Abstract
Myc oncoproteins induce genes driving aerobic glycolysis, including lactate dehydrogenase-A that generates lactate. Here we report that Myc controls transcription of the lactate transporter SLC16A1/MCT1, and that elevated MCT1 levels are manifest in premalignant and neoplastic Eμ-Myc transgenic B cells and in human malignancies with MYC or MYCN involvement. Notably, disrupting MCT1 function leads to an accumulation of intracellular lactate that rapidly disables tumor cell growth and glycolysis, provoking marked alterations in glycolytic intermediates, and reductions in glucose transport, and in levels of ATP, NADPH and glutathione. Reductions in glutathione then lead to increases in hydrogen peroxide, mitochondrial damage and, ultimately, cell death. Finally, forcing glycolysis by metformin treatment augments this response and the efficacy of MCT1 inhibitors, suggesting an attractive combination therapy for MYC/MCT1-expressing malignancies.
Introduction
Myc oncogenic transcription factors are activated in a large cast of human malignancies, and it has been estimated that that 100,000 deaths/year are associated with deregulated MYC expression (1). Myc drives continuous cell growth and division, which triggers DNA damage and apoptotic checkpoints that are then bypassed by mutations that lead to frank malignancy. Accordingly, forced expression of Myc is sufficient to provoke tumor formation in mouse models of human cancer (1, 2). Further, Myc is required to sustain the malignant state, as Myc inactivation usually provokes rapid tumor regression (3, 4).
Myc oncoproteins are basic-helix-loop-helix-leucine zipper (bHLH-Zip) transcription factors that regulate a large cast of targets to coordinate cell growth, metabolism and division (5). Myc functions require dimerization with the related bHLH-Zip partner Max, and Myc:Max dimers activate target genes by binding to E-box elements (CACGTG) (6). In addition, Myc represses transcription via inhibitory interactions with the transcriptional activator Miz-1 (7, 8). Finally, Myc oncoproteins have recently been suggested to function as universal amplifiers of active genes (9, 10), which may occur through their ability to recruit histone modifying enzymes (11) and/or by occupying pre-existing open chromatin and promoting transcription or pause release at promoters loaded with RNA polymerase II (9, 10).
Prominent targets induced by Myc include a cast of metabolic enzymes, some of which drive aerobic glycolysis, a hallmark of cancer cells (12, 13). Indeed, in cell culture models Myc oncoproteins induce many aspects of cancer cell metabolism, where Myc targets include glucose and glutamine transporters, glutaminase and glycolytic enzymes, including lactate dehydrogenase-A (LDH-A) (14–17).
Increased glycolytic flux in cancer cells leads to high levels of lactate that is exported by proton-dependent twelve-membrane pass monocarboxylic acid solute transporters coined MCT1-4 (18). Cell surface expression of MCT1 and MCT4 requires co-expression of the immunoglobulin-like molecule CD147 (19). While MCT4 transcription is regulated by hypoxia inducible factor-1α (20) and in renal clear cell cancer by promoter methylation (21), much less is known regarding the control of MCT1 transcription, other than MCT1 expression is elevated in Myc-expressing MCF10 breast epithelial cells and in some tumors (22).
Blocking lactate transport impairs tumor cell growth through several mechanisms. First, blocking lactate export leads to an accumulation of lactic acid and decreases intracellular pH (23). This response appears to contribute to growth arrest of Ras-transformed fibroblasts triggered by MCT1 inhibitors and to the effects of CD147 knockdown on tumor xenografts (24). Second, some tumor cells rely on lactate as a substrate for oxidative phosphorylation, and in this scenario blocking lactate import inhibits tumor cell growth (25, 26). However, this is the exception as most tumor cells express high levels of LDH-A, which drives the production of lactate from pyruvate (27). Finally, lactate uptake in vascular endothelial cells via MCT1 appears to promote tumor angiogenesis; thus blocking this response impairs tumorigenesis (28, 29). Given these effects, a recent AstraZeneca patent application claims the use of MCT1 inhibitors for the treatment of certain cancers (30).
Given their effects on aerobic glycolysis, we reasoned that Myc oncoproteins would control lactate transport. Here we report that Myc directly and selectively activates MCT1 transcription and that elevated MCT1 levels are a hallmark of human malignancies with MYC or MYCN involvement. Notably, we show that blocking MCT1 function rapidly disables glycolysis, leading to reductions in ATP and glutathione levels, and that co-treatment with metformin, which forces the glycolytic phenotype, augments the in vivo efficacy of MCT1 inhibitors against MYC-expressing malignances.
Materials and Methods
RNA analyses
Total RNA was prepared from B220+ B cells of the spleens or bone marrow (BM) of 4–6 week old wild type and premalignant Eμ-Myc littermates. Lymphomas were from individual animals. B cells were purified using magnetic-activated cell sorting and beads conjugated with antibodies to B220 (Miltenyi). RNA was isolated from purified B220+ B cells or tumors using RNA shredder and RNA easy kits (Qiagen) or the NucleoSpin RNA II kit (Macherey-Nagel).For qRT-PCR analyses cDNA was synthesized using iScript reverse transcriptase (BioRad), amplified with Perfecta Sybr Green (Quanta) and measured using the iCycler real-time PCR machine (BioRad). Experimental Ct values were normalized with ubiquitin Ct values and were expressed relative to wild type Ct values. Sequences of primers are provided upon request.
Proliferation, lactate and ATP assays
Proliferation was measured by counting cells daily or by MTT assay (Millipore). Lactate was measured using lactate assay kits (A-108, University of Buffalo and K607-100, BioVision) according to the manufacturers’ instructions. Cells cultured at 5 × 105 cells/ml +/− MCT1 inhibitors (100 nM) and/or metformin (1 mM) for 24 hr were washed twice in cold PBS, lysed in 100-µl cold water and lactate concentrations measured from the lysate. ATP was measured using an ATP detection kit (Perkin Elmer) according to the manufacturers’ instructions. Raji cells (5 × 104/well in 96 well plate) were treated with inhibitors (100 nM) for indicated times before assay.
siRNA knockdown
MCF7 cells (2.5 × 105 cells/well) were transfected with 50–100 nM siRNA On-TARGETplus SMARTpool siRNA against hSLC16a1 (L-007402-00), c-Myc (L-003282-00-0005) or control siRNA (D-001810-01-05) from Thermo Scientific together with 3 µl Lipofectamine-2000 (Invitrogen 11668-027) into a six well dish. Cells were incubated for 24 hr before plating for growth assays, metabolic assays, and protein and RNA isolation. For growth assays cells were plated at 10,000 cells/well into 24-well dishes and three wells/day were counted.
Metabolic analysis
ECAR and OCR were measured using the Seahorse Bioscience XF96 Analyzer. Before each measurement wells were mixed for 2 min followed by analyte measurements every 22 sec for 5 min. Each data point is the change in analyte concentration over 5 min and is reported as extracellular acidification rate (ECAR, mpH/min) and oxygen consumption rate (OCR, pMoles oxygen/min). Agents were injected into wells through pre-loaded reagent delivery chambers in the sensor probe. Final concentration of agents used was: MCT1 inhibitors 10 nM-1µM, Oligomycin 1µM (Calbiochem), FCCP 300 nM (Sigma) and Rotenone 100 nM (Sigma). Raji, Daudi and Ramos BL cells (60,000 cells/well) were adhered to poly-D-lysine coated 96-well plates by centrifuging. MCF7 cells were plated at 15,000 cells/well the day before analysis. Protein concentration in wells was measured to normalize ECAR and OCR data.
Glutathione measurements and glutathione rescue
Total glutathione (GSH/GSSG) was measured according the manufacturer’s protocol (Cayman’s Glutathione Assay Kit) from Raji cells treated with vehicle or SR13800 (1 µM) for 8 hr. Raji cells (2 × 105 cells/ml), cultured in RPMI with FBS, were pre-treated with glutathione reduced ethyl ester (3–5 mM, Sigma) for 1 hr followed by treatment with SR13800 (1 µM) and metformin (1 mM). H2O2 was measured by H2DCFDA. Viability was measured by trypan blue dye exclusion. Proliferation was measured by MTT assay (Millipore) 3 days after treatment.
Results
Myc coordinates the control of lactate homeostasis
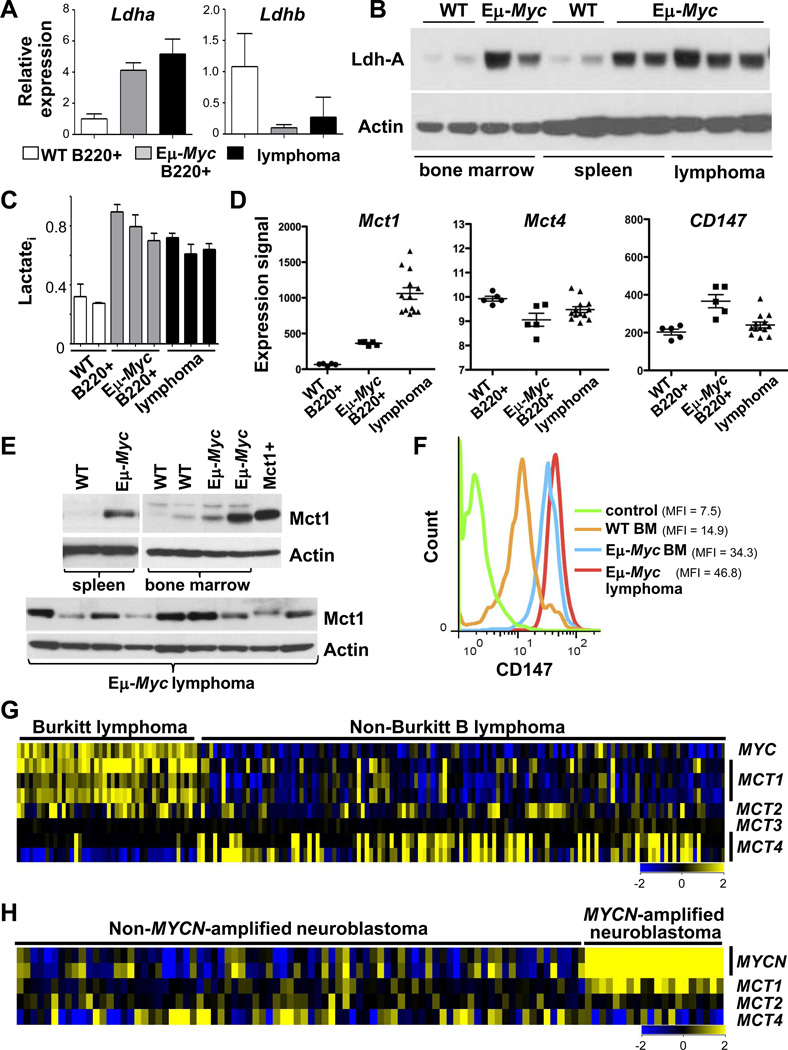
To assess if Myc controls lactate production and export in vivo we used the Eμ-Myc transgenic mouse model of human B lymphoma (1), where a protracted premalignant state allows one to identify targets that play roles in Myc-driven tumorigenesis (32). Expression analyses of B220+ splenic and bone marrow (BM) B cells from wild type and premalignant Eμ-Myc littermates established that Eμ-Myc B cells express elevated levels of several glycolytic enzymes, including Ldh-A, and that this response was augmented in neoplastic Eμ-Myc B cells (Fig. 1A and Fig. S1A–B).
Figure 1.
Myc augments lactate production and MCT1 expression. A–F, B220+ B cells isolated from bone marrow (BM) or spleen of 4–6 week old wild type (WT) or premalignant Eμ-Myc littermates or from Eμ-Myc lymphoma were analyzed: A, by qRT-PCR for Ldh-A (left) or Ldh-B (right) expression (WT BM, n = 2; premalignant Eμ-Myc,n = 2; lymphoma, n = 3; error bars are SD); B, for Ldh-A protein levels; C, intracellular lactate (Lactatei) (triplicate assays); D, by expression profiling analysis of Mct1, Mct4 and CD147 mRNA (circles, WT splenic B cells, n = 5; squares, premalignant Eμ-Myc, n = 5; triangles, lymphoma, n = 13) (significant differences in mean Mct1 levels in premalignant (p < 0.00002) and neoplastic (p < 0.000002) Eμ-Myc B cells versus WT B cells); E, for MCT1 protein levels (Mct1+, lysate from 293T cells expressing MCT1); and F, by flow cytometry for CD147 levels on B220+ BM B cells from WT (orange line) and premalignant Eμ-Myc (blue line) littermates, and of Eμ-Myc lymphoma (red line). MFI, mean fluorescence intensity. G, Gene expression profiling of MYC, MCT1, MCT2, MCT3 and MCT4 in human BL (n = 44) and non-BL (n = 129, from GSE4475, ref. 33). H, MCT1 expression in primary human MYCN-amplified neuroblastoma (n = 20) versus non-MYCN-amplified neuroblastoma (n = 81, from GSE3960, ref. 34).
In accord with these analyses, marked increases in Ldh-A protein were manifest in Eμ-Myc versus wild type B cells (Fig. 1B), and the preponderance of Ldh-A versus Ldh-B suggested that pyruvate would be converted to lactate (27). Indeed, intracellular lactate levels were elevated 2–3-fold in Eμ-Myc versus wild type B cells (Fig. 1C). Thus, Myc activates glycolysis and drives lactate production during lymphomagenesis.
We reasoned that lactate was exported from Eμ-Myc B cells by SLC16A/MCT transporters. Notably, there were marked increases in Mct1 mRNA levels, but not of Mct2, Mct3 and Mct4 transcripts, in Eμ-Myc B cells (Fig. 1D and Fig. S1C). Levels of CD147 transcripts, the obligate MCT1/MCT4 chaperone (19), were similar in wild type and Eμ-Myc B cells (Fig. 1D). Immunoblot analyses confirmed elevated MCT1 protein levels in premalignant and neoplastic Eμ-Myc B cells (Fig. 1E). MCT4 protein was also detected at low levels in some Eμ-Myc lymphomas (Fig. S1D), presumably due to hypoxic regions in these tumors (20, 25). Cell surface expression of MCT1 and CD147 are co-dependent (19); thus, FACS analyses of CD147 serves as a surrogate for cell surface MCT1. Notably, there were marked increases in the cell surface levels of CD147 in premalignant and neoplastic Eμ-Myc B cells versus wild type B cells (Fig. 1F). Thus, there are marked increases in MCT1 expression during Myc-induced lymphomagenesis.
Elevated MCT1 expression is a hallmark of MYC-driven malignancies
To determine if elevated MCT1 expression is manifest in human malignancies having MYC involvement, we first queried expression datasets of Burkitt lymphoma (BL) having MYC/Immunoglobulin chromosomal translocations and of MYCN-amplified neuroblastoma (33, 34). Notably, there are concordantly high levels of MCT1 and MYC mRNA in BL, and of MCT1 and MYCN in MYCN-amplified neuroblastoma (Fig. 1G–H). In contrast, MCT2, MCT3 and MCT4 are not specifically elevated in tumors with MYC/MYCN involvement; indeed, MCT4 transcript levels are low in MYC-expressing tumors (Fig. 1G–H). Further, the high-MCT1/low-MCT4 profile is a hallmark of MYC-expressing colon cancer (Fig. S2A) (35) (though normal colon was heterogeneous in MCT1 mRNA levels) and elevated MCT1 levels are manifest in the squamous carcinoma subtype of non-small cell lung cancer (Fig. S2C) (36). Lung adenocarcinoma and breast adenocarcinoma that express high levels of MYC also expressed high levels of MCT1, yet some of these also elevated levels of MCT4, possibly due to hypoxic regions in these tumors (Fig. S2C–D). Finally, a high-MYC-high-MCT1 signature connotes poor outcome in basal-like breast cancer and all lung cancers (Fig. S2E–G).
MCT1 is a Myc transcription target
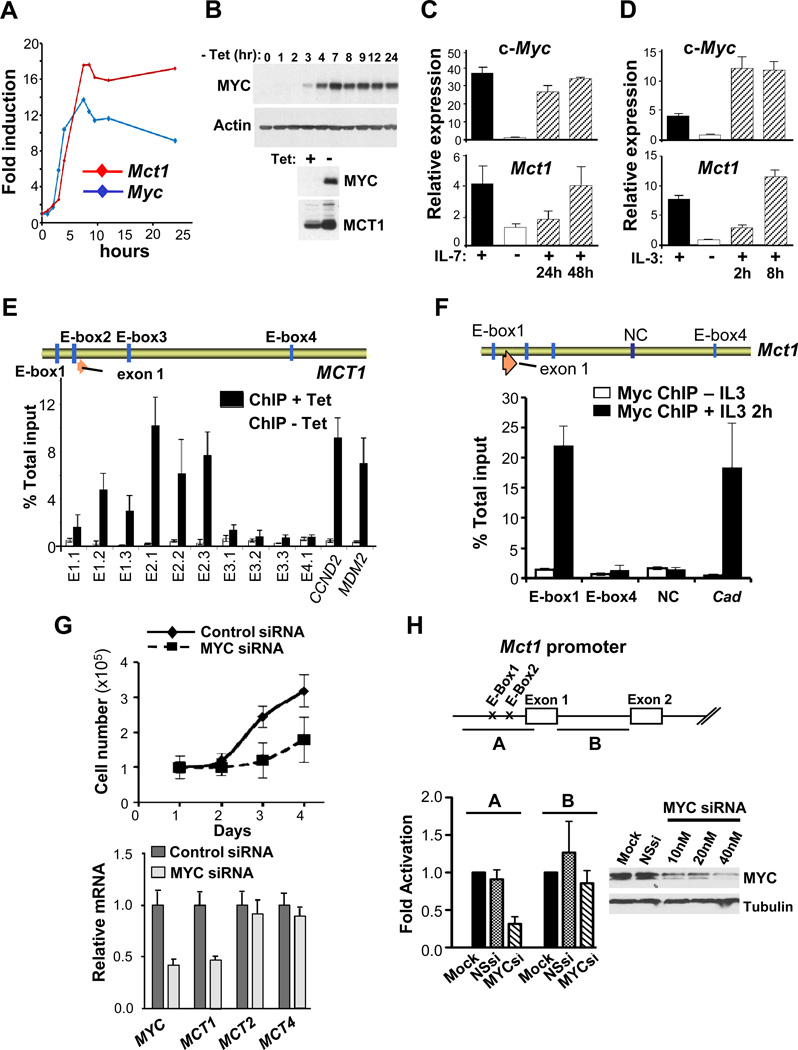
We evaluated the control of MCT1 expression in human P493-6 B lymphoma cells, which bear a tetracycline (Tet)-repressible c-Myc transgene (37). The induction of c-Myc mRNA and protein in P493-6 cells deprived of Tet was followed by marked increases in MCT1 mRNA and protein (Fig. 2A–B). These findings agree with RNA-seq and expression profiling analyses of P493-6 cells (9) (Fig. S3A).
Figure 2.
MCT1 is a Myc transcription target. A, human P493-6 B lymphoma cells bearing a Tet-repressible c-Myc transgene were cultured for 72 hr in Tet (1µg/ml, Myc-Off), washed and cultured in medium lacking Tet (Myc-On) and assessed for MYC and MCT1 mRNA levels by qRT-PCR. B, Top, c-Myc protein levels at the indicated intervals following withdrawal of Tet from P493-6 cells. Bottom, MCT1 protein levels 24 hr after removal (−) of Tet. C and D, Primary BM-derived mouse pre-B cells grown in IL7 (C) and mouse 32D.3 myeloid cells grown in IL3 (D) were deprived of ligand for 16 hr and then re-stimulated with IL7 or IL3. Levels of c-Myc and Mct1 transcripts were determined by qRT-PCR (n = 3). Expression is relative to Ub. E, Top, schematic of human SLC16A1/MCT1. Vertical blue bars indicate four E-box (CACGTG) sequences and orange arrow indicates exon 1. Bottom, P493-6 cells cultured for 72 hr with Tet (white bars, Myc-Off) then washed and cultured for 8 hr without Tet (black bars, Myc-On) was chromatin immunoprecipitated with c-Myc antibody. Designated E-boxes and controls (cyclin D2 [CCND2] and MDM2) were amplified by qRT-PCR and compared to total input. F, Top, schematic of mouse Slc16a1/Mct1. Vertical blue bars denote four E-box sequences in the mouse Mct1 gene and an intronic region (NC) used as a control for ChIP analyses. Orange arrow indicates exon 1. Bottom, 32D.3 cells cultured without IL3 for 16 hr (white bars) and then stimulated with IL3 for 2 hr (black bars) were chromatin immunoprecipitated with c-Myc antibody. Mouse Cad was amplified as a positive control for c-Myc binding. G, MCF7 cells were transfected with control siRNA or c-MYC-specific siRNA. Top, cells were counted daily (n = 3). Bottom, 48 hr after transfection cells were harvested for qRT-PCR analyses. H, Top, mouse Mct1 promoter regions used to generate firefly luciferase reporters. Bottom left, fold activation of Mct1 promoter-reporters in HEK 293T cells transfected with 40 nM scambled siRNA (NSsi) or MYC-specific siRNA (MYCsi) relative to mock-transfected cells (n=3, in quadruplicate). Bottom right, c-MYC protein knockdown in HEK293T cells.
To test if Mct1 is also induced by physiological cues that control c-Myc, we assessed if Mct1 expression is cytokine-dependent. Primary mouse bone marrow-derived pre-B cells grown in IL7 medium were deprived of and then restimulated with ligand. Like c-Myc, Mct1 expression was also IL7 dependent and its induction followed that of c-Myc (Fig. 2C). Similarly, Mct1 expression in immortal 32D.3 myeloid cells was IL3 dependent and followed the induction of c-Myc (Fig. 2D).
To test if Myc binds to the endogenous MCT1 gene via identified E-boxes, we performed chromatin immunoprecipitation (ChIP) analyses in P493-6 cells and 32D.3 myeloid cells. There was a marked and specific enrichment for c-Myc binding to human MCT1 E-box2 and E-box1 in P493-6 cells and to mouse Mct1 E-box1 in IL3-stimulated 32D.3 cells (Fig. 2E–F). In contrast, c-Myc binding to human E-box3 or E-box4, or to mouse E-box4 was not detected (Fig. 2E–F). Similar findings were evident in querying the genome-wide ChIP-seq datasets of Myc and Max target genes in P493-6 cells (9), which showed that MCT1 was transcriptionally silent until bound by Myc (Fig. S3B). In addition, Myc-directed control of MCT1 was specific, as c-Myc knockdown in MCF7 breast cancer cells impaired proliferation and reduced MCT1 expression but not levels of MCT2 or MCT4 (Fig. 2G), which are expressed in these cells (Fig. S3C). Finally, Myc knockdown selectively reduced the activity of a MCT1 promoter-reporter harboring E-box1 and E-box2 (Fig. 2H). Thus, MCT1 is a direct Myc transcription target in normal and tumor cells.
MCT1 is necessary for tumor cell proliferation
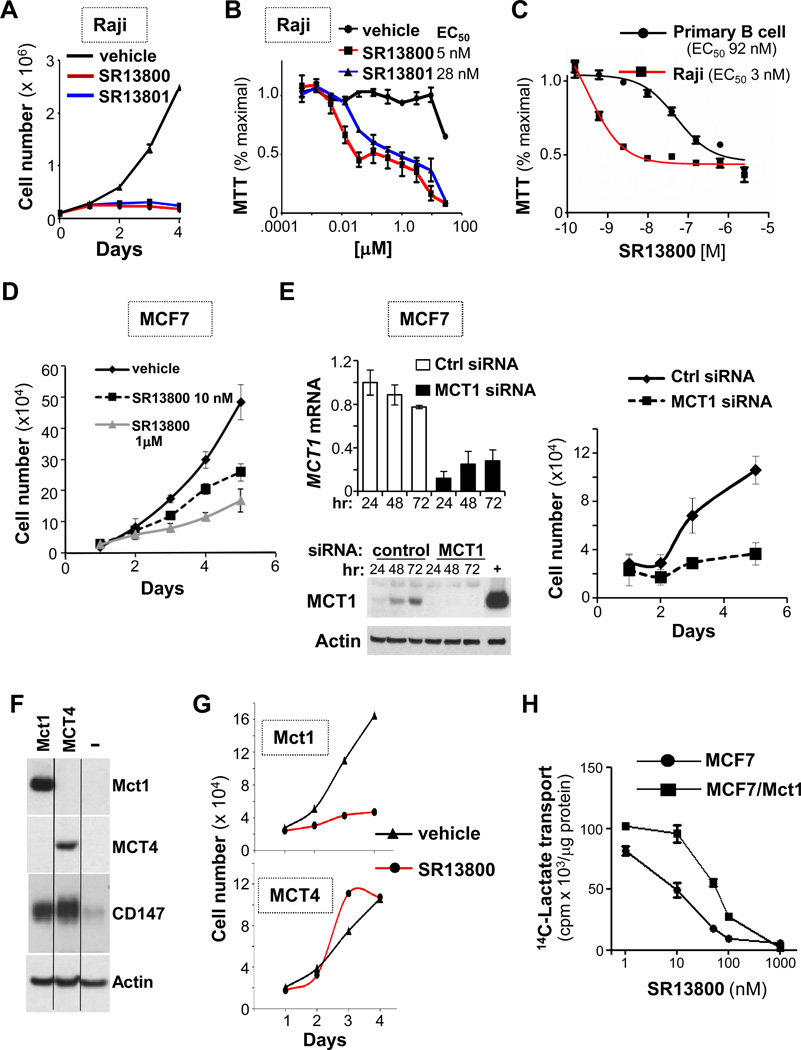
Most tumor cells produce and export excess lactate. We thus reasoned that blocking MCT1 function would impair the growth of MCT1-expressing tumor cells. Potent and specific inhibitors that disable MCT1- and MCT2-directed lactate transport (but not that of MCT4) block growth of activated T cells (38, 39). We synthesized two of these pyrole pyrimidine-based molecules (Fig. S4A, AR-C122982 [hereafter SR13800] and AR-C155858 [SR13801]) and assessed their effects on Raji Burkitt lymphoma (BL) cells that only express MCT1 (Fig. S3C). Both inhibitors blocked Raji cell proliferation (Fig. 3A–B), at doses at least 10-fold lower than those needed to impair growth of primary bone marrow-derived B cells (Fig. 3C). Growth inhibition was due to proliferative arrest, which was followed by protracted cell death (Fig. S4B–C). These inhibitors also blocked MCF7 breast cancer cell growth (Fig. 3D), and that of Raji BL and 70Z3 mouse B cell lymphoma in methylcellulose (Fig. S4D).
Figure 3.
MCT1 inhibition blocks tumor cell proliferation. A, 1 × 105 Raji cells/ml were treated with vehicle (black line), 100 nM SR13800 (red line) or SR13801 (blue line) and counted at the indicated times (n = 3, representative of three experiments). B, Raji cells were treated with indicated doses of SR13800 or SR13801 for 3 days and assayed by MTT (n = 3). C, MTT assay of Raji cells or primary mouse B cells treated with SR13800 for 3 days (n = 3). D, 1 × 105 MCF7 cells treated with vehicle (black line), or 10 nM (dashed black line) or 1 µM SR13800 (gray line) and cell numbers were counted at the indicated times (n = 3, representative of two experiments). E, siRNA knockdown of MCT1 mRNA (top left) and MCT1 protein (bottom left) in MCF7 cells. “+”control lysate from 293T cells expressing Mct1. Right, cells transfected with control siRNA (black line) or MCT1 siRNA (dashed line) and counted (n = 3, representative of three experiments). F, MCF7 cells engineered to overexpress mouse Mct1 or human MCT4 were assessed for expression of MCT1, MCT4 and CD147 proteins. G, MCF7 cells overexpressing MCT1 or MCT4 were treated with vehicle (black lines) or SR13800 (1 µM, red lines) and cell number was determined daily (n = 3, representative of two experiments). H, 14C lactate transport in MCF7 cells overexpressing MCT1 and treated with indicated doses of SR13800 (10 minutes). (n = 4, representative of three experiments).
Manipulating MCT1 expression was not feasible in Raji B lymphoma. Thus, to confirm that these inhibitors were on target MCT1 expression was silenced in MCF7 cells. Silencing MCT1 blocked the growth of MCF7 cells similar to treatment with the inhibitor (Fig. 3E). We also tested the effects of MCT1 or MCT4 overexpression in MCF7 cells. As expected (19), overexpression of either transporter increased CD147 levels (Fig. 3F), and MCT4, but not MCT1, overexpression was sufficient to confer SR13800 resistance (Fig. 3G). MCT1 overexpression augmented lactate transport (Fig. S4E) and shifted the IC50 for SR13800 in inhibiting transport of 14C-lactate (Fig. 3H). However, forced expression of MCT1 (or MCT4) did not affect MCF7 cell proliferation (Fig. S4F). Thus, in tumor cells that express MCT1, this transporter is necessary for proliferation but does not augment cell growth.
MCT1 inhibition derails lactate homeostasis, glycolysis and glutathione synthesis
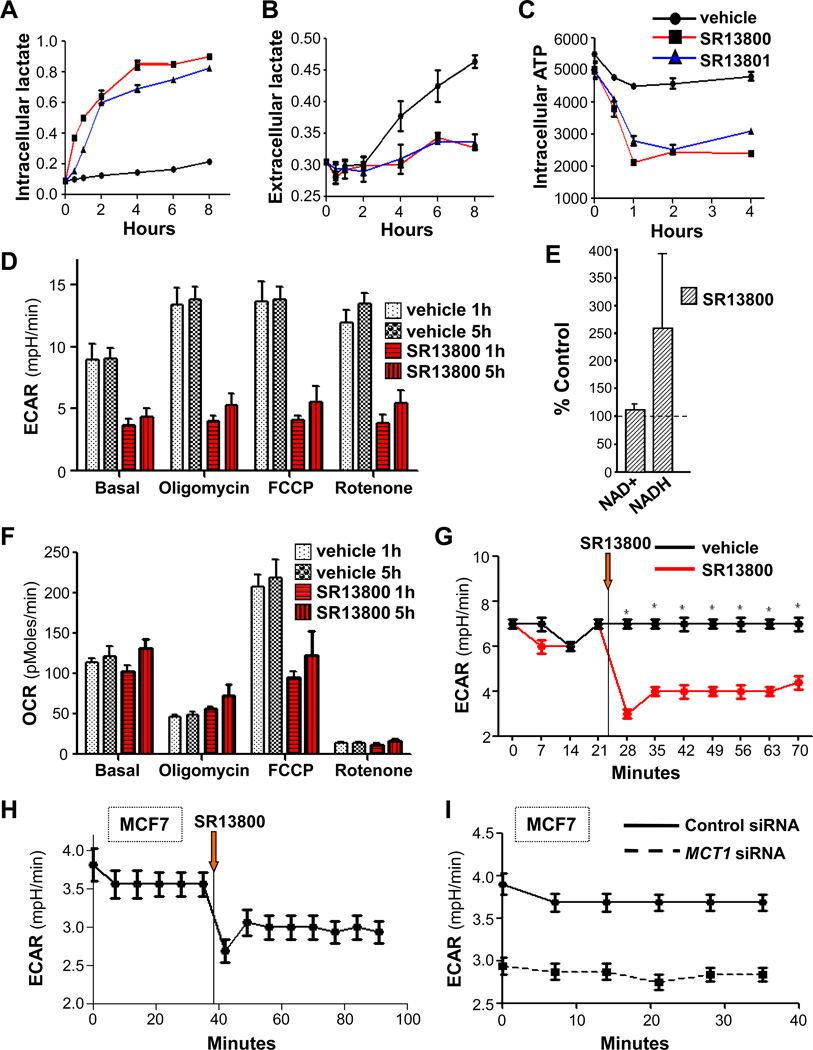
In Raji cells treated with SR13800 or SR13801 there were immediate increases in the levels of intracellular lactate and a block in lactate export (Fig. 4A–B). However, levels of intracellular lactate reached a maximum within ~4 hours (Fig. 4A), suggesting effects on metabolism. Indeed, there were rapid and marked reductions in the levels of intracellular ATP (Fig. 4C) and in the extracellular acidification rate (ECAR) (Fig. 4D, 4G). By contrast, there were little effects of SR13800 on basal oxygen consumption rate (OCR) (Fig. 4F). Rapid decreases in ECAR provoked by SR13800 were manifest in other BL lines that express MCT1 (Fig. S5B) and in MCF7 breast cancer cells (Fig. 4H). Similar decreases were observed in MCF7 cells following MCT1 knockdown (Fig. 4I). Further, oligomycin treatment, which blocks ATP synthesis and activates glycolysis, induced ECAR in vehicle- but not SR13800-treated cells (Fig. 4D). Notably, dissipating the proton gradient with FCCP revealed that MCT1 inhibition abolishes reserve mitochondrial capacity (Fig. 4F) without reducing levels of NADH (Fig. 4E), an electron donor that drives oxidative phosphorylation. Rotenone treatment, which inhibits mitochondrial complex I, abolished the majority of OCR in Raji cells, and this was unaffected by SR13800 (Fig. 4F). Thus, MCT1 inhibition rapidly disables tumor cell metabolism.
Figure 4.
MCT1 inhibition derails lactate homeostasis and cancer cell metabolism. A–C, cells treated with vehicle (black lines) or 100 nM of SR13800 (red lines) or SR13801 (blue lines) and levels of intracellular (A) and extracellular (B) lactate, and of intracellular ATP (C), were determined (n = 3, representative of three experiments). D, ECAR and F, OCR, in Raji cells treated for 1 or 5 hr with vehicle or SR13800 (1 µM). Effects of addition of oligomycin, FCCP or rotenone are also shown (n = 6). E, Raji cells were treated for 8 hr with SR13800 (100 nM) or vehicle and levels of NAD+ and NADH (n = 3) were determined. Levels are shown relative to those of vehicle treated cells (dashed line). G, ECAR in Raji BL cells treated with vehicle (black line) or SR13800 (red line) at time indicated (orange arrow) (n = 6, representative of four experiments). H, ECAR in MCF7 cells treated with SR13800 at time indicated (orange arrow) (n = 6, representative of three experiments). I, ECAR in control and MCT1 siRNA transfected MCF7 cells (n = 6).
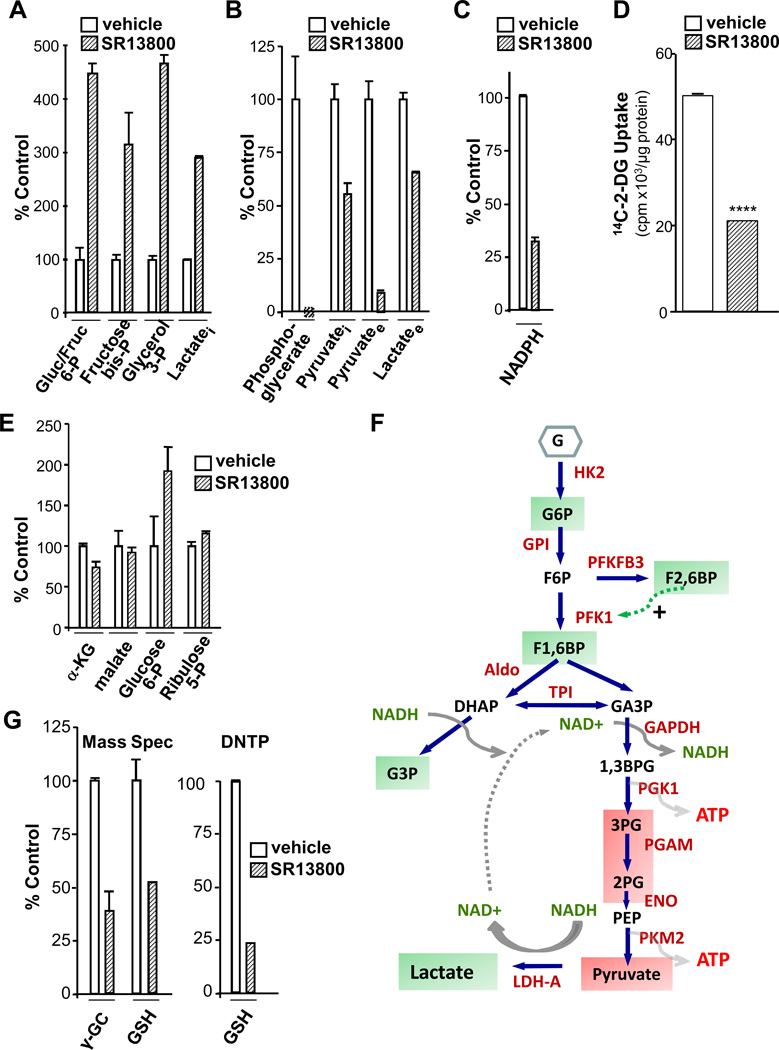
To assess the effects of MCT1 inhibition on metabolism, we measured metabolic intermediates in Raji lymphoma cells by mass spectrometry. Again, SR13800 treatment led to rapid increases in the levels of intracellular lactate and corresponding decreases in extracellular lactate (Fig. 5A–B). Further, as MCT family members transport pyruvate (18), SR13800 treatment led to marked reductions in extracellular pyruvate (Fig. 5B). Notably, MCT1 inhibition increased the levels of glucose- and fructose-6-phosphate, fructose-bisphosphate and glycerol-3-phosphate, and markedly reduced products of the ATP-generating arm of glycolysis, including phosphoglycerate and pyruvate (Fig. 5A–B, 5F). Further, there were marked reductions in NADPH levels and glucose transport (Fig. 5C–D). These changes were not associated with alterations in the levels of glycolytic enzymes (Fig. S5C), and reduced glucose transport was not due to changes in the cell surface levels of glucose transporters (not shown).
Figure 5.
MCT1 inhibition impairs glucose transport and glycolysis, and reduces glutathione pools. A–C, Raji cells were treated for 8 hr with SR13800 (100 nM) or vehicle and levels of glycolytic metabolites were determined by (A, B) mass spectrometry (n = 6, representative of four experiments) and (C) levels of NADPH were determined by NADP/NADPH Quantification Kit (BioVision) (n=3). D, Uptake of 14C-2-deoxyglucose (14C-2-DG), (0.5 µCi for 1 hr in quadruplicate) in Raji cells pre-treated with vehicle or SR13800 (100 nM). (****, p <0.0001, representative of three experiments). E, Levels of pentose phosphate pathway and TCA cycle products from Raji cells treated for 8 hr with SR13800 (100 nM) or vehicle were assessed by mass spectrometry (n = 6). F, summary of effects of inhibition of lactate transport on glycolytic intermediates. Green boxes indicate accumulations and red boxes reductions of intermediates. G, Left, mass spectrometry of γ-GC and GSH levels from Raji cells treated with SR13800 (100 nM) for 8 hr (n = 6, representative of three experiments). Right, total GSH (GSH plus GSSG) levels in Raji cells assayed by DNTP (n = 3, representative of three experiments).
Decreases in NADPH following MCT1 inhibition were not associated with changes in ribulose-5-phosphate or malate, two intermediates in NADPH production (Fig. 5E). NADPH is consumed in the conversion of glutathione disulfide (GSSG) to reduced glutathione (GSH) by glutathione reductase. The first step of GSH synthesis, the production of γ-glutamylcysteine (γ-GC), requires ATP (Fig. S5D). Thus, we reasoned that MCT1 inhibition might compromise GSH synthesis. Indeed, by two measures, MCT1 inhibition led to rapid reductions in γ-GC and GSH, and in total glutathione (GSH and GSSG, Fig 5G). Reductions in GSH did not reflect changes in the levels of the catalytic (GCLC) and modulatory (GCLM) subunits of glutamyl cysteine ligase (GCL) that catalyzes γ-GC production (Fig. S5E). Thus, the reductions in ATP levels triggered by inhibiting lactate transport (Fig. 4C) are associated with reduced levels of GSH.
Metformin augments the potency and anti-tumor activity of MCT1 inhibitors
Metformin disables oxidative phosphorylation (OXPHOS) by inhibiting mitochondrial complex I and forces a glycolytic phenotype that we reasoned may augment sensitivity to MCT1 inhibitors. Though metformin alone had little effect, co-treatment of Raji lymphoma with SR13800 and metformin led to higher levels of intracellular lactate (Fig. S6A), rapid growth arrest and cell death (Fig. S6B). Synergy of SR13800 and metformin was evident in all MCT1-expressing human and mouse tumor cell lines (Fig. S6C–D), yet had, as expected, no effects on MCT4-expressing human tumor cells nor upon Eμ-Myc lymphomas in transplant experiments (not shown), likely due to hypoxia-mediated induction of Mct4 (Fig. S1D). Thus, metformin augments the anti-cancer potency of MCT1/MCT2 inhibitors in tumor cells expressing these transporters.
The efficacy of SR13800 +/− metformin was tested in vivo using NOD/SCID mice injected intravenously with Raji lymphoma, or injected subcutaneously with MCT1-expressing human T47D estrogen receptor-positive breast cancer cells. Immunohistochemical (IHC) analyses confirmed that high levels of MCT1, but not MCT4, were expressed in both tumor models (Fig. S7A–B). In vivo DMPK analyses demonstrated that a daily intraperitoneal injection of 30 mg/kg of SR13800 was sufficient to maintain serum levels above the EC50 of the drug (~5 nM, Fig. 3B), and mice tolerated long-term daily dosing of SR13800 for >130 days without toxic effects (not shown). There were also no significant effects of SR13800 treatment on numbers of white or red blood cells that express high CD147 levels (not shown).
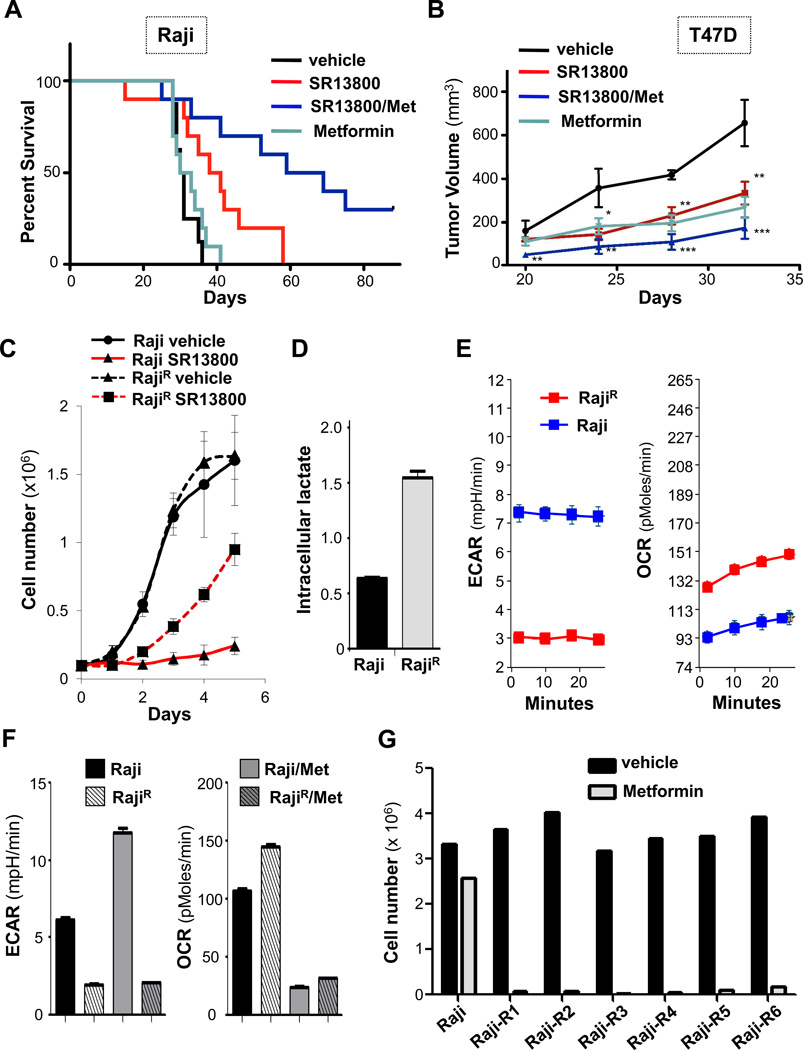
Notably, SR13800 treatment delayed the onset and penetrance of disease relative to vehicle-treated Raji lymphoma recipients (Fig. 6A and Fig. S7C). In contrast, metformin treatment (at doses not affecting blood glucose) did not affect lymphoma onset (Fig. 6A). Recipients treated with both SR13800 plus metformin had remarkably delayed disease (Fig. 6A); indeed, some recipients never developed tumors, even after being taken off drug for over 80 days (data not shown). In the T47D model, SR13800 and metformin had comparable activity in impairing tumorigenecity (Fig. 6B). However, the SR13800/metformin combination was again superior in blocking tumor growth, without affecting the health of transplant recipients (Fig. 6B, Fig. S7D–F). Thus, MCT1 inhibitors have in vivo anti-cancer activity and the MCT1 inhibitor/metformin combination is an attractive therapeutic strategy for treating MCT1-expressing malignancies.
Figure 6.
Metformin augments the anti-tumor activity of MCT1 inhibitors and overcomes resistance due to shifts to OXPHOS. A, Survival of NOD/SCID mice transplanted with Raji lymphoma. At day 3 mice were treated with water or water with 5 mg/kg metformin and daily injections of vehicle or 30 mg/kg SR13800. Median survival: vehicle (n=8), 30 days; metformin (n=10), 31 days; SR13800 (n=10), 39 days (p = 0.016); SR13800 + metformin (n=10), 64 days (p = 0.005). B, Tumor volumes measured with calipers in NOD/SCID mice injected sub-Q with T47D cells and treated as in (A). At day 32 there were significant differences in tumor volume for vehicle (n = 5) versus SR13800 (n = 9, p = 0.01), metformin (n = 8, p = 0.002) and SR13800/metformin (n = 10, p = 0.0005) cohorts. C, Parental Raji and SR13800-resistant Raji (RajiR) cells were treated with vehicle or 100 nM SR13800 and cell numbers determined (n = 4, representative of two experiments). D, levels of intracellular lactate in Raji and RajiR cells in normal growth medium (n = 3, representative of two experiments). E, ECAR (left) and OCR (right) measurements of Raji (blue lines) and RajiR cells (red lines) (n = 6, representative of two experiments). F, parental Raji and RajiR cells were assessed for ECAR and OCR +/− metformin (1 mM) for 16 hr (n = 6, representative of two experiments). Cell number of Raji or RajiR cells treated with vehicle (black bars) or metformin (1 mM, gray bars) for 24h (representative of two experiments).
Metformin overcomes resistance to MCT1 inhibitors due to shifts to OXPHOS
To assess if, in addition to MCT4 expression, there were other tumor cell intrinsic mechanisms of resistance to MCT1 inhibitors, Raji cells were cultured with low doses of SR13800 and surviving cells serially passaged into media having increased levels in SR13800, to generate independent pools of SR13800-resistant Raji (RajiR) cells (n=6, (Fig. S8A). The growth rates of RajiR cells were comparable to parental Raji cells in normal media yet they continued to proliferate in media containing SR13800, whereas parental Raji lymphoma cells underwent growth arrest (Fig. 6C).
The resistance of RajiR cells was not associated with MCT4 induction or changes in MCT1 levels (Fig. S8B). However, RajiR cells had elevated levels of intracellular lactate and low ECAR compared to parental Raji cells growing in normal media (Fig. 6D–E). Notably, RajiR cells had an increase in OCR compared to parental cells (Fig. 6E). Treatment of RajiR cells with metformin alone led to a rapid collapse in metabolism, with a marked reduction in OCR (Fig. 6F). Although metformin treatment led to reductions in OCR in parental Raji lymphoma cells, it also, as predicted, increased ECAR, thus providing energetic compensation for inhibiting OXPHOS (Fig. 6F). Finally, metformin alone led to the rapid death of RajiR but not parental Raji cells (Fig. 6G), demonstrating RajiR cells are dependent on OXPHOS. Thus, a metabolic shift to OXPHOS is a mechanism of resistance to MCT1 inhibitors and metformin overcomes this resistance.
Reductions of GSH pools trigger hydrogen peroxide-induced cell death
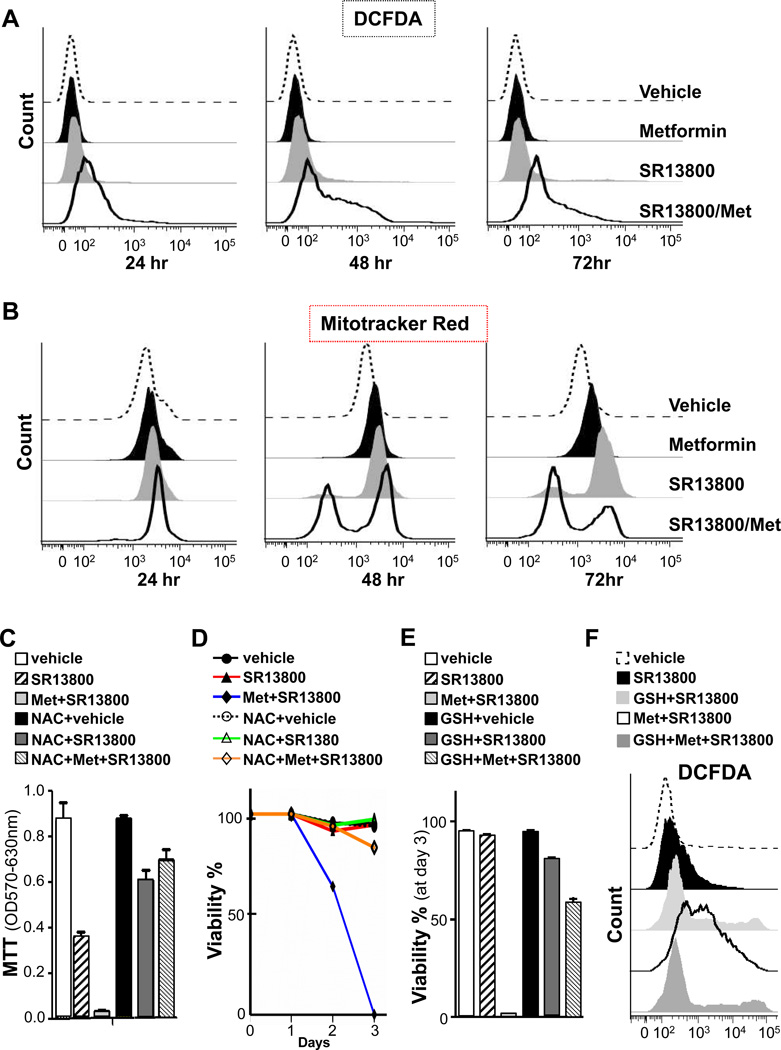
GSH is a major anti-oxidant in cells that neutralizes free radicals and reactive oxygen species (Fig. S5D). Indeed, the marked reduction in GSH in SR13800-treated Raji cells was followed by increases of hydrogen peroxide (H2O2) but not of mitochondrial or cytosolic superoxide anions (Fig. 7A and not shown). Notably, SR13800/metformin treatment led to a profound increase in H2O2 levels that were associated with the death of Raji BL.
Figure 7.
MCT1 Inhibitor/metformin combination triggers increases in hydrogen peroxide, mitochondrial damage and cell death that is blocked by glutathione. Raji cells were treated with vehicle, SR13800 (1 µM), metformin (1 mM) or SR13800 plus metformin for the indicated times and analyzed by flow cytometry for: A, DCFDA; and B, Mitotracker Red stain (representative of three experiments). C–F, NAC (C, D) and GSH (E, F) override the anti-cancer effects of SR13800 +/− metformin. Raji cells were pretreated with or without NAC (10 mM) or glutathione ester (3 mM [D] or 5 mM [E, F]) for 1 hr and then treated with vehicle or SR13800 (100 nM [C–E] or 1 µM [F]) +/− metformin (1 mM). C, MTT assays after 3 days (n = 3). D, E, Cell survival by trypan blue dye exclusion (n = 3). F, DCFDA fluorescence after 24 hr of treatment (n = 3).
ROS compromise the function of mitochondria. Analyses with MitoTracker Green, which measures mitochondrial mass, indicated that treatment of Raji BL with SR13800, metformin or SR13800/metformin did not change mitochondrial mass (Fig. S8C). However, analyses with MitoTracker Red, which measures mitochondrial membrane potential, demonstrated that SR13800 treatment, and especially SR13800/metformin, triggered marked reductions in numbers of Raji BL cells having functional mitochondria (Fig. 7B).
These findings suggested that reductions in GSH and subsequent increases in H2O2 triggered by SR13800 or SR13800/metformin compromise tumor cell survival. Indeed, pre-treatment of Raji BL cells with the anti-oxidants N-acetyl cysteine (NAC) or GSH blocked the deleterious effects of SR13800 and of SR13800/metformin on Raji cell growth and survival (Fig. 7C–E). Finally, GSH treatment impaired H2O2 production in SR13800- and SR13800/metformin-treated cells (Fig. 7F). Thus, metabolic demise triggered by the inhibition of lactate transport leads to a collapse in ATP pools, reductions in GSH and increases in H2O2 that contribute to tumor cell death (Figure S8D).
Discussion
Aerobic glycolysis is a hallmark of most tumor types and our findings have established that Myc is sufficient to drive glycolysis in vivo, where premalignant Eμ-Myc B cells express elevated levels of several glycolytic enzymes that leads to increased lactate levels, in accord with Ldh-A induction by Myc in vitro (14, 40). Further, this response is amplified in Eμ-Myc lymphoma. These findings parallel the increased dependence on glycolysis and lactate production manifest during the step-wise transformation of primary human fibroblasts (41). Finally, in rapidly expanding tumors HIF-1α also contributes to glycolysis via the induction of hypoxia (42).
In cancer cells glucose transport and glycolytic flux is high, and this drives the production of ATP and intermediates necessary for anabolic pathways (43). A consequence of this shift is the production of toxic levels of lactate. Here we show this is diverted by expression of high MCT1 levels in Myc-expressing malignancies and that MCT1 is necessary for the proliferation of these tumor cells. Further, exporting lactate into the milieu may play roles in promoting tumor progression, as lactate can trigger an inflammatory response (44) that drives tumor progression (45). Finally, lactate also plays other roles in tumorigenesis, where it can be converted into pyruvate that enters the TCA cycle (25, 26) and/or stimulate endothelial cell angiogenesis (28, 29).
Our findings establish that Myc controls lactate homeostasis by inducing MCT1 transcription. Pharmacological inhibition and knockdown of MCT1 demonstrates the essential role of this transporter in maintaining the metabolism, growth, survival and tumorigenecity of MCT1-expressing tumor cells. These findings are consistent with studies showing the anti-cancer effects of non-specific inhibitors of lactate transporters such as α-cyanocinnamate (25) and with the block in metabolism and growth of Ras-transformed fibroblasts ex vivo by MCT1 inhibitors (24). However, the mechanism by which inhibiting lactate transport compromises cancer cell growth has been unclear and we now show is due to a collapse in glycolysis and accompanying reductions in ATP and glutathione (Figure S8D).
Changes in metabolic intermediates suggest how inhibiting lactate export disables glycolysis. In particular, our findings are remarkably akin to the changes in glycolytic intermediates observed in the exercised muscle of LDH-A deficient patients, where reduced levels of NAD+ compromise GAPDH activity and favor the oxidation of NADH and the production of glycerol-3-phospate by GPDH (46). In MCT1 inhibitor-treated cells, intracellular lactate levels and NADH levels rise, suggesting that the conversion of pyruvate to lactate and the coupled oxidation of NADH to NAD+ are impeded by high intracellular lactate levels. High NADH levels favor GPDH conversion of DHAP to glycerol-3-phosphate, which diverts triosephosphates from GAPDH and the ATP generating arm of glycolysis (Figure 8SD). As a consequence this leads to reductions in phosphoglycerate and pyruvate. This chain of events, coupled with feedback inhibition by lactate on PFK1 (47), explains increased levels of more upstream intermediates such as glucose-6-phosphate (G-6-P) and the effects upon glucose transport. Specifically, G-6-P feedback inhibits hexokinase, which results in increased intracellular free glucose that then inhibits glucose transport. Collectively these events converge to markedly impair ATP levels and the synthesis of the major antioxidant of the cell, glutathione, which is a key target, as GSH is sufficient to override the acute effects of MCT1 inhibitors on tumor cell survival.
Importantly, these findings suggest that any agent targeting glycolysis will compromise ATP production and GSH levels, resulting in H2O2-induced death. In accord with this notion, LDH-A inhibition triggers oxidative stress and blocks tumor cell proliferation (48) and the antioxidants NAC and GSH block the anti-tumor activity of MCT1 inhibitors. Our findings also suggest that pro-oxidant therapies will augment the potency of lactate transport inhibitors and that other means of disrupting GSH synthesis, for example by inhibiting cysteine or cystine transporters (49), will be effective therapeutic strategies for tumors with MYC involvement.
Elevated MCT1 expression is a hallmark of human malignancies with MYC involvement. Notably, many MYC-expressing tumors express reduced MCT4 levels, suggesting MCT1 inhibitors will have a therapeutic benefit in such tumors. However, it is clear that tumor cells that express MCT4 will be refractory to MCT1 inhibitors, and that patient tumors should be assessed for MCT1 and MCT4 expression prior to using MCT1 inhibitors for treatment (25). A second mechanism of resistance identified herein is a shift of tumor cells to OXPHOS, and we show that agents such as metformin that disrupt OXPHOS disable this resistance, leading to a collapse in tumor cell metabolism and rapid death. Notably metformin use in type-2 diabetes appears to decrease cancer risk and may provide benefit as a cancer therapeutic (50). Together, these findings support the use of MCT1 inhibitors in combination with metformin, or with metformin derivatives, in therapies for MCT1- and MYC-expressing malignancies.
Supplementary Material
Acknowledgments
We thank Nancy Philp (Thomas Jefferson University) for MCT antibodies, the FACS, Genomics and ARC Cores of Scripps Florida, members of the Cleveland lab and Anke Klippel-Giese (Pfizer Oncology) for discussions, Marika Kernick for editing, Justin Lucas (Pfizer Oncology) for assistance, and Dr. George Hu (Pfizer Oncology) for pathological analyses. This study is dedicated to the memory of our beloved colleague Dr. Mark Hall.
Grant Support
This work was supported by NIH grants CA076379 and CA169142 to J.L.C., CA057341 to C.V.D., GM026782, GM038436, CA169142 and U54MH074404 to W.R.R., F32 CA134121 to A.L.A, the Jane & Leonard Korman Family Foundation Postdoctoral Fellowship to K.E.N.S., and by monies from the State of Florida to Scripps Florida.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors declare that they have no potential conflicts of interest.
Authors’ Contributions
Conception and design: J.R. Doherty, C. Yang, K.E.N. Scott, W.R. Roush, J.L. Cleveland
Development of Methodology: J.R. Doherty, C. Yang, K.E.N. Scott, M.D. Cameron, J.K. Mishra. F. Li, M. Tortosa
Acquisition of data: J.R. Doherty, C. Yang, K.E.N. Scott, M.D. Cameron, M.A. Hall., A.L. Amelio, H.M. Genau., M. Fallahi, Y. Lu, A.T. Hooper, K. Unsal-Kacmaz
Analysis and interpretation of data: J.R. Doherty, C. Yang, K.E.N. Scott, M.D. Cameron, M. Fallahi, J.L. Cleveland
Writing, review and/or revision of the manuscript: J.R. Doherty, C. Yang, K.E.N. Scott, C.V. Dang, W.R. Roush, J.L. Cleveland
Administrative, technical or material support: W. Li, J.K. Mishra, F. Li, M. Tortosa, H.M. Genau, R.J. Rounbehler, K.G. Kumar, A.A. Butler, T.D. Bannister
Study supervision: J.L. Cleveland, W.R. Roush.
References
- 1.Gardner L, Lee L, Dang C. The c-Myc Oncogenic Transcription Factor. The Encyclopedia of Cancer, Second Edition. 2002:555–561. [Google Scholar]
- 2.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 3.Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, et al. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- 4.Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 5.Ji H, Wu G, Zhan X, Nolan A, Koh C, De Marzo A, et al. Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PLoS One. 2011;6e:26057. doi: 10.1371/journal.pone.0026057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 7.Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 8.Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massagué J. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nature Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- 9.Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, et al. c-Myc Is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 13.Kroemer G, Pouyssegur J. Tumor cell metabolism: Cancer's Achilles' heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci USA. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 16.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halestrap AP, Price NT. The proton-linked monocarboxylate transporter (MCT) family: structure, function and regulation. Biochem J. 1999;343:281–299. [PMC free article] [PubMed] [Google Scholar]
- 19.Kirk P, Wilson MC, Heddle C, Brown MH, Barclay AN, Halestrap AP. CD147 is tightly associated with lactate transporters MCT1 and MCT4 and facilitates their cell surface expression. EMBO J. 2000;19:3896–3904. doi: 10.1093/emboj/19.15.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ullah MS, Davies AJ, Halestrap AP. The plasma membrane lactate transporter MCT4, but not MCT1, is up-regulated by hypoxia through a HIF-1α-dependent mechanism. J Biol Chem. 2006;281:9030–9037. doi: 10.1074/jbc.M511397200. [DOI] [PubMed] [Google Scholar]
- 21.Fisel P, Kruck S, Winter S, Bedke J, Hennenlotter J, Nies AT, et al. DNA Methylation of the SLC16A3 promoter regulates expression of the human lactate transporter MCT4 in renal cancer with consequences for clinical outcome Clin. Cancer Res. 2013;19:5170–5181. doi: 10.1158/1078-0432.CCR-13-1180. [DOI] [PubMed] [Google Scholar]
- 22.Pinheiro C, Longatto-Filho A, Azevedo-Silva J, Casal M, Schmitt FC, Baltazar F. Role of monocarboxylate transporters in human cancers: state of the art. J Bioenerg Biomembr. 2012;44:127–139. doi: 10.1007/s10863-012-9428-1. [DOI] [PubMed] [Google Scholar]
- 23.Halestrap AP, Meredith D. The SLC16 gene family-from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflugers Arch. 2004;447:619–628. doi: 10.1007/s00424-003-1067-2. [DOI] [PubMed] [Google Scholar]
- 24.Le Floch R, Chiche J, Marchiq I, Naïken T, Ilk K, Murray CM, et al. CD147 subunit of lactate/H+ symporters MCT1 and hypoxia-inducible MCT4 is critical for energetics and growth of glycolytic tumors. Proc Natl Acad Sci USA. 2011;108:16663–16668. doi: 10.1073/pnas.1106123108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118:3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiaschi T, Marini A, Giannoni E, Taddei ML, Gandellini P, De Donatis A, et al. Reciprocal metabolic reprogramming through lactate shuttle coordinately influences tumor-stroma interplay. Cancer Res. 2012;72:5130–5140. doi: 10.1158/0008-5472.CAN-12-1949. [DOI] [PubMed] [Google Scholar]
- 27.Dawson DM, Goodfriend TL, Kaplan NO. Lactic Dehydrogenases: Functions of the Two Types Rates of Synthesis of the Two Major Forms Can Be Correlated with Metabolic Differentiation. Science. 1964;143:929–933. doi: 10.1126/science.143.3609.929. [DOI] [PubMed] [Google Scholar]
- 28.Végran F, Boidot R, Michiels C, Sonveaux P, Feron O. Lactate influx through the endothelial cell monocarboxylate transporter MCT1 supports an NF-κB/IL-8 pathway that drives tumor angiogenesis. Cancer Res. 2011;71:2550–2560. doi: 10.1158/0008-5472.CAN-10-2828. [DOI] [PubMed] [Google Scholar]
- 29.Sonveaux P, Copetti T, De Saedeleer CJ, Végran F, Verrax J, Kennedy KM, et al. Targeting the lactate transporter MCT1 in endothelial cells inhibits lactate-induced HIF-1 activation and tumor angiogenesis. PLoS One. 2012;7:e33418. doi: 10.1371/journal.pone.0033418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Critchlo SE, Tate L. Use of a MCT1 inhibitor in the treament of cancers expressing MCT1 over MCT4. WO 2010/089580 A1. [Google Scholar]
- 31.Rounbehler RJ, Fallahi M, Yang C, Steeves MA, Li W, Doherty JR, et al. Tristetraprolin impairs myc-induced lymphoma and abolishes the malignant state. Cell. 2012;150:563–574. doi: 10.1016/j.cell.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, et al. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 34.Wang Q, Diskin S, Rappaport E, Attiyeh E, Mosse Y, Shue D, et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006;66:6050–6062. doi: 10.1158/0008-5472.CAN-05-4618. [DOI] [PubMed] [Google Scholar]
- 35.Matsuyama T, Ishikawa T, Mogushi K, Yoshida T, Iida S, Uetake H, et al. MUC12 mRNA expression is an independent marker of prognosis in stage II and stage III colorectal cancer. Int J Cancer. 2010;127:2292–2299. doi: 10.1002/ijc.25256. [DOI] [PubMed] [Google Scholar]
- 36.Kuner R, Muley T, Meister M, Ruschhaupt M, Buness A, Xu EC, et al. Global gene expression analysis reveals specific patterns of cell junctions in non-small cell lung cancer subtypes. Lung Cancer. 2009;63:32–38. doi: 10.1016/j.lungcan.2008.03.033. [DOI] [PubMed] [Google Scholar]
- 37.Pajic A, Spitkovsky D, Christoph B, Kempkes B, Schuhmacher M, Staege MS, et al. Cell cycle activation by c-myc in a Burkitt lymphoma model cell line. Int J Cancer. 2000;87:787–793. doi: 10.1002/1097-0215(20000915)87:6<787::aid-ijc4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 38.Murray CM, Hutchinson R, Bantick JR, Belfield GP, Benjamin AD, Brazma D, et al. Monocarboxylate transporter MCT1 is a target for immunosuppression. Nat Chem Biol. 2005;1:371–376. doi: 10.1038/nchembio744. [DOI] [PubMed] [Google Scholar]
- 39.Guile SD, Bantick JR, Cooper ME, Donald DK, Eyssade C, Ingall AH, et al. Optimization of monocarboxylate transporter 1 blockers through analysis and modulation of atropisomer interconversion properties. J Med Chem. 2007;50:254–263. doi: 10.1021/jm060995h. [DOI] [PubMed] [Google Scholar]
- 40.Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc Natl Acad Sci USA. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramanathan A, Wang C, Schreiber SL. Perturbational profiling of a cell-line model of tumorigenesis by using metabolic measurements. Proc Natl Acad Sci U S A. 2005;102:5992–5997. doi: 10.1073/pnas.0502267102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marin-Hernandez A, Gallardo-Perez JC, Ralph SJ, Rodriguez-Enriquez S, Moreno-Sanchez R. HIF-1alpha modulates energy metabolism in cancer cells by inducing over-expression of specific glycolytic isoforms. Mini Rev Med Chem. 2009;9:1084–1101. doi: 10.2174/138955709788922610. [DOI] [PubMed] [Google Scholar]
- 43.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shime H, Yabu M, Akazawa T, Kodama K, Matsumoto M, Seya T, et al. Tumor-secreted lactic acid promotes IL-23/IL-17 proinflammatory pathway. J Immunol. 2008;180:7175–7183. doi: 10.4049/jimmunol.180.11.7175. [DOI] [PubMed] [Google Scholar]
- 45.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanno T, Sudo K, Maekawa M, Nishimura Y, Ukita M, Fututake K. Lactate dehydrogenase M-subunit deficiency: a new type of hereditary exertional myopathy. Clinica Chimica Acta. 1988;173:88–98. doi: 10.1016/0009-8981(88)90359-2. [DOI] [PubMed] [Google Scholar]
- 47.Costa Leite T, Da Silva D, Guimarães Coelho R, Zancan P, Sola-Penna M. Lactate favours the dissociation of skeletal muscle 6-phosphofructo-1-kinase tetramers down-regulating the enzyme and muscle glycolysis. Biochem J. 2007;408:123–130. doi: 10.1042/BJ20070687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, et al. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci USA. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y, Dai Z, Barbacioru C, Sadee W. Cystine-glutamate transporter SLC7A11 in cancer chemosensitivity and chemoresistance. Cancer Res. 2005;65:7446–7454. doi: 10.1158/0008-5472.CAN-04-4267. [DOI] [PubMed] [Google Scholar]
- 50.Kourelis TV, Siegel RD. Metformin and cancer: new applications for an old drug. Med Oncol. 2012;29:1314–1327. doi: 10.1007/s12032-011-9846-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.