Abstract
17ß-Estradiol, an epigenetic modulator, is involved in the increased prevalence of migraine in women. Together with the prophylactic efficacy of valproate, which influences DNA methylation and histone modification, this points to the involvement of epigenetic mechanisms. Epigenetic studies are often performed on leukocytes, but it is unclear to what extent methylation is similar in other tissues. Therefore, we investigated methylation of migraine-related genes that might be epigenetically regulated (CGRP-ergic pathway, estrogen receptors, endothelial NOS, as well as MTHFR) in different migraine-related tissues and compared this to methylation in rat as well as human leukocytes. Further, we studied whether 17ß-estradiol has a prominent role in methylation of these genes. Female rats (n = 35) were ovariectomized or sham-operated and treated with 17β-estradiol or placebo. DNA was isolated and methylation was assessed through bisulphite treatment and mass spectrometry. Human methylation data were obtained using the Illumina 450k genome-wide methylation array in 395 female subjects from a population-based cohort study. We showed that methylation of the Crcp, Calcrl, Esr1 and Nos3 genes is tissue-specific and that methylation in leukocytes was not correlated to that in other tissues. Interestingly, the interindividual variation in methylation differed considerably between genes and tissues. Furthermore we showed that methylation in human leukocytes was similar to that in rat leukocytes in our genes of interest, suggesting that rat may be a good model to study human DNA methylation in tissues that are difficult to obtain. In none of the genes a significant effect of estradiol treatment was observed.
Introduction
Migraine is a neurovascular disorder affecting 8–17% of the population [1], [2]. Little is known about the cause of migraine and the prophylactic medication that is used to prevent migraine is only effective in half of the patients [3]. Family studies show a heritability of ∼40% [4], [5] and a monogenetic inheritance pattern has only been identified for familial hemiplegic migraine. Hence, there may be a major contribution of environmental (including hormonal) influences, possibly via epigenetic mechanisms. The methylene tetrahydrofolate reductase gene (MTHFR), encoding an important protein of the DNA methylation cycle, has been suggested to be involved in migraine [6], [7], although genome-wide association studies did not confirm this. However, they pointed to other genes involved in epigenetic mechanisms [8]–[10]. Moreover, the migraine prophylactic valproate, inhibits histone deacetylation as well as DNA methylation [11]. Finally, 17β-estradiol, which seems to be responsible for the 2–3 times higher prevalence of migraine in women compared to men, may exert at least part of its effects via epigenetic mechanisms [12], [13]. Thus, DNA methylation may be involved in migraine pathophysiology.
DNA methylation occurs at cytosines of CpG dinucleotides, often localized in CpG rich regions called CpG islands [14]. CpG island methylation is changed in different types of cancer, during aging, and probably also during the menstrual cycle [15]–[18]. Environmental factors that might be responsible for these epigenetic mechanisms include female hormones and nutrition [19], [20].
DNA methylation can differ greatly between tissues, so ideally it should be studied in the tissue of interest. For complex brain diseases like migraine, this is technically impossible in humans. Yet, blood can be derived easily, and thus if the DNA methylation pattern of leukocytes is correlated to that of other tissues, this would allow conclusions from methylation studies in leukocytes.
The aim of our study was to compare the methylation of genes that are probably involved in the generation of a migraine attack and might be epigenetically regulated [21], [22], as well as to examine the role of 17ß-estradiol in the methylation of our genes of interest. We focussed on the CGRP-ergic system because of its prominent role in migraine [23] We investigated DNA methylation in the Calcitonin related peptide alpha (Calca), receptor activity-modifying protein 1 (Ramp1), calcitonin receptor component protein (Crcp), calcitonin receptor-like receptor (Calcrl), upstream stimulating factor 2 (Usf2), Estrogen receptor 1 (Esr1), G-protein coupled estrogen receptor 1 (Gper), nitric oxide synthase 3 (Nos3)) and Mthfr genes in several tissues relevant to the pathophysiology of migraine (dura mater, trigeminal ganglion and trigeminal caudal nucleus). We investigated methylation in leukocytes and aorta as a peripheral control. To investigate whether DNA methylation in rats may be representative of that in humans, we also studied DNA methylation in leukocytes obtained from healthy human females.
Materials and Methods
Ethics statement
The animal experiments were performed in our laboratory with permission of the ethics committee of the Erasmus Medical Center in Rotterdam, The Netherlands (Permit Number: EMC2345(127-11-02)). All surgeries were performed under sodium pentobarbital anesthesia and all effort was made to minimize suffering.
The human blood samples were obtained from the Rotterdam Study [24], which has been approved by the institutional review board (Medical Ethics Committee) of the Erasmus Medical Center and by the review board of The Netherlands Ministry of Health, Welfare and Sports. All subjects provided written informed consent.
Animals
Female Sprague Dawley rats (Harlan Netherlands, Horst, The Netherlands) (N = 11–12 per group, weight at the start of the study ∼250 g) (See Table S1 in File S1) were kept at room temperature (22°C) at a 12/12 hours dark/light cycle with unlimited access to food and water in their home cages. The animals were anesthetized (50 mg/kg) and ovariectomized or sham-treated on day 1 of the study. After 7 days, a pellet releasing placebo or 17β-estradiol (21-day release pellet, 12 µg/day, Innovative Research, USA) was implanted subcutaneously in the neck. Blood samples were collected on day 1, 7 and 21 for CGRP and hormone measurements (See Table S2 in File S1). On day 21, the animals were sacrificed via an overdose of sodium pentobarbital (200 mg/kg). A leukocyte differentiation count was performed to verify whether the proportion of different types of leukocytes was the same for all animals. 0.5 ml whole blood, dura mater, trigeminal ganglia, caudal nuclei and a 5-mm segment of thoracic aorta were snap frozen in liquid nitrogen and stored at −80°C for DNA isolation. A vaginal smear was taken at all three time points to establish the phase of the estrous cycle according to proportions of epithelial cells, cornified cells and leukocytes present in the smear. Because an epigenetic study as in the current experiments has not been performed previously, we based the number of animals on our previous results, showing increased vascular endogenous CGRP responses after treatment with 17β-estradiol [25] and explored whether differences in DNA methylation caused by 17β-estradiol can be demonstrated in this model.
DNA methylation measurements
DNA of leukocytes, thoracic aorta, dura mater, trigeminal ganglia and caudal nuclei from all animals was isolated (DNeasy, Qiagen, Germantown, MD, USA) and quantified (Nanodrop, Thermo Scientific, Wilmington, DE, USA).
CpG islands of the genes of interest were determined using UCSC (http://genome.ucsc.edu/) and Ensemble genome browsers (http://www.ensembl.org). Primers were designed for each CpG island located in the promoter region of the genes of interest with the EpiDESIGNER primer design software (Sequenom, San Diego, CA, USA, Table 1). When the CpG island could not be covered in total, multiple primer sets were designed to cover the largest possible part of the CpG Island. With the BiSearch web server [26] primer sets were checked for having only one product.
Table 1. Primers and conditions for PCR on CpG island located in the promoter region of genes of interest.
| Amplicon | Primer Sequence | Amplicon size (bp) | Number of analyzed CpG's |
| Calca-part 1 | Forward-TTTAAATGGTGTTATTTTGTTAGATGTT Reverse-TAAACAAAAACCTCAAAACTCACCT | 282 | 7 |
| Calca-part 2 | Forward-GTAATTGTGGTTGTTGGTTTTTGTT Reverse-CACCAAATAAACCCTAAAATTCCTA | 405 | 7 |
| Ramp1 | Forward-GGGGGTTATGGTAAGTAGAGTTT Reverse-TTACAAAACAAACCCCAAAATAACT | 294 | 17 |
| Crcp | Forward-TTAGTAGTGGGTTTAGGAAGAGAGTG Reverse-CTAAAAAACAAAATTCTAAATACACAAAAC | 290 | 11 |
| Calcrl | Forward-GGGTTTTGTTGTTTGGATTTTA Reverse-TCCAAAAACTTACCTTATAACCTATTCA | 244 | 2 |
| Usf2 | Forward-GGTAGTAGTGTTGATTTTGGTGGG Reverse-TCACCTAACCTCCATTACTCTCTATAC | 411 | 15 |
| Esr1-part 1 | Forward-GGTAGTAGGGTATTTGGTGGTTATG Reverse-CAACTCAAAATACCCATAAAAAAAA | 321 | 9 |
| Esr1-part 2 | Forward-TTTTTTGATTTTTTAGAAGGGTGG Reverse-TAATCTAAAAACTTTCCCCCAACTC | 293 | 13 |
| Gper1-part 1 | Forward-GAAGATTATTTTTAGGGTTTTTTGTTTG Reverse-ACACCTTCATATCCCTCTTCCTACAA | 397 | 7 |
| Gper1-part 2 | Forward-TTGTTGTATATGTTGATTTGTAGGAAGA Reverse-CCCAAACCTATACTTCATCAACCTAA | 152 | 2 |
| Nos3 | Forward-TTTTTGTAAAGAAAAATTTTGGGTG Reverse-CACAATAAAAACTACCCCTAAACCT | 404 | 11 |
| Mthfr | Forward-GGGGTGATAGTTATTATAAGTTTATAGGTT Reverse-TTAACTAACTTCCCAAAAAACCTCC | 244 | 12 |
A 10mer (aggaagagag) sequence was added to the forward primers and a T7 sequence (cagtaatacgactcactatagggagaaggct) to the reverse primers. The primers were obtained from Invitrogen (Life Technologies Corporation, Carlsbad, CA, USA).
500 ng DNA of all tissue samples was treated with bisulfite (EZ-96 DNA-methylation kit (Shallow), Zymo Research, Irvine, CA, USA) for 16 hours to convert all non-methylated cytosines into uracil nucleotides. After bisulfite conversion PCR, reverse transcription and uracil specific cleavage was performed. For quantitative DNA methylation measurements the MassARRAY EpiTYPEr was used (Sequenom, San Diego, CA, USA). 2.5 µg of low methylated and 2.5 µg high methylated genomic rat DNA (EpigenDx, Hopkinton, MA, USA) were mixed into 0%, 25%, 50%, 75% and 100% methylated DNA as a control and also treated with bisulfite. To check the accuracy of the methylation measurements, methylation of a standard curve, of which the % of methylation is known (0–100%, increasing in steps of 10%), was measured for all amplicons (See Figure S1 in File S1). After the validation steps, a PCR was performed in triplo for all amplicons on the bisulfite-treated samples. Percentage methylation for each CpG in all genes and tissues was calculated with Sequenom EpiTYPER software (MassARRAY EpiTYPER Analyzer software v1.0, build1.0.6.88 Sequenom, Inc, San Diego, USA). Samples were excluded when the standard deviation of triplos was larger than 10%.
Genome-wide methylation study in humans
DNA methylation profiles from whole blood were assessed in a subset of 395 healthy women (age ≥45) from the Rotterdam Study-III, a population-based cohort study in the Netherlands. The design and rationale of the study has been published previously [24]. Samples were excluded when showing a low detection rate (<99%), incomplete bisulfite conversion, or gender swaps. Probes with a detection p-value >0.01 in >1% samples, were filtered out. β-values for the two assay-types on the array were corrected with SWAN (Subset-Within-Array-Normalization). Mean and standard deviation for the probes in the equivalent human genomic regions were calculated for 395 samples that passed quality control filters.
Data analysis
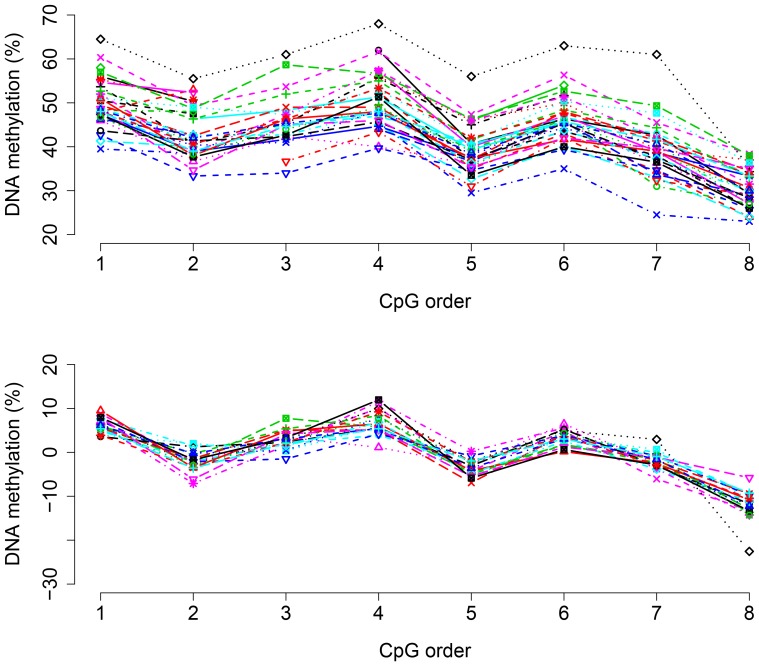
Calculations of differences in methylation and correlation between tissues were performed using SPSS software. The linear mixed model was used to estimate the size and standard errors of the effects of tissue and treatment. The model contains random effects for the individual rats, to correct for the strong correlations between CpGs within a gene. Typically, when one CpG shows a low (or high) level of methylation for a chosen rat, all the other CpGs show a low (or high) level as well. A strong example is shown in Fig. 1, for the Crcp gene in the trigeminal ganglion. The profiles for different rats run more or less in parallel, but at quite different levels. Subtracting the mean of all CpGs per rat removes the differences almost completely. The computations were done with the R system [27], version 2.15.2, using the function lmer in the library lme4.
Figure 1. Methylation of the Crcp gene in the trigeminal ganglion.
Methylation of each studied CpG of the promoter region of the Crcp gene is shown without correction for rat levels (upper panel) and with correction for rat levels (lower panel).
Results
DNA methylation measurements
Because all CpGs per CpG island that we studied were strongly correlated, we calculated the mean methylation of a CpG island and used these values for further analysis (see paragraph 2.4, Statistics). DNA methylation below 5% was observed for the Calca, Ramp1, Usf2 and Mthfr genes in all tissues (Table 2). DNA methylation higher than 90%, was observed for the Gper gene in all tissues (Table 2). DNA methylation differed substantially between tissues for the Nos3, Esr1, Crcp and Calcrl genes. Nos3 showed a two times significantly higher DNA methylation in leukocytes than in the other tissues. Methylation of the Crcp gene was high in aorta, leukocytes and trigeminal caudal nucleus, and significantly lower in dura mater and trigeminal ganglion. The Calcrl gene showed low methylation in leukocytes, intermediate methylation in dura mater and trigeminal caudal nucleus and high methylation in aorta and trigeminal ganglion. DNA methylation of Esr1 was significantly higher in the aorta than in other tissues (Fig. 2 and Table 2). Remarkable is that the variation in DNA methylation is small in some tissues and large in others. This is not a tissue-specific pattern, but it varies between tissues for the Nos3, Esr1, Crcp and Calcrl genes, as illustrated by the differences in standard deviation (Table 2).
Table 2. Mean methylation of CpG islands of candidate genes in aorta, leukocytes, dura mater, trigeminal caudal nucleus and trigeminal ganglion.
| Aorta | Leukocytes | Dura | Trigeminal Caudal Nucleus | Trigeminal Ganglion | |
| Mean ± SD (%) | Mean ± SD (%) | Mean ± SD (%) | Mean ± SD (%) | Mean ± SD (%) | |
| Calca * | 2.3±0.6 | 4.2±1.6 | 2.4±0.7 | 2.8±1.0 | 2.5±1.1 |
| Ramp1 * | 2.3±0.5 | 2.8±0.8 | 2.3±0.5 | 3.2±0.7 | 2.5±0.6 |
| Crcp * | 84.3±2.4 | 91.0±2.5 | 82.1±1.6 | 41.1±14.1 | 43.2±5.1 |
| Calcrl * | 70.6±17.6 | 2.9±2.2 | 28.2±9.6 | 15.0±13.5 | 69.6±10.3 |
| Usf2 | 1.5±0.3 | 1.7±0.6 | 1.5±0.3 | 1.5±0.3 | 1.6±0.3 |
| Esr1 * | 13.4±2.6 | 3.4±0.9 | 4.3±0.9 | 4.1±1.3 | 3.0±0.8 |
| Gper * | 93.7±0.8 | 93.4±1.7 | 92.6±0.7 | 90.2±2.2 | 92.9±0.8 |
| Nos3 * | 6.4±3.9 | 23.0±14.6 | 9.7±5.1 | 7.1±6.0 | 6.1±5.6 |
| Mthfr | 2.0±0.4 | 2.0±0.6 | 1.9±0.5 | 2.2±0.6 | 1.9±0.4 |
*) Significant differences between tissue means are present within the respective gene. P-value <0.0001.
Figure 2. DNA methylation of Crcp, Calcrl, Esr1 and Nos3 in different tissues.
Mean methylation of the promoter region of the Crcp (upper left panel), Calcrl (upper right panel), Esr1 (lower left panel) and Nos3 (lower right panel) genes is shown for the different tissues studied. Each symbol represents one animal and the different colors and symbols represent the treatment of the animals. Red circle: OVX+17β-estradiol; Green square: OVX+placebo; Blue triangle: sham+placebo.
Estradiol treatment did not induce any statistically significant differences in DNA methylation in any of the examined genes in any investigated tissue (Fig. 2). Therefore, the data of the different treatment groups were pooled for further statistical analyses, where treatment effect was not included.
The DNA methylation of leukocytes and that of the other tissues were compared to each other for all genes. No correlation was seen between DNA methylation in the leukocyte samples and samples from other tissues (Table 3). A leukocyte differentiation analysis was performed, but no influence was found of leukocyte composition on methylation of blood samples for any of the genes (data not shown).
Table 3. Relationship of methylation of leukocytes with other tissues.
| Methylation of leukocytes compared to other tissues (Pearsons r1) | ||||
| Aorta | Dura | Trigeminal Caudal Nucleus | Trigeminal Ganglion | |
| Calca | −0.23 | −0.33 | −0.38 | −0.01 |
| Ramp1 | 0.26 | 0.64 | 0.40 | 0.40 |
| Crcp | 0.04 | 0.33 | 0.09 | 0.17 |
| Calcrl | 0.08 | 0.29 | 0.52 | −0.37 |
| Usf2 | 0.07 | 0.50 | 0.07 | −0.06 |
| Esr1 | 0.43 | 0.53 | 0.18 | 0.58 |
| Gper | 0.01 | 0.02 | 0.17 | 0.07 |
| Nos3 | 0.02 | 0.18 | −0.36 | −0.38 |
| Mthfr | 0.33 | −0.09 | −0.02 | 0.13 |
) The relevance of the correlations can best be appreciated from a prediction perspective. Assume that we are interested in estimating the mean amount of methylation of some gene in some tissue. If we have no specific information, the best we can do is take the observed mean in our sample. The uncertainty is quantified by the observed standard deviation (SD). If a linear relationship with the mean amount of methylation in blood exists, a prediction model could be derived. Using basic statistical theory, one can show that the uncertainty is reduced to c * SD, where c follows form the formula c2 = 1−r2, if r is the correlation coefficient. To get c = 0.5, thus halving the uncertainty, r has to be as high as 0.87. Conversely, when r = 0.5, c = 0.87 and thus only a 13% reduction is obtained. For reliable prediction really high correlations (e.g. r = 0.97 for c = 0.25) are needed.
Concordance of DNA methylation in human and rat leukocytes
Human female DNA methylation data of our genes of interest were obtained from a genome wide methylation array. Human DNA methylation data of exactly the same genomic region of rat DNA was available for the CALCA, CALCRL, USF2, ESR1, GPER and MTHFR genes. For the other genes, RAMP1, CRCP and NOS3, the region studied in rat was very close (<100 bp) to the regions studied in human. We observed comparable values of DNA methylation in our genes of interest in human leukocytes as in rat leukocytes; the two genes with a high percentage of DNA methylation in the rat had also a high percentage of DNA methylation in the human samples, while the seven genes with a low percentage of DNA methylation were also concordant between the rat and human samples (Table 4, Fig. 3).
Table 4. Methylation of amplicons of rat genes compared to homologues regions of the human genome.
| % DNA methylation Rat | % DNA methylation human | Location of rat amplicons in human genome (Build 37) | Location Illumina probes (Build 37) | Accession Number Rat | Accession Number Human | ||
| High DNA methylation | Crcp | 91.0±2.5 | 86.0±5.8 | 7:65587384–7:65587475 | 7:65583242–7:65604635 | NM_053670.3 | NM_014478.4 |
| Gper1 | 93.4±1.7 | 98.3±0.8 | 7:1131643–7:1132074 | 7:1132036 | NM_133573.1 | NM_001505.2 | |
| Low DNA methylation | Calca | 4.2±1.6 | 30.0±3.8 | 11:14993934–11:14994000 | 11:14993929–11:14293977 | NM_017338.2 | NM_001741.2 |
| Calcrl | 2.9±2.2 | 20.8±5.7 | 2:188312541 | 2:188312833 | NM_012717.1 | NM_005795.5 | |
| Esr1 | 3.4±0.9 | 6.1±2.3 | 6:152129135–6:152129504 | 6:152129388–6:152129400 | NM_012689.1 | NM_000125.3 | |
| Mthfr | 2.0±0.6 | 1.8±1.0 | 1:11866155–1:11866365 | 1:11866155–1:11866236 | NM_005957.4 | XM_001074061.4 | |
| Nos3 | 23±14.6 | 19.7±4.3 | 7:150710828–7:10571127 | 7:150710730–7:150711138 | NM_021838.2 | NM_000603.4 | |
| Ramp1 | 2.8±0.8 | 6.1±2.3 | 2:238768163–2:238768240 | 2:238768104–2:238828641 | NM_031645.1 | NM_005855.2 | |
| Usf2 | 1.7±0.6 | 6.8±2.3 | 19:345760455–19:35760666 | 19:35760554 | NM_031139.1 | NM_003367.2 | |
Figure 3. Rat leukocyte DNA methylation compared to human leukocyte DNA methylation.
The methylation of DNA from rat leukocytes is compared to methylation of DNA from human leukocytes for different migraine-related genes. Our genes of interest that are high methylated in rat leukocytes, are also high methylated in human leukocytes and the genes that are low methylated in rat leukocytes are also low methylated in human leukocytes.
Discussion
DNA methylation and gene expression across tissues
DNA methylation and histone modifications, making the DNA accessible or locked for transcriptional regulation, are responsible for physiological processes like stem cell differentiation. Therefore, CpG island methylation may be similar between tissues but can also differ substantially. DNA methylation of the Calca, Ramp1, Usf2 and Mthfr genes was low in all tissues. Not taking into account other regulatory mechanisms, expression of these proteins would thus be expected in all investigated tissues. Notwithstanding the fact that tissue-specific information is not always available, all four genes are indeed expressed in several brain and cardiovascular tissues [28]–[33]. The expression in human leukocytes [34]–[37] suggests the expression in rat leukocytes as well.
In contrast, DNA methylation of the CpG island of the Gper gene was 90–94% in all investigated tissues, suggesting that expression of this gene might be low or absent. However, Gper mRNA is present in cells of the aorta [38] and in the central and peripheral nervous system [39] But the quantitative extent of this expression is unknown. Further, regulation of protein expression might even occur at high percentages of DNA methylation. Obviously, other regulatory mechanisms, such as histone modifications or non-epigenetic mechanisms like transcription factor binding to specific regions on the DNA, may also have an additional effect on protein expression. Hence, DNA methylation is not necessarily related to protein expression for every gene [40], [41].
Our results show that the variation in methylation differs between tissues in the Crcp, Calcrl, Esr1 and Nos3 genes, which are expressed in several brain and vascular tissues [32], [42]. ESR1 and NOS3 are also expressed in human leukocytes [43], [44] Large variations could result from contamination with other tissues, but variations would then be expected to be small in tissues that are a distinct anatomical entity, such as aorta and trigeminal ganglion. Furthermore, outliers potentially caused by contaminated tissue, should then be the same in all examined genes, which was not the case. It would be tempting to quantitatively relate differences in DNA methylation to differences in gene expression and subsequent translation into proteins in the different tissues. However, such a quantification between tissues, which is clearly beyond the scope of our study, would be hampered by the different characteristics of the tissues studied, with the consequent lack of a validated internal standard between tissues. Taken together, the substantial variations in DNA methylation between tissues for Crcp, Calcrl, Esr1 and Nos3 genes in some of the tissues (Fig. 2), suggests a regulatory role for DNA methylation in the expression of these genes in the respective tissues.
DNA methylation in leukocytes versus other tissues
DNA methylation is regularly examined in leukocytes with the aim of discovering biomarkers for several diseases [45], [46] because of the difficulty of obtaining affected tissues. For example, leukocyte DNA methylation is correlated to different types of cancer [47]–[49]. In our study, there was no correlation between DNA methylation of the candidate genes in leukocytes and that in the other tissues. Thus, it can be concluded that leukocytes are not representative for changes in DNA methylation in aorta, dura mater, trigeminal caudal nucleus and trigeminal ganglion for the migraine-related genes we investigated (Table 2).
Is the rat a valid model for human DNA methylation?
We studied whether DNA methylation of human leukocytes in our genes of interest is similar to DNA methylation of rat leukocytes. Since DNA methylation in human and rat leukocytes was found to be concordant, we hypothesize that this might also be the case for other tissues. This finding, combined with the obvious limitations in obtaining human tissues, suggests that animal models are relevant to study DNA methylation changes in specific tissues.
DNA methylation and environmental factors
DNA methylation may be influenced by environmental factors like nutrition [50] and maternal care [51]. Hormones like 17β-estradiol may also alter DNA methylation [52], [53]. Interestingly, female hormones probably play an important role in migraine as migraine is much more common in females than in males especially during the fertile part of their lives, and migraine attacks in females often start at the day before menstruation [54]. It is well known that estradiol affects epigenetic mechanisms [12], [13]. In addition, it potentiates the response to an important peptide in migraine pathophysiology, calcitonin gene-related peptide (CGRP) [24], [25], which is widely expressed in the central and peripheral nervous system [55]. We previously demonstrated that estrogen increases vascular sensitivity to CGRP and trigeminal innervation [25]. Estrogen can also modulate CGRP expression in the spinal portion of the trigeminal caudal nucleus and the cervical spinal cord [56], [57]. How estrogen causes these effects is still unknown, but a recent study showed that CGRP expression is epigenetically regulated [21]. Epigenetic mechanisms like DNA methylation may thus be involved in the differences in migraine prevalence between men and women.
When examining mean values of DNA methylation of the three different treatment groups (Fig. 2) some differences were observed between the groups, but these were not statistically significant. Because we are, to the best of our knowledge, the first to perform a study on DNA methylation of candidate genes in our field, we could not perform an adequate power calculation. We based our group sizes on our previous studies using the same animal model [25], where the same dose of and duration of treatment with 17β-estradiol as in the current study were adequate to induce changes in CGRP-ergic pathways. Furthermore, others have shown that DNA methylation patterns can change within hours or days [16], [58]. A study with a similar design as our current study showed that an environmental factor, bisphenol A, can increase methylation of the promoter region of the estrogen receptor [59], indicating that the methods we applied are suitable to detect changes in DNA methylation due to environmental factors.
The large variation in methylation in our investigated tissues is likely to have reduced the statistical power of our study when investigating the effects of 17ß-estradiol treatment. We observed a maximal difference of 8% methylation with a standard deviation of 18% between animals treated with 17β-estradiol and animals treated with placebo (data not shown). Based on a post-hoc power calculation, a group size of 318 animals would be needed to obtain sufficient statistical power to investigate these hormone effects. This is practically not feasible so another study setup, possibly an in vitro study, falling beyond the scope of the current study, is needed to investigate the effect of 17β-estradiol on DNA methylation. Thus, with our experiment, we cannot categorically exclude that the methylation of the genes that we have studied may be influenced by 17ß-estradiol.
Relevance of CpG islands
Methylation of CpG islands in and close to promoter regions of genes can cause a decrease in transcription of those genes during development and in certain types of cancer, Therefore, the focus in DNA methylation studies, including our current study, is often on CpG islands [60], [61]. CpG islands in or close to the promoter region of a gene are only present in 60% of the genes. Also in our case, there are genes that were of potential interest because they are involved in CGRP and estrogen signalling, as for example estrogen receptor 2 (Esr2) or the transient receptor potential cation channel, subfamily V, member 1 (Trpv1), but that are devoid of CpG islands in the rat genome. Therefore, we did not investigate these genes in our study. Recently, it has been shown that also methylation changes in enhancer regions of the DNA [62], as well as the CpG island shores [63], may induce differences between subjects, providing another regulatory region in which DNA methylation plays a role in the regulation of transcription. The general relevance of these findings, however, needs to be confirmed in future studies.
Overall conclusion
In conclusion, our results show that DNA methylation of the rat genes we studied is variable, tissue specific and cannot be extrapolated from leukocytes to other tissues. On the other hand, we observed a high degree of concordance between human and rat DNA methylation in leukocytes, suggesting that it is possible to study effects on DNA methylation in rat tissues that are difficult to obtain from humans, for example several types of brain tissues. The large variation of DNA methylation in the Crcp, Calcrl, Esr1 and Nos3 genes suggests that these genes are prone to changes in DNA methylation, but challenges drawing conclusions about the effect of 17β-estradiol.
Supporting Information
Figure S1. Example of standard curve, showing the distribution of methylation of the known samples for the Esr1 gene. Table S1. Body weight (g) and bodyweight changes (Δ, g) in rats after different treatments (n = 11–14). * p<0.05 compared to OVX placebo. Differences were calculated using one-way ANOVA. Table S2. Estradiol and CGRP concentrations measured at day 1, day 7 and day 21 and progesterone concentrations measured at day 21. * p<0.05 compared to both sham and OVX animals treated with placebo pellet. Differences were calculated using one-way ANOVA.
(DOCX)
Acknowledgments
We would like to thank Dr. Sandra Heil for her help with the leukocyte differentiations and Piet Kramer for his help in assessing the vaginal smears. We thank the members of the Genomics Lab and the ERGO support team for their help in sampling the data and in creating the database for the Rotterdam Study. The authors are grateful to the study participants, the staff from the Rotterdam Study and the participating general practitioners and pharmacists.
Funding Statement
The generation and management of genomics data for the Rotterdam Study is supported by the Netherlands Organisation of Scientific Research NWO investments (nr. 175.010.2005.011, 911-03-012) and the Netherlands Genomics Initiative (NGI)/NWO project nr. 050-060-810 (Netherlands Consortium for Healthy Aging; NCHA). The Rotterdam Study is funded by the Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organisation for the Health Research and development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII) and the Municipality of Rotterdam. Dr. Antoinette MaassenVanDenBrink was supported by the Netherlands Organization for Scientific Research (Vidi grant 917-11-349). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Stovner LJ, Andree C (2010) Prevalence of headache in Europe: a review for the Eurolight project. J Headache Pain 11: 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stovner L, Hagen K, Jensen R, Katsarava Z, Lipton R, et al. (2007) The global burden of headache: a documentation of headache prevalence and disability worldwide. Cephalalgia 27: 193–210. [DOI] [PubMed] [Google Scholar]
- 3. Goadsby PJ, Lipton RB, Ferrari MD (2002) Migraine–current understanding and treatment. N Engl J Med 346: 257–270. [DOI] [PubMed] [Google Scholar]
- 4. Chen CC, Mengersen KL, Keith JM, Martin NG, Nyholt DR (2009) Linkage and heritability analysis of migraine symptom groupings: a comparison of three different clustering methods on twin data. Hum Genet 125: 591–604. [DOI] [PubMed] [Google Scholar]
- 5. Nyholt DR, Gillespie NG, Heath AC, Merikangas KR, Duffy DL, et al. (2004) Latent class and genetic analysis does not support migraine with aura and migraine without aura as separate entities. Genet Epidemiol 26: 231–244. [DOI] [PubMed] [Google Scholar]
- 6. Oterino A, Toriello M, Valle N, Castillo J, Alonso-Arranz A, et al. (2009) The Relationship Between Homocysteine and Genes of Folate-Related Enzymes in Migraine Patients. Headache HED1484 [pii] 10.1111/j.1526-4610.2009.01484.x. [DOI] [PubMed] [Google Scholar]
- 7. Schurks M, Rist PM, Kurth T (2010) MTHFR 677C>T and ACE D/I polymorphisms in migraine: a systematic review and meta-analysis. Headache 50: 588–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wessman M, Terwindt GM, Kaunisto MA, Palotie A, Ophoff RA (2007) Migraine: a complex genetic disorder. Lancet Neurol 6: 521–532. [DOI] [PubMed] [Google Scholar]
- 9. Eising E, N AD, van den Maagdenberg AM, Ferrari MD (2013) Epigenetic mechanisms in migraine: a promising avenue? BMC Med 11: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Durham P, Papapetropoulos S (2013) Biomarkers associated with migraine and their potential role in migraine management. Headache 53: 1262–1277. [DOI] [PubMed] [Google Scholar]
- 11. Manev H, Uz T (2002) DNA hypomethylating agents 5-aza-2′-deoxycytidine and valproate increase neuronal 5-lipoxygenase mRNA. Eur J Pharmacol 445: 149–150. [DOI] [PubMed] [Google Scholar]
- 12. Green CD, Han JD (2011) Epigenetic regulation by nuclear receptors. Epigenomics 3: 59–72. [DOI] [PubMed] [Google Scholar]
- 13. Imamura T (2011) Epigenetic setting for long-term expression of estrogen receptor alpha and androgen receptor in cells. Horm Behav 59: 345–352. [DOI] [PubMed] [Google Scholar]
- 14. Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492. [DOI] [PubMed] [Google Scholar]
- 15. Bergman Y, Cedar H (2013) DNA methylation dynamics in health and disease. Nat Struct Mol Biol 20: 274–281. [DOI] [PubMed] [Google Scholar]
- 16. Zhao Z, Fan L, Frick KM (2010) Epigenetic alterations regulate estradiol-induced enhancement of memory consolidation. Proc Natl Acad Sci U S A 107: 5605–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gentilini D, Mari D, Castaldi D, Remondini D, Ogliari G, et al. (2012) Role of epigenetics in human aging and longevity: genome-wide DNA methylation profile in centenarians and centenarians' offspring. Age (Dordr) 10.1007/s11357-012-9463-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo SW (2012) The endometrial epigenome and its response to steroid hormones. Mol Cell Endocrinol 358: 185–196. [DOI] [PubMed] [Google Scholar]
- 19. Kvisvik EV, Stovner LJ, Helde G, Bovim G, Linde M (2011) Headache and migraine during pregnancy and puerperium: the MIGRA-study. J Headache Pain 12: 443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Novensa L, Novella S, Medina P, Segarra G, Castillo N, et al. (2011) Aging negatively affects estrogens-mediated effects on nitric oxide bioavailability by shifting ERalpha/ERbeta balance in female mice. PLoS One 6: e25335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park KY, Fletcher JR, Raddant AC, Russo AF (2011) Epigenetic regulation of the calcitonin gene-related peptide gene in trigeminal glia. Cephalalgia 31: 614–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiao J, Opal MD, Dulawa SC (2012) Gestational environment programs adult depression-like behavior through methylation of the calcitonin gene-related peptide gene. Mol Psychiatry mp2012136 [pii] 10.1038/mp.2012.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ho TW, Edvinsson L, Goadsby PJ (2010) CGRP and its receptors provide new insights into migraine pathophysiology. Nat Rev Neurol 6: 573–582. [DOI] [PubMed] [Google Scholar]
- 24. Hofman A, van Duijn CM, Franco OH, Ikram MA, Janssen HL, et al. (2011) The Rotterdam Study: 2012 objectives and design update. Eur J Epidemiol 26: 657–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gupta S, Villalon CM, Mehrotra S, de Vries R, Garrelds IM, et al. (2007) Female sex hormones and rat dural vasodilatation to CGRP, periarterial electrical stimulation and capsaicin. Headache 47: 225–235. [DOI] [PubMed] [Google Scholar]
- 26. Aranyi T, Varadi A, Simon I, Tusnady GE (2006) The BiSearch web server. BMC Bioinformatics 7: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.RCoreTeam (2012) R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 28. Finkelstein JD, Martin JJ, Kyle WE, Harris BJ (1978) Methionine metabolism in mammals: regulation of methylenetetrahydrofolate reductase content of rat tissues. Arch Biochem Biophys 191: 153–160. [DOI] [PubMed] [Google Scholar]
- 29. Carl GF (1986) Effect of chronic valproate treatment on folate-dependent methyl biosynthesis in the rat. Neurochem Res 11: 671–685. [DOI] [PubMed] [Google Scholar]
- 30. Li D, Peng J, Xin HY, Luo D, Zhang YS, et al. (2008) Calcitonin gene-related peptide-mediated antihypertensive and anti-platelet effects by rutaecarpine in spontaneously hypertensive rats. Peptides 29: 1781–1788. [DOI] [PubMed] [Google Scholar]
- 31. Park KY, Russo AF (2008) Control of the calcitonin gene-related peptide enhancer by upstream stimulatory factor in trigeminal ganglion neurons. The Journal of biological chemistry 283: 5441–5451. [DOI] [PubMed] [Google Scholar]
- 32. Eftekhari S, Edvinsson L (2011) Calcitonin gene-related peptide (CGRP) and its receptor components in human and rat spinal trigeminal nucleus and spinal cord at C1-level. BMC Neurosci 12: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Edvinsson L, Eftekhari S, Salvatore CA, Warfvinge K (2011) Cerebellar distribution of calcitonin gene-related peptide (CGRP) and its receptor components calcitonin receptor-like receptor (CLR) and receptor activity modifying protein 1 (RAMP1) in rat. Molecular and cellular neurosciences 46: 333–339. [DOI] [PubMed] [Google Scholar]
- 34. Crescenti A, Sola R, Valls RM, Caimari A, Del Bas JM, et al. (2013) Cocoa Consumption Alters the Global DNA Methylation of Peripheral Leukocytes in Humans with Cardiovascular Disease Risk Factors: A Randomized Controlled Trial. PLoS One 8: e65744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dumeaux V, Johansen J, Borresen-Dale AL, Lund E (2006) Gene expression profiling of whole-blood samples from women exposed to hormone replacement therapy. Molecular cancer therapeutics 5: 868–876. [DOI] [PubMed] [Google Scholar]
- 36. Brooks JT, Elvidge GP, Glenny L, Gleadle JM, Liu C, et al. (2009) Variations within oxygen-regulated gene expression in humans. J Appl Physiol 106: 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McHale CM, Zhang L, Lan Q, Li G, Hubbard AE, et al. (2009) Changes in the peripheral blood transcriptome associated with occupational benzene exposure identified by cross-comparison on two microarray platforms. Genomics 93: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lindsey SH, Cohen JA, Brosnihan KB, Gallagher PE, Chappell MC (2009) Chronic treatment with the G protein-coupled receptor 30 agonist G-1 decreases blood pressure in ovariectomized mRen2.Lewis rats. Endocrinology 150: 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brailoiu E, Dun SL, Brailoiu GC, Mizuo K, Sklar LA, et al. (2007) Distribution and characterization of estrogen receptor G protein-coupled receptor 30 in the rat central nervous system. J Endocrinol 193: 311–321. [DOI] [PubMed] [Google Scholar]
- 40. Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, et al. (2006) DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet 38: 1378–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, et al. (2007) Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466. [DOI] [PubMed] [Google Scholar]
- 42. Fehrenbacher JC, Loverme J, Clarke W, Hargreaves KM, Piomelli D, et al. (2009) Rapid pain modulation with nuclear receptor ligands. Brain research reviews 60: 114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scariano JK, Emery-Cohen AJ, Pickett GG, Morgan M, Simons PC, et al. (2008) Estrogen receptors alpha (ESR1) and beta (ESR2) are expressed in circulating human lymphocytes. J Recept Signal Transduct Res 28: 285–293. [DOI] [PubMed] [Google Scholar]
- 44. Jenkins NT, Landers RQ, Prior SJ, Soni N, Spangenburg EE, et al. (2011) Effects of acute and chronic endurance exercise on intracellular nitric oxide and superoxide in circulating CD34(+) and CD34(−) cells. J Appl Physiol 111: 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soubry A, Schildkraut JM, Murtha A, Wang F, Huang Z, et al. (2013) Paternal obesity is associated with IGF2 hypomethylation in newborns: results from a Newborn Epigenetics Study (NEST) cohort. BMC Med 11: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bailey KA, Wu MC, Ward WO, Smeester L, Rager JE, et al. (2013) Arsenic and the epigenome: interindividual differences in arsenic metabolism related to distinct patterns of DNA methylation. J Biochem Mol Toxicol 27: 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM (2011) DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics 6: 828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Summers T, Langan RC, Nissan A, Brucher BL, Bilchik AJ, et al. (2013) Serum-based DNA methylation biomarkers in colorectal cancer: potential for screening and early detection. J Cancer 4: 210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bijron JG, Bol GM, Verheijen RH, van Diest PJ (2012) Epigenetic biomarkers in the diagnosis of ovarian cancer. Expert Opin Med Diagn 6: 421–438. [DOI] [PubMed] [Google Scholar]
- 50. Mathers JC (2006) Nutritional modulation of ageing: genomic and epigenetic approaches. Mech Ageing Dev 127: 584–589. [DOI] [PubMed] [Google Scholar]
- 51. Zhang TY, Labonte B, Wen XL, Turecki G, Meaney MJ (2013) Epigenetic mechanisms for the early environmental regulation of hippocampal glucocorticoid receptor gene expression in rodents and humans. Neuropsychopharmacology 38: 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lenz KM, Nugent BM, Haliyur R, McCarthy MM (2013) Microglia are essential to masculinization of brain and behavior. J Neurosci 33: 2761–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kolodkin MH, Auger AP (2011) Sex difference in the expression of DNA methyltransferase 3a in the rat amygdala during development. J Neuroendocrinol 23: 577–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Silberstein S, Merriam G (1999) Sex hormones and headache 1999 (menstrual migraine). Neurology 53: S3–13. [PubMed] [Google Scholar]
- 55. Eftekhari S, Edvinsson L (2010) Possible sites of action of the new calcitonin gene-related peptide receptor antagonists. Ther Adv Neurol Disord 3: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pardutz A, Multon S, Malgrange B, Parducz A, Vecsei L, et al. (2002) Effect of systemic nitroglycerin on CGRP and 5-HT afferents to rat caudal spinal trigeminal nucleus and its modulation by estrogen. Eur J Neurosci 15: 1803–1809. [DOI] [PubMed] [Google Scholar]
- 57. Moussaoul S, Duval P, Lenoir V, Garret C, Kerdelhue B (1996) CGRP in the trigeminal nucleus, spinal cord and hypothalamus: effect of gonadal steroids. Neuropeptides 30: 546–550. [DOI] [PubMed] [Google Scholar]
- 58. Byun HM, Nordio F, Coull BA, Tarantini L, Hou L, et al. (2012) Temporal stability of epigenetic markers: sequence characteristics and predictors of short-term DNA methylation variations. PLoS One 7: e39220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Doshi T, Mehta SS, Dighe V, Balasinor N, Vanage G (2011) Hypermethylation of estrogen receptor promoter region in adult testis of rats exposed neonatally to bisphenol A. Toxicology 289: 7482. [DOI] [PubMed] [Google Scholar]
- 60. Lamson G, Stockdale FE (1989) Developmental and muscle-specific changes in methylation of the myosin light chain LC1f and LC3f promoters during avian myogenesis. Dev Biol 132: 62–68. [DOI] [PubMed] [Google Scholar]
- 61. Momparler RL, Bovenzi V (2000) DNA methylation and cancer. J Cell Physiol 183: 145–154. [DOI] [PubMed] [Google Scholar]
- 62. Zhang B, Zhou Y, Lin N, Lowdon RF, Hong C, et al. (2013) Functional DNA methylation differences between tissues, cell types, and across individuals discovered using the M&M algorithm. Genome Res gr.156539.113 [pii] 10.1101/gr.156539.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Portela A, Esteller M (2010) Epigenetic modifications and human disease. Nat Biotechnol 28: 1057–1068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Example of standard curve, showing the distribution of methylation of the known samples for the Esr1 gene. Table S1. Body weight (g) and bodyweight changes (Δ, g) in rats after different treatments (n = 11–14). * p<0.05 compared to OVX placebo. Differences were calculated using one-way ANOVA. Table S2. Estradiol and CGRP concentrations measured at day 1, day 7 and day 21 and progesterone concentrations measured at day 21. * p<0.05 compared to both sham and OVX animals treated with placebo pellet. Differences were calculated using one-way ANOVA.
(DOCX)