Abstract
The solution conformation behavior of the macrolide core of microtubule-stabilizing agents (−)-zampanolide and (−)-dactylolide has been determined through a combination of high-field NMR experiments and computational modeling. Taken together, the results demonstrate that in solution both molecules exist as a mixture of three interconverting conformational families, one of which bears strong resemblance to zampanolide’s tubulin-bound conformation.
Tubulin-binding agents that disrupt microtubule dynamics represent an important class of chemotherapeutic agents for the treatment of multiple cancers.1 However, several key microtubule-stabilizing and -destabilizing agents possess significant clinical limitations in the form of tubulin resistance2, dose-dependent toxicities, and peripheral neuropathy3. As a result, there has been widespread interest in investigating the clinical potential of natural and synthetic compounds with this mode of action. Recently, our laboratory has become interested in the marine polyketide (−)-zampanolide 1, a potent microtubule-stabilizing agent that exhibits low nanomolar cytotoxicity (1–5 nM) against both normal and multi-drug resistant cell lines. Field and co-workers have shown via H/D-exchange mass spectrometry that zampanolide binds tubulin irreversibly via a covalent interaction with the amino acid H229 in the paclitaxel-binding site.4 Interestingly, a structurally related polyketide natural product, (+)-dactylolide, was isolated in 2001 off the coast of the Vanatu islands and found to possess the same macrolactone core as zampanolide, albeit in the opposite absolute configuration.5 Dactylolide displays modest cytotoxicity in the low μM range against numerous human cancer cell lines and binds tubulin through the same covalent binding mechanism as zampanolide.4
The potent biological activity of zampanolide and its relatively few stereocenters has attracted significant attention from the synthetic and biological communities. However, despite isolation from two marine organisms6 and several total syntheses,7 including our own,8 zampanolide remains in scarce supply and only a few analogues have been reported to date. While previous studies with these compounds have centered on determining their mode of action and molecular interactions with tubulin4 there’s been little focus on understanding their conformational behavior in solution, which we believe is crucial in the process of designing bioactive analogues.9 Herein, we report the conformational analysis of zampanolide and dactylolide’s shared macrolide core.
Structurally, the core of zampanolide consists of a 20-membered macrolactone containing a cis-2,6-tetrahydropyran, three (E)- and one (Z)-olefinic bonds, 11 rotatable σ bonds, and three stereocenters. Additionally, zampanolide contains an unusual N-acyl hemiaminal side chain at C19 while dactylolide possesses an aldehyde at the same position. Though zampanolide is regarded as the more attractive of the two from a clinical standpoint, dactylolide was chosen for molecular modeling studies and NMR experiments because the side chain complicates both spectral and computational analyses without offering additional significant insight into the macrolactone’s conformational preferences.10
To identify the major solution conformations, we first conducted conformational searches using Macromodel 9.9. In order to completely explore the molecules conformational space, 50,000-step Monte Carlo searches were performed using the MM3*, MMFF*, and OPLS2005* force fields, which were run in conjunction with the GB/SA solvation model for water to prevent hydrophilic collapse of the ring.11 Structures that fell within 20 kJ/mol (4.8 kcal/mol) of the global minimum were saved from each search, and duplicate conformers across the three runs were eliminated to yield a single library.
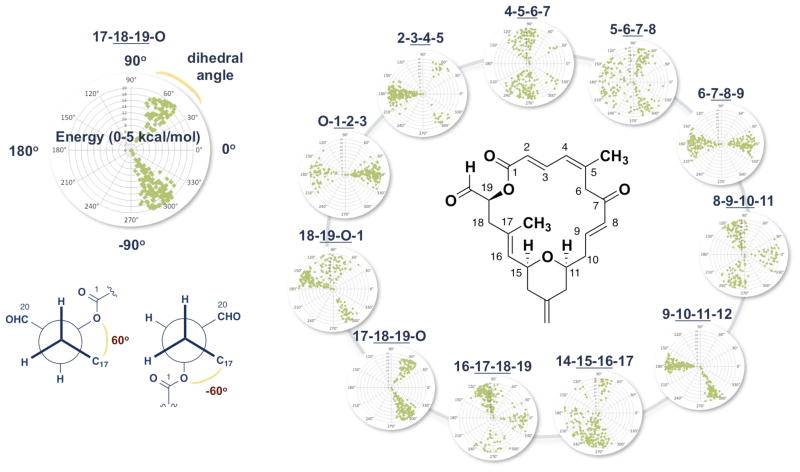
The flexible portions of the molecule were identified employing the same methodology previously used by our group in the conformational analysis of the epothilones,11 extracting the backbone torsional angles from each conformation and plotting them onto polar coordinate graphs (Figure 2). The resulting polar maps revealed a mixture of flexible and rigid torsions throughout the macrolactone core. Of particular note was the enone-containing eastern fragment from C6–C11, which appears to be rotating on a plane perpendicular to the pyran ring in either an s-cis or s-trans orientation. While the northern fragment possesses a certain amount of rigidity in the form of the s-trans diene, the O-C1-C2-C3 and 18-19-O-C1 dihedrals indicate that the exclusively s-trans lactone linkage readily flips back and forth relative to the plane of the macrolide ring. Additionally, the flexibility in the C5–C7 region suggests that the nothern fragment as a whole oscillates relative to the rest of the macrolide. Finally, the western region showed some preference at lower energies for a conformation that minimizes A1,3- and A1,2-strain involving the C17Me, but otherwise the relative lack of stereogenic centers prevents most of the molecule from adopting a single conformational structure.
Figure 2.
Dactylolide dihedral angle distribution for Monte Carlo conformational search.
To evaluate the solution behavior of the shared macrolide core, we used NMR experiments in conjuction with molecular modeling studies to determine its major conformational preferences. Our first task was the complete assignment of dactylolide’s nonoverlapping protons with the correlation spectroscopy (COSY)12 technique at 600 MHz, using DMSO as the solvent to simulate a biological environment relative to the previously reported solvent, chloroform. Serendipitously, use of DMSO also allowed for the unambiguous identification of a nearly complete set of 1H coupling constants, including ones in the C9–C11 region that were unresolved in chloroform. To complete our NMR studies, rotating-frame Overhauser effect (ROESY)13 experiments were undertaken next. In order to avoid spin diffusion effects, buildup curves were generated at various mixing times to determine the optimum mixing time (800 ms). In total, 39 cross-peaks were observed from the spectra.
Overall, the flexibility demonstrated in our computational studies indicated that the core of dactylolide and zampanolide exists as an ensemble of conformations in solution. While there are several different methods for deconvoluting time-averaged NMR data into individual structures, we decided to use the DISCON (Distribution of Solution Conformations) software package developed by the Smith laboratory,14 which is similar to the NAMFIS15 methodology but uses clustering based on NMR variables to avoid overfitting a given set of conformers. In order to utilize DISCON, the ROESY cross-peaks obtained from our dactylolide NMR experiments were normalized and integrated using the C10 methylene protons as reference to yield quantitative peak volumes. Of the 39 cross-peaks previously obtained, 11 were identified for use in deconvolution, along with 6 of the 14 3JH-H coupling constants from the newly-resolved 1H DMSO spectrum (see Supporting Information).
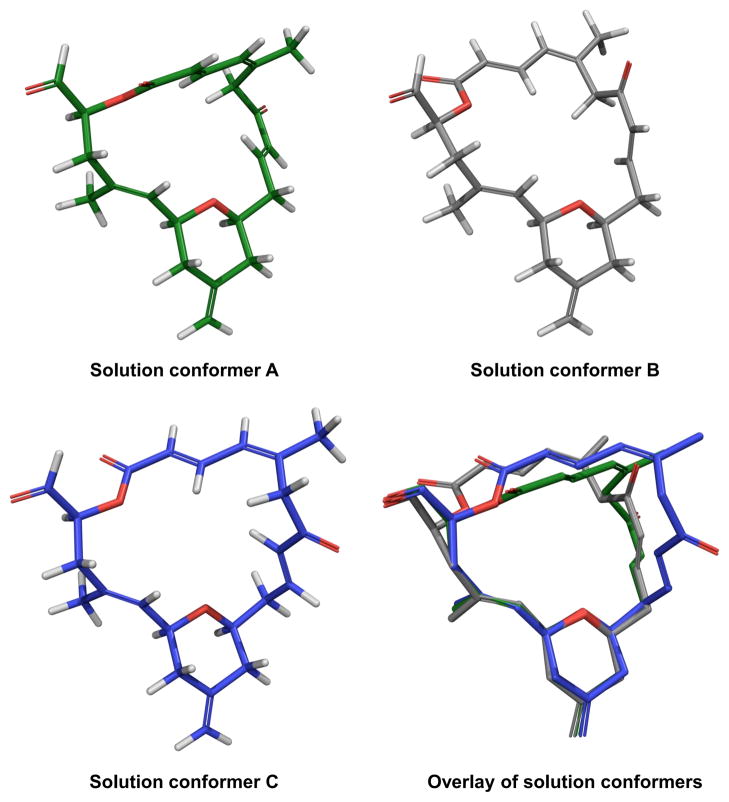
Repeated evaluation of the conformational library using DISCON’s clustering analysis revealed an ensemble containing three conformational families that fit the observed NMR data better than any individual structure (Figure 3). Two of the families were identified as the majority with the primary difference being the orientation of the mostly inflexible northern fragment relative to the plane of the macrolide ring, changing the overall profile from hooked (conformer A, 33%) to flat (conformer B, 35%). The third conformational family (conformer C, 20%) was largely similar to the other two families, save for a 180° rotation around the C6–C7 bond that contributes to a twisted shape. The remaining conformers were insignificant with respect to the NMR data, with the next most populated family contributing less than 5%.
Figure 3.
DISCON-derived solution conformations
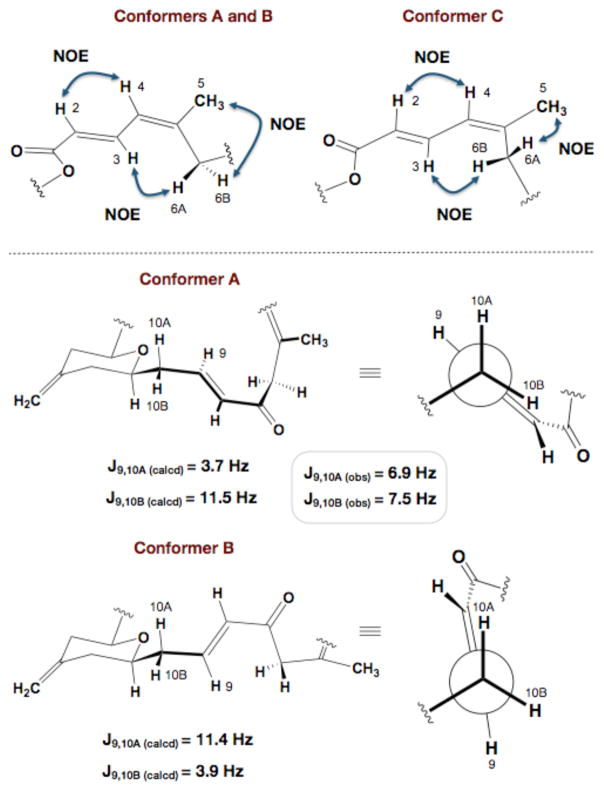
One of the interesting features shared by the three conformational families was a suprisingly rigid C15–C19 region, which was only vaguely suggested by the polar map analysis of the computational results. The rigidity present within this region was supported by strong C17Me-H15 and C17Me-H19 ROESY correlations along with a well-defined anti H18–H19 coupling constant (3JH18–H19 = 11.0 Hz). This conformational preference can be rationalized by considering the destabilizing allylic strain interactions between the C17Me-pyran oxygen and the C15 stereogenic center within the pyran. In addition, the northern diene’s preference for an s-trans conjugated system was also represented in all three conformers and supported by a strong H2–H4 NOE cross-peak and a large 3JH3–H4 coupling constant of 11.2 Hz (Figure 4). Though the western and diene segments were confirmed to be conformationally rigid, the macrolide exhibited a high degree of flexibility within the C6–C11 region of the molecule. A portion of this flexibility arose from the ability of the C5-C6-C7-C8 torsion to adopt 60° and 300° dihedral angles. Support for this torsional rotation and thus the existence of minor conformer C resulted from observed NOE correlations between both H3-H6A and H3-H6B. Interestingly, it appears that the conformational preferences of the C6–C11 region are primarily controlled by rapid rotation around the C9–C10 bond, which was observed in the computational polar map (Figure 2; 8-9-10-11) and is experimentally supported by time-averaged 3JH9-H10A and 3JH9-H10B coupling constant values of 6.9 and 7.5 Hz, respectively. This particular preference for torsions at 0°, 120°, and −120° results from the minimization of A-strain through exclusively low-energy eclipsed conformations and nicely represents the conformational distribution displayed by 1-butene (Figure 4).16
Figure 4.
Conformational averaging of enone and northern diene region
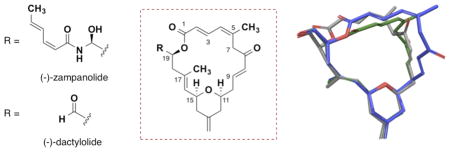
During the course of this work, Prota and co-workers reported a high resolution co-crystal structure of zampanolide bound to α,β-tubulin.17 The structure not only confirms the unique covalent binding exhibited by both zampanolide and dactylolide but also provided the bioactive conformation of zampanolide, which bears a strong resemblance to major solution conformer A (Figure 5). However, based on the stereochemistry of C9 in the bound complex, the C7–C9 enone must adopt an s-cis orientation that more closely resembles solution conformer C prior to conjugate addition.
Figure 5.
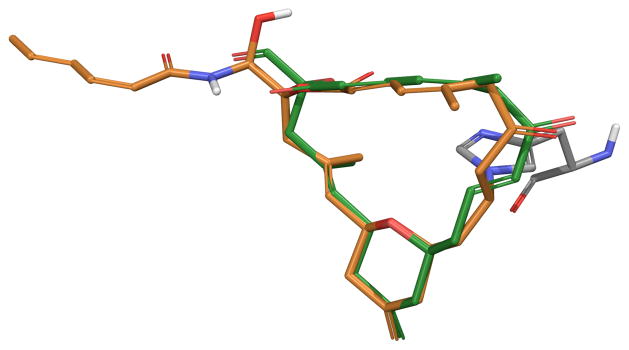
Overlay of zampanolide co-crystal structure (orange) bound to tubulin HIS229 (gray) with solution conformer A (green)
In summary, we have determined the solution conformational preferences of the macrolactone core shared by dactylolide and zampanolide. Through a combination of high-field NMR experiments and molecular modeling, we found that the macrolide displays a preference for three interconverting conformational families while in solution. Given its low-nanomolar cytotoxicity and relatively few observed hydrogen-bonding interactions with tubulin, we believe that mimicking the bioactive conformation of zampanolide is an important aspect to consider in the design of potent analogues. Studies involving the synthesis and evaluation of such conformationally-designed analogues are ongoing in our laboratory.
Supplementary Material
Figure 1.
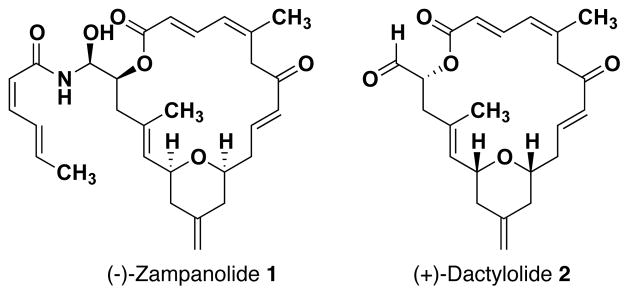
Zampanolide and Dactylolide
Acknowledgments
Financial support by the National Institutes of Health is gratefully acknowledged (GM084922). We also thank Eric Stefan (Broad Institute) for assisting with the synthesis of both dactylolide and zampanolide.
Footnotes
Supporting Information Available: NMR characterization data and computational protocols. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Dumontet C, Jordan MA. Nat Rev Drug Discov. 2010;9:790–803. doi: 10.1038/nrd3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kavallaris M. Nat Rev Cancer. 2010;10:194–204. doi: 10.1038/nrc2803. [DOI] [PubMed] [Google Scholar]
- 3.Lee JJ, Swain SM. J Clin Oncol. 2006;24:1633–1642. doi: 10.1200/JCO.2005.04.0543. [DOI] [PubMed] [Google Scholar]
- 4.Field JJ, Pera B, Calvo E, Canales A, Zurwerra D, Trigili C, Rodríguez-Salarichs J, Matesanz R, Kanakkanthara A, Wakefield SJ, Singh AJ, Jiménez-Barbero J, Northcote P, Miller JH, López JA, Hamel E, Barasoain I, Altmann KH, Díaz JF. Chem Biol. 2012;19:686–698. doi: 10.1016/j.chembiol.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutignano A, Bruno I, Bifulco G, Casapullo A, Cécile D, Gomez-Paloma L, Riccio R. Eur J Org Chem. 2001:775–778. [Google Scholar]
- 6.a) Tanaka J, Higa T. Tetrahedron Lett. 1996;37:5535–5538. [Google Scholar]; b) Field JJ, Singh AJ, Kanakkanthara A, Halafihi T, Northcote PT, Miller JH. J Med Chem. 2009;52:7328–7332. doi: 10.1021/jm901249g. [DOI] [PubMed] [Google Scholar]
- 7.a) Smith AB, III, Safonov IG, Corbett RM. J Am Chem Soc. 2001;123:12426–12427. doi: 10.1021/ja012220y. [DOI] [PubMed] [Google Scholar]; b) Hoye TR, Hu M. J Am Chem Soc. 2003;125:9576–9577. doi: 10.1021/ja035579q. [DOI] [PubMed] [Google Scholar]; c) Uenishi J, Iwamoto T, Tanaka J. Org Lett. 2009;11:3262–3265. doi: 10.1021/ol901167g. [DOI] [PubMed] [Google Scholar]; d) Ghosh AK, Cheng X. Org Lett. 2011;13:4108–4111. doi: 10.1021/ol201626h. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Zurwerra D, Glaus F, Betschart L, Schuster J, Gertsch J, Ganci W, Altmann KH. Chem Eur J. 2012;18:16868–16883. doi: 10.1002/chem.201202553. [DOI] [PubMed] [Google Scholar]
- 8.a) Wilson MR, Taylor RE. Org Lett. 2012;14:3408–3411. doi: 10.1021/ol301383a. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wilson MR, Larsen EM, Stefan E, Sackett DL, Taylor RE. manuscript in preparation. [Google Scholar]
- 9.a) Taylor RE, Chen Y, Galvin GM, Pabba PK. Org Biomol Chem. 2004;2:127–132. doi: 10.1039/b312213c. [DOI] [PubMed] [Google Scholar]; b) Taylor RE, Chen Y, Beatty A, Myles DC, Zhou Y. J Am Chem Soc. 2003;125:26–27. doi: 10.1021/ja028196l. [DOI] [PubMed] [Google Scholar]; c) Zhao Z, Taylor RE. Org Lett. 2012;14:669–671. doi: 10.1021/ol203268t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Initial computational searches were complicated by the presence of the side chain as the modeling software will treat changes to the freely-rotating chain as new conformations, vastly increasing the number of Monte Carlo steps necessary for a complete search of the conformational space. b) For an analysis of the side chain conformation of zampanolide and the importance of intramolecular hydrogen bonding networks, see Troast DM, Porco JA., Jr Org Lett. 2002;4:991–994. doi: 10.1021/ol025558l.
- 11.Taylor RE, Zajicek J. J Org Chem. 1999;64:7224–7228. [Google Scholar]
- 12.Rance M, Sorensen OW, Bodenhausen G, Wagner G, Ernst RR, Wuthrich K. Biochem Biophys Res Commun. 1983;117:479. doi: 10.1016/0006-291x(83)91225-1. [DOI] [PubMed] [Google Scholar]
- 13.a) Bothner-By AA, Stephens RL, Lee JM, Warren CD, Jeanloz RW. J Am Chem Soc. 1984;106:811–813. [Google Scholar]; b) Bax A, Davis DG. J Magn Reson. 1985;63:207–213. [Google Scholar]
- 14.a) Atasoylu O, Furst G, Risatti C, Smith AB., III Org Lett. 2010;12:1788–1791. doi: 10.1021/ol100417d. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Smith AB, III, Risatti CA, Atasoylu O, Bennett CS, Liu J, Cheng H, TenDyke Xu Q. [accessed Nov 12, 2012];J Am Chem Soc. 2011 133:14042–14053. doi: 10.1021/ja2046167. DISCON software and documentation is available for download at http://discon.sourceforge.net/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cicero DO, Barbato G, Bazzo R. J Am Chem Soc. 1995;117:1027–1033. [Google Scholar]
- 16.a) Wiberg KB, Martin E. J Am Chem Soc. 1985;107:5035–5041. [Google Scholar]; b) Broeker JL, Hoffmann RW, Houk KN. J Am Chem Soc. 1991;113:5006–5017. [Google Scholar]
- 17.Prota AE, Bargsten K, Zurwerra D, Field JJ, Díaz JF, Altmann KH, Steinmetz MO. Science. 2013;339:587–590. doi: 10.1126/science.1230582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.