Abstract
Purpose
To elucidate molecular pathways contributing to metastatic cancer progression and poor clinical outcome in serous ovarian cancer.
Experimental Design
Poor survival signatures from three different serous ovarian cancer datasets were compared and a common set of genes was identified. The predictive value of this gene signature was validated in independent datasets. The expression of the signature genes was evaluated in primary, metastatic, and/or recurrent cancers using qPCR and in situ hybridization. Alterations in gene expression by TGFβ1 and functional consequences of loss of COL11A1 were evaluated using pharmacologic and knockdown approaches, respectively.
Results
We identified and validated a 10-gene signature (AEBP1, COL11A1, COL5A1, COL6A2, LOX, POSTN, SNAI2, THBS2, TIMP3, VCAN) that is associated with poor overall survival in patients with high-grade serous ovarian cancer. The signature genes encode extracellular matrix proteins involved in collagen remodeling. Expression of the signature genes is regulated by TGFβ1 signaling and is enriched in metastases in comparison to primary ovarian tumors. We demonstrate that levels of COL11A1, one of the signature genes, continuously increase during ovarian cancer disease progression, with the highest expression in recurrent metastases. Knockdown of COL11A1 decreases in vitro cell migration and invasion and tumor progression in mice.
Conclusion
Our findings suggest that collagen-remodeling genes regulated by TGFβ1 signaling promote metastasis and contribute to poor overall survival in patients with serous ovarian cancer. Our 10-gene signature has both predictive value and biological relevance and thus may be useful as a therapeutic target.
Keywords: serous ovarian cancer, microarray, gene signature, TGF-beta, collagen, stroma
INTRODUCTION
Ovarian cancer is the fifth leading cause of cancer death among women and the most lethal gynecologic cancer in the United States with survival rates for advanced stage disease that have not changed in several decades (1, 2). Despite similarities in initial presentation, patients with high-grade serous ovarian cancer display a broad range of survival end points. Some patients develop a chronic-type disease and can be maintained on chemotherapy for more than 5 years while others are either intrinsically chemo-resistant or rapidly become chemo-resistant following an initial period of chemosensitivity. Once chemoresistance develops in these patients, their response rate to other second-line agents is very low. A reliable method that identifies poor outcome patients early in the course of their disease would facilitate timely inclusion into clinical trials or personalized treatment strategies. The OncotypeDX® and Mammaprint® assays for breast cancer are successful examples of this approach and have become the standard of care for individualized treatment decision-making in breast cancer (3, 4). Currently, there is no fully-validated and clinically-applied test to guide treatment decisions in ovarian cancer.
Several research groups have utilized expression profile data to develop signatures that predict clinical outcomes in ovarian cancer. Although each signature has an associated predictive ability, the gene signatures described to date exhibit little overlap and lack apparent biological relevance to poor outcome. The mechanisms by which individual genes or a group of genes contribute to poor clinical outcome are also not well understood. Consequently, there is not only a critical need for diagnostic classifiers that can assess the risk of poor survival in patients with serous ovarian cancer but also for a better understanding of the molecular mechanisms that are involved in tumor progression and can be used to develop new treatment strategies.
MATERIALS AND METHODS
Patients and samples
Snap-frozen and paraffin-embedded patient samples and paired clinical information were retrieved from the Department of Pathology and Laboratory Medicine and from the Women’s Cancer Program Biorepository at Cedars-Sinai Medical Center. All patients signed the consent form for bio-banking, clinical data extraction, and molecular analysis and received adjuvant chemotherapy. This study was approved by the Institutional Review Board at Cedars-Sinai Medical Center.
Microarray data analyses for the identification of a poor outcome gene signature
To identify poor outcome gene signatures, we used three datasets which primarily included high-grade, advanced-stage serous ovarian cancer samples: the Cancer Genome Atlas (TCGA) dataset (n=403) (5), the GSE26712 dataset (N=185) (6), and our own microarray dataset, the Karlan dataset (n=122). Expression data and clinical data for the TCGA and GSE26712 datasets were downloaded from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp) and the Gene Expression Omnibus (GEO) website (http://www.ncbi.nlm.nih.gov/gds/), respectively. Expression data and clinical data for the Karlan dataset have been deposited in GEO (GSE51088).
For the TCGA dataset, 403 samples of high-grade serous ovarian cancer that were profiled on the Affymetrix U133A platform were pre-processed with dChip (version 12/05/2011) software (7) as described in the manual. The genes were filtered using 2 criteria: 1) the signal intensity of each gene should be 1.00 < Standard Deviation / Mean < 10.00; and 2) the P call %, the percentage of gene identified as present (P) in the whole array should be ≥ 20%. The 1,458 genes that remained after filtration were used for unsupervised clustering. The Pearson correlation was used to calculate the similarity between genes and samples. The samples were clustered into four major groups. Survival analyses were performed using the dChip survival analysis module (7). In the group that was associated with the worst survival outcome in comparison to all other groups (P=0.01869), a cluster of 86 genes was identified (Supplementary Table S1A).
For the analysis of the GSE26712 dataset, the probe level raw expression data in the ovarian cancer samples (n=185) were analyzed using GeneSpring GX 11.5 (Agilent Technologies). The Robust Multi-chip Averaging (RMA) algorithm was applied to the Affymetrix human U133A microarray data. Background correction, normalization, and summarization were performed and a baseline transformation to the median of all samples was applied. Samples were grouped into 2 distinct survival groups: short survivors (overall survival (OS) < 2.0 years, n=52) and long survivors (OS > 4.0 years, n=67). A gene list was made by selecting genes with a fold change ≥1.5 between short survivors and long survivors and an independent t-test was applied to compare short and long survivors. The Benjamini-Hochberg multiple comparison adjustment was applied and the corrected P value <0.05 was considered statistically significant. The 68 genes that satisfy these criteria are listed in Supplementary Table S1B.
In the Karlan dataset, periostin (POSTN) was selected as a gene of interest because our previous studies demonstrated that POSTN expression is significantly increased in ovarian tumors in comparison to normal ovarian epithelia (8) and that POSTN is required for ovarian cancer progression (9, 10). The Rosetta Similarity Search Tool (ROAST) was used in 122 serous ovarian cancer patients to identify a cluster of 188 genes (Supplementary Table S1C) that were highly correlated (r range=0.701–0.919; P<0.0002) with POSTN expression and associated with poor survival (Karlan et al., manuscript submitted).
Functional annotation of the genes associated with poor survival in each dataset was accomplished using the Database for Annotation, Visualization and Integrated Discovery (DAVID) tool (Supplementary Table S2).
Validation of the 10-gene signature
Three discovery datasets (TCGA, GSE26712, and Karlan) and one independent dataset (Tothill (11)) were used to validate the predictive value of the 10-gene signature. Microarray data and clinical data from the Tothill validation dataset were downloaded from the GEO website. Expression values for the 10 genes were extracted from the microarray data. The ‘risk index’ of the 10 genes was calculated from a linear combination of the gene expression values and their estimated multivariable Cox proportional hazard regression coefficients. The median risk index was used to define two patient groups – one characterized by high expression of the 10 genes and the other by low expression of the 10 genes. The Kaplan-Meier method was used to estimate overall survival (OS) and the log-rank test was applied to compare OS across groups. All analyses were performed using R packages (http://www.r-project.org/). Additional validation of the gene signature using the web-based Kaplan-Meier Plotter tool (http://kmplot.com/) (12) is described in the Supplementary Figure 3 legend.
Microarray data comparison in primary and metastatic tumors
The Oncomine expression analysis tool (www.oncomine.org) was used to examine the expression of the signature genes in primary (P) and metastatic (M) serous ovarian cancers in three large datasets: Anglesio (P=74, M=16), Bittner (P=166, M=75), and Tothill (P=189, M=54). For the GSE30587 dataset, which contained nine pairs of matched primary and metastatic serous ovarian cancers, the GEO2R tool (www.ncbi.nlm.nih.gov/gds/) was used to identify the top 250 gene probes that are most differentially expressed between paired primary and metastatic tumors.
RNA isolation and qRT-PCR analysis
OpenArray® Real-Time PCR was used to measure mRNA expression in patient samples obtained from the Women’s Cancer Program Biorepository. Total RNA (2 µg) was extracted from snap-frozen tumors using the TRI reagent (Molecular Research Center, Inc) and reverse-transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). cDNA (120 ng) was mixed with TaqMan OpenArray Real-time Mix (Applied Biosystems) and loaded onto OpenArray® Real-Time PCR plates containing the following probes for 9 of the 10 signature genes: Hs00937468_m1 (AEBP1), Hs01097664_m1 (COL11A1), Hs00609088_m1 (COL5A1), Hs00942480_m1 (LOX), Hs00170815_m1 (POSTN), Hs00950344_m1 (SNAI2), Hs01568063_m1 (THBS2), Hs00165949_m1 (TIMP3), Hs00171642_m1 (VCAN) as well as the large ribosomal protein P0 internal control Hs99999902_m1 (RPLP0). The probe for COL6A2 was not available on this array. The qRT-PCR reaction was performed by the Cedars-Sinai Medical Center Genomics Core using the BioTrove OpenArray® NT Cycler System and the data were analyzed by the 2(−ΔC(T)) method. For other qRT-PCR analyses, total RNA was extracted using the RNeasy mini kit (Qiagen) and was reverse-transcribed to cDNA using the Quantitect Reverse Transcription Kit (Qiagen). A total of 50 ng of cDNA was mixed with primers and iQSYBR-Green Supermix (BioRad) in a 96-well plate format. The qRT-PCR reaction was performed using an iCycler thermo cycler (BioRad) and the data were analyzed by the 2(−ΔC(T)) method. Primers for human AEBP1 (VHPS–207), COL6A2 (VHPS-2115), LOX (VHPS-5341), SNAI2 (VHPS-8686), and TIMP3 (VHPS-9288) were purchased from realtimeprimers.com (http://www.realtimeprimers.com). Primer sequences for human COL5A1, COL11A1, POSTN, THBS2 and VCAN, and the ribosomal protein L32 (RPL32) which served as an internal control in this assay, are shown in Supplementary Table S3. Expression levels of the 10 genes in the two designated groups were plotted and analyzed by an unpaired t-test using GraphPad PRISM (version 6.0; Graph-Pad Software).
Analysis of the molecular pathway and upstream regulators
Selected gene lists obtained from the microarray analyses were uploaded to Ingenuity Pathway Analysis (Ingenuity® Systems) and a core biological pathway analysis was performed to identify molecular networks and upstream regulators.
Cell culture
The OVCAR3 cell line was obtained from Dennis Slamon (University of California, Los Angeles) in 2011 and the A2780 ovarian cancer cell line was purchased from Sigma. The authenticity of the OVCAR3 and A2780 cell lines was confirmed by Laragen using the Short Tandem Repeat (STR) method. The TRS3 cell line was generated by immortalizing stromal cells isolated from a normal ovary with SV40 T Antigen. The OVCAR3 and A2780 cells were cultured in DMEM (Corning) and RPMI 1640 media (Corning), respectively, supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. The TRS3 cells were cultured in MCDB 105 (Sigma) and 199 (GIBCO) (1:1) media supplemented with 15% fetal bovine serum and 1% penicillin-streptomycin. For TGFβ1 treatment, 105 cells were plated in 6-well plates, serum-starved overnight, pre-treated with 10 µM A83-01 (Sigma) for 30 minutes, then incubated with 10 ng/ml TGFβ1 (Sigma) for a designated time before harvesting. For the COL11A1 knockdown, 3×105 A2780 cells were incubated with 5×104 transduction units of MISSION shRNA lentiviral transduction particles specific for human COL11A1 or scrambled control (Sigma-Aldrich) with polybrene (8 µg/ml) for 24 hours and allowed to recover for 24 hours in fresh medium. Puromycin (5 µg/ml) selection was performed 72 hours after infection and polyclonal populations of infected cells were used for phenotype analysis. The COL11A1 knockdown was confirmed by real-time PCR and Western blotting. To assess cell proliferation, cells were plated in 96-well plates at 103 cells per well in quadruplicate and cell viability was measured on days 1, 2, 3 and 4 by the CellTiter-Glo luminescent cell viability assay (Promega), according to the manufacturer’s instructions. For the migration and invasion assays, 105 of A2780 cells with sh-COL11A1 or scrambled control were seeded in serum-free media onto 24-well inserts with 8 µm PET membrane (BD Biosciences) or a BD Matrigel invasion chamber (BD Biosciences). Medium with 10% FBS was placed in the wells beneath the chambers as a chemoattractant. Migrated cells at 24 hours and invasive cells at 48 hours were stained with the Diff-Quik stain set (Siemens Healthcare Diagnostics) and counted in four different fields under an Olympus BX43 upright microscope (Olympus). The average numbers of migratory/invasive cells in the two cell lines were compared by an unpaired t-test using GraphPad PRISM (version 6.0; Graph-Pad Software).
In situ hybridization
Probes for COL11A1, the housekeeping gene HPRT (positive control), the bacterial gene dapB (negative control) and the hybridization kit (RNAscope® 2.0 FFPE Assay) were purchased from Advanced Cell Diagnostics, Inc. Paraffin slides were processed by the Cedars-Sinai Pathology Core as recommended by the manufacturer. Slides were examined using the Olympus BX43 upright microscope (Olympus). Matched primary ovarian cancers, concurrent metastases and recurrent/persistent disease were scored using the Leica Systems imaging hub. The H score was determined as % positive stromal cells × intensity (0, 1+, 2+, 3+) under 10× objective in nine intratumoral fields.
Immunohistochemistry
Immunohistochemical detection of COL11A1 was performed on 4-µm formalin-fixed, paraffin-embedded tissue sections using the monoclonal COL11A1 antibody DMTX InvaScan (Oncomatrix) at 1:100 dilution. Staining was done on the Ventana Benchmark Ultra (Tucson, AZ) automated slide stainer using an onboard heat-induced epitope retrieval method in high pH CC1 buffer. The staining was visualized using the Ventana optiview DAB Detection System. The slides were subsequently counterstained with Mayer’s hematoxylin.
Tumor xenografts
Mouse injections were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of Cedars-Sinai Medical Center. Six week-old female nude (nu/nu) mice (Charles River Laboratories) were intraperitoneally injected with 107 cells in 100 µl of PBS and combined with 100 µl of Matrigel.
RESULTS
Identification of a 10-gene signature associated with poor overall survival in patients with serous ovarian cancer
With the goal of identifying genes that can predict poor overall survival, we analyzed three large microarray datasets that primarily included high-grade, advanced-stage, primary serous ovarian carcinoma samples: TCGA (n=403) (5), the GSE26712 dataset (N=185) (6), and the Karlan dataset (n= 122; GSE51088). The workflow and methods for the identification of genes associated with poor survival in each dataset are outlined in Supplementary Fig. S1. Comparison of the three resultant gene signatures of poor survival (Supplementary Table S1A–C) showed that a total of 61 genes were present in at least two of the three signatures (19 genes in TCGA and GSE26712; 38 genes in TCGA and Karlan; and 24 genes in GSE26712 and Karlan) and that 10 of these 61 genes were present in all three datasets (Fig. 1A). These 10 genes (AEBP1, COL11A1, COL5A1, COL6A2, LOX, POSTN, SNAI2, THBS2, TIMP3, VCAN) are known to be localized in the extracellular matrix and are involved in cell adhesion and collagen remodeling (Table 1). A Pearson correlation showed that expression of the 10 genes is highly correlated (Supplementary Fig. S2), suggesting their involvement in similar biological processes. The identification of 10 collagen-remodeling genes as a poor outcome gene signature suggests that collagen-remodeling might be a common biological process that contributes to poor overall survival among patients with serous ovarian carcinoma.
Fig. 1.
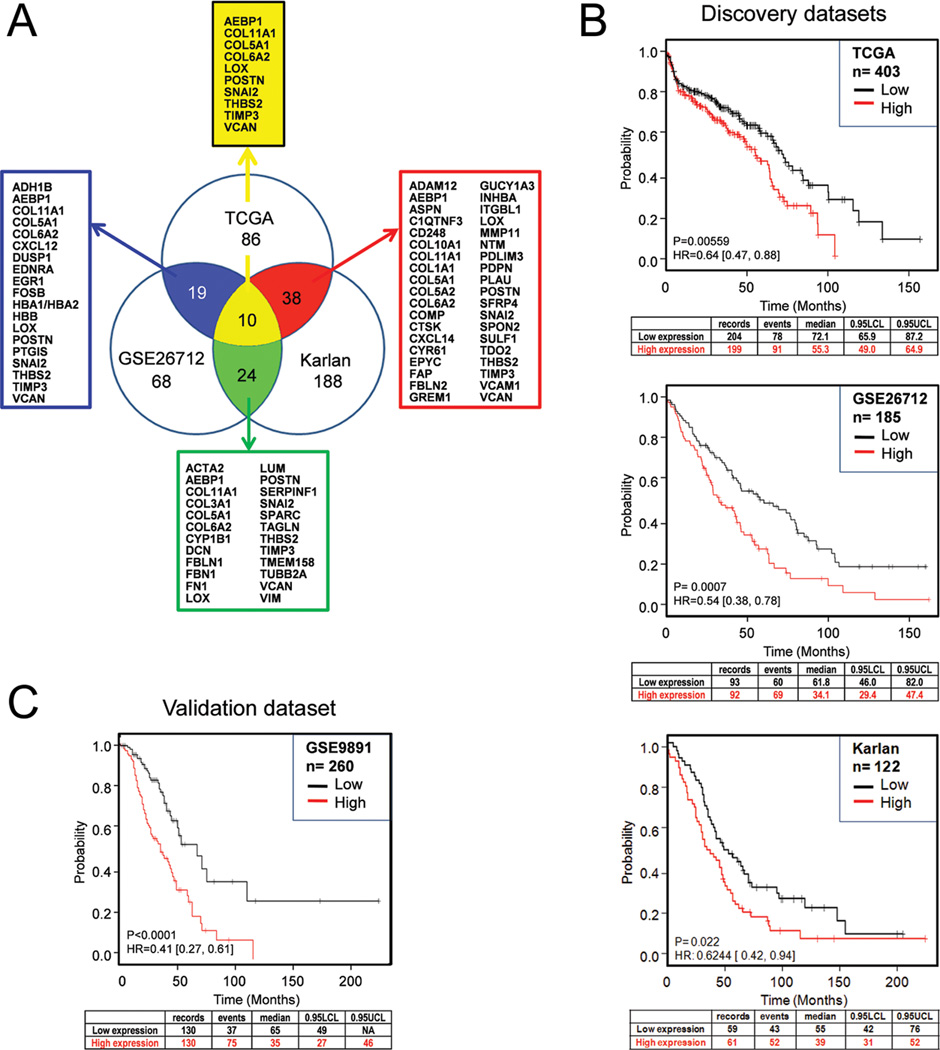
Identification and validation of the 10-gene signature associated with poor overall survival. (A) Venn diagram of poor outcome gene signatures identified from three microarray datasets (TCGA, GSE26712, and Karlan). The number of overlapping genes is indicated and arrows point to the corresponding lists of overlapping genes. The 10 genes present in all three signatures are listed at the top. (B) Validation of the predictive value of the 10-gene signature from three discovery datasets (TCGA, GSE26712, and Karlan) and (C) one independent validation dataset (Tothill). Kaplan-Meier curves, log-rank P values and hazard ratios (HR) are shown to compare overall survival between two patient groups with ‘high’ (indicated by the red line) and ‘low’ (indicated by the black line) expression of the 10-gene signature. The cutoff for the risk index is the median of the continuous risk factor. 0.95 LCL, the 95% lower confidence limit interval for the median time; UCL, upper confidence limit.
Table 1.
Ten genes associated with poor overall survival in all 3 datasets.
| Gene Symbol |
Affymetrix Probe ID |
Gene Name | Functions |
|---|---|---|---|
| AEBP1 | 201792_at | Adipocyte enhancer binding protein 1 | Transcriptional repressor with carbo-xypeptidase activity; Enhances adipocyte proliferation and reduces adipocyte differentiation |
| COL11A1 | 204320_at | Collagen, type XI, alpha 1 | Encodes one of the two alpha chains of type XI collagen, a minor fibrillar collagen |
| COL5A1 | 203325_s_at | Collagen, type V, alpha 1 | Encodes an alpha chain for one of the low abundance fibrillar collagens |
| COL6A2 | 213290_at | Collagen, type VI, alpha 2 | Encodes one of the three alpha chains of type VI collagen, a beaded filament collagen found in most connective tissues |
| LOX | 215446_s_at | Lysyl oxidase | An extracellular copper enzyme that initiates the crosslinking of collagens and elastin |
| POSTN | 210809_s_at | Periostin, osteoblast specific factor | Plays a role in cell adhesion and collagen remodeling; Induces cell attachment and spreading |
| SNAI2 | 213139_at | Snail homolog 2 (Drosophila) | A transcriptional repressor that binds to E-box motifs; Involved in epithelial-mesenchymal transition |
| THBS2 | 203083_at | Thrombospondin 2 | Encodes a disulfide-linked homotrimeric glycoprotein that mediates cell-to-cell and cell-to-matrix interactions |
| TIMP3 | 201147_s_at | Tissue inhibitor of metalloproteinase 3 | Encodes an inhibitor of the matrix metalloproteinases, a group of peptidases involved in the degradation of the extracellular matrix |
| VCAN | 204619_s_at | Versican | Encodes a large chondroitin sulfate proteoglycan, a major component of the extracellular matrix |
Validation of the 10-gene signature in predicting poor overall survival
Since the 10 signature genes were selected based on overlap among the three survival signatures rather than on predictive efficiency, we evaluated the potential predictive value of the 10-gene signature in the three discovery datasets and one independent validation dataset by comparing survival in patient groups with ‘high’ and ‘low’ expression of the 10 genes. In each of the 3 discovery datasets, the patient group with ‘high’ expression of the 10-gene signature had poor overall survival: TCGA (log-rank P=0.00559; HR=0.64 [0.47, 0.88]), GSE26712 (log-rank P=0.0007; HR=0.54 [0.38, 0.78]), and Karlan (log-rank P=0.022; HR=0.6244 [0.42, 0.94]) (Fig. 1B). For validation, we used the Tothill dataset (GSE9891) since it comprised a large number of serous ovarian cancer samples (n=260) with well-defined clinical outcome data (11). In this validation dataset, the 10-gene signature predicted poor overall survival with statistical significance (log-rank P<0.0001; HR=0.41 [0.27, 0.61]) (Fig. 1C). Similar results were obtained after adjusting for cancer stage (not shown). The 10 gene-signature also predicted poor overall survival with statistical significance (log-rank P=2.2E-05; HR=1.46 [1.22, 1.74]) when applied to a large combined ovarian cancer dataset (n=1,058) consisting of 10 publicly available datasets (Supplementary Fig. S3).
Regulation of poor outcome gene expression by the TGFβ signaling pathway
Ingenuity Pathway Analysis (IPA) using the 61 poor outcome signature genes that were present in at least two of the three initial discovery datasets (Fig. 1A) revealed that many of these genes form a network centered around TGFβ and collagens (Fig. 2A). Identification of upstream regulators by IPA also indicated TGFβ1 as the top molecule regulating expression of the 61-gene poor outcome signature (Fig. 2B). In addition to TGFβ1, other members of the TGFβ signaling pathway (TGFβ2, TGFβ3, SMAD3, and SMAD7) were also identified as top transcription factors regulating expression of the 61 poor outcome genes (Fig. 2B), suggesting that the TGFβ signaling pathway may be the main upstream regulator of these genes.
Fig 2.
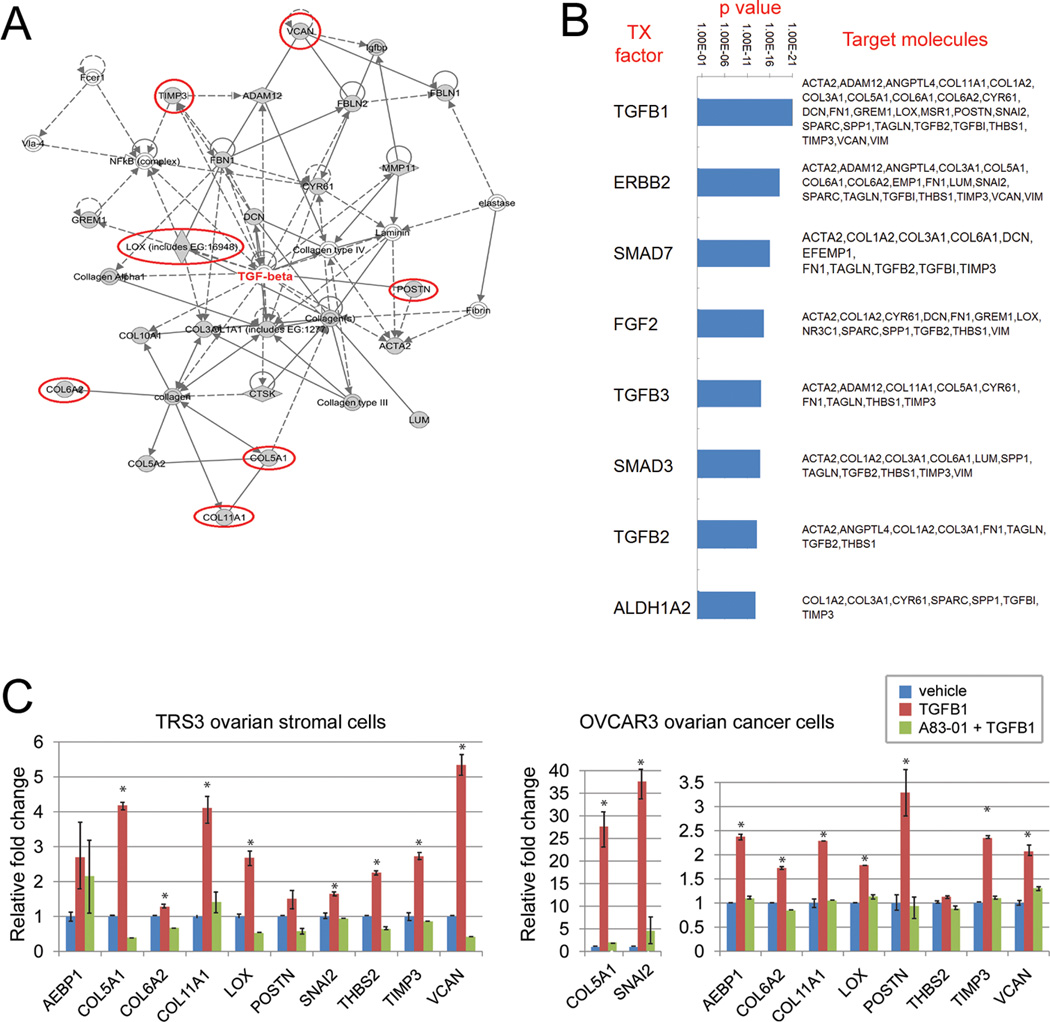
Regulation of the poor outcome signature genes by TGFβ signaling. (A) Ingenuity Pathway Analysis of the 61 genes present in at least two of the three discovery signatures of poor outcome. Genes that are present in all three discovery signatures are circled in red. (B) Top transcription factors regulating the 61 poor survival genes are ranked by p values. Downstream target genes are listed. (C) Induction of the 10 poor outcome signature genes by TGFβ1 in the ovarian stromal cell line TRS3 and the ovarian cancer cell line OVCAR3. Cells were treated with TGFβ1 (10 ng/ml) for 48 hours (TRS3) or 1–3 hours (OVCAR3) with or without pre-treatment with the TGFβ1 receptor inhibitor, A83-01. Shown is the relative fold change of the mRNA levels compared to untreated control cells. Data are presented as the mean +/− SEM in triplicate samples. * indicates P<0.05. Data are representative of at least three independent experiments.
To validate the predicted regulation of the poor outcome genes by TGFβ1, we treated the human ovarian stromal cell line TRS3 and the ovarian cancer cell line OVCAR3 with TGFβ1 and measured mRNA expression of the 10 poor outcome genes before and after TGFβ1 treatment. We included the ovarian stromal cell line since many of our poor outcome genes are thought to be expressed in stromal cells. Most of the 10 genes were found to be induced by TGFβ1 in both ovarian stromal and cancer cell lines (Fig. 2C). To further validate that this induction was mediated by TGFβ1 signaling, we measured expression of the 10 genes in cells pre-treated with the TGFβ1 receptor inhibitor A83-01 before adding TGFβ1. We found that the TGFβ1-induced expression of the 10 genes was abrogated by A83-01 (Fig. 2C).
Enrichment of the 10-gene signature in metastatic ovarian cancer
To identify the underlying biological mechanism that could explain the observed association of poor survival with high expression of the collagen-remodeling genes, we evaluated the expression of the 10 signature genes in primary and metastatic serous ovarian tumors in three Oncomine datasets that contained primary ovarian tumors (P) and metastatic tumors (M): Anglesio (P=74, M=16), Bittner (P=166, M=75), and Tothill (P=189, M=54). The probes used for the analysis are listed in Supplementary Table S4. Markedly higher expression levels of the 10 genes were observed in the metastatic tumors in all three datasets (Fig. 3A). The minimal difference in the expression of the epithelial marker, EPCAM, and the stromal marker, vimentin, in primary and metastatic tumors (Fig. 3A) indicates that the epithelium-to-stroma ratio is not significantly different between samples.
Fig 3.
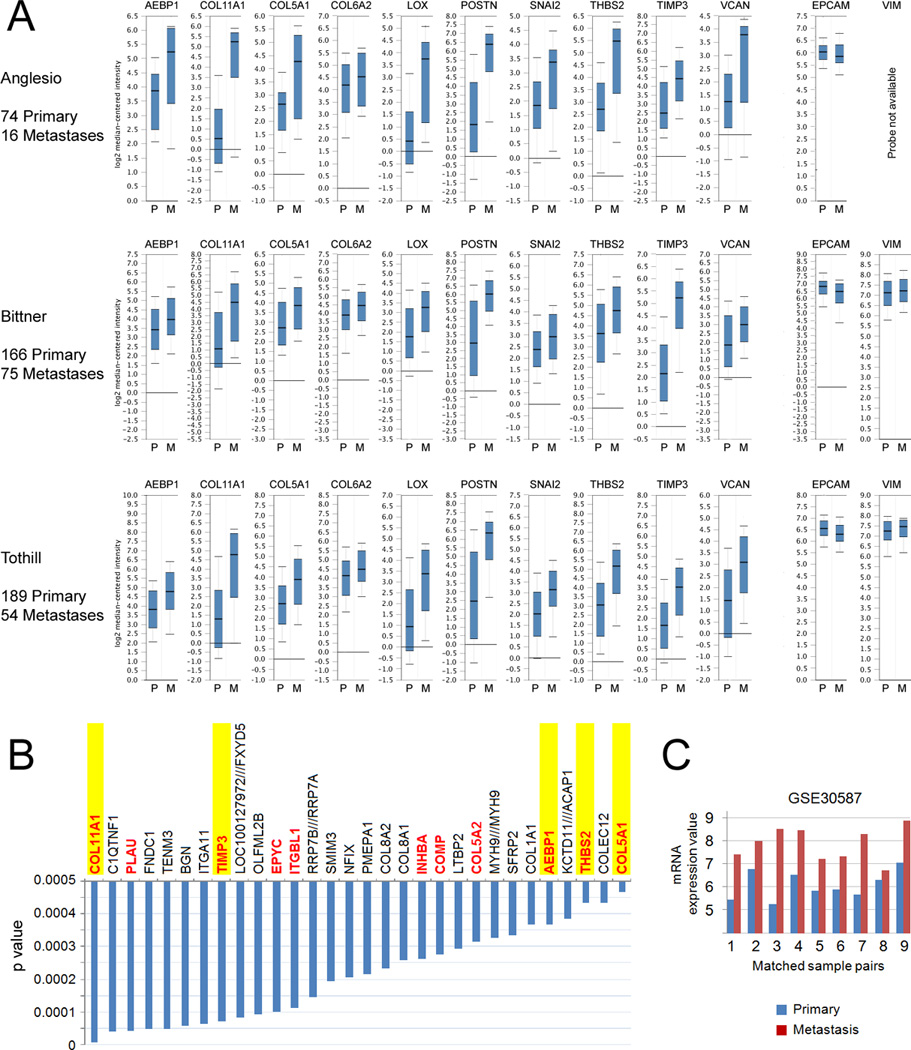
Enrichment of the 10-gene signature in metastatic ovarian cancer. (A) Oncomine mRNA expression analysis of the 10 poor outcome genes in three public ovarian cancer microarray datasets. Expression of the poor outcome genes are shown in primary (P) and metastatic (M) ovarian tumor samples using whisker plots with log2 median-centered intensity. EPCAM and VIM were used as markers of the relative content of epithelial and stromal cells, respectively. (B) List of genes enriched in metastases compared to primary tumors in the GSE30587 microarray dataset, which consists of nine matched pairs of primary and metastatic tumors. Genes that are present in at least two of the three poor prognosis signatures are in red font. Genes that overlap with the 10-gene signature are highlighted in yellow. (C) COL11A1 mRNA expression in nine matched primary and metastatic ovarian tumor samples in the GSE30587 microarray dataset.
We further validated that the signature genes are enriched during ovarian cancer progression by using another method of mRNA detection (qPCR) in an independent patient cohort that included 8 normal ovaries, 30 primary serous ovarian tumors, and 29 metastatic serous ovarian tumors from the Women’s Cancer Program Biorepository (Supplementary Fig. S4).
Next, we conducted an unbiased global identification of genes that are differentially expressed between primary tumors and metastases using nine matched pairs of primary ovarian tumors and omental metastases (GSE30587 dataset). The top 20 gene probes that exhibited increased expression in metastases are ranked according to statistical significance in Fig 3B. This analysis showed a marked overlap between our poor prognosis signature genes and genes that are enriched in metastases (Fig. 3B). One of our signature genes, COL11A1, was identified as the most statistically significant differentially expressed gene in the nine matched pairs of primary and metastatic tumor samples (Fig. 3B). Fig. 3C shows COL11A1 mRNA expression values in matched pairs of primary ovarian tumors and omental metastases in the nine matched tumor pairs.
Enrichment of COL11A1 during ovarian cancer disease progression
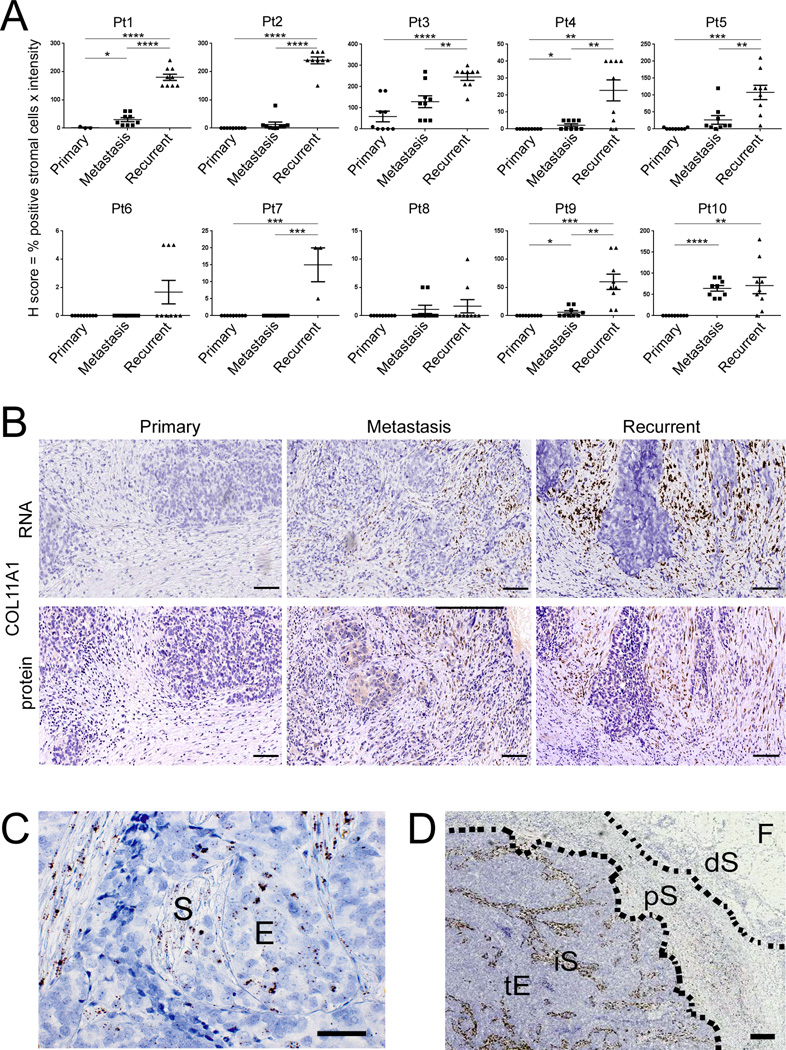
To test whether COL11A1 is a marker of tumor progression, we used in situ hybridization in 10 patients with “triplet” samples (primary ovarian cancer, concurrent metastasis, and recurrent/persistent metastasis) and eight additional patients with matched primary ovarian cancer and recurrent/persistent metastatic tumor samples (Supplementary Table S5). In each of the 18 patients, COL11A1 expression increased in the recurrent/persistent metastasis compared to the matched primary ovarian tumor (Fig. 4A,B and Supplementary Fig. S5). In the 10 patients with “triplet” samples, COL11A1 exhibited the lowest levels in primary ovarian cancer samples, medium levels in concurrent metastases, and highest levels in recurrent/persistent metastases (Fig. 4A). Representative in situ hybridization images for Patient 1 are shown in Fig. 4B. To correlate RNA and protein expression, serial sections from primary ovary, concurrent metastatic, and recurrent/persistent metastatic tumors were stained for the COL11A1 protein using immunohistochemistry. The COL11A1 protein levels and pattern of expression in these serial sections were consistent with COL11A1 RNA levels and pattern of expression (Fig. 4B); however, in situ hybridization provided a higher-resolution signal at a cellular level. COL11A1 expression was predominantly confined to stromal cells although rare clusters of positive epithelial cells were observed in some tumors (Fig. 4C). Interestingly, COL11A1 was specifically expressed in the intra/peri-tumoral stromal cells while stromal cells >1 mm from the epithelial tumor cells were always negative (Fig. 4D and Supplementary Fig. S6).
Fig 4.
Increase in COL11A1 expression during ovarian cancer progression. (A) Quantification of COL11A1 in situ hybridization signal in matched triplets of primary ovarian cancer, concurrent metastasis, and recurrent/persistent metastasis from 10 patients. H score = % positive stromal cells × intensity (0, 1+, 2+, 3+) under 10× objective. Each point represents the H score in a single field. Nine intratumoral fields were scored in each sample except for two samples in which only three fields were scored due to a minimal amount of tumor tissue. Data are presented as the mean +/− SEM. *P<0.05; **P<0.005; ***P<0.0005; ****P<0.0001. (B) Representative COL11A1 in situ hybridization and COL11A1 immunohistochemistry in serial sections of samples from Patient 1. (C) Detection of a positive focal COL11A1 in situ hybridization signal in cells exhibiting stromal (S) and epithelial (E) morphology. (D) Representative image of COL11A1 distribution in intra- and peri-tumoral areas. tE, tumor epithelium; iS, intratumoral stroma; pS, peritumoral stroma; dS, distant stroma; F, fat. Hematoxylin counterstain. Size bar is 100 µm in all panels.
Attenuation of tumor progression upon COL11A1 knockdown
To determine whether COL11A1 has a functional role in tumor progression, we used a mouse tumor xenograft model with A2780 human ovarian cancer cells. Despite their epithelial morphology, these cells exhibit a mesenchymal-like expression profile, including low levels of E-cadherin and high levels of N-cadherin proteins (Ruby Huang, Cancer Science Institute of Singapore, personal communication). A2780 cells have relatively high levels of endogenous COL11A1 and thus may represent the small subset of COL11A1-expressing epithelial tumor cells that we observed in patient tumors (Fig. 4C). COL11A1 expression in A2780 cells was silenced using shRNA lentiviral particles. Effective silencing of COL11A1 was confirmed by real-time PCR and Western blotting (Supplementary Fig. S7). COL11A1 knockdown resulted in decreased cell migration and invasion (Fig. 5A,B). To assess the effect of COL11A1 on tumor progression in vivo, we intraperitoneally injected nude mice with 107 sh-scr A2780 cells or sh-COL11A1 A2780 cells. The experiment was first conducted with 5 mice per group (Fig. 5C) and then replicated with 10 mice per group (Fig. 5D). In both sets of experiments, tumor growth was significantly reduced in mice injected with sh-COL11A1 A2780 cells compared to the mice injected with sh-scr A2780 cells (Fig. 5C,D).
Fig 5.
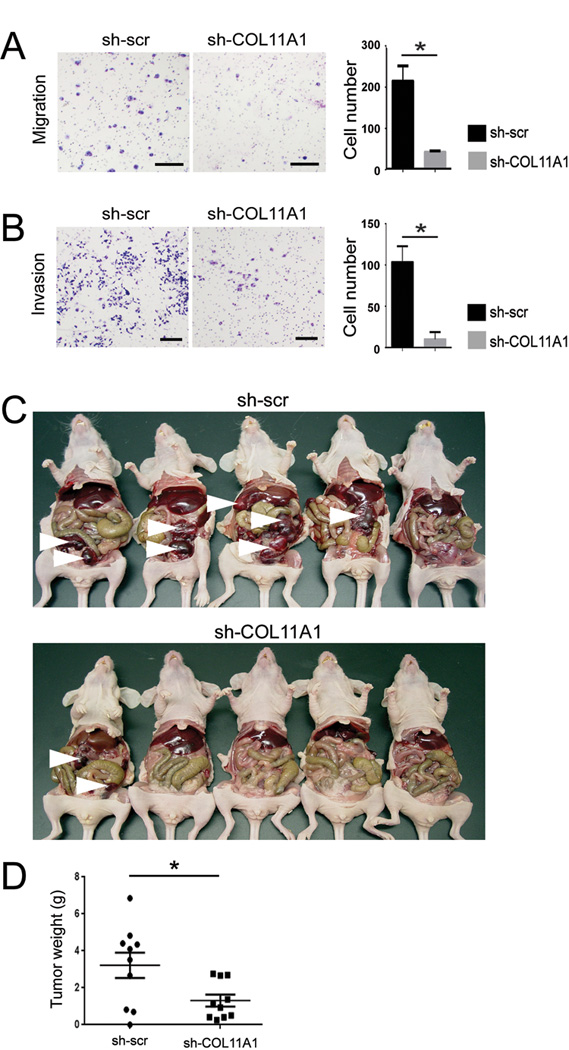
COL11A1 knockdown results in decreased cell migration, invasion, and tumor progression. (A) Migration and (B) invasion assays of A2780 cells with scrambled shRNA (sh-scr) or shRNA specific to COL11A1 (sh-COL11A1). Shown are representative images of migrated cells after 24 hours and invasive cells after 48 hours. Size bar, 25 µm. The bar graph shows the quantification of migrated cells in four different fields at 10× magnification and invasive cells in four different fields at 4× magnification. Data are presented as the mean +/− standard deviation. *P<0.05. (C) Photograph of nude mice with tumors that formed 14 days after intraperitoneal injection of A2780 cells transduced with scrambled shRNA control (sh-scr; 5 mice) or shRNA specific to COL11A1 (sh-COL11A1; 5 mice). White arrowheads indicate large tumor nodules. (D) Quantification of wet tumor weight after resection of tumor nodules from 20 mice in the replication experiment of intraperitoneal injection of A2780 cells transduced with sh-scr (10 mice) or sh-COL11A1 (10 mice). Each dot indicate an individual mouse. Data are presented as the mean +/− SEM, *P=0.02.
DISCUSSION
Expression profile data have been used extensively in efforts to develop gene signatures that relate to clinical outcomes in ovarian cancer. A key advantage of our 10-gene signature is that gene selections were based on overlap among three individual signatures of poor outcome, each of which had been derived using entirely different patient populations and statistical methods for microarray analyses. Thus, this signature should be independent of technical variations associated with microarray analyses and should be associated with poor survival in diverse patient populations. Indeed, these 10 genes are highly enriched in patient subgroups with the worst clinical outcome in published datasets, including the discovery and validation datasets used in this study. For example, among patient subgroups identified in the original ovarian TCGA study, our 10 signature genes exhibit the highest expression in the mesenchymal subgroup described by Verhaak et al. (13) or subtype 2 described by Zhang et al. (14), which has the worst survival in that dataset. Furthermore, genes involved in ‘cell adhesion’, ‘TGFβ binding’, and ‘epithelial mesenchymal transition’ were significantly upregulated in subtype 2 (14), similar to our observation. In the study by Tothill et al., our 10 genes are most highly enriched in the C1 subtype, which has the worst survival in that dataset (11).
Our 10-gene signature is robust in its ability to predict poor survival as demonstrated in two large validation datasets consisting of 260 ovarian cancer patient samples and 1,058 pooled ovarian cancer patient samples. Interestingly, individual genes or groups of genes from our 10-gene signature, including COL11A1, POSTN, SNAI2, THBS2 and TIMP3, have also been associated with poor survival in other solid tumors including breast, colorectal, lung, oral, and head and neck carcinomas as well as melanoma (15–23), suggesting that expression of this signature is not specific to ovarian cancer but might characterize aggressive behavior across cancer types.
Another major strength of our 10-gene signature is its clear biological relevance to cancer progression. Previously identified gene signatures in ovarian cancer consist of genes that are involved in many diverse biological processes, making it difficult to assess their biological relevance or functional role in cancer progression. All 10 of our signature genes are present in the 351-gene signature that was identified as upregulated in invasive ductal carcinoma (IDC) when compared to noninvasive ductal carcinoma in situ (DCIS) (24), supporting the important role of these genes in early local invasion. We further showed that the 10 signature genes are highly enriched in metastases and that knockdown of one of these genes, COL11A1, results in reduced cell migration, invasion, and tumor progression, suggesting that collagen remodeling could be important in ovarian cancer progression and metastasis. The higher expression of 10 genes in metastasis does not appear to be due to a higher stroma-to-tumor ratio in metastatic tumors for several reasons. First, tumor samples from the TCGA dataset were selected to have >70% tumor cells (5). Second, we did not observe different expression levels of the epithelial marker, EPCAM, and the stromal marker, vimentin, in metastatic tumors compared to primary tumors (Fig. 3A), indicating that the stroma-to-tumor ratio is not significantly different between samples of primary tumors and metastases. Third, our in situ hybridization results showed that regardless of the overall amount of stroma in tumor sections, COL11A1 expression was confined to intra-/peri-tumoral stromal cells and rare foci of tumor epithelial cells, while stromal cells that were >1 mm from epithelial tumor cells were completely negative (Fig. 4B-D). This indicates that COL11A1 is a specific marker of carcinoma-associated fibroblasts (CAFs) and possibly cancer cells that are undergoing EMT. The in situ hybridization analysis of COL11A1 in matched triplets of primary ovarian cancer, concurrent metastasis, and recurrent/persistent metastasis, demonstrated a marked increase in COL11A1 during cancer progression in all patients (Fig. 4A), indicating that COL11A1 could serve as a marker of cancer progression.
Collagen-rich stroma is thought to maintain tissue architecture and, under normal conditions, serve as a barrier to epithelial cell migration. However, when modified by cancer cells, collagen-rich stroma can promote tumor progression (25). Enhanced collagen deposition and cross-linking has been shown to increase breast cancer risk (26, 27). Increased levels of LOX, an enzyme responsible for collagen cross-link formation (28), result in increased collagen stiffness (29). POSTN also promotes collagen cross-linking by interacting with BMP-1 to enhance the proteolytic activity of LOX (30), which results in the reorganization of loose connective tissue into linear tracks of fibers that promote chemotaxis of tumor cells (27, 31). Furthermore, increased collagen deposition and remodeling increases interstitial pressure, thereby severely compromising the efficacy of drug delivery (25). Of particular interest, an increase in collagen expression and remodeling has been associated with cisplatin resistance in ovarian cancer (32–34), suggesting that cisplatin resistance might be one of the factors contributing to poor survival.
During the revision of this manuscript, Yeung and colleagues published an in vitro study demonstrating that VCAN and POSTN are induced by TGFβ and involved in ovarian cancer invasion (35), while Wu and colleagues showed that COL11A1 is not only a predictor of ovarian cancer recurrence and poor clinical outcome but also plays a role in ovarian cancer invasion in vitro and in mouse xenografts (36). We anticipate that future studies will reveal that most of our signature genes play critical functional roles in ovarian cancer progression.
Finally, the clinically-relevant strength of the 10-gene signature is that it can be not only used as a biomarker to identify patients with poor outcome but also as a guide to individualize their therapy. In fact, several of our 10 signature genes have been validated as promising therapeutic targets in mouse models. POSTN, an extracellular matrix protein that is highly expressed in late-stage ovarian cancer (9), is thought to play a role in metastatic colonization by forming a niche for cancer stem cells (37). Treatment with a POSTN-neutralizing antibody led to a significant decrease in ovarian tumor growth and metastasis in a mouse model (10). Similarly, inhibiting LOX by treatment with β-aminopropionitrile, neutralizing antibodies, or RNA interference inhibited tumor metastasis in xenograft and transgenic mouse models (38). Our COL11A1 knockdown result suggests that targeting collagen might be an effective approach to preventing ovarian cancer progression and metastasis. A recent study of collagen mimetic peptides (CMPs), 2–3 kDa small peptides that bind to type I collagen, showed that they can specifically bind to tumors with high matrix metalloproteinase (MMP) activity in xenograft models (39). This is a promising approach to treating tumors associated with excessive collagen remodeling and high MMP activity. Furthermore, we showed that collagen-remodeling genes are regulated by TGFβ1, suggesting that targeting TGFβ1 signaling might be an efficient way to impede metastatic progression. High TGFβ1 signaling activity was reported in patients with metastatic ovarian cancer (40) and the antibody against TGFβ was shown to be effective in suppressing metastasis in a preclinical model of ovarian cancer (41). Currently, there are several TGFβ1 inhibitors in phase I/II clinical trials (42). It will be important to test the effectiveness of these agents as inhibitors of ovarian cancer progression and metastasis as single agents or in combination with chemotherapy.
In conclusion, we identified a gene signature that is correlated with ovarian cancer progression and poor outcome. The signature has strong predictive value, biological relevance, and translational potential. Future studies are warranted to optimize the gene signature for its predictive power and develop a quantitative assay that is appropriate for use in the clinical setting. Using the validated gene signature to identify patients who are unlikely to respond to standard treatment will provide opportunities to deliver individualized therapies that target the underlying mechanism of the poor outcome signature genes. Furthermore, a better understanding of how collagen remodeling contributes to ovarian cancer progression and metastasis could reveal the “Achilles heel” of these tumors and thus have a major impact on the development of improved therapies for advanced ovarian cancer.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Ovarian cancer is the most lethal gynecologic cancer. Despite clinical and histopathologic similarities at diagnosis, survival outcomes differ due to the development of chemo-resistant tumors. Early identification of these tumors could help direct the use of tailored or experimental therapies. In this study, we report a 10-gene signature that is associated with poor overall survival in patients with serous ovarian cancer. The 10 validated signature genes are enriched in metastatic serous carcinomas and are primarily involved in collagen remodeling. The signature genes are regulated by TGFβ1 signaling, suggesting that TGFβ1 inhibitors might be an effective adjunct in treating these patients and may play a role in the biology of tumor metastasis. Our poor outcome signature provides a novel therapeutic strategy that targets collagen-remodeling genes as a means to improve survival in women with serous ovarian cancer.
ACKNOWLEDGMENTS
We are grateful to Alexander Brodsky (Brown University) for depositing his unpublished data for matched primary and metastatic ovarian tumor samples into the GEO website, which facilitated our initial identification of the metastasis-associated genes. We thank Michael Birrer, Marc Goodman, Neil Bhowmick, and members of the Women’s Cancer Program at Cedars-Sinai Medical Center for insightful discussions and critical readings of the manuscript; Hang Tran for technical assistance; the Cedars-Sinai Genomics Core for ABI Open Array; Steven Swartwood in the Biobank and Translational Research Core for in situ hybridization analysis; and Kristy Daniels for assistance in the preparation of the manuscript.
Grant support: S.O. is supported by grants from the American Cancer Society (RSG-10-252-01-TBG), Sandy Rollman Ovarian Cancer Foundation, Pacific Ovarian Cancer Research Consortium (Developmental Grant, P50 CA083636), and the National Center for Advancing Translational Sciences (UL1TR000124) as well as funds from the Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute at Cedars-Sinai Medical Center. Y.T. is supported by the career award K99CA138914. X.C. is supported by the National Institutes of Health grant CA151610. B.Y.K. is supported by an American Cancer Society Clinical Research Professorship (SIOP-06-258-01-COUN).
Footnotes
COI: All authors declare no conflict of interest
REFERENCES
- 1.Bast RC, Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9:415–428. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 4.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, Bogomolniy F, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478–5486. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ismail RS, Baldwin RL, Fang J, Browning D, Karlan BY, Gasson JC, et al. Differential gene expression between normal and tumor-derived ovarian epithelial cells. Cancer Res. 2000;60:6744–6749. [PubMed] [Google Scholar]
- 9.Zhu M, Fejzo MS, Anderson L, Dering J, Ginther C, Ramos L, et al. Periostin promotes ovarian cancer angiogenesis and metastasis. Gynecol Oncol. 2010;119:337–344. doi: 10.1016/j.ygyno.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Zhu M, Saxton RE, Ramos L, Chang DD, Karlan BY, Gasson JC, et al. Neutralizing monoclonal antibody to periostin inhibits ovarian tumor growth and metastasis. Mol Cancer Ther. 2011;10:1500–1508. doi: 10.1158/1535-7163.MCT-11-0046. [DOI] [PubMed] [Google Scholar]
- 11.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 12.Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 13.Verhaak RG, Tamayo P, Yang JY, Hubbard D, Zhang H, Creighton CJ, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest. 2013;123:517–525. doi: 10.1172/JCI65833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W, Liu Y, Sun N, Wang D, Boyd-Kirkup J, Dou X, et al. Integrating genomic, epigenomic, and transcriptomic features reveals modular signatures underlying poor prognosis in ovarian cancer. Cell Rep. 2013;4:542–553. doi: 10.1016/j.celrep.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 15.Albinger-Hegyi A, Stoeckli SJ, Schmid S, Storz M, Iotzova G, Probst-Hensch NM, et al. Lysyl oxidase expression is an independent marker of prognosis and a predictor of lymph node metastasis in oral and oropharyngeal squamous cell carcinoma (OSCC) Int J Cancer. 2010;126:2653–2662. doi: 10.1002/ijc.24948. [DOI] [PubMed] [Google Scholar]
- 16.Helleman J, Jansen MP, Ruigrok-Ritstier K, van Staveren IL, Look MP, Meijer-van Gelder ME, et al. Association of an extracellular matrix gene cluster with breast cancer prognosis and endocrine therapy response. Clin Cancer Res. 2008;14:5555–5564. doi: 10.1158/1078-0432.CCR-08-0555. [DOI] [PubMed] [Google Scholar]
- 17.Kim H, Watkinson J, Varadan V, Anastassiou D. Multi-cancer computational analysis reveals invasion-associated variant of desmoplastic reaction involving INHBA, THBS2 and COL11A1. BMC Med Genomics. 2010;3:51. doi: 10.1186/1755-8794-3-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kornfeld JW, Meder S, Wohlberg M, Friedrich RE, Rau T, Riethdorf L, et al. Overexpression of TACE and TIMP3 mRNA in head and neck cancer: association with tumour development and progression. Br J Cancer. 2011;104:138–145. doi: 10.1038/sj.bjc.6606017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Liu X, Huang H, Ma H, Cai W, Hou J, et al. Deregulation of Snai2 is associated with metastasis and poor prognosis in tongue squamous cell carcinoma. Int J Cancer. 2012;130:2249–2258. doi: 10.1002/ijc.26226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilgus ML, Borczuk AC, Stoopler M, Ginsburg M, Gorenstein L, Sonett JR, et al. Lysyl oxidase: a lung adenocarcinoma biomarker of invasion and survival. Cancer. 2011;117:2186–2191. doi: 10.1002/cncr.25768. [DOI] [PubMed] [Google Scholar]
- 21.Farmer P, Bonnefoi H, Anderle P, Cameron D, Wirapati P, Becette V, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15:68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- 22.Soikkeli J, Podlasz P, Yin M, Nummela P, Jahkola T, Virolainen S, et al. Metastatic outgrowth encompasses COL-I, FN1, and POSTN up-regulation and assembly to fibrillar networks regulating cell adhesion, migration, and growth. Am J Pathol. 2010;177:387–403. doi: 10.2353/ajpath.2010.090748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen JL, Espinosa I, Lin AY, Liao OY, van de Rijn M, West RB. Stromal Responses among Common Carcinomas Correlated with Clinicopathologic Features. Clin Cancer Res. 2013;19:5127–5135. doi: 10.1158/1078-0432.CCR-12-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuetz CS, Bonin M, Clare SE, Nieselt K, Sotlar K, Walter M, et al. Progression-specific genes identified by expression profiling of matched ductal carcinomas in situ and invasive breast tumors, combining laser capture microdissection and oligonucleotide microarray analysis. Cancer Res. 2006;66:5278–5286. doi: 10.1158/0008-5472.CAN-05-4610. [DOI] [PubMed] [Google Scholar]
- 25.Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev Cell. 2010;18:884–901. doi: 10.1016/j.devcel.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ursin G, Hovanessian-Larsen L, Parisky YR, Pike MC, Wu AH. Greatly increased occurrence of breast cancers in areas of mammographically dense tissue. Breast Cancer Res. 2005;7:R605–R608. doi: 10.1186/bcr1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Provenzano PP, Inman DR, Eliceiri KW, Knittel JG, Yan L, Rueden CT, et al. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6:11. doi: 10.1186/1741-7015-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barker HE, Cox TR, Erler JT. The rationale for targeting the LOX family in cancer. Nat Rev Cancer. 2012;12:540–552. doi: 10.1038/nrc3319. [DOI] [PubMed] [Google Scholar]
- 29.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maruhashi T, Kii I, Saito M, Kudo A. Interaction between periostin and BMP-1 promotes proteolytic activation of lysyl oxidase. J Biol Chem. 2010;285:13294–13303. doi: 10.1074/jbc.M109.088864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Helleman J, Jansen MP, Span PN, van Staveren IL, Massuger LF, Meijer-van Gelder ME, et al. Molecular profiling of platinum resistant ovarian cancer. Int J Cancer. 2006;118:1963–1971. doi: 10.1002/ijc.21599. [DOI] [PubMed] [Google Scholar]
- 33.Jazaeri AA, Awtrey CS, Chandramouli GV, Chuang YE, Khan J, Sotiriou C, et al. Gene expression profiles associated with response to chemotherapy in epithelial ovarian cancers. Clin Cancer Res. 2005;11:6300–6310. doi: 10.1158/1078-0432.CCR-04-2682. [DOI] [PubMed] [Google Scholar]
- 34.Sherman-Baust CA, Weeraratna AT, Rangel LB, Pizer ES, Cho KR, Schwartz DR, et al. Remodeling of the extracellular matrix through overexpression of collagen VI contributes to cisplatin resistance in ovarian cancer cells. Cancer Cell. 2003;3:377–386. doi: 10.1016/s1535-6108(03)00058-8. [DOI] [PubMed] [Google Scholar]
- 35.Yeung TL, Leung CS, Wong KK, Samimi G, Thompson MS, Liu J, et al. TGF-beta Modulates Ovarian Cancer Invasion by Upregulating CAF-Derived Versican in the Tumor Microenvironment. Cancer Res. 2013;73:5016–5028. doi: 10.1158/0008-5472.CAN-13-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu YH, Chang TH, Huang YF, Huang HD, Chou CY. COL11A1 promotes tumor progression and predicts poor clinical outcome in ovarian cancer. Oncogene. 2013 doi: 10.1038/onc.2013.307. [DOI] [PubMed] [Google Scholar]
- 37.Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delaloye JF, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2012;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 38.Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT, et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Foss CA, Summerfield DD, Doyle JJ, Torok CM, Dietz HC, et al. Targeting collagen strands by photo-triggered triple-helix hybridization. Proc Natl Acad Sci U S A. 2012;109:14767–14772. doi: 10.1073/pnas.1209721109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamura S, Matsumura N, Mandai M, Huang Z, Oura T, Baba T, et al. The activated transforming growth factor-beta signaling pathway in peritoneal metastases is a potential therapeutic target in ovarian cancer. Int J Cancer. 2012;130:20–28. doi: 10.1002/ijc.25961. [DOI] [PubMed] [Google Scholar]
- 41.Nam JS, Terabe M, Mamura M, Kang MJ, Chae H, Stuelten C, et al. An anti-transforming growth factor beta antibody suppresses metastasis via cooperative effects on multiple cell compartments. Cancer Res. 2008;68:3835–3843. doi: 10.1158/0008-5472.CAN-08-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Connolly EC, Freimuth J, Akhurst RJ. Complexities of TGF-beta targeted cancer therapy. Int J Biol Sci. 2012;8:964–978. doi: 10.7150/ijbs.4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.