Abstract
Macrophages are central to essential physiological processes including the regulation of innate and adaptive immunity, but they are also central to a number of inflammatory disease states. These immune cells also possess remarkable plasticity and display various shades of functionalities based on changes in the surrounding molecular environment. Macrophage biology has defined various phenotypes and roles in inflammation based primarily on cytokine and chemokine profiles of cells in different activation states. Importantly, macrophages are elite producers of eicosanoids and other related lipid mediators during inflammation, but specific roles of these molecules have not generally been incorporated into the larger context of macrophage biology. In this review, we discuss the current classification of macrophage types and their roles in inflammation and disease, along with the practical challenges of studying biologically relevant phenotypes ex vivo. Using the latest advances in eicosanoid lipidomics, we highlight several key studies from our laboratory that provide a comprehensive understanding of how eicosanoid metabolism differs between macrophage phenotypes, along with how this metabolism is altered by changes in membrane fatty acid distribution and varied durations of Toll-like receptor (TLR) priming. In conclusion, we summarize several examples of the benefit of macrophage plasticity to develop accurate cellular mechanisms of lipid metabolism, and insights from lipidomic analyses about the differences in eicosanoid pathway enzyme activity in vitro vs. ex vivo cell studies. Examples of new techniques to further understand the role of macrophage eicosanoid signaling in vivo are also discussed.
Introduction
From vertebrate systems, macrophages can be evolutionarily traced to the ancient invertebrate mononuclear phagocyte system (Mellor and Munn, 2004; Ottaviani and Franceschi, 1997). The macrophage has recently been suggested as a potential relative of the protozoan Acanthamoeba (Siddiqui and Khan, 2012), based in part on their analogous proficiencies for engulfing large particles and cells through phagocytosis. Phagocytosis was the original hallmark leading to the macrophage’s discovery by Elie Metchnikoff in 1866; and consequentially, his discovery of the first immune cell (Chang, 2009).
Immune cells exist in multicellular organisms largely to protect the host from general traumas and invasion by pathogens in part, by summoning inflammation. The orchestration of innate and adaptive immunity, including inflammatory processes, requires the actions of myriad immune cells including macrophages, neutrophils, T- and B-lymphocytes, and other white blood cells, in a cooperative fashion. The macrophage has received particular focus for understanding immunity and inflammation because of its central role and dynamic functionality.
Along with being an efficient phagocytic cell, macrophages express numerous receptors that recognize foreign molecular motifs. They can respond to these danger signals through upregulation of proteins and peptides and synthesis of eicosanoids and other lipid molecules that altogether act to recruit other immune cells to a site of attack, along with other functions. Additionally, macrophages possess the ability to promote tissue repair once infection has been thwarted. Aside from general characterizations, macrophages actually represent a diverse range of unique phenotypes existing throughout the body, with specialized functions unique to their site of residence. This review will discuss the current understanding of macrophage biology and our recent work to understand the macrophage’s roles in inflammation associated with eicosanoid signaling.
Macrophage origins and phenotypic variability
Haematopoiesis and macrophage lineages
Haematopoietic stem cells (HSC) are the precursors to blood-derived mature macrophages and precursor macrophages, called monocytes. HSCs reside and multiply in bone marrow where specific molecular cues promote their differentiation into a range of mature cell types.
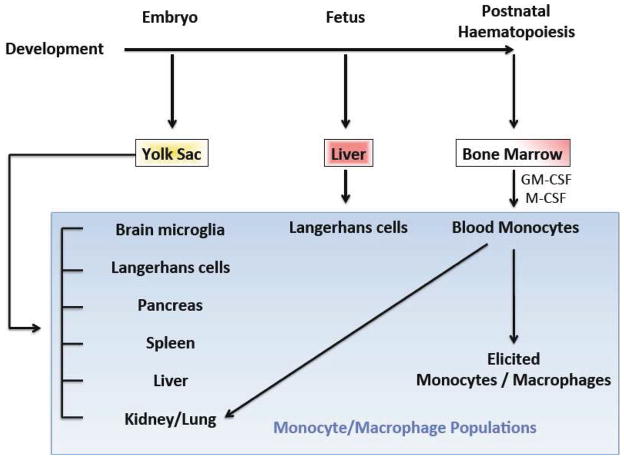
Only a few years ago, it was widely viewed that all resident tissue macrophages were derived from peripheral blood monocytes (Mosser and Edwards, 2008). However, the most recent understanding (in mice (Wynn et al., 2013)) proposes that all macrophages and precursors indirectly stem from the yolk sac; and a few directly (Figure 1). Macrophages in the brain (microglia), pancreas, spleen, liver (Kupffer cells), kidney, lung, and some Langerhans cells (a dendritic cell subset) derive from the yolk sac directly (Ginhoux et al., 2010; Hoeffel et al., 2012; Wynn et al., 2013). The remaining precursors are seeded from the yolk sac into the fetal liver, which is the predominant source of Langerhans cells (Hoeffel et al., 2012; Wynn et al., 2013). In the adult, bone marrow takes over as the source for circulating blood monocytes and macrophages (Schulz et al., 2012; Wynn et al., 2013) that can be elicited upon stimulation to various physiological sites, including the peritoneum. Some macrophages in the kidney and lung stem from blood monocytes (Wynn et al., 2013).
Figure 1. Macrophage origins and haematopoiesis.
A number of macrophage populations originate from the yolk sac, while blood monocytes and macrophages that originate from progenitors are derived from bone marrow stem cells. Langerhans cells have some overlapping functions with macrophages but are actually dendritic cells found in skin and mucosa. Figure is based on a previously published illustration (Wynn et al., 2013).
Monocytes
Monocytes represent a heterogeneous population of circulating cells that are precursors of macrophages as well as other white blood cells, including dendritic cells (Auffray et al., 2009). To become monocytes, HSCs must first commit to the myeloid lineage, and can then differentiate to one of at least two monocyte lineages. The initial steps in the commitment to precursor monocytes (monoblasts and pro-monocytes) involve cytokines, granulocyte/macrophage colony stimulating factor (GM-CSF) and further stimulation with macrophage-colony stimulating factor (M-CSF) (Mosser and Edwards, 2008). This is, of course, only a model of the differentiation process and other factors are sure to be involved in vivo. How monocytes are further differentiated to the multitude of distinct macrophages found in inflammation is also unresolved and complex.
In addition to being differentiated into macrophage phenotypes, monocytes themselves can be recruited to specific sites and play roles in host defense (Serbina et al., 2008). They can also be recruited to sites of tumors to inhibit tumor-specific defense mechanisms (Shi and Pamer, 2011). Expression of chemokine receptors allow monocytes to be recruited, and differential expression of these receptors along with other cell surface markers are used to distinguish subsets that are often characterized as either “inflammatory” or “patrolling” (Shi and Pamer, 2011); “resident” (Mosser and Edwards, 2008) instead of “patrolling” is sometimes used but this is mostly a semantic difference. These terms also refer to their actions, defining inflammatory monocytes as cells that can leave the blood, and patrolling/resident monocytes as cells that remain in blood circulation.
Macrophage functions and phenotypes
The “janitor” role: phagocytosis
During homeostasis, red blood cells are produced in large quantities that must be controlled to avoid excessive accumulation. Macrophages are well equipped to handle this due to expression of phosphatidylserine, thrombospondin, complement, and scavenger receptors, as well as integrins (Erwig and Henson, 2007). The ability to seek out and phagocytize cells and debris allow macrophages to clear about 200 billion red blood cells per day, which recycles some 3 kg of iron and haemoglobin yearly (Mosser and Edwards, 2008). This house-keeping function appears to be independent of immune signaling, since clearance of apoptotic cells by unstimulated macrophages does not lead to production of immune mediators (Kono and Rock, 2008). Phagocytosis is also an important function of macrophages during inflammation. Pathogens and elicited cells create a significant mass that must be removed by macrophages in order to resume normal tissue functions.
During development, cells of the macrophage lineage (containing the PU.1 transcription factor) are the most adept at recognizing and clearing apoptotic cells from the webbing of hand and foot digits. In mice null for PU.1, other mesenchymal cells are able to account for the loss of macrophages, but require 3 times as many cells to accomplish the same amount of neonatal cell clearance (Wood et al., 2000).
The “sentinel” role: responding to danger signals
Pathogenic assault can lead to necrosis of host cells. Pathogen-associated molecular patterns are shed from bacteria, fungi, and viruses and can be identified in necrotized host cells by Toll-like receptors (TLRs) expressed in macrophages (Chen et al., 2007; Kono and Rock, 2008; Park et al., 2004). Additionally, intracellular pattern-recognition receptors (PRRs) and the interleukin-1 receptor (IL-1R), along with TLRs, largely signal through the adaptor molecule myeloid differentiation primary-response gene 88 (MyD88) (Chen et al., 2007). Macrophages and other haematopoietic cells rather uniquely express the purinergic receptor, P2X7, at high levels. This ionotropic receptor is activated by concentrations of adenosine 5′-triphosphate (ATP) above 100 μM and up to low mM concentrations, and can promote killing of infectious organisms, mediate cell death, and regulate immune responses (Bulanova et al., 2009; Gavala et al., 2008; Jiang, 2009; Khakh, 2001; Miller et al., 2011). Complex signaling by macrophages commences from all of these receptor stimuli in order to neutralize injury and infection before rebuilding the affected site.
Activation states: M1 vs. M2
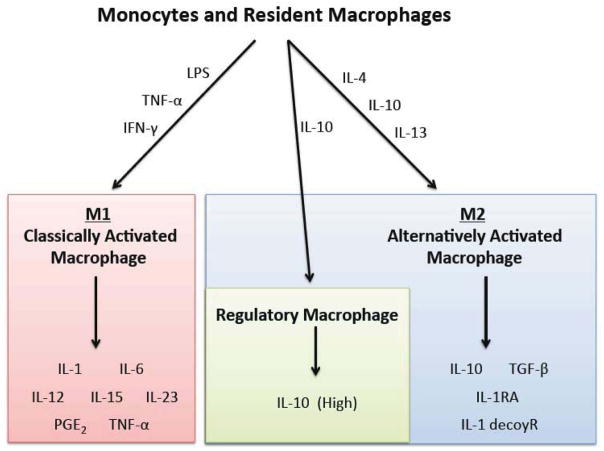
During immune responses, macrophages respond differently depending on the specific agonists presented (Figure 2). Classically, two types of responses, M1 and M2, have been described in macrophages and result from different sets of stimuli (Sica and Mantovani, 2012), mirroring the Th-1 and Th-2 states of T-lymphocytes (Biswas and Mantovani, 2010; Mantovani et al., 2002).
Figure 2. Activation states of macrophages.
Monocytes and macrophages commit to several activated phenotypes in response to specific stimuli. M1 “classically activated macrophages” release pro-inflammatory cytokines and have bactericidal activity; M2 “alternatively activated macrophages” release anti-inflammatory cytokines and certain subsets display wound healing properties, while a distinct subset termed “regulatory macrophages” have an anti-inflammatory phenotype primarily characterized by production of high levels of IL-10.
M1 activation is defined by high expression levels of proinflammatory cytokines, significant production of reactive oxygen and nitrogen species, commitment to microbicidal and tumoricidal actions, as well as the promotion of the Th-1 response (Sica and Mantovani, 2012). They manifest from stimulation by TLR, TNF, and IFNγ signals (Mosser and Edwards, 2008; Sica and Mantovani, 2012). A combination of transcription factors promote the M1 phenotype: signal transducer and activation of transcription (STAT) molecules are activated by IFNγ, while nuclear factor-κB (NFκB) and mitogen-activated protein kinases (MAPKs) are activated by TLR ligands and TNF (Mosser and Edwards, 2008; O’Shea and Murray, 2008). Macrophages in the M1 state promote destruction of tissues and initiate pro-inflammatory responses (Biswas and Mantovani, 2010; Gordon and Martinez, 2010), and are commonly referred to as “classically activated”. They produce pro-inflammatory signals, including IL-1, IL-6, IL-12, IL-15, IL-23, and TNF-α cytokines, and prostaglandin E2 (PGE2) along with other eicosanoids.
The M2 activation state is characterized by a commitment to pathogen containment, promotion of tissue remodeling (and tumor progression), and regulation of immune responses (Sica and Mantovani, 2012). Promotion of cells to this state involves stimulation with IL-4 or IL-13 (Mosser and Edwards, 2008; Sica and Mantovani, 2012). More specifically, M2 macrophages possess significant phagocytic activity, and high expression of scavenging molecules, mannose and glucose receptors, IL-10, IL-1decoyR, IL-1RA, and TGF-β, low expression of IL-12 and other characterizations (Gordon and Martinez, 2010; Mantovani et al., 2002; Sica and Mantovani, 2012). M2 macrophages are generally associated with tissue repair (Lucas et al., 2010; Sica and Mantovani, 2012). The M2 state is also commonly referred to as “alternatively activated”, and essentially serves as a catchall term for macrophage phenotypes that do not resemble the M1, classically activated, macrophage (Martinez et al., 2008).
A more recently described state of macrophage activation has been coined as a “regulatory macrophage”, or “anti-inflammatory” macrophage, that requires activation with IL-10; stimulation with prostaglandins, GPCR ligands, immune complexes, and other stimuli have also been shown to induce this phenotype (Mosser and Edwards, 2008). These cells produce high levels of IL-10 and can further promote wound healing, or M2-like macrophage states, and overall suppress inflammatory signaling. While some immune cells, like T-cells, undergo considerable epigenetic modifications during differentiation to limit phenotypic variation, macrophages possess a marked difference in this respect (Stout et al., 2005), which very likely underlies their tremendous plasticity.
Wound healing
Wound healing properties of macrophages largely appear to be induced by IL-4, a signal produced by the innate immune system (Mosser and Edwards, 2008) that is considered a major ligand involved in the switch from innate to adaptive immunity. The mannose receptor was originally found to be upregulated by IL-4 in macrophages, prompting the naming of the phenotype as “alternatively activated” (Stein et al., 1992).
IL-4 stimulation also causes resident macrophages to contribute in the production of the extracellular matrix via arginase-mediated conversion of arginine to ornithine, a precursor of polyamines and collagen (Kreider et al., 2007). Chitinase, and chitinase-like molecules including YM1 and YM2, acidic mammalian chitinase (AMCase), and stabilin-interacting chitinase-like protein are also produced by “alternatively activated macrophages” and have been found to be involved in wound healing based on carbohydrate and matrix-binding activity (Bleau et al., 1999; Fusetti et al., 2002; Kzhyshkowska et al., 2006; Raes et al., 2002; Zhu et al., 2004). Concomitantly, macrophages stimulated with IL-4 or IL-13 are inefficient producers of pro-inflammatory cytokines, reactive oxygen and nitrogen species, fail to present antigen to T-cells, and are less able to kill intracellular pathogens, compared to classically activated macrophages (Edwards et al., 2006; Mosser and Edwards, 2008). Altogether, these results demonstrate that macrophages can be programmed to reduce cytotoxic functions and increase tissue regeneration processes.
Primary and immortalized macrophages for ex vivo studies
Murine macrophages, but not other cells, attach to tissue culture-grade polystyrene plates due to unique expression of divalent cation-independent receptors: murine scavenger receptors (MSRs) (Fraser et al., 1993). This allows for efficient attachment of macrophages and removal of contaminating cells by simple aspiration. Macrophages can also adhere to non-tissue culture-treated petri dishes via complement receptor 3 (CR3; CD11b/CD18) integrins (Rosen and Gordon, 1987).
A large majority of cell culture studies utilize only a few macrophage phenotypes for practical reasons (Figure 3). The peritoneum provides a fairly clean and simple source for macrophage isolation that involves a quick lavage with phosphate-buffered saline. Resident peritoneal macrophages (RPM) can be removed directly, though only ~1 million cells can be removed from one mouse. This is a relatively low yield that makes large-scale experiments quite difficult (and expensive) to attempt. Alternatively, thioglycollate-elicited macrophages (TGEM) can be harvested from the peritoneum in quantities of ~30–40 million cells/mouse but are a significantly different phenotype relative to RPM cells. Still, they are relevant for studying blood-elicited macrophages initiated by sterile peritonitis.
Figure 3. Macrophage phenotypes.
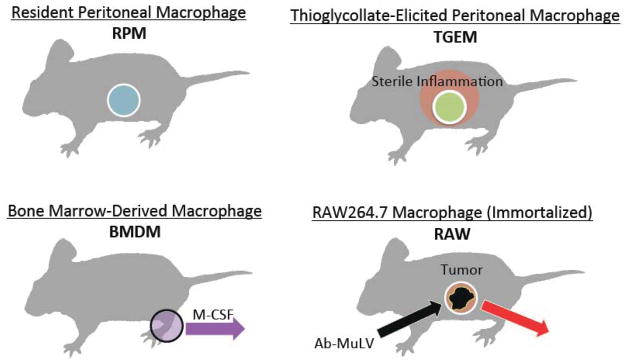
Resident peritoneal macrophages (RPM) are present during homoestasis and maintain other cell populations before and during sterile or pathogenic assaults. Thioglycollate-elicited peritoneal macrophages (TGEM) are derived from monocytes elicited from the blood and are likely alternatively activated macrophages involved with cell clearance, wound healing, and resolution of inflammation. Bone marrow-derived macrophages (BMDM) arise from the differentiation of stem cells stimulated with monocyte colony-stimulating factor (M-CSF); BMDM are generated artificially for study in cell culture and have lost some biological relevance in this context. RAW264.7 (RAW) macrophages are immortalized cells obtained from mouse tumors induced by peritoneal administration of the Abelson Murine Leukemia Virus (Ab-MULV).
Bone marrow-derived macrophages (BMDM) represent another primary macrophage phenotype, which is a highly homogeneous population that requires only a few mice for most experiments. However, BMDM require isolation of stem cells that are further cultured for ~1 week in the presence of M-CSF for differentiation, which represents a fairly artificial environment.
RAW264.7 (RAW) cells are one of the most widely used macrophage phenotypes in cell culture due to their ability to replicate rapidly; they can also be removed from tissue-culture plates via simple scraping rather than via trypsin or EDTA, which makes for simple handling and passaging. RAW cells represent an immortalized phenotype derived from peritoneal tumors induced in BALB/c male mice by the Abelson murine leukemia virus. These cells have been passaged for over 20 years ex vivo, thus relating information from RAW cells to a biological context requires some direct comparison with primary macrophages.
Advances in understanding macrophage eicosanoid signaling using lipidomics
Inflammatory eicosanoid metabolism in macrophages
Eicosanoids are 20-carbon oxidized fatty acids derived from arachidonic acid (AA; 20:4). During inflammation, cytosolic phospholipase A2 (cPLA2) is activated by calcium-dependent and independent mechanisms leading to hydrolysis of AA from phospholipid membranes (Dennis et al., 2011). Calcium independent (i) and secretory (sPLA2) may also contribute to AA release in some contexts. AA is further metabolized by the three major enzymatic pathways: cyclooxygenase (COX), lipoxygenase (LOX), and cytochrome P450 (CYP450), which also include numerous downstream enzymes that collectively form many distinct structures. Additionally, non-enzymatic pathways can oxidize AA. Refer to (Buczynski et al., 2009) for an extensive review of eicosanoids and pathway enzymes. COX-1 and COX-2 convert AA to prostaglandins and thromboxane (with COX-2 being upregulated in many tissues during inflammation), while 5-LOX converts AA to leukotrienes. These enzymes are the primary drug targets for alleviating pain, swelling, fever, and asthmatic symptoms associated with inflammation, however metabolism by these enzymes help regulate important physiological processes like vascular tone and platelet aggregation.
Macrophage roles in inflammatory signaling are mostly understood only in the context of cytokine and chemokine production. Though a large body of studies focused on the production of eicosanoids in macrophages now exists in the literature, not much of this can be easily incorporated into the greater context of macrophage biology. At most, one function particular to macrophages involves prostaglandin E2 (PGE2) and PGI2, which can be produced by resident peritoneal macrophages for autoregulatory signaling through IP, EP2 and EP4 receptors that downregulates pro-inflammatory TNF-α and upregulates anti-inflammatory IL-10 (Shinomiya et al., 2001). Still, this study elegantly demonstrates the tight connection between inflammatory eicosanoid and cytokine signaling in macrophages, and further provides a key example of how widely used non- steroidal anti-inflammatory drugs (NSAIDS) inhibit pro-inflammatory prostaglandin formation while enhancing pro-inflammatory cytokine signaling.
Quantitative eicosanoid profiling and predictive modeling
Eicosanoid metabolism has been well characterized using purified enzymes and overexpression studies. Currently, most interest in the field has shifted to defining the pathway in cellular inflammatory contexts. Understanding the complex networks of eicosanoid metabolism and signaling at the physiological level requires dissection of individual cell contributions and applying the information to whole tissues. Lipidomic mass spectrometry methodologies have been significantly advanced to the point where nearly complete and quantitative eicosanoid profiles in different cells can be generated to define specific signaling roles. Ultimately, creating predictive models of eicosanoid metabolism at the cell level is a major goal from a systems biology standpoint because therapeutic outcomes could be more rapidly and accurately tested with computational tools before clinical trials.
Our lab has previously generated a complete lipidomic quantitative dataset of eicosanoids produced after stimulation with the TLR4 ligand Kdo2 lipid A (KLA) in RAW264.7 cells that was used to develop a kinetic model (Gupta et al., 2009). This model accurately predicts changes in RAW cells quite well, but does not account for phenotypic differences in eicosanoid metabolism.
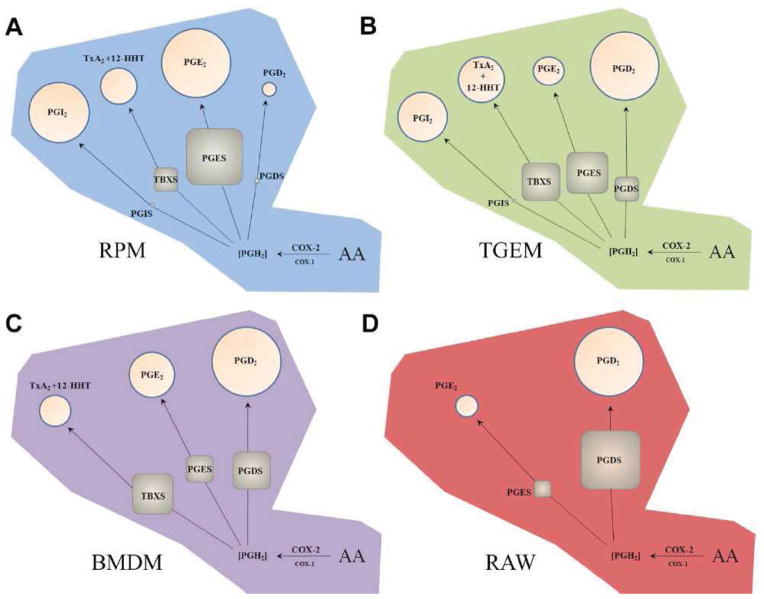
More recently, we have compared temporal quantitative differences in eicosanoid levels and related enzyme transcript expression levels generated after stimulation of TLR4 with KLA in the four types of macrophages illustrated in Figure 3, namely: RPM, TGEM, BMDM, and RAW (Figure 4) (Norris et al., 2011). In this study, all of these macrophage phenotypes were found to produce almost exclusively COX pathway metabolites where total quantities of COX metabolites directly correlated with COX-2 transcript and protein expression. Primary peritoneal macrophages (RPM and TGEM) expressed significant levels of prostaglandin I2 synthase (PGIS) and produced significant levels of PGI2, while BMDM and RAW cells neither expressed PGIS nor produced PGI2. Additionally, RPM and TGEM cells exclusively produced exponentially lower levels of PGE2 vs. PGD2 relative to PGES and PGDS transcript expression. BMDM and RAW cells did not express PGIS or produce PGI2 at detectable levels, and produced remarkably similar ratios of PGE2:PGD2 vs. PGES:PGDS.
Figure 4. Phenotypic differences in quantitative COX pathway enzyme transcript expression and eicosanoid production after TLR4 stimulation.
(a) RPM, (b) TGEM, (c) BMDM, and (d) RAW macrophages express different proportions of TBXS, PGIS, PGDS (H-PGDS), and induced PGES (mPGES-1) that result in a close correlation with metabolite proportions, but only in cells that lack PGIS. Area of circles and squares represent percentage of total metabolites and enzyme transcripts after 8 hours KLA stimulation, respectively. COX-2 is dominant over COX-1 in this scenario, where PGIS possesses the most efficient coupling. Figure is from (Norris et al., 2011) and reprinted with permission.
These results are connected to the model of functional coupling (Murakami and Kudo, 2004), which suggests that PGIS activity is the prostaglandin synthase most tightly dependent on COX-2 metabolism. Activity of prostaglandin synthases rely on PGH2 substrate concentration derived from either COX-1 expressed at the ER and to a lesser extent at the perinuclear membrane, or COX-2 that is expressed almost exclusively at the perinuclear membrane. PGIS is primarily expressed at the perinuclear membrane along with COX-2.
These studies suggest that predictive modeling of eicosanoid metabolism in macrophages requires additional work to incorporate the complications of PGIS expression. Preferential coupling of enzymes to COX-1 or COX-2 likely require additional functions that account for topographical distances and site-specific substrate concentrations. Alternatively, PGIS protein translation may be more efficient compared to other prostaglandin and thromboxane synthases, or an unidentified adaptor protein may yield more efficient enzyme activity. Ultimately, COX metabolism in macrophages and other cells requires more elaborate models for accurate predictions.
To account for disparities between transcript and protein expression, a mass spectrometry-based proteomic methodology has recently been developed to accompany our lipidomic methodology for eicosanoid metabolism (Sabido et al., 2012). This methodology has only been utilized for analyzing protein levels in RAW cells, though analysis of RPM or TGEM cells (which contain the full cassette of prostaglandin and thromboxane synthases) would provide a more complete understanding of eicosanoid metabolism connected directly to COX pathway enzyme expression.
Eicosanoid metabolism and fatty acid distribution in membrane phospholipids
Eicosanoid pathway enzymes are generally thought to possess strict specificity for AA vs. other fatty acid substrates. Evidence from in vitro studies supports specificity of AA for cPLA2, COX-1, and COX-2 (Wada et al., 2007). Lipoxygenases, however, appear to be equally efficient enzymes against other poly-unsaturated fatty acids (PUFAs) such as omega-3 eicosapentaenoic acid (EPA; 20:5) and docosahexaenoic acid (DHA; 22:6). In RAW cells, we have shown that 5-LOX is not inhibited by EPA or DHA, and actually produce similar levels of hydroxy-EPA, and hydroxy-DHA compared with hydroxy-AA (5-hydroxyeicosatetraenoic acid; 5-HETE) (Norris and Dennis, 2012). More significantly, we have also demonstrated that cPLA2 activated by P2X7 stimulation can actually hydrolyze phospholipid-esterified EPA at a rate roughly equal to that of AA.
In this study, we also found that resident peritoneal macrophages elongate AA and EPA quite proficiently, meaning that supplementation of fish oil omega-3 fatty acids used for cardioprotection and anti-inflammatory effects may function inadvertently through the 2-carbon elongation product of EPA, docosapentaenoic acid (DPA), along with DHA. Ultimately, lipidomic profiling of total fatty acid levels in membrane phospholipids can offer significant insights about the source of downstream signaling effects in cells. While significant mass spectrometry advancements have been made to efficiently profile phospholipid species, quantitative standards for phospholipids are very limited and a total accounting of fatty acid quantities using these methods cannot be reliably obtained. We have found that using the older method of saponification, to hydrolyze all of the fatty acids from membranes, and total lipidomic quantitation of eicosanoids and fatty acids provides the best starting point for addressing the complexity of phospholipids. Ultimately, there appear to be significant differences in substrate specificities of eicosanoid pathway enzymes in cells vs. purified enzymes in vitro.
Macrophages in health and inflammatory-based diseases
Effects of macrophage ablation
A significant advance in understanding macrophage biology in vivo comes from the development of a selective strategy for ablating macrophages (Cailhier et al., 2005). While diphtheria toxin (DT) poorly binds to the murine DT receptor (DTR; also known as hbEGF), the human form of DTR is quite sensitive to DT. Thus, transgenic mice (CD11bDTR) expressing the human DTR under control of the CD11b promotor yield selective macrophage ablation with DT administration via inhibition of protein synthesis. Using this strategy, resident peritoneal macrophages were shown to be critical for initiating CXC chemokine-mediated neutrophil infiltration after thioglycollate administration (Cailhier et al., 2005).
This same transgenic model was also used to investigate the role of macrophages in wound healing. Depletion of macrophages with DT resulted in delayed re-epithelialization, diminished collagen deposition, compromised angiogenesis, and inhibition of cell proliferation in healing wounds. The absence of macrophages overall retarded wound closure and dermal healing that was associated with increased levels of TNF-α and diminished levels of TGF-1β (Mirza et al., 2009).
Role in atherosclerosis: macrophage foam cells
Formation of atherosclerotic plaques appears to be centrally mediated by monocytes and macrophages. Activated endothelial cells at lesion prone sites within large arteries promote adherence of monocytes, their migration to the subendothelial space, and further differentiation to macrophages (Li and Glass, 2002). Atherosclerotic plaques progress when macrophages increase uptake of oxidized-LDL and cholesterol/cholesterol esters that are stored as lipid droplets (Yu et al., 2013). The same scavenger receptors that macrophages use to recognize pathogens and apoptotic cells also sense oxidized lipid patterns present in oxidized LDL, which can lead to significant accumulation of lipid droplets in macrophages resulting in formation of foam cells, a major component of atherosclerotic plaques (de Villiers and Smart, 1999; Linton and Fazio, 2001). The most profound example of the macrophage’s role in atherosclerosis comes from a study demonstrating that hypercholesterolemic mice gain extreme resistance to atherosclerosis when bred to macrophage-deficient mice (Smith et al., 1995).
Role in insulin resistance: Type-2 diabetes
Macrophages are also considered as a major central component that drives progression of type-2 diabetes mellitus (Olefsky and Glass, 2010). In obese subjects, macrophages are present at significantly higher levels in adipose tissue than in lean subjects, and are putatively a major source for pro-inflammatory mediators linked to insulin resistance, like TNF-α (Heilbronn and Campbell, 2008; Weisberg et al., 2003; Xu et al., 2003). Obese mouse and human tissues have been found to contain increased numbers of infiltrated macrophages (Weisberg et al., 2003; Xu et al., 2003). Insulin resistance appears to be promoted by a switch from M2/alternatively activated macrophages, to M1/classically activated macrophages, that is driven by NFκB, AP-1, and possibly other transcription factors, creating a feed-forward loop in pro-inflammatory mediator production (Olefsky and Glass, 2010).
Macrophages and cancer
A general view of macrophage involvement in cancer suggests that the M1 phenotype can be either antagonistic or cooperative toward early tumor progression, while prolonged tumor progression is mostly augmented by macrophages with alternatively activated/regulatory phenotypes (Mosser and Edwards, 2008). Classically activated macrophages in vitro can kill transformed cells, although some of their products can also promote tumorigenesis (Klimp et al., 2002). Macrophages associated with progressed tumors have been shown to display regulatory phenotypes that produce relatively high levels of IL-10 and low levels of IL-12 and TNF, and can ultimately suppress actions of neighboring macrophages and antigen presenting cells, along with the ability to promote tumor growth via angiogenesis (Biswas et al., 2006; Lin et al., 2006; Mosser and Edwards, 2008; Pollard, 2008). Roles for macrophages in cancer are largely generalized, but some differences are likely to be found in specific cases, along with differences in age, sex, and genetic variations.
Conclusion
The macrophage has now been studied for more than 100 years and is still one of the most highly studied immune cells because of its central importance in immune processes and disease. Its ability to shift through an array of phenotypes has presented a great challenge in classifying specific roles, yet we have embraced macrophage plasticity as a useful tool to probe a deeper understanding of cellular eicosanoid signaling. By comparing macrophage phenotypes directly using “omics” strategies, the need for genetic manipulation or inhibitors can be avoided to study unadulterated biological systems. The complete picture of eicosanoid signaling can now be accurately viewed using LC-MS/MS lipidomic and proteomic strategies (Dumlao et al., 2011; Quehenberger et al., 2008; Sabido et al., 2012). Our application of eicosanoid lipidomics has provided important insights about signaling roles in various diseases and inflammatory conditions (Gregus et al., 2012; Harmon et al., 2010; Tam et al., 2013). Conditional knockout strategies have also been recently developed to probe the roles of enzymes in specific cells (Ishikawa and Herschman, 2006; Ishikawa et al., 2011; Lao et al., 2012), which we envision will provide useful information about the roles of eicosanoids in macrophages and other cells in vivo that could be coupled with ex vivo studies to provide sound and relevant mechanisms.
Acknowledgments
This work was supported by the Lipid Metabolites and Pathways Strategy (LIPID MAPS) Large Scale Collaborative Grant U54 GM069338 and Grant R01 GM64611 from the National Institutes of Health; and University of California at San Diego Graduate Training Program in Cellular and Molecular Pharmacology National Institutes of Health Grant T32 GM007752 (to P.C.N.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annual review of immunology. 2009;27:669–692. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- 2.Biswas SK, Gangi L, Paul S, Schioppa T, Saccani A, Sironi M, Bottazzi B, Doni A, Vincenzo B, Pasqualini F, et al. A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-kappaB and enhanced IRF-3/STAT1 activation) Blood. 2006;107:2112–2122. doi: 10.1182/blood-2005-01-0428. [DOI] [PubMed] [Google Scholar]
- 3.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nature immunology. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 4.Bleau G, Massicotte F, Merlen Y, Boisvert C. Mammalian chitinase-like proteins. Exs. 1999;87:211–221. doi: 10.1007/978-3-0348-8757-1_15. [DOI] [PubMed] [Google Scholar]
- 5.Buczynski MW, Dumlao DS, Dennis EA. Thematic Review Series: Proteomics. An integrated omics analysis of eicosanoid biology. Journal of lipid research. 2009;50:1015–1038. doi: 10.1194/jlr.R900004-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulanova E, Budagian V, Orinska Z, Koch-Nolte F, Haag F, Bulfone-Paus S. ATP induces P2X7 receptor-independent cytokine and chemokine expression through P2X1 and P2X3 receptors in murine mast cells. Journal of leukocyte biology. 2009;85:692–702. doi: 10.1189/jlb.0808470. [DOI] [PubMed] [Google Scholar]
- 7.Cailhier JF, Partolina M, Vuthoori S, Wu S, Ko K, Watson S, Savill J, Hughes J, Lang RA. Conditional macrophage ablation demonstrates that resident macrophages initiate acute peritoneal inflammation. J Immunol. 2005;174:2336–2342. doi: 10.4049/jimmunol.174.4.2336. [DOI] [PubMed] [Google Scholar]
- 8.Chang ZL. Recent development of the mononuclear phagocyte system: in memory of Metchnikoff and Ehrlich on the 100th Anniversary of the 1908 Nobel Prize in Physiology or Medicine. Biology of the cell / under the auspices of the European Cell Biology Organization. 2009;101:709–721. doi: 10.1042/BC20080227. [DOI] [PubMed] [Google Scholar]
- 9.Chen CJ, Kono H, Golenbock D, Reed G, Akira S, Rock KL. Identification of a key pathway required for the sterile inflammatory response triggered by dying cells. Nature medicine. 2007;13:851–856. doi: 10.1038/nm1603. [DOI] [PubMed] [Google Scholar]
- 10.de Villiers WJ, Smart EJ. Macrophage scavenger receptors and foam cell formation. Journal of leukocyte biology. 1999;66:740–746. doi: 10.1002/jlb.66.5.740. [DOI] [PubMed] [Google Scholar]
- 11.Dennis EA, Cao J, Hsu YH, Magrioti V, Kokotos G. Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chemical reviews. 2011;111:6130–6185. doi: 10.1021/cr200085w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumlao DS, Buczynski MW, Norris PC, Harkewicz R, Dennis EA. High-throughput lipidomic analysis of fatty acid derived eicosanoids and N-acylethanolamines. Biochimica et biophysica acta. 2011;1811:724–736. doi: 10.1016/j.bbalip.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards JP, Zhang X, Frauwirth KA, Mosser DM. Biochemical and functional characterization of three activated macrophage populations. Journal of leukocyte biology. 2006;80:1298–1307. doi: 10.1189/jlb.0406249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erwig LP, Henson PM. Immunological consequences of apoptotic cell phagocytosis. The American journal of pathology. 2007;171:2–8. doi: 10.2353/ajpath.2007.070135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser I, Hughes D, Gordon S. Divalent cation-independent macrophage adhesion inhibited by monoclonal antibody to murine scavenger receptor. Nature. 1993;364:343–346. doi: 10.1038/364343a0. [DOI] [PubMed] [Google Scholar]
- 16.Fusetti F, von Moeller H, Houston D, Rozeboom HJ, Dijkstra BW, Boot RG, Aerts JM, van Aalten DM. Structure of human chitotriosidase. Implications for specific inhibitor design and function of mammalian chitinase-like lectins. The Journal of biological chemistry. 2002;277:25537–25544. doi: 10.1074/jbc.M201636200. [DOI] [PubMed] [Google Scholar]
- 17.Gavala ML, Pfeiffer ZA, Bertics PJ. The nucleotide receptor P2RX7 mediates ATP-induced CREB activation in human and murine monocytic cells. Journal of leukocyte biology. 2008;84:1159–1171. doi: 10.1189/jlb.0907612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Gregus AM, Doolen S, Dumlao DS, Buczynski MW, Takasusuki T, Fitzsimmons BL, Hua XY, Taylor BK, Dennis EA, Yaksh TL. Spinal 12-lipoxygenase-derived hepoxilin A3 contributes to inflammatory hyperalgesia via activation of TRPV1 and TRPA1 receptors. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6721–6726. doi: 10.1073/pnas.1110460109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta S, Maurya MR, Stephens DL, Dennis EA, Subramaniam S. An integrated model of eicosanoid metabolism and signaling based on lipidomics flux analysis. Biophysical journal. 2009;96:4542–4551. doi: 10.1016/j.bpj.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harmon GS, Dumlao DS, Ng DT, Barrett KE, Dennis EA, Dong H, Glass CK. Pharmacological correction of a defect in PPAR-gamma signaling ameliorates disease severity in Cftr-deficient mice. Nature medicine. 2010;16:313–318. doi: 10.1038/nm.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heilbronn LK, Campbell LV. Adipose tissue macrophages, low grade inflammation and insulin resistance in human obesity. Current pharmaceutical design. 2008;14:1225–1230. doi: 10.2174/138161208784246153. [DOI] [PubMed] [Google Scholar]
- 24.Hoeffel G, Wang Y, Greter M, See P, Teo P, Malleret B, Leboeuf M, Low D, Oller G, Almeida F, et al. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. The Journal of experimental medicine. 2012;209:1167–1181. doi: 10.1084/jem.20120340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa TO, Herschman HR. Conditional knockout mouse for tissue-specific disruption of the cyclooxygenase-2 (Cox-2) gene. Genesis. 2006;44:143–149. doi: 10.1002/gene.20192. [DOI] [PubMed] [Google Scholar]
- 26.Ishikawa TO, Oshima M, Herschman HR. Cox-2 deletion in myeloid and endothelial cells, but not in epithelial cells, exacerbates murine colitis. Carcinogenesis. 2011;32:417–426. doi: 10.1093/carcin/bgq268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang LH. Inhibition of P2X(7) receptors by divalent cations: old action and new insight. European biophysics journal: EBJ. 2009;38:339–346. doi: 10.1007/s00249-008-0315-y. [DOI] [PubMed] [Google Scholar]
- 28.Khakh BS. Molecular physiology of P2X receptors and ATP signalling at synapses. Nature reviews Neuroscience. 2001;2:165–174. doi: 10.1038/35058521. [DOI] [PubMed] [Google Scholar]
- 29.Klimp AH, de Vries EG, Scherphof GL, Daemen T. A potential role of macrophage activation in the treatment of cancer. Critical reviews in oncology/hematology. 2002;44:143–161. doi: 10.1016/s1040-8428(01)00203-7. [DOI] [PubMed] [Google Scholar]
- 30.Kono H, Rock KL. How dying cells alert the immune system to danger. Nature reviews Immunology. 2008;8:279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreider T, Anthony RM, Urban JF, Jr, Gause WC. Alternatively activated macrophages in helminth infections. Current opinion in immunology. 2007;19:448–453. doi: 10.1016/j.coi.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kzhyshkowska J, Mamidi S, Gratchev A, Kremmer E, Schmuttermaier C, Krusell L, Haus G, Utikal J, Schledzewski K, Scholtze J, et al. Novel stabilin-1 interacting chitinase-like protein (SI-CLP) is up-regulated in alternatively activated macrophages and secreted via lysosomal pathway. Blood. 2006;107:3221–3228. doi: 10.1182/blood-2005-07-2843. [DOI] [PubMed] [Google Scholar]
- 33.Lao HC, Akunda JK, Chun KS, Flake GP, Yuspa SH, Langenbach R. Genetic ablation of cyclooxygenase-2 in keratinocytes produces a cell-autonomous defect in tumor formation. Carcinogenesis. 2012;33:2293–2300. doi: 10.1093/carcin/bgs267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li AC, Glass CK. The macrophage foam cell as a target for therapeutic intervention. Nature medicine. 2002;8:1235–1242. doi: 10.1038/nm1102-1235. [DOI] [PubMed] [Google Scholar]
- 35.Lin EY, Li JF, Gnatovskiy L, Deng Y, Zhu L, Grzesik DA, Qian H, Xue XN, Pollard JW. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer research. 2006;66:11238–11246. doi: 10.1158/0008-5472.CAN-06-1278. [DOI] [PubMed] [Google Scholar]
- 36.Linton MF, Fazio S. Class A scavenger receptors, macrophages, and atherosclerosis. Current opinion in lipidology. 2001;12:489–495. doi: 10.1097/00041433-200110000-00003. [DOI] [PubMed] [Google Scholar]
- 37.Lucas T, Waisman A, Ranjan R, Roes J, Krieg T, Muller W, Roers A, Eming SA. Differential roles of macrophages in diverse phases of skin repair. J Immunol. 2010;184:3964–3977. doi: 10.4049/jimmunol.0903356. [DOI] [PubMed] [Google Scholar]
- 38.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends in immunology. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 39.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Frontiers in bioscience: a journal and virtual library. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 40.Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nature reviews Immunology. 2004;4:762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- 41.Miller CM, Boulter NR, Fuller SJ, Zakrzewski AM, Lees MP, Saunders BM, Wiley JS, Smith NC. The role of the P2X(7) receptor in infectious diseases. PLoS pathogens. 2011;7:e1002212. doi: 10.1371/journal.ppat.1002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mirza R, DiPietro LA, Koh TJ. Selective and specific macrophage ablation is detrimental to wound healing in mice. The American journal of pathology. 2009;175:2454–2462. doi: 10.2353/ajpath.2009.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nature reviews Immunology. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murakami M, Kudo I. Recent advances in molecular biology and physiology of the prostaglandin E2-biosynthetic pathway. Progress in lipid research. 2004;43:3–35. doi: 10.1016/s0163-7827(03)00037-7. [DOI] [PubMed] [Google Scholar]
- 45.Norris PC, Dennis EA. Omega-3 fatty acids cause dramatic changes in TLR4 and purinergic eicosanoid signaling. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8517–8522. doi: 10.1073/pnas.1200189109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Norris PC, Reichart D, Dumlao DS, Glass CK, Dennis EA. Specificity of eicosanoid production depends on the TLR-4-stimulated macrophage phenotype. Journal of leukocyte biology. 2011;90:563–574. doi: 10.1189/jlb.0311153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Shea JJ, Murray PJ. Cytokine signaling modules in inflammatory responses. Immunity. 2008;28:477–487. doi: 10.1016/j.immuni.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annual review of physiology. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 49.Ottaviani E, Franceschi C. The invertebrate phagocytic immunocyte: clues to a common evolution of immune and neuroendocrine systems. Immunology today. 1997;18:169–174. doi: 10.1016/s0167-5699(97)84663-4. [DOI] [PubMed] [Google Scholar]
- 50.Park JS, Svetkauskaite D, He Q, Kim JY, Strassheim D, Ishizaka A, Abraham E. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. The Journal of biological chemistry. 2004;279:7370–7377. doi: 10.1074/jbc.M306793200. [DOI] [PubMed] [Google Scholar]
- 51.Pollard JW. Macrophages define the invasive microenvironment in breast cancer. Journal of leukocyte biology. 2008;84:623–630. doi: 10.1189/jlb.1107762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quehenberger O, Armando A, Dumlao D, Stephens DL, Dennis EA. Lipidomics analysis of essential fatty acids in macrophages. Prostaglandins, leukotrienes, and essential fatty acids. 2008;79:123–129. doi: 10.1016/j.plefa.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raes G, De Baetselier P, Noel W, Beschin A, Brombacher F, Hassanzadeh GhG. Differential expression of FIZZ1 and Ym1 in alternatively versus classically activated macrophages. Journal of leukocyte biology. 2002;71:597–602. [PubMed] [Google Scholar]
- 54.Rosen H, Gordon S. Monoclonal antibody to the murine type 3 complement receptor inhibits adhesion of myelomonocytic cells in vitro and inflammatory cell recruitment in vivo. The Journal of experimental medicine. 1987;166:1685–1701. doi: 10.1084/jem.166.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabido E, Quehenberger O, Shen Q, Chang CY, Shah I, Armando AM, Andreyev A, Vitek O, Dennis EA, Aebersold R. Targeted proteomics of the eicosanoid biosynthetic pathway completes an integrated genomics-proteomics-metabolomics picture of cellular metabolism. Molecular & cellular proteomics: MCP. 2012;11:M111. doi: 10.1074/mcp.M111.014746. 014746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 57.Serbina NV, Jia T, Hohl TM, Pamer EG. Monocyte-mediated defense against microbial pathogens. Annual review of immunology. 2008;26:421–452. doi: 10.1146/annurev.immunol.26.021607.090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nature reviews Immunology. 2011;11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shinomiya S, Naraba H, Ueno A, Utsunomiya I, Maruyama T, Ohuchida S, Ushikubi F, Yuki K, Narumiya S, Sugimoto Y, et al. Regulation of TNFalpha and interleukin-10 production by prostaglandins I(2) and E(2): studies with prostaglandin receptor-deficient mice and prostaglandin E-receptor subtype-selective synthetic agonists. Biochemical pharmacology. 2001;61:1153–1160. doi: 10.1016/s0006-2952(01)00586-x. [DOI] [PubMed] [Google Scholar]
- 60.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. The Journal of clinical investigation. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siddiqui R, Khan NA. Acanthamoeba is an evolutionary ancestor of macrophages: a myth or reality? Experimental parasitology. 2012;130:95–97. doi: 10.1016/j.exppara.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 62.Smith JD, Trogan E, Ginsberg M, Grigaux C, Tian J, Miyata M. Decreased atherosclerosis in mice deficient in both macrophage colony-stimulating factor (op) and apolipoprotein E. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:8264–8268. doi: 10.1073/pnas.92.18.8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. The Journal of experimental medicine. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stout RD, Jiang C, Matta B, Tietzel I, Watkins SK, Suttles J. Macrophages sequentially change their functional phenotype in response to changes in microenvironmental influences. J Immunol. 2005;175:342–349. doi: 10.4049/jimmunol.175.1.342. [DOI] [PubMed] [Google Scholar]
- 65.Tam VC, Quehenberger O, Oshansky CM, Suen R, Armando AM, Treuting PM, Thomas PG, Dennis EA, Aderem A. Lipidomic profiling of influenza infection identifies mediators that induce and resolve inflammation. Cell. 2013;154:213–227. doi: 10.1016/j.cell.2013.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wada M, DeLong CJ, Hong YH, Rieke CJ, Song I, Sidhu RS, Yuan C, Warnock M, Schmaier AH, Yokoyama C, et al. Enzymes and receptors of prostaglandin pathways with arachidonic acid-derived versus eicosapentaenoic acid-derived substrates and products. The Journal of biological chemistry. 2007;282:22254–22266. doi: 10.1074/jbc.M703169200. [DOI] [PubMed] [Google Scholar]
- 67.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. The Journal of clinical investigation. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wood W, Turmaine M, Weber R, Camp V, Maki RA, McKercher SR, Martin P. Mesenchymal cells engulf and clear apoptotic footplate cells in macrophageless PU.1 null mouse embryos. Development. 2000;127:5245–5252. doi: 10.1242/dev.127.24.5245. [DOI] [PubMed] [Google Scholar]
- 69.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. The Journal of clinical investigation. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu XH, Fu YC, Zhang DW, Yin K, Tang CK. Foam cells in atherosclerosis. Clinica chimica acta; international journal of clinical chemistry. 2013 doi: 10.1016/j.cca.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 72.Zhu Z, Zheng T, Homer RJ, Kim YK, Chen NY, Cohn L, Hamid Q, Elias JA. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 2004;304:1678–1682. doi: 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]