transactivation
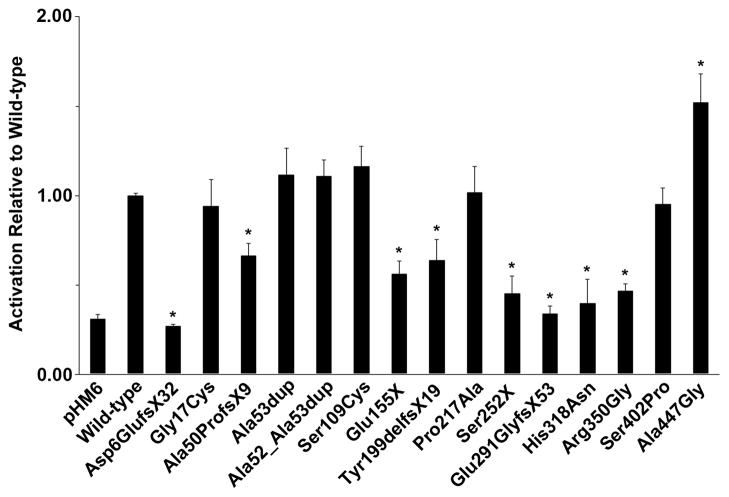
As a transcription factor, ZIC3 function is dependent on both its localization to the nucleus and its subsequent ability to activate target genes. Previous studies have demonstrated the importance for the zinc-finger domains in fulfilling these functions (Ware, et al., 2004; Bedard, et al., 2007; Hatayama, et al., 2008); however, the impact of mutations in upstream and downstream regions, including the polyalanine tract, remain less clearly defined. We, therefore, sought to assess the pathogenic significance of each of the 11 ZIC3 variants identified in our cohort. To maximize the breadth of our analyses, we supplemented these with 4 recently published variants for which functional assessments were lacking. Site-directed mutagenesis was used to generate mutant ZIC3 constructs from a previously described expression vector encoding the wild-type ZIC3-A open reading frame fused to an N-terminal hemagglutinin (HA) epitope tag (Ware, et al., 2004). Wild-type and mutant ZIC3 constructs were subsequently co-transfected into NIH/3T3 cells with an SV40 promoter-driven firefly luciferase reporter (Ware, et al., 2004). In agreement with previous studies (Ware, et al., 2004; Bedard, et al., 2007; Hatayama, et al., 2008), ZIC3 mutations in the zinc-finger regions significantly disrupted reporter gene transactivation relative to wild-type controls (Figure 2). Included in this group were 2 missense variants affecting highly conserved amino acid residues (His318Asn, Arg350Gly), as well as 6 truncating variants individually producing proteins missing at least 4 out of 5 zinc-fingers (Asp6GlufsX32, Ala50ProfsX9, Glu155X, Tyr199delfsX19, Ser252X, Glu291GlyfsX53). Of the remaining variants, Ala447Gly was exceptional, demonstrating significant and reproducible increase in luciferase reporter transactivation. As the most C-terminal ZIC3 variant identified to date, Ala447Gly lies downstream of any mapped functional domains and affects only the larger ZIC3-A isoform, which preferentially splices in the Ala447Gly-containing exon 3 at the expense of the ZIC3-B specific exon 4 (Bedard, et al., 2011).
Figure 2. Mutations in ZIC3 alter transactivation of an SV40 luciferase reporter.
Wild-type (HA-ZIC3 WT) or mutant ZIC3 constructs were co-transfected into NIH/3T3 cells with pGL3-SV40 firefly and pRL-TK renilla luciferase reporters. Luciferase activities were measured 24 hours post-transfection. Mean-fold activations relative to wild-type are shown. Results represent average luciferase activations across a minimum of 4 individual experiments. Standard errors are indicated by vertical lines. Asterisks denote statistical significance (p<0.05) by two-tailed, unpaired Student’s T-tests assuming unequal variance.
Zinc-finger domain mutations demonstrate aberrant cytoplasmic subcellular localization
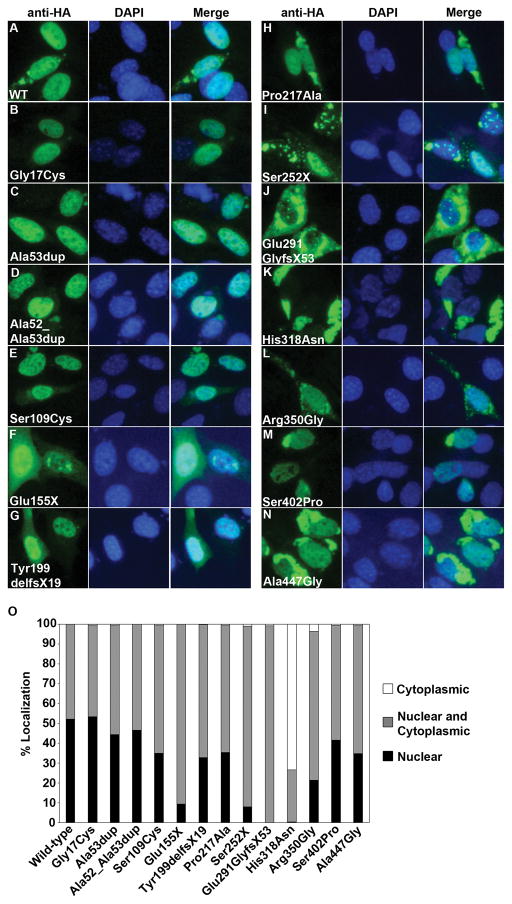
As an additional assessment of the functional impact of ZIC3 variants, transient transfections of HA-tagged ZIC3 constructs and immunofluorescence analyses were performed to determine the effect of each variant on ZIC3 protein subcellular localization. Localization experiments for the current study were carried out in the same NIH/3T3 cells used for the luciferase transactivation studies. Cells were counted and scored as having “nuclear”, “cytoplasmic” or “nuclear and cytoplasmic” ZIC3, as described (see Materials and Methods). Results from these experiments are summarized in Figure 3.
Figure 3. Mutations in ZIC3 alter subcellular localization.
(A–N) Representative subcellular localization for wild-type (WT) or mutant HA-tagged ZIC3 constructs transfected into NIH/3T3 cells. Anti-HA and DAPI staining are shown individually and merged. (O) Cells transfected with each ZIC3 construct were classified as having either nuclear or cytoplasmic. A minimum of 250 cells were scored for each construct.
Gli superfamily protein subcellular localization has previously been shown to be cell line dependent (Koyabu, et al., 2001). In NIH/3T3 cells, exogenously expressed ZIC3 localizes to the nucleus in approximately 50–55% of cells with the remaining 45–50% demonstrating mixed nuclear and cytoplasmic ZIC3 localization (Figure 3). Similar localization patterns were observed following transfection with the Gly17Cys construct, encoding the most N-terminal missense mutation reported to date (this study, De Luca, et al., 2010). Slightly fewer cells (40–45%) exhibited nuclear localization with either of the polyalanine expansions or with the C-terminal Ser402Pro variant; each of which lie downstream from Gly17Cys in non-zinc-finger domains. Three similarly positioned missense mutations (Ser109Cys, Pro217Ala, Ala447Gly) also yielded large proportions of cells with nuclear ZIC3 (30–35%), albeit at decreased levels relative to wild-type transfections. In contrast, ZIC3 was exclusively nuclear in <20% of cells transfected with mutant constructs affecting the zinc-finger domains (Figure 3O). Notably, each of these 5 variants (Glu155X, Ser252X, Glu291GlyfsX53, His318Asn, Arg350Gly) are predicted to disrupt one or more amino acids in previously mapped nuclear localization (NLS) and nuclear export (NES) signals (Hatayama, et al., 2008; Bedard, et al., 2007). Most striking was His318Asn, which affects a single highly conserved histidine residue in overlapping zinc-finger 2 NLS and NES sequences (Bedard, et al., 2007; Hatayama, et al., 2008). Finally, it should be noted that no detectable fluorescent signal was observed for 2 out-of-frame mutations (Asp6GlufsX32 and Ala50ProfsX9), presumably due to protein instability resulting from early truncation. Results from all functional assays are summarized in Table 3. Collectively, they support previous findings that ZIC3 subcellular localization and transcriptional activity is most sensitive to mutations affecting amino acids in zinc-finger domains and suggest the likelihood of alternative pathogenic mechanisms for variants affecting the N- and C-terminal domains (Ware, et al., 2004; Bedard, et al., 2007; Hatayama, et al., 2008).
Table 3.
Summary of variant analyses
| Variant | Variant Type | 1KG / ESP (MAF)1 | Pathogenicity Predicitons2 | Reporter Gene Transactivation | Subcellular Localization | Variant Interpretation3 | Reference |
|---|---|---|---|---|---|---|---|
| p.Asp6GlufsX32 (c.18_19delinsA) | Frameshift | na / na | na | Decreased | na | Disease causing | This study |
| p.Gly17Cys (c.49G>T) | Missense | 0.002 / 0.003 | + + + + − | Normal | Normal | Variant of uncertain significance | This study; DeLuca et al. 2010 |
| p.Ala50ProfsX9 (c.148delG) | Frameshift | na / na | na | Decreased | na | Disease causing | Ma et al. 2011 |
| p.Ala53dup (c.157_159dupGCC) | Polyalanine expansion | na / na | na | Normal | Normal | Variant of uncertain significance | This study; D’Alessandro et al. 2013 |
| p.Ala52_Ala53dup (c.154_159dupGCCGCC) | Polyalanine expansion | na / na | na | Normal | Normal | Variant of uncertain significance | Wessels et al. 2010 |
| p.Ser109Cys (c.326C>G) | Missense | na / 0.0002 | + + na − − | Normal | Abnormal | Likely disease associated | This study |
| p.Glu155X (c.463G>T) | Nonsense | na / na | na | Decreased | Abnormal | Disease causing | This study |
| p.Tyr199delfsX19 (c.595-610del TACCGCCCAGTGGCCA) | Frameshift | na / na | na | Decreased | Abnormal | Disease causing | This study |
| p.Pro217Ala (c.649C>G) | Missense | 0.003 / 0.008 | − − + − + | Normal | Abnormal | Likely disease associated | This study; Ware et al. 2004 |
| p.Ser252X (c.755C>A) | Nonsense | na / na | na | Decreased | Abnormal | Disease causing | This study |
| p.Glu291GlyfsX53 (c.866dupG) | Frameshift | na / na | na | Decreased | Abnormal | Disease causing | This study |
| p.His318Asn (c.952C>A) | Missense | na / na | + + + + − | Decreased | Abnormal | Disease causing | This study |
| p.Arg350Gly (c.1048A>G) | Missense | na / na | + + + + + | Decreased | Abnormal | Disease causing | D’Alessandro et al. 2011 |
| p.Ser402Pro (c.1204T>C) | Missense | na / na | + + + + + | Normal | Abnormal | Disease causing | Ma et al. 2011 |
| p.Ala447Gly (c.1340C>G) | Missense | na / na | + + + + − | Increased | Abnormal | Disease causing | This study |
MAFs represent allele frequencies for the minor allele (in all populations) from the 1000 Genomes Project (1KG, http://www.1000genomes.org/) and NHLBI Exome Sequencing Project (ESP, http://evs.gs.washington.edu/EVS/) databases. 1KG data encompass whole genome indels and SNP variants from 1092 individuals representing 14 world-wide populations (20110521-V3), while ESP data encompass exome SNP variants from 6503 individuals of European American and African American descent (ESP6500SI-V2).
Missense variant pathogenicity predictions were generated using (from left to right) Polyphen (genetics.bwh.harvard.edu/pph2/), SIFT (sift.jcvi.org/), PANTHER (http://www.pantherdb.org/tools/csnpScoreForm.jsp), Mutation Taster (http://www.mutationtaster.org/), and PMut (http://mmb2.pcb.ub.es:8080/PMut/) algorithms. Variants were predicted to be damaging (+), or benign/neutral (−). Analyses which were not possible are also indicated (na).
Based on clinical molecular diagnostic laboratory guidelines (see Materials and Methods)
DISCUSSION
To date, sequencing efforts have identified 23 novel mutations affecting ZIC3 coding regions (Stenson, et al., 2003; D’Alessandro, et al., 2013), establishing ZIC3 loss of function as an important cause of both familial (~75%) and sporadic (~1%) heterotaxy (Ware, et al., 2004). In the present study, we report sequencing results from an additional 440 patients with heterotaxy and/or heterotaxy-spectrum CHD, reassess mutation prevalence, and describe functional analyses for 15 previously uncharacterized ZIC3 variants, 8 of which were novel to our cohort.
In agreement with previous findings (Gebbia, et al., 1997; Ware, et al., 2004; De Luca, et al., 2010; Chung, et al., 2011; Ma, et al., 2012), ZIC3 variants were detected in both males and females with familial and sporadic disease. Notably, the observed 3.8% (10/264) incidence in sporadic cases was higher than expected based on our previous analyses of a similar patient population (Ware, et al., 2004). The majority of this increase is attributable to a higher than anticipated proportion of mutation carriers among males with sporadic disease (8/153 = 5.2%). These results have important implications for the diagnostic evaluation and risk-based counseling of sporadic heterotaxy.
Pathogenicity determinations and relevant data for all ZIC3 variants are summarized in Table 3 and reflect clinical molecular diagnostic laboratory standards for mutation interpretation (see Materials and Methods). Ten of the 15 analyzed ZIC3 variants were interpreted as disease causing mutations, including 4 missense mutations (His318Asn, Arg350Gly, Ser402Pro, Ala447Gly), 2 nonsense mutations (Glu155X, Ser252X), and 4 out-of-frame mutations predicted to cause premature protein truncation upstream of all zinc-finger domains (Asp6GlufsX32, Ala50ProfsX9, Tyr199delfsX19, Glu291GlyfsX53 ). Each mutation was absent from 1KG and ESP public databases, was predicted to be pathogenic by relevant predictive software algorithms, and disrupted ZIC3 subcellular localization and nuclear activity in vitro. Two additional missense variants (Ser109Cys, Pro217Ala) were identified as ultrarare variants incontrol populations and had abnormal in vitro functional results. Bioinformatic prediction programs differed in their prediction of pathogenicity. An increased prevalence of the Pro217Ala variant has been observed among obstensively healthy individuals of African American descent [MAFAA = 0.02 (EVS), 0.01 (1KG)]; however, our functional results suggest that Pro217Ala and Ser109Cys may act as susceptibility alleles alongside other genetic or environmental modifying factors. Lastly, one missense variant (Gly17Cys) and both polyalanine expansions (Ala53dup, Ala52_Ala53dup) were interpreted to be variants of uncertain significance based on lack of effect on tested functional parameters. Future experimentation will be required to definitively determine the pathogenic potential of these variants; however, support for potential pathogenicity of Gly17Cys is provided by the fact that Gly17Cys affects a highly conserved residue, was predicted to be functionally deleterious by 4/5 mutation pathogenicity programs, and is represented in <1% of 1KG and ESP control populations.
Although pathogenic mutations have been identified throughout the ZIC3 gene, most have clustered in the latter portion of exon 1, a sequence encoding 3 of 5 highly conserved Cys2His2 zinc-finger domains. Pathogenic significance of this DNA-binding region has been demonstrated by cell-based analyses of human heterotaxy mutations (Ware, et al., 2004; Hatayama, et al., 2008) and by observations of L-R patterning defects in Xenopus embryos expressing zinc-fingerless ZIC3 protein (Kitaguchi, et al., 2000). Furthermore, recent studies have identified NLS and NES signals overlapping amino acids in these regions (Bedard, et al., 2007; Hatayama, et al., 2008), indicating their importance for not only DNA-binding, but also ZIC3 subcellular trafficking.
In contrast to ‘classical’ NLSs, which comprise one or more short stretches of basic amino acids (reviewed in Lange, et al., 2007), the ZIC3 NLS is formed through juxtaposition of physically separated (interspersed) basic residues throughout the zinc-finger domain (Hatayama, et al., 2008). Similar interspersed NLSs have been described in other zinc-finger transcription factors (Pandya and Townes, 2002). Systematic mutagenesis of ZIC3 coding sequences has mapped the functional ZIC3 NLS to a long stretch of amino acids (292–356) in zinc-fingers 2 + 3 (ZF2+3) (Hatayama, et al., 2008) and to a bipartite sequence (367–382,403–412) in zinc-fingers 4 and 5 (ZF4+5) (Bedard, et al., 2007). Surprisingly, the ZF2+3 NLS was also found to overlap a cryptic CRM-1 dependent NES spanning amino acids 313–325 (Bedard, et al., 2007). Structural changes in ZIC3 protein structure, including those caused by heterotaxy mutations, can expose the export motif and facilitate cytoplasmic localization of properly imported ZIC3 protein (Bedard, et al., 2007).
We have previously reported significant decreases in ZIC3 nuclear localization and luciferase reporter transactivation for both missense (His286Arg, Thr323Met) and nonsense (Cys268X, Gln292X) mutations affecting the ZF2+3 NLS (Ware, et al., 2004). In the present study, a missense mutation in zinc-finger 2 (His318Asn), as well as several truncation mutants lacking these and all other downstream zinc-finger domains (Glu155X, Ser252X, Glu291GlyfsX53), resulted in similarly severe functional abnormalities. The His318Asn mutation uniquely maps to amino acids overlapping both the ZF2+3 NLS and the cryptic NES, which may account for the striking lack of nuclear ZIC3 in nearly 75% of His318Asn-transfected cells (Figure 3). In contrast, ZIC3 nuclear localization and luciferase transactivation levels were considerably less affected by non-truncating mutations in regions outside of NLS/NES regions, including those upstream of (Gly17Cys, Ala53dup, Ala52_Ala53dup, Ser109Cys, Pro217Ala), between (Ser402Pro), and downstream of (Ala447Gly) the ZF2+3 and ZF4+5 localization signals (Ware, et al., 2004, present study). Collectively, these analyses underline the functional relevance of ZIC3 mutations affecting the zinc-finger, NLS, and NES domains and lend support to heterotaxy pathogenesis models whereby altered subcellular trafficking of mutant ZIC3 prevents downstream transcriptional activation of as yet unidentified developmentally critical genes (Ware, et al., 2004; Bedard, et al., 2007; Hatayama, et al., 2008).
The N- and C-terminal domains of the ZIC3 protein are highly conserved, but have yet to be ascribed specific cellular functions. Only 9 N-terminal variants have been previously reported, including an out-of-frame deletion (Ala50ProfsX9) (Ma, et al., 2012), 3 polyalanine tract expansions (Wessels, et al., 2010; D’Alessandro, et al., 2013), a pair of nonsense mutations (Ser43X, Gln249X) (Ware, et al., 2004), and 3 missense mutations (Ala33Val, Gly17Cys, and Pro217Ala) (Ware, et al., 2004; De Luca, et al., 2010; D’Alessandro, et al., 2013). Three of these variants (Gly17Cys, Ala53dup, Pro217Ala) were additionally identified in patients from our cohort. We have significantly expanded this mutation spectrum through identification of 5 additional N-terminal variants (Asp6GlufsX32, Ser109Cys, Glu155X, Tyr199delfsX19, and Ser252X). Collectively, 7 of the 14 known N-terminal variants (Gly17Cys, Ala33Val, Ser109Cys, Pro217Ala, and the 3 polyalanine expansions) preserve the ZIC3 reading frame and, presumably, maintain structural and functional integrity of all downstream zinc-finger sites. Our functional analyses indicate that amino acids in the N-terminal domain, including those in the polyalanine tract, have little to no effect on ZIC3 subcellular trafficking and transcriptional activation. Although it remains a possibility that the identified N-terminal variants lack functional significance, high levels of amino acid conservation throughout the domain suggest the potential for alternative pathogenic mechanism(s). It is noteworthy that the polyalanine expansions, if definitely established as pathogenic, would represent the shortest disease-causing mutations of this type yet reported.
A comparison of phenotypic characteristics among ZIC3 mutation carriers and non-carriers in our heterotaxy and CHD cohort revealed few significant differences (Supp. Table S2), with the exception of an over-representation of pulmonary atresia in the ZIC3 mutation cohort (40% vs. 15.29%, p = 0.0215). In general, in-frame N-terminal ZIC3 variants were associated with isolated and sporadic CHD (Patients 2, 4, 6, 7, and 10). These findings echo previously published data for Gly17Cys and Pro217Ala variants (Ware, et al., 2004; De Luca, et al., 2010) and suggest potentially milder phenotypes for N-terminal mutations relative to those in zinc-finger regions, which are more often associated with familial heterotaxy, situs inversus, and/or other situs abnormalities (Gebbia, et al., 1997; Ware, et al., 2004; Chhin, et al., 2007; Wessels, et al., 2010; D’Alessandro, et al., 2011; Ma, et al., 2012, this study). These findings are also in agreement with existing in vitro data, which almost invariably indicates more severe abrogation of ZIC3 function by mutations in zinc-finger domains (Ware, et al., 2004; Hatayama, et al., 2008; D’Alessandro, et al., 2013; this study). Nevertheless, more severe phenotypes in 3 patients with N-terminal mutations and (Patients 3, 5, and 11) and recent identification of polyalanine tract expansions in patients with VACTERL and heterotaxy (Wessels, et al., 2010; D’Alessandro, et al., 2013) suggest that definitive genotype-phenotype correlations are unlikely to be so simplistic and may be influenced by other modifying factors. Future genetic and functional studies will undoubtedly help to further delineate the phenotypic and functional consequences of N-terminal variants, including those affecting the polyalanine tract.
We recently reported the existence of a second, shorter ZIC3 splice variant, which preferentially includes a downstream fourth exon at the expense of exon 3 (Bedard, et al., 2011). Consequently, the C-terminal domain is alternatively encoded by exon 3 (in ZIC3-A) or 4 (in ZIC3-B), depending on post-transcriptional splicing. Our mutation analyses represent the first broad scale sequencing efforts to include full sequencing coverage of exon 4, although no ZIC3-B specific mutations were identified in our cohort. We did, however, identify a novel C to G transversion in exon 3, resulting in substitution of a highly conserved alanine for a glycine (Ala447Gly) in a female patient with isolated CHD. This mutation uniquely and reproducibly increased activation of the luciferase reporter, despite a 10% decrease in cells with nuclear localization of ZIC3 (Figure 2). As of the time of writing, no other C-terminal missense mutations have been reported, precluding genotype-phenotype comparisons. The functional relationship between ZIC3-A and ZIC3-B remains uncertain, with both isoforms expressed in similar tissues and at similar time-points during embryonic development (Bedard, 2011). Nevertheless, although both ZIC3-A and ZIC3-B are capable of transcriptionally activating a Gli-binding site reporter, only ZIC3-A does so synergistically with Gli3, suggesting some level of functional distinction between the two isoforms (Bedard, 2011). Because all subcellular localization and luciferase reporter analyses were carried out using the ZIC3-A ORF, we cannot exclude the possibility that individual mutations may have different effects on ZIC3-B function. Future analyses utilizing both isoforms will, consequently, be beneficial in determining the pathophysiological relevance N- and C-terminal variants of uncertain functional impact, as well as the extent to which activating mutations like Ala447Gly contribute to CHD and heterotaxy.
In summary, we have described in vitro functional significance of 15 ZIC3 variants, including 8 novel variants identified in a cohort of 440 patients with heterotaxy and related CHD. Twelve of these 15 variants were considered to be disease causing or disease associated, while the significance of 3 variants, including both analyzed polyalanine tract expansions, remains uncertain. Collectively, these analyses greatly expand the known ZIC3 mutation spectrum, particularly in the relatively less characterized N-terminal protein domain, and suggest that ZIC3 mutations are more common in sporadic heterotaxy than previously anticipated. This information is essential for providing accurate risk-based counseling to families with heterotaxy and associated cardiovascular disease. Finally, our data confirm the importance of the zinc-finger domains for ZIC3 cellular trafficking and nuclear function, and suggest the potential existence of alternative pathogenic mechanisms for variants affecting N- and C-terminal regions, including the polyalanine tract.
Supplementary Material
Acknowledgments
We thank the patients and families for their participation. The authors have no conflicts to disclose.
Grant Sponsors: This work was supported by National Institutes of Health grants RO1 HL088639 and HL088639-03S1 (to S.M.W.) and the Burroughs Wellcome Clinical Scientist Awards in Translational Research #1008496 (to S.M.W.).
Footnotes
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
References
- Aruga J, Kamiya A, Takahashi H, Fujimi TJ, Shimizu Y, Ohkawa K, Yazawa S, Umesono Y, Noguchi H, Shimizu T, et al. A wide-range phylogenetic analysis of Zic proteins: implications for correlations between protein structure conservation and body plan complexity. Genomics. 2006;87:783–92. doi: 10.1016/j.ygeno.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Bedard JE, Purnell JD, Ware SM. Nuclear import and export signals are essential for proper cellular trafficking and function of ZIC3. Hum Mol Genet. 2007;16:187–98. doi: 10.1093/hmg/ddl461. [DOI] [PubMed] [Google Scholar]
- Bedard JE, Haaning AM, Ware SM. Identification of a novel ZIC3 isoform and mutation screening in patients with heterotaxy and congenital heart disease. PLoS One. 2011;6:e23755. doi: 10.1371/journal.pone.0023755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont JW, Mohapatra B, Towbin JA, Ware SM. Molecular genetics of heterotaxy syndromes. Curr Opin Cardiol. 2004;19:216–20. doi: 10.1097/00001573-200405000-00005. [DOI] [PubMed] [Google Scholar]
- Casey B, Devoto M, Jones KL, Ballabio A. Mapping a gene for familial situs abnormalities to human chromosome Xq24–q27.1. Nat Genet. 1993;5:403–7. doi: 10.1038/ng1293-403. [DOI] [PubMed] [Google Scholar]
- Cast AE, Gao C, Amack JD, Ware SM. An essential and highly conserved role for Zic3 in left-right patterning, gastrulation and convergent extension morphogenesis. Dev Biol. 2012;364:22–31. doi: 10.1016/j.ydbio.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhin B, Hatayama M, Bozon D, Ogawa M, Schon P, Tohmonda T, Sassolas F, Aruga J, Valard AG, Chen SC, et al. Elucidation of penetrance variability of a ZIC3 mutation in a family with complex heart defects and functional analysis of ZIC3 mutations in the first zinc finger domain. Hum Mutat. 2007;28:563–70. doi: 10.1002/humu.20480. [DOI] [PubMed] [Google Scholar]
- Chung B, Shaffer LG, Keating S, Johnson J, Casey B, Chitayat D. From VACTERL-H to heterotaxy: variable expressivity of ZIC3-related disorders. Am J Med Genet A. 2011;155A:1123–8. doi: 10.1002/ajmg.a.33859. [DOI] [PubMed] [Google Scholar]
- D’Alessandro LC, Casey B, Siu VM. Situs Inversus Totalis and a Novel ZIC3 Mutation in a Family with X-linked Heterotaxy. Congenit Heart Dis. 2011 doi: 10.1111/j.1747-0803.2011.00602.x. [DOI] [PubMed] [Google Scholar]
- D’Alessandro LC, Latney BC, Paluru PC, Goldmuntz E. The phenotypic spectrum of ZIC3 mutations includes isolated d-transposition of the great arteries and double outlet right ventricle. Am J Med Genet A. 2013;161:792–802. doi: 10.1002/ajmg.a.35849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca A, Sarkozy A, Consoli F, Ferese R, Guida V, Dentici ML, Mingarelli R, Bellacchio E, Tuo G, Limongelli G, et al. Familial transposition of the great arteries caused by multiple mutations in laterality genes. Heart. 2010;96:673–7. doi: 10.1136/hrt.2009.181685. [DOI] [PubMed] [Google Scholar]
- Gebbia M, Ferrero GB, Pilia G, Bassi MT, Aylsworth A, Penman-Splitt M, Bird LM, Bamforth JS, Burn J, Schlessinger D, et al. X-linked situs abnormalities result from mutations in ZIC3. Nat Genet. 1997;17:305–8. doi: 10.1038/ng1197-305. [DOI] [PubMed] [Google Scholar]
- Hatayama M, Tomizawa T, Sakai-Kato K, Bouvagnet P, Kose S, Imamoto N, Yokoyama S, Utsunomiya-Tate N, Mikoshiba K, Kigawa T, et al. Functional and structural basis of the nuclear localization signal in the ZIC3 zinc finger domain. Hum Mol Genet. 2008;17:3459–73. doi: 10.1093/hmg/ddn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman GE, El-Hodiri HM. The role of ZIC3 in vertebrate development. Cytogenet Genome Res. 2002;99:229–35. doi: 10.1159/000071598. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Zhu L, Hu L, Slesnick TC, Pautler RG, Justice MJ, Belmont JW. Zic3 is required in the extra-cardiac perinodal region of the lateral plate mesoderm for left-right patterning and heart development. Hum Mol Genet. 2013;22:879–89. doi: 10.1093/hmg/dds494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaguchi T, Nagai T, Nakata K, Aruga J, Mikoshiba K. Zic3 is involved in the left-right specification of the Xenopus embryo. Development. 2000;127:4787–95. doi: 10.1242/dev.127.22.4787. [DOI] [PubMed] [Google Scholar]
- Klootwijk R, Franke B, van der Zee CE, de Boer RT, Wilms W, Hol FA, Mariman EC. A deletion encompassing Zic3 in bent tail, a mouse model for X-linked neural tube defects. Hum Mol Genet. 2000;9:1615–22. doi: 10.1093/hmg/9.11.1615. [DOI] [PubMed] [Google Scholar]
- Koyabu Y, Nakata K, Mizugishi K, Aruga J, Mikoshiba K. Physical and functional interactions between Zic and Gli proteins. J Biol Chem. 2001;276:6889–92. doi: 10.1074/jbc.C000773200. [DOI] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LS, Hong FH, Kunarso G, Stanton LW. The pluripotency regulator Zic3 is a direct activator of the Nanog promoter in ESCs. Stem Cells. 2010;28:1961–9. doi: 10.1002/stem.527. [DOI] [PubMed] [Google Scholar]
- Ma L, Selamet Tierney ES, Lee T, Lanzano P, Chung WK. Mutations in ZIC3 and ACVR2B are a common cause of heterotaxy and associated cardiovascular anomalies. Cardiol Young. 2012;22:194–201. doi: 10.1017/S1047951111001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AR, Merzdorf CS. Expression of the zic1, zic2, zic3, and zic4 genes in early chick embryos. BMC Res Notes. 2010;3:167. doi: 10.1186/1756-0500-3-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megarbane A, Salem N, Stephan E, Ashoush R, Lenoir D, Delague V, Kassab R, Loiselet J, Bouvagnet P. X-linked transposition of the great arteries and incomplete penetrance among males with a nonsense mutation in ZIC3. Eur J Hum Genet. 2000;8:704–8. doi: 10.1038/sj.ejhg.5200526. [DOI] [PubMed] [Google Scholar]
- Messaed C, Rouleau GA. Molecular mechanisms underlying polyalanine diseases. Neurobiol Dis. 2009;34:397–405. doi: 10.1016/j.nbd.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Mizugishi K, Aruga J, Nakata K, Mikoshiba K. Molecular properties of Zic proteins as transcriptional regulators and their relationship to GLI proteins. J Biol Chem. 2001;276:2180–8. doi: 10.1074/jbc.M004430200. [DOI] [PubMed] [Google Scholar]
- Nagai T, Aruga J, Takada S, Gunther T, Sporle R, Schughart K, Mikoshiba K. The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev Biol. 1997;182:299–313. doi: 10.1006/dbio.1996.8449. [DOI] [PubMed] [Google Scholar]
- Nakata K, Nagai T, Aruga J, Mikoshiba K. Xenopus Zic3, a primary regulator both in neural and neural crest development. Proc Natl Acad Sci U S A. 1997;94:11980–5. doi: 10.1073/pnas.94.22.11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya K, Townes TM. Basic residues within the Kruppel zinc finger DNA binding domains are the critical nuclear localization determinants of EKLF/KLF-1. J Biol Chem. 2002;277:16304–12. doi: 10.1074/jbc.M200866200. [DOI] [PubMed] [Google Scholar]
- Purandare SM, Ware SM, Kwan KM, Gebbia M, Bassi MT, Deng JM, Vogel H, Behringer RR, Belmont JW, Casey B. A complex syndrome of left-right axis, central nervous system and axial skeleton defects in Zic3 mutant mice. Development. 2002;129:2293–302. doi: 10.1242/dev.129.9.2293. [DOI] [PubMed] [Google Scholar]
- Quinn ME, Haaning A, Ware SM. Preaxial polydactyly caused by Gli3 haploinsufficiency is rescued by Zic3 loss of function in mice. Hum Mol Genet. 2012;21:1888–96. doi: 10.1093/hmg/dds002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai-Kato K, Ishiguro A, Mikoshiba K, Aruga J, Utsunomiya-Tate N. CD spectra show the relational style between Zic-, Gli-, Glis-zinc finger protein and DNA. Biochim Biophys Acta. 2008;1784:1011–9. doi: 10.1016/j.bbapap.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577–81. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- Sutherland MJ, Ware SM. Disorders of left-right asymmetry: heterotaxy and situs inversus. Am J Med Genet C Semin Med Genet. 2009;151C:307–17. doi: 10.1002/ajmg.c.30228. [DOI] [PubMed] [Google Scholar]
- Sutherland MJ, Wang S, Quinn ME, Haaning A, Ware SM. Zic3 is required in the migrating primitive streak for node morphogenesis and left-right patterning. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzschach A, Hoeltzenbein M, Hoffmann K, Menzel C, Beyer A, Ocker V, Wurster G, Raynaud M, Ropers HH, Kalscheuer V, et al. Heterotaxy and cardiac defect in a girl with chromosome translocation t(X;1)(q26;p13.1) and involvement of ZIC3. Eur J Hum Genet. 2006;14:1317–20. doi: 10.1038/sj.ejhg.5201707. [DOI] [PubMed] [Google Scholar]
- Vandenberg LN, Levin M. Far from solved: a perspective on what we know about early mechanisms of left-right asymmetry. Dev Dyn. 2010;239:3131–46. doi: 10.1002/dvdy.22450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale E, Brancolini V, De Rienzo A, Bird L, Allada V, Sklansky M, Chae CU, Ferrero GB, Weber J, Devoto M, et al. Suggestive linkage of situs inversus and other left-right axis anomalies to chromosome 6p. J Med Genet. 2001;38:182–5. doi: 10.1136/jmg.38.3.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware SM, Peng J, Zhu L, Fernbach S, Colicos S, Casey B, Towbin J, Belmont JW. Identification and functional analysis of ZIC3 mutations in heterotaxy and related congenital heart defects. Am J Hum Genet. 2004;74:93–105. doi: 10.1086/380998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware SM, Harutyunyan KG, Belmont JW. Zic3 is critical for early embryonic patterning during gastrulation. Dev Dyn. 2006a;235:776–85. doi: 10.1002/dvdy.20668. [DOI] [PubMed] [Google Scholar]
- Ware SM, Harutyunyan KG, Belmont JW. Heart defects in X-linked heterotaxy: evidence for a genetic interaction of Zic3 with the nodal signaling pathway. Dev Dyn. 2006b;235:1631–7. doi: 10.1002/dvdy.20719. [DOI] [PubMed] [Google Scholar]
- Wessels MW, Kuchinka B, Heydanus R, Smit BJ, Dooijes D, de Krijger RR, Lequin MH, de Jong EM, Husen M, Willems PJ, et al. Polyalanine expansion in the ZIC3 gene leading to X-linked heterotaxy with VACTERL association: a new polyalanine disorder? J Med Genet. 2010;47:351–5. doi: 10.1136/jmg.2008.060913. [DOI] [PubMed] [Google Scholar]
- Zhu L, Belmont JW, Ware SM. Genetics of human heterotaxias. Eur J Hum Genet. 2006;14:17–25. doi: 10.1038/sj.ejhg.5201506. [DOI] [PubMed] [Google Scholar]
- Zhu L, Peng JL, Harutyunyan KG, Garcia MD, Justice MJ, Belmont JW. Craniofacial, skeletal, and cardiac defects associated with altered embryonic murine Zic3 expression following targeted insertion of a PGK-NEO cassette. Front Biosci. 2007a;12:1680–90. doi: 10.2741/2180. [DOI] [PubMed] [Google Scholar]
- Zhu L, Harutyunyan KG, Peng JL, Wang J, Schwartz RJ, Belmont JW. Identification of a novel role of ZIC3 in regulating cardiac development. Hum Mol Genet. 2007b;16:1649–60. doi: 10.1093/hmg/ddm106. [DOI] [PubMed] [Google Scholar]
- Zhu L, Zhou G, Poole S, Belmont JW. Characterization of the interactions of human ZIC3 mutants with GLI3. Hum Mutat. 2008;29:99–105. doi: 10.1002/humu.20606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.