Abstract
Background
Dual antiplatelet therapy is the cornerstone in the management of acute coronary syndromes (ACS) and prevention of stent thrombosis (ST). Genetic polymorphisms in CYP2C19 gene involved in hepatic activation of clopidogrel leads to clopidogrel non-responsiveness and may influence clinical outcomes. These polymorphisms in CYP2C19 gene and their impact on clinical outcome in coronary artery disease (CAD) have not been studied in Indian population.
Methods
We studied 110 consecutive patients (mean age 55.7 ± 10.7 years; 90% male) taking clopidogrel with angiographically proven CAD for various genetic polymorphisms in CYP2C19 gene. Relationship between loss of function mutation and clinical presentation with recurrent ACS including ST was analyzed.
Results
Out of 110 patients, 26 (23.64%) had normal genotype, 52 (47.23%) had loss of function mutation *2 and 39 (35.45%) had a gain of function mutation *17, 7 (6.36%) patients were undefined metabolizers (*2/*17) which were excluded from analyses. Final analyses included 103 patients, with 45 (40.90%) having loss of function. Overall 51 patients had ACS, with 27 developing recurrence while on clopidogrel. The prevalence of loss of function mutation was no different between the group with recurrences and those without recurrences (55.6% vs. 50%, p = 0.7). Two patients developed ST while on clopidogrel; both had loss of function mutation.
Conclusion
CYP2C19 gene polymorphisms are common in Indian population. Loss of function mutation status did not affect the clinical outcomes. A larger study also considering P2Y12 receptor polymorphisms together with platelet activity testing, may be required to establish the role of CYP2C19 gene polymorphisms in clinical practice.
Keywords: Clopidogrel, CYP2C19 polymorphism, Coronary artery disease
1. Introduction
Dual antiplatelet therapy with aspirin and clopidogrel is the cornerstone in the management of STEMI,1 acute coronary syndromes (UA/NSTEMI),2 prevention of stent thrombosis (ST) in patients undergoing percutaneous coronary intervention (PCI)3 and patients with high risk chronic stable angina.4 In spite of significant benefits of dual antiplatelet treatment proven in various trials, adverse ischemic events, including stent thrombosis, do occur and continue to haunt clinicians worldwide. It is well established that the antiplatelet response to clopidogrel shows tremendous individual variation in the level of platelet inhibition achieved.5, 6 Patients displaying little attenuation (<30%) of platelet reactivity under clopidogrel therapy are recognized as low or non-responders, while those with <10% attenuation are termed as clopidogrel resistant.7, 8 Clopidogrel is a pro-drug that requires hepatic bioactivation involving 2steps, which are regulated by cytochromes (CYP).9 The genes encoding the enzymes are characterized by several polymorphisms, some of which are able to modify the activity of proteins, reducing the concentration of active metabolite.10, 11 Carriers of at least one CYP2C19 reduced-function allele had a relative reduction of 32.4% in plasma exposure to the active metabolite as compared to non-carriers, whereas the carriers of other CYP reduced-function alleles did not have significant differences in platelet inhibition as compared to non-carriers.12
Several studies have associated these gene polymorphisms with inadequate platelet inhibition resulting in adverse clinical outcomes, including recurrent ischemic cardiovascular events, stent thrombosis (ST) and peri-procedural myocardial infarction.12, 13, 14, 15, 16, 17 Based on these studies, ACC (American College of Cardiology) has issued an advisory stating that, “ genetic testing to determine if a patient is predisposed to poor clopidogrel metabolism (“poor metabolizers”) may be considered before starting clopidogrel therapy in patients believed to be at moderate or high risk for poor outcomes”.18
The frequency of CYP2C19 polymorphism shows variations among different ethnic groups, which translates into significantly variable metabolic phenotypes across the populations. CYPC19 polymorphisms and their impact on clinical outcomes in CAD patients have not been studied in Indian population. We conducted this study to identify and describe the distribution of genetic polymorphisms in CYP2C19 gene in Indian patients and study relationship between loss of function mutation and presentation with recurrent ACS including ST while taking adequate dose of clopidogrel.
2. Material and methods
We conducted a prospective cohort study in a tertiary hospital in north India. We studied 110 patients of angiographically proven CAD taking clopidogrel. Patients with history of major bleeding, recent history of stroke or transient ischemic attack (TIA), clopidogrel/aspirin sensitivity, renal dysfunction, hepatic dysfunction, malignancy, patients having non-coronary indication for clopidogrel (e.g. renal/carotid stenting), patients on other P2Y12 antagonists within 4 weeks of inclusion and those taking omeprazole were excluded. The study is approved by Institute's ethics committee.
Written and informed consent was taken from all eligible patients prior to inclusion into the study. All patients were questioned regarding any of the above mentioned exclusion criteria, coronary risk factors, history of medications they have been taking, past history of STEMI, ACS and percutaneous coronary interventions. The patients were divided into 2 groups.
2.1. Recurrent ACS group
Patients who developed an episode of stent thrombosis or acute coronary syndrome while they were taking adequate dose of dual antiplatelet agents.
2.2. No recurrence group
All other patients on clopidogrel initiated for the first event or prior to PCI without clinical recurrence of ACS.
The baseline demographic and coronary details were recorded in a standard proforma. Standard definitions of ACS as defined by ACC guidelines were used in the study. The patients were managed as per current guideline by their physicians. A blood sample (10 ml) was collected in EDTA anti-coagulated vacutainer and stored for genetic analysis of CYP2C19 polymorphisms at −80 °C.
2.3. Method of genetic testing
The method followed was based on the Sequenom MassARRAY platform (Sequenom Inc, San Diego, CA). The assay was based on primer extension and offers two levels of specificity. First, a locus-specific amplification PCR reaction took place, followed by a locus-specific primer extension reaction, in which an oligonucleotide primer annealed immediately upstream of the polymorphic site being genotyped. In the iPLEX assay, the primer and amplified target DNA were incubated with mass-modified dideoxynucleotide terminators. The primer extension was made according to the sequence of the variant site, and was a single complementary mass-modified base. Through the use of MALDI–TOF mass spectrometry (Dynamo, Thermo Bioanalysis, Santa Fe, NM) the mass of the extended primer was determined. The primer's mass indicated the sequence and, therefore, the alleles present at the polymorphic site of interest. Sequenom supplied software (TYPER) that automatically translated the mass of the observed primers into a genotype for each reaction.
The assay reported the alleles as *1, *2, *3, *4, *5 and *17 and reported genotypes as normal extensive metabolizers (*1/*1), poor metabolizers (*2–*5/*2–*5), intermediate metabolizers (*1/*2–*5), undefined phenotype (*17/*2–*5) and ultra-rapid metabolizers (*17/*17 and *1/*17). Based on this two groups were made, one with loss of function allele (homozygous or heterozygous for *2–*5 allele, which includes *2–5/*2–5 and *1/*2–5) and another without loss of function allele (not having any *2–*5 allele, which included *1/*1, *1/*17, and *17/*17). The allele *17/*2–5 was labelled as undefined phenotype. There are various publications based on the currently used assay.19, 20, 21, 22
3. Statistical analysis
The description of genetic polymorphism profile of consecutive Indian patients is reported as a percentage. Continuous values are expressed as mean ± SD. For normally distributed continuous variables, the Student's t-test was applied for comparison between two groups. Pearson's Chi-square test was used for evaluating dichotomous variables. Number of patients with reduced-function mutations in recurrent ACS versus first time ACS group were compared using Pearson's Chi-square test with two tailed significance level. A p value of <0.05 was considered significant. Analysis was done on SPSS statistics 17.0 (IBM corp.).
4. Results
Out of the 125 patients with diagnosed coronary artery disease taking clopidogrel, 15 patients were excluded for not giving consent.
4.1. Characteristics of study population
Baseline characteristics of study population are shown in Table 1. Majority of 57 (51.8%) patients presented with CSA (Chronic stable angina), while 22 (20%) and 29 (26.4%) presented with STEMI or UA/NSTEMI respectively. There were two instances of ST (1.8%). In all, 53 (48.2%) presented with acute coronary events, 28 patients of these presented with recurrence of ACS while taking adequate dose of clopidogrel.
Table 1.
Demographic features of the study population.
| Characteristics | Total n = 110 | n% | |
|---|---|---|---|
| Age (years) | 55.7 ± 10.7 | ||
| Sex | Male | 90 | 81.8% |
| Coronary Risk factors | Hypertension | 68 | 61.8% |
| Diabetes | 39 | 35.5% | |
| Dyslipidemia | 36 | 32.7% | |
| Smoker | 29 | 26.4% | |
| Tobacco chewer | 30 | 27.3% | |
| Obese | 27 | 24.5 | |
| Positive family history | 22 | 20% | |
| Medical treatment | Aspirin | 110 | 100% |
| Clopidogrel | 110 | 100% | |
| Beta-blocker | 110 | 100% | |
| ACEI/ARB | 78 | 70.9% | |
| Statins | 110 | 100% | |
| Presentation | CSA | 57 | 51.8% |
| STEMI | 22 | 20% | |
| UA/NSTEMI | 29 | 26.4% | |
| Stent thrombosis | 2 | 1.8% | |
| Coronary angiography | Normal | 14 | 12.7% |
| SVD | 41 | 37.3% | |
| DVD | 30 | 27.3% | |
| TVD | 21 | 19.1% | |
| LMCA disease | 4 | 3.6% | |
| LV function | Normal | 64 | 58.2% |
| LV dysfunction (LVEF <55%) | 46 | 41.8% | |
ACEI: Angiotensin converting enzyme inhibitor, ARB: angiotensin receptor blocker, CSA: chronic stable angina, STEMI: ST elevation myocardial infarction, NSTEMI: non-ST elevation myocardial infarction, UA: unstable angina, ACS: acute coronary syndrome; SVD: single vessel disease, DVD: double vessel disease, TVD: triple vessel disease, LMCA: left main coronary artery, LV: left ventricle, LVEF: LV ejection fraction.
4.2. Genetic analysis
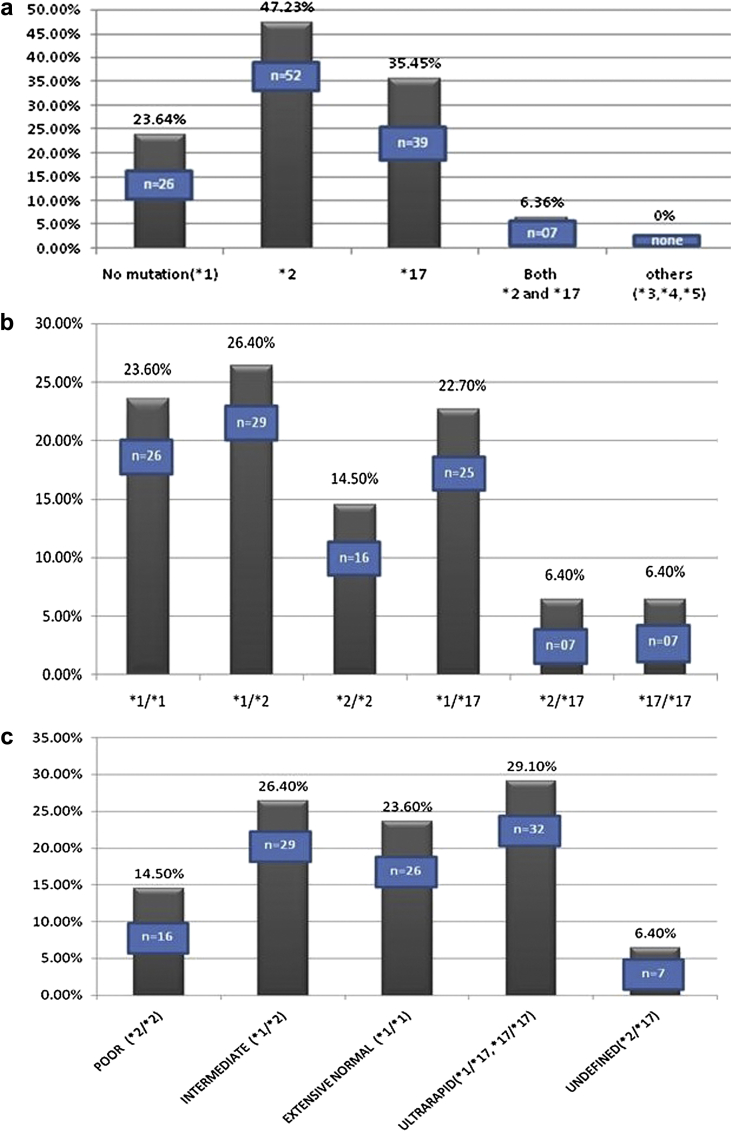
The frequency of CYP2C19 mutant alleles in study population is shown in Fig. 1a. Genetic analysis revealed presence of loss of function allele *2 and gain of function allele *17 in the study population, while there were no *3, *4 or *5 loss of function alleles. Only one fourth (26 patients) carried the normal wild type allele *1 at both (homozygous) loci, 52 patients (47.27%) carried loss of function allele *2 at both (homozygous) or one (heterozygous) loci, 39 patients (35.46%) carried gain of function allele *17 at both (homozygous) or one (heterozygous) loci and 7 patients (6.36%) carried both *2 and *17 mutations.
Fig. 1.
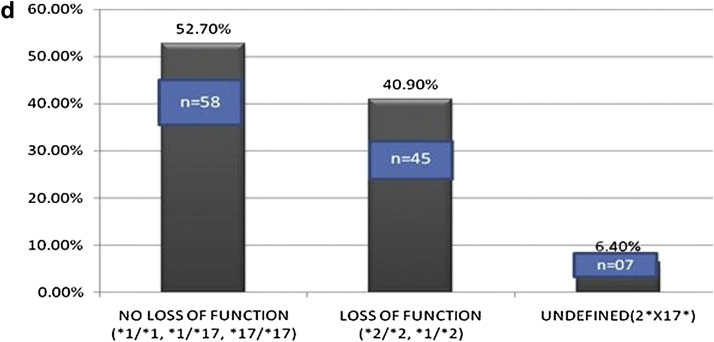
a – Frequency of CYP2C19 mutant alleles in study population. b – Frequency of clopidogrel genotypes in study population. c – Frequency of CYP2C19 alleles, according to metabolizer status in the study population. d – Patient groups according to loss of function versus no loss of function genotype in the study population.
Twenty-six patients (23.60%) had normal genotype and were homozygous for the wild allele *1, 16 patients (14.50%) were homozygous for loss of function allele *2, 7 patients (6.40%) were homozygous for gain of function allele *17, 29 patients (26.40%) were heterozygous with one loss of function allele *2 and other normal allele, 25 patients (22.70%) were heterozygous with one gain of function allele *17 and other normal allele while 7 patients (6.40%) formed the undefined group which were heterozygous with both loss of function allele *2 and gain of function allele *17 (Fig. 1b).
Patients with *2 mutation are either homozygous (*2/*2) or heterozygous (*1/*2), and are termed as poor metabolizers and intermediate metabolizers respectively. Patients with *17 mutation are either homozygous (*17/*17) or heterozygous (*1/*17), both are classified as metabolically ultra-rapid metabolizers. Patients homozygous for normal wild type allele *1 i.e. (*1/*1) are metabolically normal and are extensive normal metabolizers. The genotype heterozygous for both *2 and *17 mutation (*2/*17) is termed as undefined metabolizer, because metabolizer status of this genotype is unknown. This group of undefined metabolizer patients has been uniformly excluded from analysis in clinical trials. Twenty-six patients (23.60%) were normal extensive metabolizers, 16 patients (14.50%) were poor metabolizers, 29 patients (26.40%) were intermediate metabolizers, 32 patients (29.10%) were ultra-rapid metabolizers and 7 patients (6.40%) formed the undefined group (Fig. 1c).
4.3. Distribution of CYP2C19 genotypes within ACS and CSA group
Distribution of CYP2C19 genotypes individually in patients presenting with CSA and ACS is shown in Table 2. More number of patients in ACS group had loss of function mutation (27/53, 50.94%) as compared to patients in chronic stable angina group (18/57, 31.58%). Numerically more patients in CSA group had normal CYP2C19 phenotype as compared to ACS group (18/57, 31.58% versus 8/53, 15.09%). Gain of function mutation was no different between both groups (16/57 versus 16/53). However these differences were not statistically significant (p = 0.079).
Table 2.
Distribution of CYP2C19 genotypes according to the mode of presentation (chronic stable angina versus acute coronary syndrome including stent thrombosis).
| Chronic stable angina (n = 57) | n% | Acute coronary syndrome (n = 53)# | n% | p value | ||
|---|---|---|---|---|---|---|
| Genotypes | *1 × *1 | 18 | 31.58 | 8 | 15.09 | |
| *1 × *2 | 13 | 22.81 | 16 | 30.19 | ||
| *2 × *2 | 5 | 8.77 | 11 | 20.75 | ||
| *1 × *17 | 14 | 24.56 | 11 | 20.75 | 0.101 | |
| *17 × *17 | 2 | 3.51 | 5 | 9.43 | ||
| *2 × *17 | 5 | 8.77 | 2 | 3.77 | ||
| Status of mutation | Normal (*1 × *1) | 18 | 31.58 | 8 | 15.09 | |
| Loss of function (*1 × *2, *2 × *2) | 18 | 31.58 | 27 | 50.94 | ||
| Gain of function (*1 × *17, *17 × *17) | 16 | 28.07 | 16 | 30.19 | 0.079 | |
| Undefined (*2 × *17) | 5 | 8.77 | 2 | 3.77 | ||
# Includes two patients with stent thrombosis.
4.4. Analyses based on loss of function versus no loss of function genotype
For analysis of loss of function genotype relationship with coronary events the patients were analyzed in two groups, one with loss of function and the other without loss of function allele. Out of 110 patients, 58 (52.70%), 45 (40.90%) and 7 patients (6.40%) were in no loss of function, loss of function and undefined groups respectively (Fig. 1d). The undefined group (n = 7) was excluded from the analysis as their metabolizer status is unknown and final analysis was done in 103 patients. The undefined metabolizer group with genotype *2/*17 had 2 males and 5 females, 5 patients presented with CSA while 2 presented with ACS, only one patient from this group had recurrent ACS while on clopidogrel.
More patients in loss of function group (n = 27 of 45, 60%) presented with an acute coronary syndrome (STEMI/NSTEMI/USA) as compared to 41.4% (n = 24 of 58) patients in no loss of function group, however this difference didn't reach statistical significance (p = 0.061). There were two cases of stent thrombosis and both were in loss of function group. There was no significant difference between both groups in any of the baseline characteristics (Table 3).
Table 3.
Comparison between patient with loss of function and no loss of function mutations.
| Characteristics | Loss of function mutation (n = 45) | No loss of function mutation (n = 58) | Total n = 103 | Total % | p value | |||
|---|---|---|---|---|---|---|---|---|
| Age (years) | 54.71 ± 10.72 | 55.34 ± 10.25 | 55.07 ± 10.41 | 0.188 | ||||
| Sex | Male | 41 | 91.1% | 47 | 81% | 88 | 85.4% | 0.150 |
| Female | 4 | 8.9% | 11 | 19% | 15 | 14.6% | ||
| Coronary risk factors | Hypertension | 30 | 66.7% | 32 | 55.2% | 62 | 60.2% | 0.237 |
| Diabetes | 16 | 35.6% | 19 | 32.8% | 35 | 34.0% | 0.766 | |
| Dyslipidemia | 14 | 31.1% | 18 | 31% | 32 | 31.1% | 0.993 | |
| Smoking | 13 | 22.4% | 15 | 33.3% | 28 | 27.18% | 0.440 | |
| Presentation | CSA | 18 | 40% | 34 | 58.6% | 52 | 50.5% | 0.132 |
| STEMI | 12 | 26.7% | 10 | 17.2% | 22 | 21.4% | ||
| USA/NSTEMI | 13 | 28.9% | 14 | 24.1% | 27 | 26.2% | ||
| Stent thrombosis | 2 | 4.4% | 0 | 0% | 2 | 1.9% | ||
| Presentation (CSA v/s ACS) | CSA | 18 | 40% | 34 | 58.6% | 52 | 50.5% | 0.061 |
| ACS | 27 | 60% | 24 | 41.4% | 51 | 49.5% | ||
| Coronary angiography | Normal | 5 | 11.1% | 6 | 10.3% | 11 | 10.7% | 0.505 |
| SVD | 21 | 46.7% | 18 | 31% | 39 | 37.9% | ||
| DVD | 10 | 22.2% | 19 | 32.8% | 29 | 28.2% | ||
| TVD | 8 | 17.8% | 12 | 20.7% | 20 | 19.4% | ||
| LMCA disease | 1 | 2.2% | 3 | 5.2% | 4 | 3.9% | ||
| Left ventricular dysfunction (LVEF <55%) | 16 | 35.6% | 29 | 50% | 45 | 43.7% | 0.143 | |
CSA: chronic stable angina, STEMI: ST elevation myocardial infarction, USA: unstable angina, NSTEMI: non-ST elevation myocardial infarction, ACS: acute coronary syndrome, SVD: single vessel disease, DVD: double vessel disease, TVD: triple vessel disease, LMCA: left main coronary artery, LVEF: left ventricular ejection fraction.
4.5. CYP2C19 loss of function and recurrent coronary events on clopidogrel within ACS group
Out of total 103 patients (after excluding 7 patients with undefined metabolizer status), 51 presented with ACS. Of these 51 patients, 27 patients (52.94%) presented with second or subsequent (recurrent) episode of acute coronary syndrome while on adequate dose of clopidogrel, while 24 patients (47.05%) presented with first episode of acute coronary syndrome.
Baseline characteristics between recurrent ACS group and no recurrence group are shown in Table 4. There was no significant difference between both groups in terms of age, gender, coronary risk factors, extent of coronary artery disease and LV dysfunction. All patients in both groups were receiving aspirin, clopidogrel, beta-blockers and statins. Significantly more number of patients 88.9% (24 out of 27) in recurrent ACS group were receiving ACEI/ARB as compared to no recurrence (first event of ACS) group 62.5% (15 out of 24), p = 0.027.
Table 4.
Comparison between patients with recurrent ACS on clopidogrel group versus no recurrence group.
| Characteristic | Recurrent ACS group (n = 27) | No recurrence group (n = 24) | Total n = 51 | Total % | p value | |||
|---|---|---|---|---|---|---|---|---|
| Age (years) | 56.74 ± 10.39 | 53.38 ± 11.86 | 55.16 ± 11.124 | 0.426 | ||||
| Sex | Male | 22 | 81.5% | 23 | 95.8% | 45 | 88.2% | 0.112 |
| Female | 5 | 18.5% | 1 | 4.2% | 6 | 11.8% | ||
| Coronary Risk factors | Hypertension | 19 | 70.4% | 13 | 54.2% | 32 | 62.7% | 0.232 |
| Diabetes | 7 | 25.9% | 8 | 33.3% | 15 | 29.4% | 0.562 | |
| Dyslipidemia | 8 | 29.6% | 6 | 25.0% | 14 | 27.5% | 0.137 | |
| Smoker | 12 | 44.4% | 7 | 29.2% | 19 | 37.3% | 0.260 | |
| Tobacco chewer | 8 | 29.6% | 5 | 20.8% | 13 | 25.5% | 0.472 | |
| Obese | 8 | 29.6% | 5 | 20.8% | 13 | 25.5% | 0.472 | |
| Family history | 6 | 22.2% | 5 | 20.8% | 11 | 21.6% | 0.904 | |
| Medical treatment | Aspirin | 27 | 100% | 24 | 100% | 51 | 100% | |
| Clopidogrel | 27 | 100% | 24 | 100% | 51 | 100% | ||
| Beta blocker | 27 | 100% | 24 | 100% | 51 | 100% | ||
| ACEI/ARB | 24 | 88.9% | 15 | 62.5% | 39 | 76.5% | 0.027 | |
| Statins | 27 | 100% | 24 | 100% | 51 | 100% | ||
| Coronary angiography | Normal | 1 | 3.7% | 2 | 8.3% | 3 | 5.9% | 0.541 |
| SVD | 10 | 37.0% | 13 | 54.2% | 23 | 45.1% | ||
| DVD | 9 | 33.3% | 5 | 20.8% | 14 | 27.5% | ||
| TVD | 6 | 22.2% | 4 | 4% | 10 | 19.6% | ||
| LMCA disease | 1 | 3.7% | 0 | 0% | 1 | 2.0% | ||
| LV dysfunction (LVEF <55%) | 16 | 59.3% | 15 | 62.5% | 31 | 60.8% | 0.813 | |
ACS: acute coronary syndrome, SVD: single vessel disease, DVD: double vessel disease, TVD: triple vessel disease, LMCA: left main coronary artery, LV: left ventricle, ACEI: angiotensin converting enzyme inhibitor, ARB: angiotensin receptor blocker, LVEF: LV ejection fraction.
Indicated bold p values are significant.
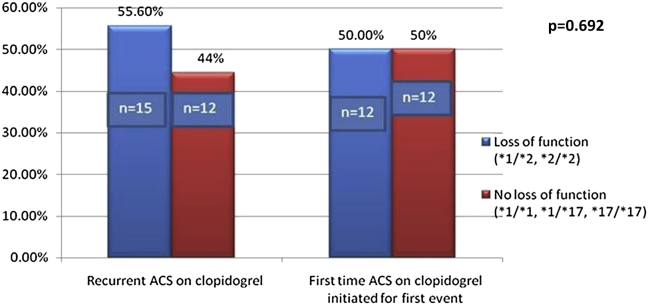
Fifteen patients (55.6%) of recurrent ACS Group had a loss of function mutation in CYP2C19 (*1/*2 or *2/*2) while 12 patients (50%) of no recurrence group had loss of function mutation (Fig. 2). This difference was not statistically significant (p = 0.692).
Fig. 2.
Relationship of loss of function mutation with recurrence of acute coronary events.
5. Discussion
Polymorphisms in cytochromes which modify the activity of hepatic enzymes, leading to reduce concentration of active metabolite, is one of the key factor involved in clopidogrel resistance.10, 11 Various loss of function alleles of CYP2C19 have been found and studied. The CYP2C19*2 allele is the most common type among the reduced-function genes and has been shown as a prime indicator of low response to clopidogrel in many studies.23, 24 Recently studies have shown that CYP2C19*3, *4, *5, *6, *7 and *8 alleles may also affect clopidogrel metabolism in the same way as CYP2C19*2, however their frequency in population is negligible as compared to CYP2C19*2 allele.25 An allele CYP2C19*17 is associated with increased activity and is been found to be having higher platelet inhibition as compared to the normal.10, 11
Even though our results cannot be compared with studies involving general population, certain inferences can be made. Our study showed a high prevalence of CYP2C19 polymorphism in a specific group of CAD population, than reported in earlier epidemiological studies conducted in general population. The frequency of CYP2C19*2 allele associated with poor metabolizer type was observed to be 47.23% in CAD patients which is higher than that in general population as previously reported by Adithan et al26 in south Indian Tamil population (37.9%) and by Kavita et al27 in western Indian population (35.2%). We did not find any loss of function CYP2C19*3 allele in our study population, which is similar to as observed in western Indian population.27 However this allele though rare (2.2%) was detected in Tamilian population.26 Other loss of function alleles CYP2C19*4 and *5, were not detected in our study population. These rare alleles have not been previously studied in Indian population, and probably are very rare like in other worldwide populations.10 Unlike CYP2C19*2 allele, the gain of function allele CYP2C19*17 associated with rapid metabolism has not been adequately studied in Indian population. Kavita et al27 and Chan et al28 reported the frequency of CYP2C19*17 allele to be 18% and 16.5% respectively in the Indian population. We observed higher allelic frequency of CYP2C19*17 at 35.45% in our study population involving CAD patients. Frequency of CYP2C19 alleles studied in varied populations is shown in Table 5. CYP2C19*2 loss of function allele is very common in Indian population as compared to the other populations studied worldwide. There is also disparity among the Indian population in terms of CYP2C19*17 allele, north Indian population had a higher frequency of this mutation as compared to south Indian population. This difference is attributed to different ethnic population with varied genetic pool among the studied groups.
Table 5.
Reported frequencies of CYP2C19 alleles in varied population and comparison with present study.
| Population | *1 allele | *2 allele | *3 allele | *4 allele | *5 allele | *17 allele |
|---|---|---|---|---|---|---|
| African10 | 68% | 15% | 0.5% | 0.09% | NA | 16% |
| American10 | 69% | 12% | 0.03% | 0.24% | 0% | 18% |
| European10 | 63% | 15% | 0.4% | 0.25% | 0.017% | 21% |
| South/Central Asian10 | 62% | 35% | 2.4% | 0% | 0% | NA |
| South Indian26 | NA | 37.9% | 2.2% | NA | NA | NA |
| Western Indian27 | NA | 35.2% | 0% | NA | NA | 18% |
| Indian28 | NA | 37.5% | 1% | NA | NA | 16.5% |
| Present study | 23.64% | 47.23% | 0% | 0% | 0% | 35.45% |
NA: not analyzed.
Various studies and meta-analyses have associated CYP2C19 loss of function mutation with adverse clinical outcomes including stent thrombosis (ST).12, 13 A genetic substudy of TRITON-TIMI 38 trial12 studied this hypothesis in a cohort of 1477 subjects with ACS, all of whom were on clopidogrel and were managed invasively with coronary artery stenting. 27.1% of the patients in this study group had at least one reduced-function CYP2C19 allele out of either *2, *3, *4, *5, *6, *7, *8, *9, *10, *12, *13 or *14 allele. The primary outcome of composite death from cardiovascular causes, myocardial infarction, or stroke was compared between two dichotomous groups, one with loss of function allele and the other not carrying such a mutation. Primary outcome was reached in 12.1% of patients carrying at least one reduced-function mutation as compared to 8.0% in those not harboring it, HR for carriers 1.53; 95% CI, 1.07–2.19 (p = 0.01). The secondary outcome of ST was also significantly high in carriers (2.6%) than non-carriers (0.8%), p = 0.02. Similarly a meta-analysis of nine studies involving patients predominantly managed invasively with PCI, showed significantly higher rates of adverse clinical events (death from cardiovascular cause, myocardial infarction or stroke), HR 1.55 (95% CI, 1.11–2.27, p = 0.01) and stent thrombosis, HR 2.67 (95% CI, 1.69–4.22, p < 0.0001), among carriers of reduced-function clopidogrel allele.13
In our study we did not found any significant interaction between the loss of function group (*2/*2, *1/*2) and no loss of function group (*1/*1, *1/*17, *17/*17) in clinical presentation. Two of our patients had stent thrombosis while on adequate dose of clopidogrel, both had loss of function mutation, one being poor metabolizer (*2/*2) and other intermediate (*1/*2). Analysis regarding this finding could not be made due to less number of events. Loss of function allele was not associated with any significant increase in acute coronary events in patients on clopidogrel.
Our results, which refute clinical significance of genetic polymorphisms in CYP2C19 gene are supported by recent studies. A Genetic substudy of CURE trial29 studied whether the benefits of clopidogrel varies according to the CYP2C19 metabolizer status. The study population consisted of patients with ACS, most of whom were managed conservatively with only 18% receiving percutaneous coronary interventions. The metabolizer status of clopidogrel didn't influence any of the primary or secondary efficacy outcomes in this study. A recent meta-analyses involving more than 25,000 patients (30% carrying reduced-function allele) from genetic substudy of TRITON-TIMI 38, CURE, PLATO, CHARISMA, and ACTIVE-A studies also echoed similar trends.30 The meta-analyses found that patients with a loss of function allele, did not have increased risk of a cardiovascular events, HR = 1.23 (95% CI 0.97–1.55), however they were prone to increased risk of ST, HR = 2.24 (95% CI 1.52–3.30). The authors concluded that effects of CYP2C19 reduced-function alleles on clopidogrel, when applied to all users as a whole may not be clinically significant, however a link to stent thrombosis remains.
Thus the role of CYP2C19 polymorphisms influencing clinical outcomes differs according to the study group. When applied to invasively managed population, these genetic polymorphisms influence the clinical outcomes, however their role in influencing clinical outcomes in patient population managed conservatively is at the best modest.
All the clinical studies of CYP2C19 polymorphisms overwhelmingly represented the Caucasian population. Though there are studies in Indian population showing attenuated platelet inhibition to clopidogrel in poor metabolizers27, 31; no study has evaluated clinical significance of this entity. Our study is the first study in the Indian population to study clinical significance of CYP2C19 reduced-function allele. Findings of this study, which had many (50.5%) patients with stable coronary artery disease supports the notion that role of CYP2C19 polymorphisms in dictating clinical outcome in patients of coronary artery disease is undermined when applied to conservatively managed patients or the entire population of clopidogrel users as a whole. P2Y12 receptor gene polymorphisms are also associated with clopidogrel resistance.32, 33 Platelet activity testing could have taken this factor into account. However we did not conduct platelet activity testing in our study population, hence co-existence of these polymorphisms could have influenced the results. May be a similar study involving only high risk patients and those receiving PCI, together with platelet activity testing show a positive association of genetic polymorphisms in CYP2C19 gene and clinical outcomes.
6. Limitations
Considering the high prevalence of genetic polymorphisms in the population, this relatively small number of patients in the study limits statistical inferences. This was a point study and follow-up of patients with loss of function mutation for coronary events has not been done. Clopidogrel CYP2C19 polymorphisms have a clinical implication in setting of PCI or high-risk patients with USA/NSTEMI/STEMI; however our study population included a large number of stable coronary artery disease patients, which might have diluted the clinical effect of these polymorphisms. Moreover, we did not conduct platelet activity testing in our study population; hence co-existence of other factors influencing clopidogrel induced platelet inhibition such as P2Y12 gene polymorphisms could have influenced the results. However, this is the first study of CYP2C19 gene polymorphisms and its clinical implication in Indian patients.
7. Conclusions
Loss of function CYP2C19*2 and gain of function CYP2C19*17 polymorphisms in Indian population are very common as compared to other populations. Other loss of function alleles *3, *4 and *5 were not found, and even if present form a very small group. Loss of function status does not affect the clinical outcomes. A larger study involving high risk patients with PCI, together with, platelet activity testing, which would also take into account effect of P2Y12 receptor polymorphisms, may be required to establish the role of CYP2C19 gene polymorphisms in clinical practice.
Conflicts of interest
All authors have none to declare.
Acknowledgment
The genetic analysis of the study was done free of cost by AceProbe Technologies (India) Pvt. Ltd., New Delhi.
References
- 1.Kushner F.G., Hand M., Smith S.C. 2009 focused updates: ACC/AHA guidelines for the management of patients with ST-elevation myocardial infarction (updating the 2004 guideline and 2007 focused update) and ACC/AHA/SCAI guidelines of percutaneous coronary intervention (updating the 2005 guideline and 2007 focused update): a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2009;120:2271–2306. doi: 10.1161/CIRCULATIONAHA.109.192663. [DOI] [PubMed] [Google Scholar]
- 2.Jneid H., Anderson J.L., Wright R.S. 2012 ACCF/AHA focused update of the guideline for the management of patients with unstable angina/non-ST-elevation myocardial infarction (updating the 2007 guideline and replacing the 2011 focused update): a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2012;126:875–910. doi: 10.1161/CIR.0b013e318256f1e0. [DOI] [PubMed] [Google Scholar]
- 3.Levine G.N., Bates E.R., Blankenship J.C. 2011 ACCF/AHA/SCAI guideline for PCI: executive summary: a report of the ACCF/AHA Task Force on Practice Guidelines and the SCAI. Circulation. 2011;124:2574–2609. doi: 10.1161/CIR.0b013e31823a5596. [DOI] [PubMed] [Google Scholar]
- 4.Bhatt D.L., Fox K.A., Hacke W. Clopidogrel and aspirin versus aspirin alone for the prevention of atherothrombotic events. N Engl J Med. 2006;354:1706. doi: 10.1056/NEJMoa060989. [DOI] [PubMed] [Google Scholar]
- 5.Dupont A.G., Gabriel D.A., Cohen M.G. Antiplatelet therapies and the role of antiplatelet resistance in acute coronary syndrome. Thromb Res. 2009;124:6–13. doi: 10.1016/j.thromres.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Gurbel P.A., Tantry U.S. Clopidogrel resistance? Thromb Res. 2007;120:311–321. doi: 10.1016/j.thromres.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Gurbel P.A., Bliden K.P., Hiatt B.L. Clopidogrel for coronary stenting: response variability, drug resistance, and the effect of pre-treatment platelet reactivity. Circulation. 2003;107:2908–2913. doi: 10.1161/01.CIR.0000072771.11429.83. [DOI] [PubMed] [Google Scholar]
- 8.Muller I., Besta F., Schulz C., Massberg S., Schomig A., Gawaz M. Prevalence of Clopidogrel non-responders among patients with stable angina pectoris scheduled for elective coronary stent placement. Thromb Haemost. 2003;89:783–787. [PubMed] [Google Scholar]
- 9.Kazui M., Nishiya Y., Ishizuka T. Identification of the human cytochrome p450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab Dispos. 2010;38:92–99. doi: 10.1124/dmd.109.029132. [DOI] [PubMed] [Google Scholar]
- 10.Scott S.A., Sangkuhl K., Gardner E.E. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther. 2011;90:328–332. doi: 10.1038/clpt.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price M.J., Tantry U.S., Gurbel P.A. The influence of CYP2C19 polymorphisms on the pharmacokinetics, pharmacodynamics, and clinical effectiveness of P2Y12 inhibitors. Rev Cardiovasc Med. 2011;12:1–12. doi: 10.3909/ricm0590. [DOI] [PubMed] [Google Scholar]
- 12.Mega J.L., Close S.L., Wiviott Cytochrome P-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360:354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 13.Mega J.L., Simon T., Collet J.P. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304:1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuster V., Sweeny J.M. Clopidogrel and the reduced-function CYP2C19 genetic variant: a limited piece of the overall therapeutic puzzle. JAMA. 2010;304:1839–1840. doi: 10.1001/jama.2010.1566. [DOI] [PubMed] [Google Scholar]
- 15.Hwang S.J., Jeong Y.H., Kim I.S. The cytochrome 2C19*2 and *3 alleles attenuate response to clopidogrel similarly in East Asian patients undergoing elective percutaneous coronary intervention. Thromb Res. 2011;127:23–28. doi: 10.1016/j.thromres.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 16.Collet J.P., Hulot J.S., Pena A. CYP 450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet. 2009;373:309–317. doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 17.Simon T., Verstuyft C., Mary-Krause M. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360:363–375. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 18.Holmes David R., Dehmer Gregory J., Kaul Sanjay, Leifer Dana, O'Gara Patrick T., Michael Stein C. ACCF/AHA clopidogrel clinical alert: approaches to the FDA “Boxed Warning”. J Am Coll Cardiol. 2010;56:321–341. doi: 10.1016/j.jacc.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 19.Falzoi M., Mossa A., Congeddu E., Saba L., Pani L. Multiplex genotyping of CYP3A4, CYP3A5, CYP2C9 and CYP2C19 SNPs using MALDI-TOF mass spectrometry. Pharmacogenomics. 2010;11:559–571. doi: 10.2217/pgs.09.172. [DOI] [PubMed] [Google Scholar]
- 20.Blievernicht J.K., Schaeffeler E., Klein K., Eichelbaum M., Schwab M., Zanger U.M. MALDI-TOF mass spectrometry for multiplex genotyping of CYP2B6 single-nucleotide polymorphisms. Clin Chem. 2007;53:24–33. doi: 10.1373/clinchem.2006.074856. [DOI] [PubMed] [Google Scholar]
- 21.Meyer K., Ueland P.M. Use of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for multiplex genotyping. Adv Clin Chem. 2011;53:1–29. doi: 10.1016/b978-0-12-385855-9.00001-1. [DOI] [PubMed] [Google Scholar]
- 22.Misra A., Hong J.Y., Kim S. Multiplex genotyping of cytochrome p450 single-nucleotide polymorphisms by use of MALDI–TOF mass spectrometry. Clin Chem. 2007;53:933–939. doi: 10.1373/clinchem.2006.080739. [DOI] [PubMed] [Google Scholar]
- 23.Shuldiner A.R., O'Connell J.R., Bliden K.P. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jinnai T., Horiuchi H., Makiyama T. Impact of CYP2C19 polymorphisms on the antiplatelet effect of clopidogrel in an actual clinical setting in Japan. Circ J. 2009;73:1498–1503. doi: 10.1253/circj.cj-09-0019. [DOI] [PubMed] [Google Scholar]
- 25.Gladding P., Webster M., Zeng I. The pharmacogenetics and pharmacodynamics of clopidogrel response: an analysis from the PRINC (Plavix Response in Coronary Intervention) trial. JACC Cardiovasc Interv. 2008;1:620–627. doi: 10.1016/j.jcin.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Adithan C., Gerard N., Vasu S., Rosemary J., Shashindran C.H., Krishnamoorthy R. Allele and genotype frequency of CYP2C19 in a Tamilian population. Br J Clin Pharmacol. 2003;56:331–333. doi: 10.1046/j.1365-2125.2003.01883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shalia Kavita K., Shah Vinod K., Pawar Poonam, Divekar Siddhi S., Payannavar Satchidanand. Polymorphisms of MDR1, CYP2C19 and P2Y12 genes in Indian population: effects on clopidogrel response. Indian Heart J. 2013;65:158–167. doi: 10.1016/j.ihj.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan M.Y., Tan K., Tan H.C. CYP2C19 and PON1 polymorphisms regulating clopidogrel bioactivation in Chinese, Malay and Indian subjects. Pharmacogenomics. 2012 Apr;13:533–542. doi: 10.2217/pgs.12.24. [DOI] [PubMed] [Google Scholar]
- 29.Paré Guillaume, Mehta Shamir R., Yusuf Salim. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med. 2010;363:1704–1714. doi: 10.1056/NEJMoa1008410. [DOI] [PubMed] [Google Scholar]
- 30.Zabalza Michel, Subirana Isaac, Sala Joan. Meta-analyses of the association between cytochrome CYP2C19 loss- and gain-of-function polymorphisms and cardiovascular outcomes in patients with coronary artery disease treated with clopidogrel. Heart. 2012;98:100–108. doi: 10.1136/hrt.2011.227652. [DOI] [PubMed] [Google Scholar]
- 31.Subraja K., Dkhar S.A., Priyadharsini R. Genetic polymorphisms of CYP2C19 influences the response to clopidogrel in ischemic heart disease patients in the South Indian Tamilian population. Eur J Clin Pharmacol. 2013 Mar;69:415–422. doi: 10.1007/s00228-012-1381-8. [DOI] [PubMed] [Google Scholar]
- 32.Fontana P., Dupont A., Gandrille S. Adenosine diphosphate induced platelet aggregation is associated with P2Y12 gene sequence variations in healthy subjects. Circulation. 2003;108:989–995. doi: 10.1161/01.CIR.0000085073.69189.88. [DOI] [PubMed] [Google Scholar]
- 33.Malek L.A., Kisiel B., Spiewak M. Coexisting polymorphisms of P2Y12 and CYP2C19 genes as a risk factor for persistent platelet activation with clopidogrel. Circ J. 2008;72:1165–1169. doi: 10.1253/circj.72.1165. [DOI] [PubMed] [Google Scholar]