Abstract
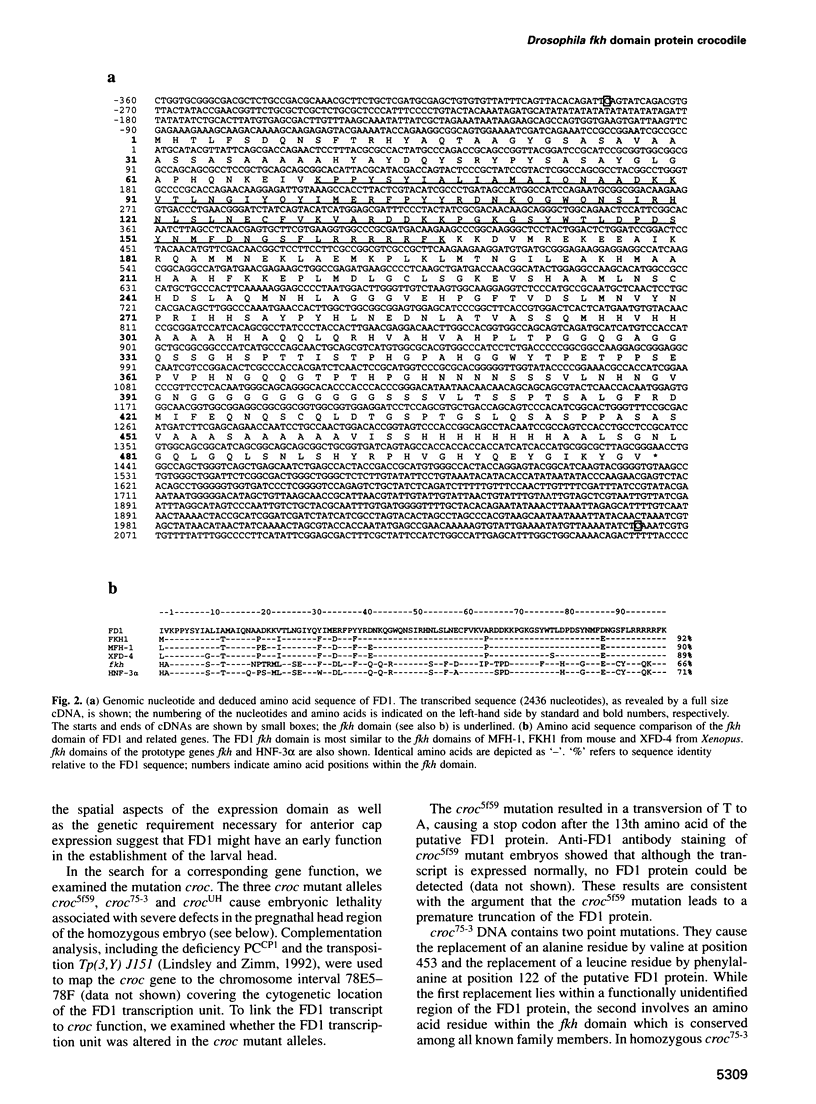
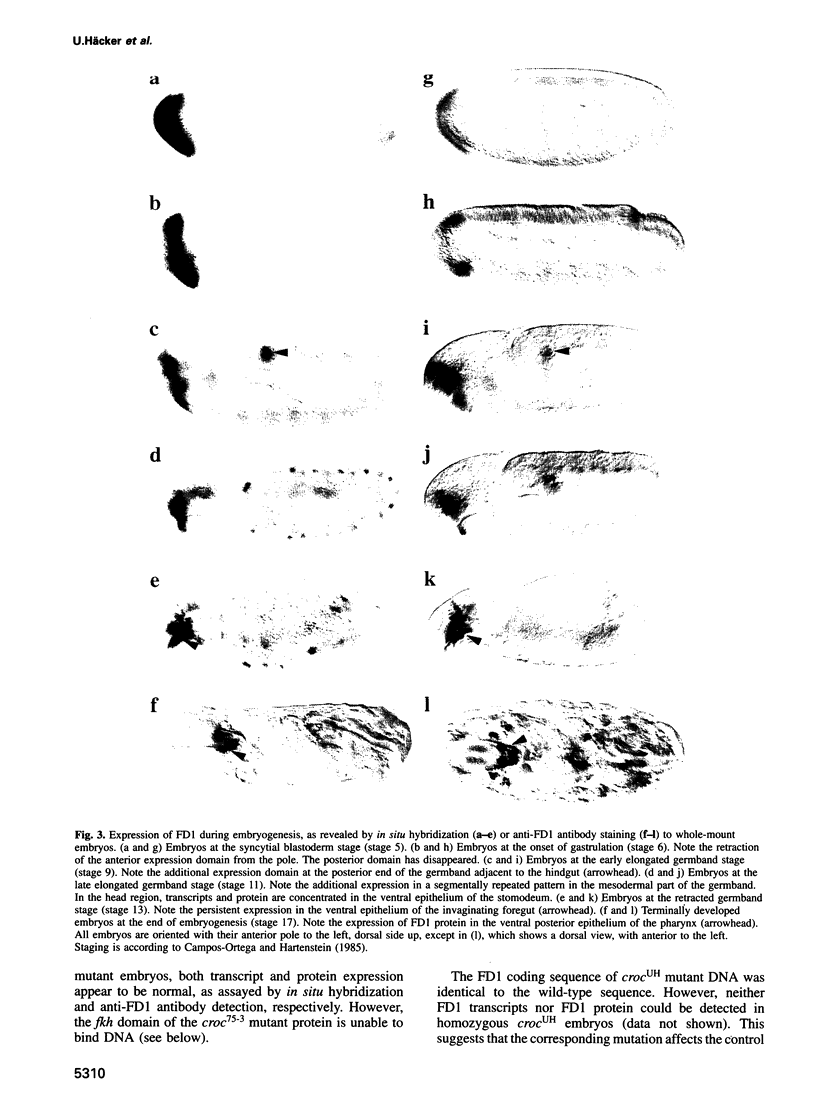
The fork head (fkh) domain defines the DNA-binding region of a family of transcription factors which has been implicated in regulating cell fate decisions across species lines. We have cloned and molecularly characterized the crocodile (croc) gene which encodes a new family member from Drosophila. croc is expressed in the head anlagen of the blastoderm embryo under the control of the anterior, the dorsoventral and the terminal maternal organizer systems. The croc mutant phenotype indicates that the croc wild-type gene is required to function as an early patterning gene in the anterior-most blastoderm head segment anlage and for the establishment of a specific head skeletal structure that derives from the non-adjacent intercalary segment at a later stage of embryogenesis. As an early patterning gene, croc exerts unusual properties which do not allow it to be grouped among the established segmentation genes. A single-site mutation within the croc fkh domain, which causes a replacement of the first out of four conserved amino acid residues thought to be involved in the coordinate binding of Mg2+, abolishes the DNA binding of the protein in vitro. In view of the resulting lack-of-function mutant phenotype, it appears likely that metal binding by the affected region of the fkh domain is crucial for proper folding of the DNA-binding structure.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987 Sep;101(1):1–22. [PubMed] [Google Scholar]
- Ang S. L., Rossant J. HNF-3 beta is essential for node and notochord formation in mouse development. Cell. 1994 Aug 26;78(4):561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990 Nov 23;250(4984):1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Brown N. H., Kafatos F. C. Functional cDNA libraries from Drosophila embryos. J Mol Biol. 1988 Sep 20;203(2):425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- Brönner G., Chu-LaGraff Q., Doe C. Q., Cohen B., Weigel D., Taubert H., Jäckle H. Sp1/egr-like zinc-finger protein required for endoderm specification and germ-layer formation in Drosophila. Nature. 1994 Jun 23;369(6482):664–668. doi: 10.1038/369664a0. [DOI] [PubMed] [Google Scholar]
- Clark K. L., Halay E. D., Lai E., Burley S. K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993 Jul 29;364(6436):412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Clevidence D. E., Overdier D. G., Tao W., Qian X., Pani L., Lai E., Costa R. H. Identification of nine tissue-specific transcription factors of the hepatocyte nuclear factor 3/forkhead DNA-binding-domain family. Proc Natl Acad Sci U S A. 1993 May 1;90(9):3948–3952. doi: 10.1073/pnas.90.9.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S., Jürgens G. Drosophila headlines. Trends Genet. 1991 Aug;7(8):267–272. doi: 10.1016/0168-9525(91)90327-M. [DOI] [PubMed] [Google Scholar]
- Dirksen M. L., Jamrich M. A novel, activin-inducible, blastopore lip-specific gene of Xenopus laevis contains a fork head DNA-binding domain. Genes Dev. 1992 Apr;6(4):599–608. doi: 10.1101/gad.6.4.599. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring W. J. Homeo boxes in the study of development. Science. 1987 Jun 5;236(4806):1245–1252. doi: 10.1126/science.2884726. [DOI] [PubMed] [Google Scholar]
- Gogos J. A., Hsu T., Bolton J., Kafatos F. C. Sequence discrimination by alternatively spliced isoforms of a DNA binding zinc finger domain. Science. 1992 Sep 25;257(5078):1951–1955. doi: 10.1126/science.1290524. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U., Cadigan K. M., Gehring W. J. Three maternal coordinate systems cooperate in the patterning of the Drosophila head. Development. 1994 Nov;120(11):3155–3171. doi: 10.1242/dev.120.11.3155. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U., Pearson R. K., Gehring W. J. The Drosophila sloppy paired locus encodes two proteins involved in segmentation that show homology to mammalian transcription factors. Genes Dev. 1992 Jun;6(6):1030–1051. doi: 10.1101/gad.6.6.1030. [DOI] [PubMed] [Google Scholar]
- Hoch M., Jäckle H. Transcriptional regulation and spatial patterning in Drosophila. Curr Opin Genet Dev. 1993 Aug;3(4):566–573. doi: 10.1016/0959-437x(93)90092-4. [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Roeder R. G. Purification of his-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res. 1991 Nov 25;19(22):6337–6338. doi: 10.1093/nar/19.22.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häcker U., Grossniklaus U., Gehring W. J., Jäckle H. Developmentally regulated Drosophila gene family encoding the fork head domain. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8754–8758. doi: 10.1073/pnas.89.18.8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kaestner K. H., Lee K. H., Schlöndorff J., Hiemisch H., Monaghan A. P., Schütz G. Six members of the mouse forkhead gene family are developmentally regulated. Proc Natl Acad Sci U S A. 1993 Aug 15;90(16):7628–7631. doi: 10.1073/pnas.90.16.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G. H., Spradling A. C. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics. 1992 Nov;132(3):737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann E., Müller D., Knöchel W. DNA recognition site analysis of Xenopus winged helix proteins. J Mol Biol. 1995 Apr 28;248(2):239–254. doi: 10.1016/s0022-2836(95)80047-6. [DOI] [PubMed] [Google Scholar]
- Knöchel S., Lef J., Clement J., Klocke B., Hille S., Köster M., Knöchel W. Activin A induced expression of a fork head related gene in posterior chordamesoderm (notochord) of Xenopus laevis embryos. Mech Dev. 1992 Aug;38(2):157–165. doi: 10.1016/0925-4773(92)90007-7. [DOI] [PubMed] [Google Scholar]
- Lai E., Prezioso V. R., Smith E., Litvin O., Costa R. H., Darnell J. E., Jr HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 1990 Aug;4(8):1427–1436. doi: 10.1101/gad.4.8.1427. [DOI] [PubMed] [Google Scholar]
- Lewin B. Commitment and activation at pol II promoters: a tail of protein-protein interactions. Cell. 1990 Jun 29;61(7):1161–1164. doi: 10.1016/0092-8674(90)90675-5. [DOI] [PubMed] [Google Scholar]
- Li J., Vogt P. K. The retroviral oncogene qin belongs to the transcription factor family that includes the homeotic gene fork head. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4490–4494. doi: 10.1073/pnas.90.10.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald P. M., Struhl G. A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature. 1986 Dec 11;324(6097):537–545. doi: 10.1038/324537a0. [DOI] [PubMed] [Google Scholar]
- Miura N., Wanaka A., Tohyama M., Tanaka K. MFH-1, a new member of the fork head domain family, is expressed in developing mesenchyme. FEBS Lett. 1993 Jul 12;326(1-3):171–176. doi: 10.1016/0014-5793(93)81785-x. [DOI] [PubMed] [Google Scholar]
- Nehls M., Pfeifer D., Schorpp M., Hedrich H., Boehm T. New member of the winged-helix protein family disrupted in mouse and rat nude mutations. Nature. 1994 Nov 3;372(6501):103–107. doi: 10.1038/372103a0. [DOI] [PubMed] [Google Scholar]
- O'Connell P. O., Rosbash M. Sequence, structure, and codon preference of the Drosophila ribosomal protein 49 gene. Nucleic Acids Res. 1984 Jul 11;12(13):5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overdier D. G., Porcella A., Costa R. H. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol Cell Biol. 1994 Apr;14(4):2755–2766. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrou S., Hellqvist M., Samuelsson L., Enerbäck S., Carlsson P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J. 1994 Oct 17;13(20):5002–5012. doi: 10.1002/j.1460-2075.1994.tb06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F., Baldarelli R. M., Steingrímsson E., Diaz R. J., Patapoutian A., Merriam J. R., Lengyel J. A. The Drosophila gene tailless is expressed at the embryonic termini and is a member of the steroid receptor superfamily. Cell. 1990 Jul 13;62(1):151–163. doi: 10.1016/0092-8674(90)90249-e. [DOI] [PubMed] [Google Scholar]
- Roeder R. G. The complexities of eukaryotic transcription initiation: regulation of preinitiation complex assembly. Trends Biochem Sci. 1991 Nov;16(11):402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- Ronchi E., Treisman J., Dostatni N., Struhl G., Desplan C. Down-regulation of the Drosophila morphogen bicoid by the torso receptor-mediated signal transduction cascade. Cell. 1993 Jul 30;74(2):347–355. doi: 10.1016/0092-8674(93)90425-p. [DOI] [PubMed] [Google Scholar]
- Rothe M., Pehl M., Taubert H., Jäckle H. Loss of gene function through rapid mitotic cycles in the Drosophila embryo. Nature. 1992 Sep 10;359(6391):156–159. doi: 10.1038/359156a0. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A., Jessell T. M. Pintallavis, a gene expressed in the organizer and midline cells of frog embryos: involvement in the development of the neural axis. Development. 1992 Sep;116(1):81–93. doi: 10.1242/dev.116.Supplement.81. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A., Prezioso V. R., Darnell J. E., Jessell T. M. Sequential expression of HNF-3 beta and HNF-3 alpha by embryonic organizing centers: the dorsal lip/node, notochord and floor plate. Mech Dev. 1993 Dec;44(2-3):91–108. doi: 10.1016/0925-4773(93)90060-b. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Ott U., Technau G. M. Expression of en and wg in the embryonic head and brain of Drosophila indicates a refolded band of seven segment remnants. Development. 1992 Sep;116(1):111–125. doi: 10.1242/dev.116.1.111. [DOI] [PubMed] [Google Scholar]
- Scott M. P., Tamkun J. W., Hartzell G. W., 3rd The structure and function of the homeodomain. Biochim Biophys Acta. 1989 Jul 28;989(1):25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Simpson-Brose M., Treisman J., Desplan C. Synergy between the hunchback and bicoid morphogens is required for anterior patterning in Drosophila. Cell. 1994 Sep 9;78(5):855–865. doi: 10.1016/s0092-8674(94)90622-x. [DOI] [PubMed] [Google Scholar]
- Sprenger F., Nüsslein-Volhard C. Torso receptor activity is regulated by a diffusible ligand produced at the extracellular terminal regions of the Drosophila egg. Cell. 1992 Dec 11;71(6):987–1001. doi: 10.1016/0092-8674(92)90394-r. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Nüsslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 1992 Jan 24;68(2):201–219. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- Strähle U., Blader P., Henrique D., Ingham P. W. Axial, a zebrafish gene expressed along the developing body axis, shows altered expression in cyclops mutant embryos. Genes Dev. 1993 Jul;7(7B):1436–1446. doi: 10.1101/gad.7.7b.1436. [DOI] [PubMed] [Google Scholar]
- Tautz D., Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989 Aug;98(2):81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Tjian R., Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994 Apr 8;77(1):5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- Volk T., VijayRaghavan K. A central role for epidermal segment border cells in the induction of muscle patterning in the Drosophila embryo. Development. 1994 Jan;120(1):59–70. doi: 10.1242/dev.120.1.59. [DOI] [PubMed] [Google Scholar]
- Weigel D., Jäckle H. The fork head domain: a novel DNA binding motif of eukaryotic transcription factors? Cell. 1990 Nov 2;63(3):455–456. doi: 10.1016/0092-8674(90)90439-l. [DOI] [PubMed] [Google Scholar]
- Weigel D., Jürgens G., Küttner F., Seifert E., Jäckle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989 May 19;57(4):645–658. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- Weinstein D. C., Ruiz i Altaba A., Chen W. S., Hoodless P., Prezioso V. R., Jessell T. M., Darnell J. E., Jr The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell. 1994 Aug 26;78(4):575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]